Abstract
Cancer cells exist in harsh microenvironments which are governed by various factors including hypoxia and nutrient deprivation. These microenvironmental stressors activate signaling pathways that affect a cancer cell’s survival. While others have previously measured microenvironmental stressors in tumors, it remains difficult to detect the real time activation of these downstream signaling pathways in primary tumors. Here, we developed transgenic mice expressing an X-box binding protein 1 (XBP1)-luciferase construct that served as a reporter for endoplasmic reticulum (ER) stress and as a downstream response for the tumor microenvironment. We find that primary mammary tumors arising in these mice possessed luciferase activity in vivo. Multiple tumors arising in the same mouse had distinct XBP1-luciferase signatures, reflecting either higher or lower levels of ER stress. Primary tumors with undetectable XBP1-luciferase activity gained bioluminescent signal after transplantation indicating that the level of XBP1-luciferase activity reflected unique microenvironments of individual primary tumors. Furthermore, variations in ER stress reflected metabolic and hypoxic differences between tumors. Finally, XBP1-luciferase activity correlated with tumor growth rates. Thus, we can visualize distinct signaling pathways in primary tumors that reflect unique tumor microenvironments, distinct metabolic signatures and may predict for tumor growth.
Keywords: Tumor microenvironment, ER stress, XBP1
Introduction
The tumor microenvironment remains a poorly characterized stress on cancer cells reflecting several variables including stromal cell interactions, hypoxia and nutrient deprivation(1–4). These microenvironmental pressures interact during tumorigenesis to promote or inhibit tumor growth. Stromal cells may receive or transmit signals from or to cancer cells to promote angiogenesis, invasion and metastasis(5). Hypoxia activates pro-growth signaling pathways and destabilizes a cancer cell’s genome to enhance tumorigenesis(6, 7). By contrast, nutrients may become limited in the tumor microenvironment potentially inhibiting cell growth and/or activating cell death pathways(7). Many groups have developed ways to quantitate these stresses in the tumor microenvironment. However, it remains unclear how much these microenvironmental stressors activate downstream signaling pathways in cancer cells. By understanding how the tumor microenvironment may impact signaling pathways among similar primary tumors, we can gain insights into cancer biology and individualizing cancer therapy.
One putative marker for the tumor microenvironment is ER stress which can be triggered within cancer cells by hypoxia or nutrient deprivation (8, 9). These stressors lead to the abnormal accumulation of proteins within the ER. Cells respond by activating the unfolded protein response (UPR) that regulates transcription and translation of genes in order to maintain ER homeostasis(10). The UPR encompasses a three pronged signaling network which includes of the Inositol-requiring 1α (Ire1α)-X-box binding protein 1 (XBP1) pathway. The UPR interacts to promote cell survival or, if the ER stress is too great or too long, to activate cell death. Several groups have shown that the UPR is activated in tumors including high grade breast, colorectal and pancreatic carcinomas(11–14). In addition, UPR proteins can promote tumor growth in both transplantable and primary tumor models(15–18). Thus, the UPR may link the tumor microenvironment to cancer cell death and survival as well as serve as a marker for the microenvironmental stressors occurring in primary tumors.
Here, we monitored the microenvironment in primary tumors by developing a transgenic mouse model to report XBP1-luciferase (XBP1-luc) activity as a marker for ER stress. XBP1-luc activity was heterogeneous among similarly sized primary tumors within the same mouse indicating that primary tumors had unique microenvironments with variable levels of ER stress. This unique XBP1-luc activity inversely correlated with the glucose avidity and directly correlated with hypoxia and tumor growth. Together, these distinct microenvironments may explain differences in growth and metabolism in similar primary tumors.
Materials and Methods
Mice
FVB/N-Tg(MMTV-PyVT)634 Mul/J (Tag), FVB/N-Tg(MMTVneu)202Mul/J (Her2), FVB/NJ and SCID mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were housed at Stanford University Research Animal Facility and maintained according to Stanford Institutional Care and Use Committee guidelines.
Generation of XBP1-Luc transgenic mice
The XBP1-Luc transgene was injected into FVB/N oocytes by Stanford Transgenic Animal Core Facility and a single founder line was maintained.
Bioluminescent imaging
Luciferase activity was measured noninvasively using the IVIS imaging system (Caliper LifeSciences, Hopkinton, MA). Mice were injected i.p. with luciferin (300mg/kg dissolved in PBS; Caliper) and anesthetized via inhaled 3% isoflurane (Abbott Laboratories Ltd, Abbott Park, IL). Exposure time ranged from 10s to 10 min. All images were analyzed using Living Image software (Caliper) with a binning of 10. In vivo bioluminescent signal was quantified by taking the average bioluminescent signal which is photon count per second per cm2. For comparisons between mice, the bioluminescent signal of the tumors was normalized to adjacent skin.
RGD and 2-DG avidity was determined using spectral fluorescence imaging via the Maestro In-Vivo imaging system (CRi, Woburn, MA). IRDye 800CW 2-DG, RGD and the unlabeled dye was obtained from Li-Cor (Lincoln, NE) and injected i.v. into tumor bearing mice. 24 to 48h later, mice were then imaged using near-infrared excitation/emission filter sets and images were obtained from 780nm to 900nm wavelengths at 10nm intervals. Exposure time ranged from 250 to 1500ms.
Additional Material and Methods are presented in the Supplemental Methods.
Results
Primary tumors that developed in transgenic mice had increased XBP1-luc activity
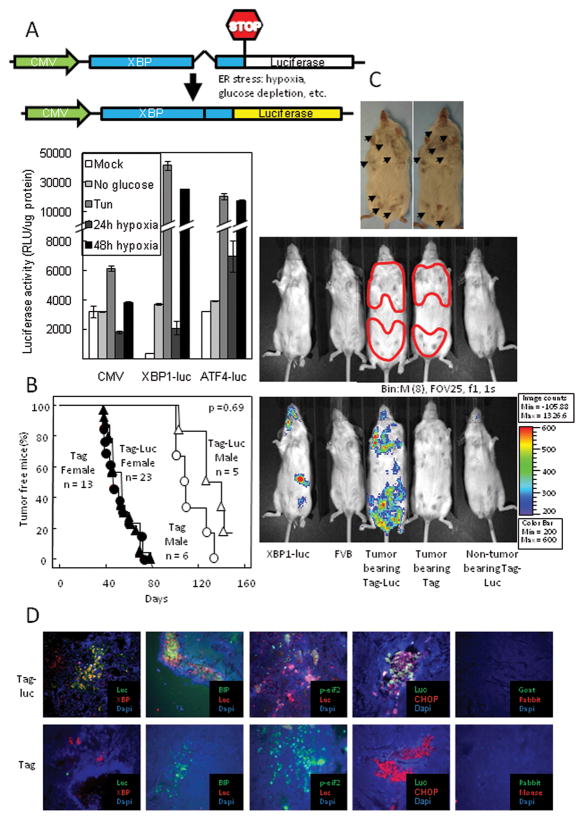
During the UPR, activation of XBP1 occurs through a novel splicing mechanism where Ire1α removes 26 nucleotides from the XBP1 mRNA(19–22). To study ER stress in tumors, we generated an XBP1-luciferase (XBP1-luc) reporter construct (Fig. 1A, upper panel). We fused the cDNA for the first 208 amino acids of the human XBP1 to firefly luciferase (XBP1-luc). This XBP1 sequence has been used in similar XBP1 reporter models and contained the hydrophobic domain-2 that regulates XBP1 activation(23–32). Under non-ER stress conditions, the XBP1-luc mRNA remains unspliced with an in-frame stop codon preventing expression of the downstream luciferase gene. Under ER stress conditions, the XBP1-luc transcript is spliced shifting the stop codon out of frame and allowing expression of the luciferase gene. We stably expressed this construct in HT1080 cells (Fig. 1A, lower panel). Similar to the UPR reporter ATF4-luc, XBP1-luc cells strongly induced luciferase activity under conditions of ER stress such as tunicamycin treatment, glucose depletion or hypoxia. XBP1-luc activity required IRE1α consistent with its requirement for splicing to report ER stress (Sup. Fig. 1).
Figure 1. Primary breast carcinomas had increased XBP1-luc activity.
(A) Upper Panel: Schematic of the XBP1-luc reporter vector. Lower panel: The XBP1-luc reporter vector induced luciferase activity in response to ER stress. HT 1080 cells expressing the XBP1-luc construct, an ATF4 or a CMV driven luciferase were subjected to 10 μg/ml Tun, glucose depletion or severe hypoxia for 48h to induce ER stress. (B) Incidence of breast carcinomas in Tag mice was similar to Tag-Luc double transgenic mice. (C) In Tag-Luc transgenic mice, primary breast tumors had increased bioluminescence. Upper panel: Photograph of breast tumors with arrows indicating each tumor along mammary chain. Middle panel: Photograph of Luc, FVB, tumor bearing Tag-Luc, tumor bearing Tag and non-tumor bearing Tag-Luc mice. Tumor areas were outlined in red. Lower panel: Overlay of bioluminescence. Note that the bioluminescence activity was detected only in tumor bearing Tag-Luc mice. (D) Luciferase localized to XBP1 and other ER stress proteins in Tag-Luc mice tumors. Frozen sections were stained with anti-luciferase and anti-XBP1, anti-BiP, anti-phopho-eIF2α or anti-CHOP.
We generated XBP1-luc transgenic mice and bred them to breast cancer prone Tag mice to generate the double transgenic Tag-Luc mouse (Fig. 1B&C & Sup. Fig. 2A). Tag mice express the middle T antigen under the control of the MMTV promoter and develop multiple primary mammary carcinomas at a mean interval of 42d (33). Normal skin fibroblasts isolated from Tag-Luc mice constitutively expressed the XBP1-luc fusion mRNA transcript (Sup. Fig. 2B). Under hypoxia, the XBP1-luc transcript was spliced similarly to the endogenous XBP1 transcript and elicited luciferase activity (Sup. Fig. 2C&D).
Tag-Luc double transgenic mice developed primary tumors at similar rates as the control Tag mice (Fig. 1B; for females, p = 0.72; for males, p = 0.07). Imaging of tumor bearing Tag-Luc mice revealed that regions corresponding to the mammary tumors possessed bioluminescent signal (Fig. 1C). By contrast, primary tumors arising in control Tag mice possessed background signal. The single transgenic XBP1-luc mice (Fig. 1C) had basal bioluminescent signal in the spleen which was confirmed with dissection (Sup. Fig. 3) and consistent with previous reports on the activity in that organ using a nearly identical XBP1-GFP transgenic reporter mouse (26). Similar to the XBP1-GFP transgenic reporter mouse, basal bioluminescent activity in our XBP1-luciferase mouse was also detectable in the pancreas and liver (data not shown). Non-tumor bearing Tag-Luc mice did not have appreciable bioluminescent signal along the mammary chains. In tumors from Tag-Luc mice, luciferase co-localized with XBP1, as well as other UPR proteins including BiP, phospho-eIF2α, and CHOP whereas, in control Tag tumors, luciferase was not detected (Fig. 1D). Therefore, primary tumors possessed XBP1-luc activity that correlated to regions of ER stress in primary tumors.
Individual primary tumors had unique levels of XBP1-luc activity that were modulated by the tumor microenvironment
Overall, Tag-Luc tumors possessed bioluminescent signal that was 9.36 ± 0.08-fold greater than the background skin and significantly greater than control tumors (Fig. 2A, left panel). Tumors isolated ex vivo by dissection from Tag-Luc transgenic mice had 14.5-fold greater luciferase activity than tumors isolated from control Tag transgenic mice (Fig. 2A, middle panel). Furthermore, in vivo bioluminescence signal significantly correlated with in vitro luciferase activity of each individual tumor (Fig. 2A, right panel). Overall, primary breast tumors possessed XBP1-luc activity that was significantly increased above background. However, we observed that each tumor within the same mouse possessed a distinct bioluminescent signal (Fig. 2B). The heterogeneity of XBP1-luc activity in different primary tumors growing in the same mouse remained a consistent phenomenon in vivo and ex vivo (Fig. 2C; Sup. Fig. 4). By contrast to Tag-luc mice, tumors arising in Tag mice (Fig. 2B, left panel and Fig. 2C) did not possess greater bioluminescence signal than non-neoplastic tissues.
Figure 2. Tumors arising in the same mouse possessed different levels of XBP1-luc activity.
(A) Left panel: Quantitation of bioluminescence activity in tumor bearing Tag-Luc and Tag mice. Middle panel: In vitro luciferase activity of mammary tumors. Mammary tumors were excised from the indicated mice, diced into fine fragments and lysed. Luciferase activity of each tumor was assessed and normalized to the luciferase activity of the autologous liver in the same mouse. Data are represented as the mean ± SEM. Right panel: Luciferase activity in vitro correlated with bioluminescence activity in vivo. Transgenic mice were imaged and bioluminescence activity quantitated. In vitro luciferase activity of each corresponding tumor was then assessed and compared to its in vivo bioluminescence. (B) Left panel: Mammary tumors in Tag-Luc mice had different levels of bioluminescence. Right panel: Heterogeneity of bioluminescence in vivo correlated with luciferase activity in vitro. Outlined tumors were excised, finely minced, lysed and analyzed for luciferase activity. (C) In vitro analysis of luciferase activity in multiple tumors from individual mice showed heterogeneity of luciferase activity. Chart legend in (C) is the same as in (B, right panel). * represent p <0.05 by Student’s t test.
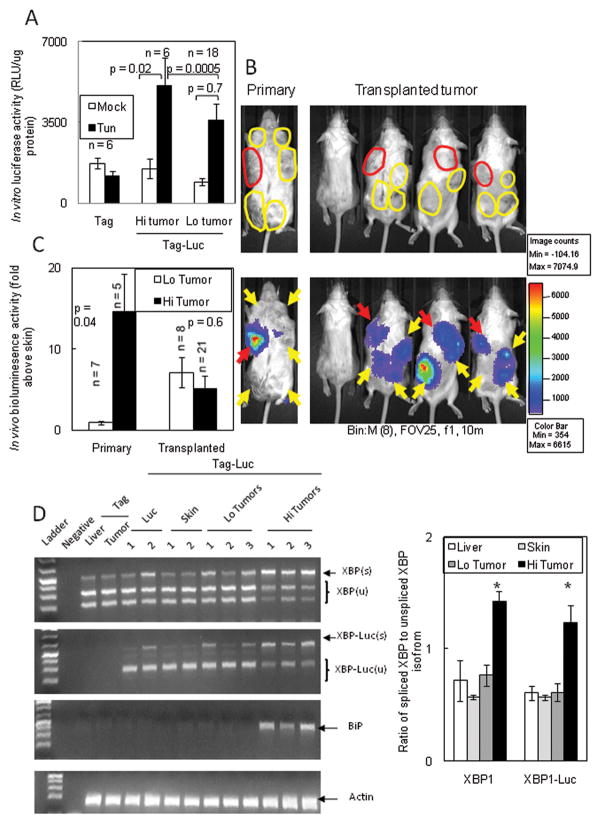
It remained unclear whether the heterogeneous XBP1-luc activity in each primary tumor reflected the variability of the tumor microenvironment or was a hereditary feature of the tumor. We defined tumors with bioluminescence signal above background skin as “Hi tumors” and those with bioluminescence signal similar to background skin as “Lo tumors”. In vivo differences in XBP1-luc activity was not maintained in cell culture as both Hi and Lo cultured tumor cells demonstrated background bioluminescent activity (Fig. 3A). After tunicamycin treatment to induce ER stress, XBP1-luc activity in Hi and Lo cultured tumor cells was induced to similar levels, indicating that both tumor phenotypes retained similar responsiveness to ER stress.
Figure 3. Tumors with low XBP1-luc activity develop increased signal after transplantation.
(A) High in vivo XBP1-luc activity was not maintained in vitro. Primary tumors with Hi or Lo XBP1-luc activity were cultured in vitro causing luciferase activity to return to background levels. Tun induced XBP1-luc activity to similar levels in cultured Hi and Lo tumors. (B) Primary tumors with low XBP1-luc activity increased bioluminescence after transplantation. Left panels: Tag-Luc mice bearing synchronous Hi (red arrows) and Lo (yellow arrows) tumors. Right panel: Primary tumors with Hi activity were transplanted into the left upper flank (red arrows) and primary tumors with Lo activity were transplanted into the remaining quadrants (yellow arrows). Bioluminescence were assessed 14d after transplantation. (C) Quantitation of primary and transplanted tumors in (B). Similar results were seen in four independent experiments using 20 mice with 71 transplanted tumors. (D) Left panel: mRNA from Hi and Lo tumors was isolated and subjected to RT-PCR analysis for the spliced endogenous XBP1, XBP1-luc and BiP. Right panel: Comparison of the ratio of spliced:unspliced isoforms of endogenous XBP1 and XBP1-luc. * represent p <0.05 by Student’s t test.
To confirm that primary tumors developed variable and non-hereditable levels of ER stress, we harvested primary tumors from mice bearing both Hi and Lo tumors and transplanted them subcutaneously into the flanks of syngeneic mice (Fig. 3B). The Hi primary tumor (indicated in red) was transplanted into the upper left flank while the Lo primary tumors (indicated in yellow) were transplanted in the remaining three quadrants of a single mouse. 14d after transplantation, we observed that both Hi and Lo tumors now had similar levels of XBP1-luc bioluminescent signal (Fig. 3B&C). Furthermore, the differences in luciferase signal between Hi and Lo tumors was not due to differences in reporter mRNA expression (Sup. Fig. 5A). Finally, Hi tumors had increased endogenous XBP1 splicing paralleling the splicing of the reporter XBP1-luc as well as increased expression of BiP confirming increased ER stress in tumors with higher bioluminescent signal (Fig. 4D & Sup. Fig. 5B). Therefore, ER stress caused by the tumor microenvironment was unique to each primary tumor and reflected the variation of microenvironmental stress.
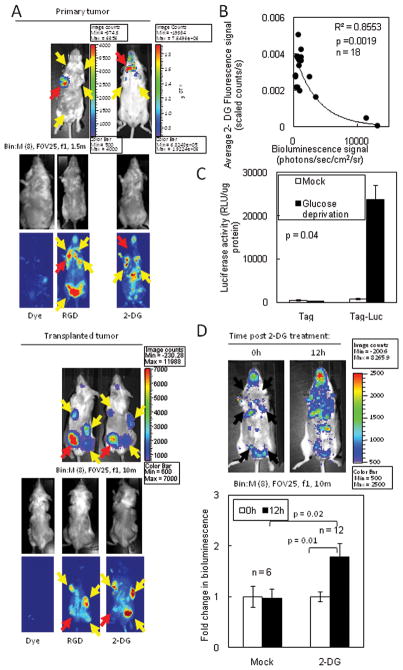
Figure 4. Loss of glucose uptake and utilization resulted in increased XBP1-luc activity in primary and transplanted tumors.
(A) XBP1-luc activity in primary tumors correlated with low glucose avidity. Tag-Luc mice bearing primary tumors (upper panels) or SCID mice (lower panels) bearing transplants of the same tumor were injected with fluorescent 2-DG or control dye and, 24h later, mice were imaged for XBP1-luc activity and 2-DG or control dye uptake. Red arrows, Hi tumors; Yellow arrows, Lo tumors. (B) For mice imaged in (A), the XBP1-luc signal was plotted against the corresponding 2-DG signal. (C) XBP-luc activity in primary tumor cells increased during glucose deprivation in vitro. (D) Upper panel: Chemical inhibition of glucose utilization in vivo increased ER stress in tumors. Tag-Luc mice were treated with vehicle or 2 mg of 2-DG i.p. and bioluminescence was assessed after 12h. Arrows indicate palpable tumors. Lower panel: Quantitation of bioluminescence. Data are represented as the mean ± SEM.
Increased XBP1-luc activity was regulated by the loss of glucose utilization in vivo
To elucidate what factors may mediate these differences of XBP1-luc activity, we injected tumor bearing Tag-Luc mice with labeled RGD peptide or 2-deoxy-d-glucose (2-DG) probes to measure angiogenesis or glucose uptake, respectively. Compared to mice injected with the RGD probe or the dye alone, primary tumors with higher XBP1-luc activity (red arrows) possessed significantly lower 2-DG uptake (Fig. 4A, upper panels and 4B). Conversely, tumors with lower XBP1-luc activity (yellow arrows) had higher 2-DG uptake. By contrast, the level of XBP1-luc activity did not significantly correlate with RGD uptake or microvessel density in tumors (Sup. Fig. 6A&B).
To confirm that differences in 2-DG uptake was not due to genetic differences or tumor size, cells from a single Tag-Luc tumor were injected at multiple sites in a single mouse. Tumors grew to similar sizes and demonstrated heterogeneous XBP1-luc signal (Fig. 4A, lower panels). As with primary tumors, transplanted tumors with higher XBP1-luc activity (red arrows) had lower 2-DG uptake. Tumors with lower XBP1-luc activity (yellow arrows) had higher 2-DG uptake. Thus, XBP1-luc activity in the microenvironment inversely correlated with the glucose uptake in tumors.
Since the XBP1-luc activity in tumors inversely correlated with glucose uptake, we determined whether inhibition of glucose utilization could modulate XBP1-luc activity in primary tumors. Indeed, tumor cells cultured in glucose depleted media had 27.2-fold more luciferase activity than control treated cells (Fig. 4C). We treated mice with 2-DG to mimic a state of glucose deprivation. Tag-Luc tumors treated with 2-DG had a 1.78-fold increase in bioluminescent compared to control mice (Fig. 4D). Therefore, the heterogeneity of ER stress in primary tumors correlated with the loss of glucose uptake as inhibition of glucose utilization increased ER stress.
Hypoxia modulated XBP1-luc signal in spontaneous tumors
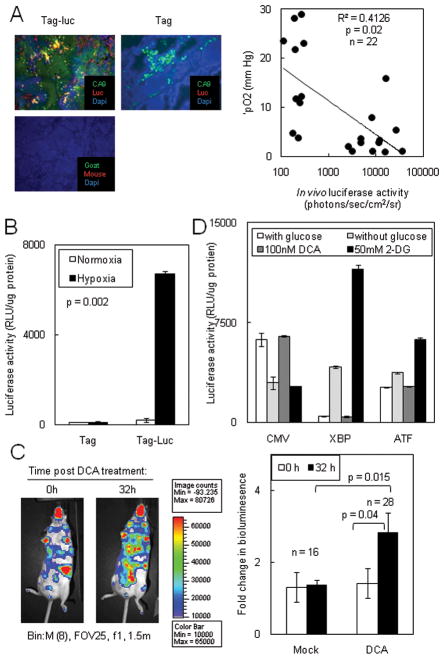
To determine if XBP1-luc activity was regulated by hypoxia, we analyzed Tag-Luc tumors for XBP1-luc protein in hypoxic areas of the tumor microenvironment. Indeed, luciferase activity co-localized with the hypoxic marker CA-9 and with lower pO2 levels indicating that XBP1 splicing occurred in hypoxic areas (Fig. 5A). To confirm that XBP1 splicing could occur in the hypoxic microenvironments, we isolated primary Tag-Luc breast tumors and cultured the cells for 5–10d. After 48h of hypoxia, Tag-Luc tumor cells had 58.0-fold more luciferase activity than normoxic cells (Fig. 5B) indicating that Tag-Luc tumors spliced XBP1 under conditions of hypoxia.
Figure 5. Hypoxia increased XBP1-luc activity.
(A) Left panel: Luciferase co-localized with the hypoxic marker CA-IX. Frozen sections from Tag-Luc and Tag mice were stained with anti-luciferase and anti-CA-IX antibodies and localization of antibodies were detected. Right panel: XBP1-luc activity was plotted against the corresponding pO2 of each tumor. (B) XBP1-luc activity in spontaneous tumors increased under hypoxia. Primary Tag-Luc tumors were cultured in vitro, subjected to normoxia or hypoxia for 48h and luciferase activity was assessed. (C) Left panel: Chemical exacerbation of hypoxia increased ER stress in spontaneous tumors. Tag-Luc mice were treated with vehicle or 50 mg/kg DCA b.i.d. for 2 days and bioluminescence was assessed after 32h. Right panel: Quantitation of bioluminescence in C. (D) DCA alone did not induce ER stress in HT1080 XBP1-luc or ATF4-luc cells.
To confirm that hypoxia regulated XBP1-luc activity, we treated mice with repeated DCA (dichloroacetic acid) injections which increases tumor oxygen consumption to mimic a state of increasing hypoxia (34). Primary Tag-Luc tumors increased bioluminescent signal by 2.0-fold after repeated DCA injections confirming that hypoxia can regulate XBP1-luc activity (Fig. 5C). DCA alone did not directly induce ER stress as HT1080 cells expressing XBP1-luc or ATF4-luc did not induce luciferase activity in response to DCA (Fig. 5D). Therefore, primary tumors in Tag-Luc mice could induce ER stress in response to hypoxic microenvironments.
Higher XBP1-luc activity correlated with faster doubling times
Various UPR proteins are important to the growth of primary tumors (15, 17, 18). Since XBP1-luc activity was heterogeneous among similar primary tumors, we determined if differences in XBP1-luc activity served as a marker for differences in tumor growth rates. Indeed, tumors with higher XBP1-luc signal had significantly faster doubling times compared to tumors with lower XBP1-luc signal (Fig. 6A). We then serially imaged Tag-Luc mice over time to detect changes in XBP1-luc signal. We observed that changes in XBP1-luc signal either increased, remained stable or decreased (Fig. 6B). Compared to tumors with stable or increasing signal, tumors with decreasing signal also had slower doubling times (Fig. 6C). By contrast, XBP1-luc activity did not correlate with tumor size indicating that XBP1-luc activity was not simply a reflection of tumor mass (Fig. 6D). Even when tumors with XBP1-luc signal that was more than 2-fold, 5-fold or 15-fold above background were only included in analysis, the lack of correlation between bioluminescent signal and tumor size remained (data not shown).
Figure 6. Tag-Luc tumors with high XBP1-luc activity possessed faster doubling times.

(A) Doubling times for tumors were calculated and plotted against bioluminescent signal. Tumors with higher XBP1-luc activity had significantly faster doubling times. (B) Tag-Luc mice were serially imaged for XBP1-luc signal. Changes in XBP1-luc signal were divided into three separate categories of increasing, stable or decreasing signal. (C) Increased or stable XBP1-luc activity was associated with faster doubling times. Doubling times were plotted against maximum change in XBP1-luc signal. (D) Bioluminescence did not correlate with tumor size.
To confirm that primary tumors possessed XBP1-luc activity, we also bred the breast cancer prone Her2 (FVB/N-Tg(MMTVneu)202Mul/J) (35) mice to the XBP1-luc reporter mice (Her2-Luc). As with Tag-Luc cells, cells from Her2-Luc tumors spliced the XBP1-luc reporter similar to the endogenous XBP1 mRNA and induced luciferase activity under ER stress (Sup. Fig 2B–2D). Her2-Luc mice developed tumors at similar rates as the Her2 controls (Sup. Fig. 6A & Sup. Table 1). Tumors arising in Her2-Luc mice possessed bioluminescence signal that co-localized to areas of ER stress as indicated by XBP1 and BiP (Sup. Fig. 6B&C). However, compared to Tag-Luc mice, XBP1 signal in Her2 tumors was less frequent and intense (Sup. Fig. 6D). Ex vivo luciferase activity of Her2-Luc tumors was statistically greater than control tumors (Sup. Fig. 6E). Therefore, in vivo and in vitro XBP1-luc activity was detected in two different primary breast tumor models.
Discussion
We observed that primary tumors possessed unique levels of ER stress that were regulated by distinct microenvironmental factors. Furthermore, these differences in ER stress reflected the metabolic profile of tumors as determined by glucose uptake and hypoxia. Finally, XBP1-luc activity also predicted for faster tumor growth. In our model, tumorigenesis was driven by the expression of the middle T antigen in mammary epithelium, which may not reflect the natural history of human breast cancers. Nevertheless, XBP1-luc activity was detected in a second Her2/neu mammary tumor model possessing a more physiologic oncogenic event and was consistent with previous clinical findings (11–13). While our reporter likely does not detect extremely low levels of XBP1 splicing that may occur in Lo tumors and normal tissues, we did observe XBP1-luc signal under conditions of ER stress sufficient for endogenous XBP1 splicing. Thus, the limitation in the sensitivity of this model did not preclude the relative comparison of ER stress between tumors. Since we examined tumors of the same histological subtype driven by the same oncogenic event, we expect that any differences in XBP1-luc signal were likely due to the biologic properties of the tumors and not an artifact of our model.
We observed that XBP1 splicing reflected ER stress imposed by the tumor microenvironment based on several findings. First, tumors with increased XBP1-luc bioluminescence signal in vivo possessed only baseline in vitro luciferase activity under non-stressed conditions. Second, XBP1-luc activity was gained or lost upon in vivo passage of primary tumors and, therefore, reflected the situational environment to which the cancer cells were subjected. Finally, treatment of mice with agents known to modulate the microenvironmental stresses of hypoxia or glucose deprivation also regulated XBP1-luc activity. Previous investigators have manipulated UPR pathways to show an effect on tumor growth in vivo but not in vitro suggesting an interplay between ER stress and the tumor microenvironment (15, 18). However, it remained unclear whether the activation of the UPR in primary tumors was a hereditary change acquired by tumor cells or a response to microenvironmental stress. By studying XBP1-luc activity under different tumor conditions, our data demonstrated that XBP1 is a critical marker of ER stress occurring in the tumor microenvironment.
Using XBP1 splicing as a surrogate for ER stress in the tumor microenvironment, we observed that primary tumors displayed different levels of XBP1-luc activity that were not maintained after in vivo or in vitro passage. It is likely that certain aspects of the tumor microenvironment remain similar between the primary and the transplanted tumor settings (3–5, 36). In contrast, our data showed that other aspects of the tumor microenvironment are unique to each primary tumor and were not maintained once tumors were introduced into a new environment. It has previously been shown that similar tumors possessed distinct metabolic (37–39), hypoxic (40) and interstitial pressure (41) profiles in their tumor microenvironments. While the exact mechanisms that regulate these differences remain to be elucidated, our data suggested that ER stress caused by the tumor microenvironment reflected glucose avidity and hypoxia within tumors. Previous reports may not have observed this because the majority of these systems dealt with a single primary tumor in a single mouse. Thus, our unique model in which several tumors arise in a single mouse allowed us to show that stress pathways important for tumor growth may be distinct among very similar tumors.
The relationship of low glucose avidity in tumors correlating with increased ER stress paralleled recent clinical observations in human tumors. Our results were consistent with the in vitro and in vivo observations that glucose deprivation is a potent inducer of ER stress. Moreover, other tumor microenvironmental stresses such as hypoxia, in combination with low glucose would further exacerbate underlying ER stress within tumors. Our results suggested that specific biological pathways may be detected and targeted by measuring glucose uptake in tumors, which is done clinically through positron emission tomography (PET) imaging. Since PET imaging has correlated with treatment response (42, 43), it is interesting to speculate how ER stress and the associated differences in glucose uptake correlate with tumor biology and response to therapy.
In addition to glucose deprivation, we also observed that prolonged hypoxia induced XBP1-luc activity in vitro and correlated with bioluminescent signal in vivo. However, the relative contributions of hypoxia and glucose avidity to ER stress remained difficult to dissect. Furthermore, increased XBP1-luc activity within tumors is a global indication of ER stress reflecting the contribution of not only hypoxia but also glucose and nutrient deprivation. Finally, since luciferase activity is dependent on the presence of O2, increasingly low O2 content in tumors may have underestimated true XBP1-luc activity and reduced the significance of this in vivo correlation. Therefore, it is difficult to determine the relative contribution of a single stressor to XBP1-luc signal.
We observed that the doubling time of tumors directly correlated with XBP1-luc activity. First, within the same mammary carcinoma model, tumors with higher XBP1-luc activity had faster doubling times whereas tumors with lower XBP1-luc activity had slower doubling times. Since it is difficult to genetically manipulate the growth rates of primary tumors in vivo, we can only correlate XBP1-luc activity to tumor growth in Tag-Luc mice. Nevertheless, in a second mammary tumor model Her2, tumors that contained inherently slower doubling times possessed lower XBP1-luc activity compared to Tag-Luc mice (Sup. Table 1; Sup. Fig. 7D&E). While the relationship between lower luciferase signal and tumor growth in the Her2 model compared with the Tag model is an over interpretation of the data, these observations are consistent with other reports that tumors with intact ER stress responses grow faster. Furthermore, our data suggests that faster growing tumors likely have less time to adapt to their blood supply leading to increased hypoxia and nutrient deprivation, a microenvironment consistent with increased ER stress.
Although tumor growth correlated with higher XBP1-luc signal, our observations did not address whether increased XBP1 activity caused increased tumor growth, or did increased tumor growth caused more ER stress. Our data more likely suggests the latter as increased tumor growth caused more ER stress associated with increased hypoxia and less glucose uptake as the tumor outgrows its blood and nutrient supply. Given these microenvironmental stresses, tumors are also more likely to have increased apoptotic proteins such as CHOP and increased cell death. Indeed, we observed that XBP1-luc co-localized to CHOP expression and tumors with higher XBP1-luc signal had increased cell death (data not shown). This parallels previous experimental and clinical observations where faster growing tumors had more necrosis and high cell loss factors(44). It is important to note that XBP1 activation, per se, did not likely lead to cell death. Rather, such an environment may have activated a separate ATF4-CHOP pathway resulting in cell death. Nevertheless, intact UPR pathways are important for cell growth as loss of the ER stress protein BiP in spontaneous mammary carcinomas resulted in slower growing tumors (17). In addition, cancer cells deficient in ATF4 or XBP1 grew slower as xenograft tumor models (15, 18). Finally, ER stress and the UPR activation was associated with higher grade breast carcinomas consistent with the observation that faster growing tumors are under more ER stress (45). Therefore, ER stress is likely a marker for faster growing tumors experiencing increased microenvironmental stressors. Thus, increased XBP1 activation may be a consequence of faster tumor growth rate rather than an enabling event for increased tumor proliferation.
In conclusion, we have developed a novel transgenic model to study ER stress caused by the tumor microenvironment of spontaneous breast tumors. This aspect of the tumor microenvironment is distinct between primary tumors and the reflects metabolic properties and growth rates of tumors. Understanding the fundamental biological processes that contribute to tumor microenvironmental stresses will result in the development of cancer therapies that target the tumor microenvironment.
Supplementary Material
Acknowledgments
We thank the Stanford Small Animal Imaging Facility including Tim Doyle, the Stanford Department of Comparative Medicine and the Stanford Transgenic Animal Core Facility for help with experiments. We thank Xiaoli Shi, Jing Wang, Marianne Powell members of the Giaccia, Koong and Le labs for assistance.
Abbreviations
- 2-DG
2-deoxy-d-glucose
- UPR
unfolded protein response
- XBP
X-box binding protein-1
Footnotes
The authors report no conflict of interest.
This work was supported by PO1 CA67166 (ACK, QTL, NCD), RSNA Research Grant RR0812 (M.T.S.). M.T.S. is a recipient of the Holman Research Pathway.
References
- 1.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 2.Hu M, Polyak K. Molecular characterisation of the tumour microenvironment in breast cancer. Eur J Cancer. 2008;44:2760–5. doi: 10.1016/j.ejca.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polyak K, Haviv I, Campbell IG. Co-evolution of tumor cells and their microenvironment. Trends Genet. 2009;25:30–8. doi: 10.1016/j.tig.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg RA. Coevolution in the tumor microenvironment. Nat Genet. 2008;40:494–5. doi: 10.1038/ng0508-494. [DOI] [PubMed] [Google Scholar]
- 5.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Denko NC, Fontana LA, Hudson KM, et al. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene. 2003;22:5907–14. doi: 10.1038/sj.onc.1206703. [DOI] [PubMed] [Google Scholar]
- 7.Graeber TG, Osmanian C, Jacks T, et al. Hypoxia-mediated selection of cells with diminished apoptotic potential in solid tumours. Nature. 1996;379:88–91. doi: 10.1038/379088a0. [DOI] [PubMed] [Google Scholar]
- 8.Feldman DE, Chauhan V, Koong AC. The unfolded protein response: a novel component of the hypoxic stress response in tumors. Mol Cancer Res. 2005;3:597–605. doi: 10.1158/1541-7786.MCR-05-0221. [DOI] [PubMed] [Google Scholar]
- 9.Koumenis C, Wouters BG. “Translating” tumor hypoxia: unfolded protein response (UPR)-dependent and UPR-independent pathways. Mol Cancer Res. 2006;4:423–36. doi: 10.1158/1541-7786.MCR-06-0150. [DOI] [PubMed] [Google Scholar]
- 10.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–58. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 11.Fujimoto T, Onda M, Nagai H, Nagahata T, Ogawa K, Emi M. Upregulation and overexpression of human X-box binding protein 1 (hXBP-1) gene in primary breast cancers. Breast Cancer. 2003;10:301–6. doi: 10.1007/BF02967649. [DOI] [PubMed] [Google Scholar]
- 12.Fujimoto T, Yoshimatsu K, Watanabe K, et al. Overexpression of human X-box binding protein 1 (XBP-1) in colorectal adenomas and adenocarcinomas. Anticancer Res. 2007;27:127–31. [PubMed] [Google Scholar]
- 13.Romero-Ramirez L, Cao H, Regalado MP, et al. X box-binding protein 1 regulates angiogenesis in human pancreatic adenocarcinomas. Transl Oncol. 2009;2:31–8. doi: 10.1593/tlo.08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee E, Nichols P, Spicer D, Groshen S, Yu MC, Lee AS. GRP78 as a novel predictor of responsiveness to chemotherapy in breast cancer. Cancer Res. 2006;66:7849–53. doi: 10.1158/0008-5472.CAN-06-1660. [DOI] [PubMed] [Google Scholar]
- 15.Bi M, Naczki C, Koritzinsky M, et al. ER stress-regulated translation increases tolerance to extreme hypoxia and promotes tumor growth. EMBO J. 2005;24:3470–81. doi: 10.1038/sj.emboj.7600777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carrasco DR, Sukhdeo K, Protopopova M, et al. The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell. 2007;11:349–60. doi: 10.1016/j.ccr.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong D, Ni M, Li J, et al. Critical role of the stress chaperone GRP78/BiP in tumor proliferation, survival, and tumor angiogenesis in transgene-induced mammary tumor development. Cancer Res. 2008;68:498–505. doi: 10.1158/0008-5472.CAN-07-2950. [DOI] [PubMed] [Google Scholar]
- 18.Romero-Ramirez L, Cao H, Nelson D, et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004;64:5943–7. doi: 10.1158/0008-5472.CAN-04-1606. [DOI] [PubMed] [Google Scholar]
- 19.Calfon M, Zeng H, Urano F, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–6. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 20.Lee AH, Iwakoshi NN, Glimcher LH. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol. 2003;23:7448–59. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen X, Ellis RE, Lee K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 23.Back SH, Lee K, Vink E, Kaufman RJ. Cytoplasmic IRE1alpha-mediated XBP1 mRNA splicing in the absence of nuclear processing and endoplasmic reticulum stress. J Biol Chem. 2006;281:18691–706. doi: 10.1074/jbc.M602030200. [DOI] [PubMed] [Google Scholar]
- 24.Brunsing R, Omori SA, Weber F, et al. B- and T-cell development both involve activity of the unfolded protein response pathway. J Biol Chem. 2008;283:17954–61. doi: 10.1074/jbc.M801395200. [DOI] [PubMed] [Google Scholar]
- 25.Iwawaki T, Akai R. Analysis of the XBP1 splicing mechanism using endoplasmic reticulum stress-indicators. Biochem Biophys Res Commun. 2006;350:709–15. doi: 10.1016/j.bbrc.2006.09.100. [DOI] [PubMed] [Google Scholar]
- 26.Iwawaki T, Akai R, Kohno K, Miura M. A transgenic mouse model for monitoring endoplasmic reticulum stress. Nat Med. 2004;10:98–102. doi: 10.1038/nm970. [DOI] [PubMed] [Google Scholar]
- 27.Kamimura D, Bevan MJ. Endoplasmic reticulum stress regulator XBP-1 contributes to effector CD8+ T cell differentiation during acute infection. J Immunol. 2008;181:5433–41. doi: 10.4049/jimmunol.181.8.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morimoto N, Oida Y, Shimazawa M, et al. Involvement of endoplasmic reticulum stress after middle cerebral artery occlusion in mice. Neuroscience. 2007;147:957–67. doi: 10.1016/j.neuroscience.2007.04.017. [DOI] [PubMed] [Google Scholar]
- 29.Ron D. Targeting of mRNAs to their sites of unconventional splicing in the unfolded protein response. Mol Cell. 2009;34:133–4. doi: 10.1016/j.molcel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 30.Yanagitani K, Imagawa Y, Iwawaki T, et al. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- 31.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–75. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshida H, Uemura A, Mori K. pXBP1(U), a negative regulator of the unfolded protein response activator pXBP1(S), targets ATF6 but not ATF4 in proteasome-mediated degradation. Cell Struct Funct. 2009;34:1–10. doi: 10.1247/csf.06028. [DOI] [PubMed] [Google Scholar]
- 33.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol Cell Biol. 1992;12:954–61. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairns RA, Papandreou I, Sutphin PD, Denko NC. Metabolic targeting of hypoxia and HIF1 in solid tumors can enhance cytotoxic chemotherapy. Proc Natl Acad Sci U S A. 2007;104:9445–50. doi: 10.1073/pnas.0611662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci U S A. 1992;89:10578–82. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu M, Yao J, Carroll DK, et al. Regulation of in situ to invasive breast carcinoma transition. Cancer Cell. 2008;13:394–406. doi: 10.1016/j.ccr.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kidd EA, Grigsby PW. Intratumoral metabolic heterogeneity of cervical cancer. Clin Cancer Res. 2008;14:5236–41. doi: 10.1158/1078-0432.CCR-07-5252. [DOI] [PubMed] [Google Scholar]
- 38.Kidd EA, Spencer CR, Huettner PC, et al. Cervical cancer histology and tumor differentiation affect 18F-fluorodeoxyglucose uptake. Cancer. 2009;115:3548–54. doi: 10.1002/cncr.24400. [DOI] [PubMed] [Google Scholar]
- 39.Lee SW, Nam SY, Im KC, et al. Prediction of prognosis using standardized uptake value of 2-[(18)F] fluoro-2-deoxy-d-glucose positron emission tomography for nasopharyngeal carcinomas. Radiother Oncol. 2008;87:211–6. doi: 10.1016/j.radonc.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 40.Rajendran JG, Mankoff DA, O’Sullivan F, et al. Hypoxia and glucose metabolism in malignant tumors: evaluation by [18F]fluoromisonidazole and [18F]fluorodeoxyglucose positron emission tomography imaging. Clin Cancer Res. 2004;10:2245–52. doi: 10.1158/1078-0432.ccr-0688-3. [DOI] [PubMed] [Google Scholar]
- 41.Lunt SJ, Kalliomaki TM, Brown A, Yang VX, Milosevic M, Hill RP. Interstitial fluid pressure, vascularity and metastasis in ectopic, orthotopic and spontaneous tumours. BMC Cancer. 2008;8:2. doi: 10.1186/1471-2407-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kidd EA, Siegel BA, Dehdashti F, Grigsby PW. The standardized uptake value for F-18 fluorodeoxyglucose is a sensitive predictive biomarker for cervical cancer treatment response and survival. Cancer. 2007;110:1738–44. doi: 10.1002/cncr.22974. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz DL, Rajendran J, Yueh B, et al. FDG-PET prediction of head and neck squamous cell cancer outcomes. Arch Otolaryngol Head Neck Surg. 2004;130:1361–7. doi: 10.1001/archotol.130.12.1361. [DOI] [PubMed] [Google Scholar]
- 44.Pavelic ZP, Porter CW, Allen LM, Mihich E. Cell population kinetics of fast- and slow-growing transplantable tumors derived from spontaneous mammary tumors of the DBA/2 Ha-DD mouse. Cancer Res. 1978;38:1533–8. [PubMed] [Google Scholar]
- 45.Davies MP, Barraclough DL, Stewart C, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–8. doi: 10.1002/ijc.23479. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.