Abstract
Transforming growth factor-β (TGF-β) regulates epithelial tissue homeostasis by activating processes that control cell cycle arrest, differentiation and apoptosis. Disruption of TGF-β signaling pathway often occurs in colorectal cancers. Previously, we have shown that TGF-β induces apoptosis through the transcription factor Smad3. Affymetrix oligonucleotide microarrays were used to identify TGF-β/Smad3 target genes that regulate apoptosis in rat intestinal epithelial cells (RIE-1). We found that TGF-β repressed the expression of the inhibitor of differentiation (Id) gene family. Knockdown of Id1 and Id2 gene expression induced apoptosis in RIE cells, whereas over-expression of Id2 attenuated TGF-β-induced apoptosis. TranSignal™ Protein/DNA arrays were used to identify hypoxia-inducing factor-1 (HIF-1) as a downstream target of TGF-β. HIF-1 is a bHLH protein, and over-expression of Id2 blocked HIF-1 activation by TGF-β. Furthermore, knockdown of HIF-1 blocked TGF-β-induced apoptosis. Thus, we have identified HIF-1 as a novel mediator downstream of Id2 in the pathway of TGF-β-induced apoptosis.
Keywords: Apoptosis, Affymetrix oligonucleotide microarrays, Inhibitor of differentiation, TranSignal™, Protein/DNA arrays, Hypoxia-inducing factor
INTRODUCTION
The gastrointestinal epithelium is one of the most dynamic tissues in the adult organism. Renewal of gut epithelium is characterized by a rapid turnover, requiring three to eight days for complete replacement of the epithelium. Renewal of the epithelium depends upon several critical cellular processes, including the cell proliferation within the epithelial crypts, differentiation of transit amplifying cells into mature epithelial enterocytes, and cellular elimination due to apoptosis and exfoliation. These processes are tightly regulated by a number of homeostatic mechanisms, among which TGF-β plays a major role by controlling the relative rates of proliferation and elimination of epithelial cells (Babyatsky and Podolsky, 1991; Ko et al., 1997). TGF-β exerts its biological effects through a cell surface receptor complex, the TGF-β type I and type II receptors TβRI and TβRII. Upon ligand binding, TβRII phosphorylates TβRI, which subsequently phosphorylates Smad2 and Smad3. Phosphorylated Smad2 and Smad3 form a heteromeric complex with Smad4, translocate into the nucleus, and regulate transcription of target genes (Heldin et al., 1997; Massague, 1998), including Id (inhibitor of differentiation or inhibitor of DNA-binding) genes.
The Id proteins are a family of helix-loop-helix (HLH) proteins, which lack the basic DNA-binding domain that is characteristic of other members of this superfamily. Ids function in a dominant negative manner by binding and sequestering basic HLH (bHLH) transcription factors, thereby blocking the binding of bHLH proteins to DNA (Norton, 2000). Four mammalian Ids, Id1, Id2, Id3 and Id4, have been identified, which are expressed in undifferentiated and proliferating cells (Hasskarl and Munger, 2002). Through binding bHLH proteins, Id proteins regulate a variety of cellular processes, including cellular growth, senescence, differentiation, apoptosis, angiogenesis, and neoplastic transformation.
Hypoxia-inducible factor HIF-1, a bHLH protein, was initially identified as a transcription factor that regulates erythropoietin gene expression in response to anemia or hypoxia (Semenza et al., 1991). HIF-1 is a heterodimeric transcription factor consisting of an oxygen-sensitive α subunit and a constitutive β subunit, with apparent molecular masses of 120–130 kD and 91–94 kD, respectively (Wang and Semenza, 1995). Nuclear sequence analysis revealed that both subunits contain bHLH and PAS domains. The bHLH domain mediates dimerization and DNA binding in a large number of transcription factors, the PAS domain provides additional dimerization motif (Semenza, 1999). Three α subunits, HIF-1α, HIF-2α and HIF-3α, have been identified, all of which can dimerize with HIF-1β (or ARNT, aryl hydrocarbon receptor nuclear translocator), ARNT2 or ARNT3 (Semenza, 1999). The heterodimeric complex constitutes the transcription factor HIF, which binds the hypoxia response element (HRE) containing the consensus sequence 5’ -RCGTG- 3’ to regulate transcription of least 70 effector genes (Wenger et al., 2005). The HIFα subunits differ in expression profiles. HIF-1α is expressed ubiquitously, whereas HIF-2α expression is restricted to endothelial, kidney, heart, lungs, and small intestine (Gordan et al., 2007). HIF-1 and HIF-2 αβ heterodimers function as transcriptional activators of oxygen-regulated target genes, whereas the role of HIF-3α is less clear, and a short splicing form of HIF-3α functions as a transcriptional repressor (Semenza, 1999; Wenger, 2002).
TGF-β has been shown to inhibit Id1, Id2 and Id3 expression in several cell lines (Lasorella et al., 2000; Ling et al., 2002). Repression of these three Id genes constitutes a TGF-β cytostatic program shared by human epithelial cell lines of different tissue origins (Kang et al., 2003). In this study, we used Affymetrix oligonucleotide microarrays to identify Id1, Id2 and Id3 as TGF-β targets in rat intestinal epithelial cells (RIE). Small interfering RNAs (siRNAs) were used to investigate the role of Id proteins in TGF-β-induced apoptosis. We report here for the first time that knockdown of Id1 and Id2 caused apoptosis in RIE cells, whereas over-expression of Id2 blocked TGF-β-induced apoptosis. Since Id proteins are generally believed to act as dominant negative inhibitors of bHLH transcription factor activity, we used TranSignal™ Protein/DNA arrays to identify bHLH transcription factors that were regulated by TGF-β. We found that TGF-β activated HIF-1, and over-expression of Id2 blocked this effect. Furthermore, knockdown of HIF-1 blocked TGF-β-induced apoptosis. Thus, we have identified HIF-1 as a novel mediator downstream of Id2 in the pathway of TGF-β-induced apoptosis.
RESULTS
TGF-β induces apoptosis in RIE-1 cells, which is Smad3-dependent
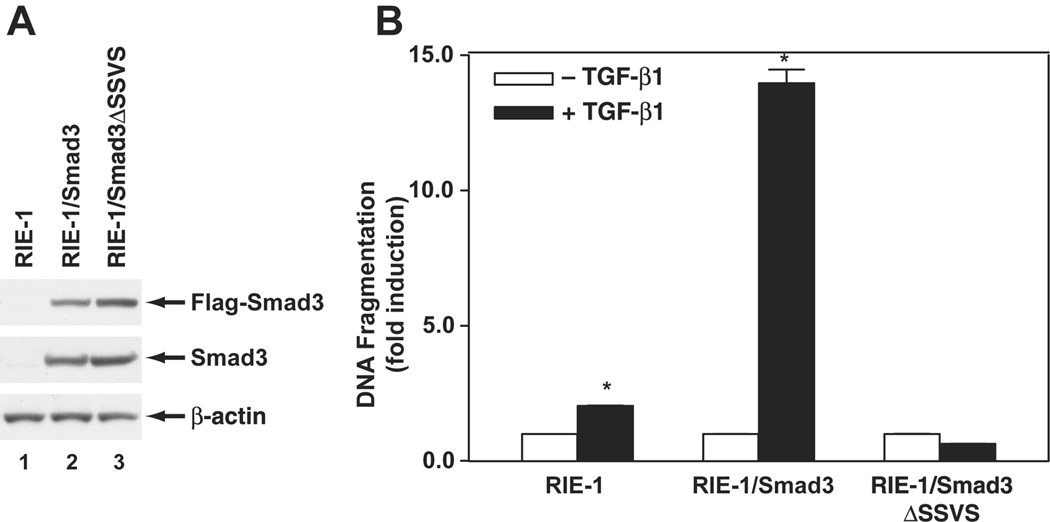
Previously, we have shown that cellular response to TGF-β-induced apoptosis is dependent on the relative activities of Smad3 and Akt (Conery et al., 2004). RIE-1 cells express low levels of Smad3 and are relatively resistant to TGF-β-induced apoptosis when maintained in normal growth medium with 5% serum. However, Akt activity is reduced when RIE-1 cells are maintained at low serum concentrations (0.5%), and apoptosis was induced when TGF-β was added to such cells (Fig. 1). Conversely, a robust apoptotic response was observed in TGF-β-treated RIE/Smad3 cells, which express higher level of Smad3. Furthermore, no apoptosis was detected in RIE cells that express a dominant negative form of Smad3 (RIE-1/Smad3ΔSSVS). These data indicated that Smad3 is the key mediator in apoptosis induction by TGF-β.
Figure 1. Smad3 mediated TGF-β-induced apoptosis.
A, Expression of Smad3 proteins was detected by Western blotting analysis using Flag antibody and Smad3 antibody, β-actin as a loading control. B, Parental RIE-1 cells, RIE-1/Smad3, and RIE-1/Smad3ΔSSVS cells were treated with TGF-β1 at 40 pmol/L in DMEM supplemented with 0.5% dFBS for 24 h. The apoptosis was quantified and expressed as mean ± SEM from triplicates. *p<0.05 compared to the group without TGF-β1 treatment.
TGF-β represses Id expression
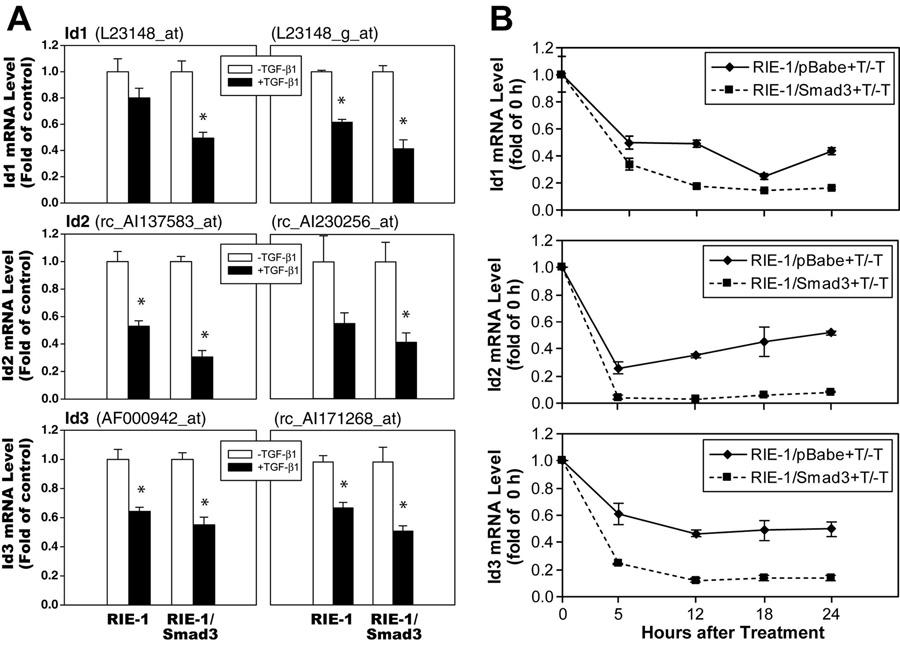
A genomic approach was utilized to compare gene expression profiles of RIE-1 and RIE-1/Smad3 cells with and without TGF-β treatment as previously described (Cao et al., 2007). Gene expression profiles were examined using Affymetrix oligonucleotide microarrays for the rat (RG-U34A, B and C), which include approximately 26,000 rat probe sets with 7,000 known genes and 19,000 ESTs. The microarray analyses revealed that TGF-β treatment decreased the abundance of mRNAs corresponding to Id1, Id2 and Id3 in RIE-1 cells and RIE-1/Smad3 cells. Downregulation of Ids by TGF-β was observed with two probe sets, and was consistently observed in three independent experiments (Fig. 2A). To validate the results of the microarray analyses, we examined the expression of Id mRNA expression by real-time PCR as shown in Fig. 2B. Consistent with the microarray results, TGF-β decreased mRNA expression of Id1, Id2, and Id3 in control RIE-1 cells infected with empty vector retrovirus (RIE-1/pBabe). This inhibitory effect was observed at 5 h and persisted to 24 h. Furthermore, in cells overexpressing Smad3 (RIE-1/Smad3), there was a further decrease of Id mRNA levels in response to TGF-β treatment. These results indicated that TGF-β represses Id mRNA expression in RIE-1 cells and that the response was enhanced by the overexpression of Smad3.
Figure 2. TGF-β repressed Id mRNA expression.
A. Cells were treated with TGF-β1 (120 pmol/L) for 6 h. Total RNA was extracted and Affymetrix Oligonucleotide Microarrays were applied to examine Id1 gene expression with two probes: L23148_at and L23148_g_at; Id2 gene expression with two probes: rc_AI137583_at and rc_AI230256_at; and Id3 gene expression with two probes: AF000942_at and rc_AI171268_at. Three independent experiments were performed. Id mRNA levels were represented as fold of control in TGF-β1 treated groups and expressed as means ± SEM. B. Cells were treated with TGF-β1 (40 pmol/L) and harvested at different time points. Total RNA was extracted and real-time PCR was applied to examine Id expression to confirm Microarray results. Id mRNA levels were represented as fold of vehicle control in TGF-β-treated groups.
Knockdown of Id1 and Id2 induces apoptosis
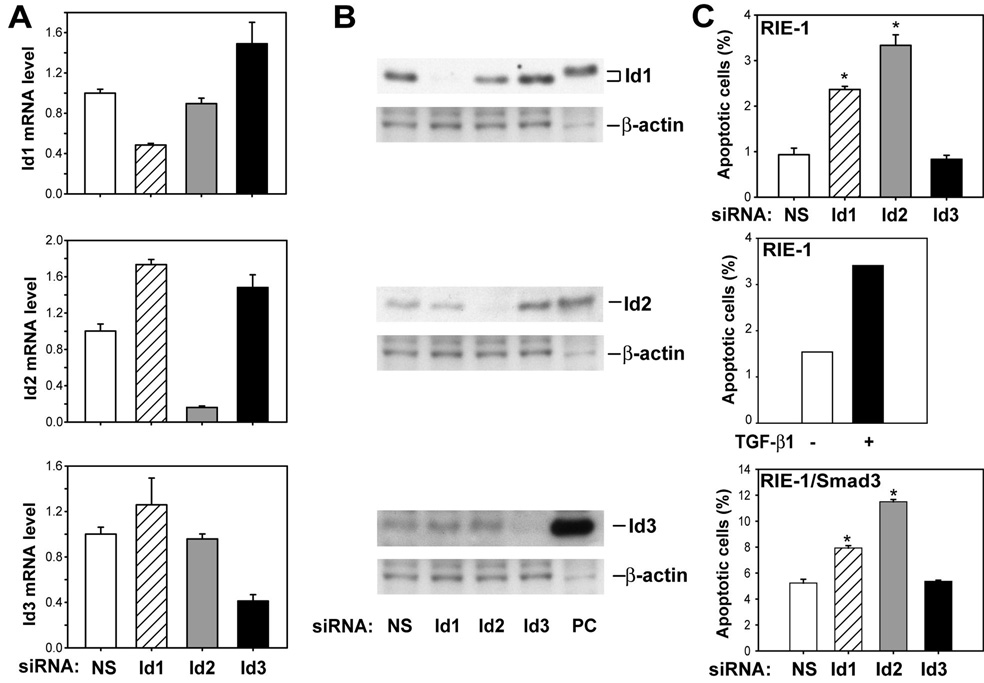
TGF-β induces apoptosis and represses Id expression in gut epithelial cells, suggesting that repression of Id gene expression by TGF-β results in apoptosis induction. This hypothesis predicts that inhibition of Id expression should be sufficient to induce apoptosis. To test this prediction, we used siRNAs specific to rat Id1, Id2, and Id3 to knockdown respective Id gene expression. The specificity and efficiency of Id siRNAs were monitored by real-time PCR (Fig. 3A) and Western blotting (Fig. 3B). Our results showed that Id siRNAs specifically knocked down respective Id gene expression compared to non-specific siRNA. Furthermore, the apoptotic effects of Id1 and Id2 siRNAs were comparable to that of TGF-β in RIE-1 cells (Fig. 3C top and middle panel). Id2 siRNA consistently induced greater apoptosis than that of Id1 siRNA; whereas Id3 siRNA did not induce apoptosis, compared to non-specific siRNA control. Similar effects of Id1 and Id2 siRNAs were observed in RIE-1/Smad3 cells (Fig. 3C). The observation that knockdown of Id1 and Id2 induces apoptosis in RIE-1 and RIE-1/Smad3 suggests that downregulation of Id expression by TGF-β is sufficient to induce apoptosis in RIE cells.
Figure 3. Knockdown of Id1 and Id2 gene expression caused apoptosis.
RIE-1 cells were transfected with rat Id1, Id2 and Id3 siRNA (50 nmol/L) using DharmaFECT3 in DMEM supplemented with 5% dFBS for 20 h and then changed fresh medium supplemented with 0.5% dFBS for 4 h after siRNA transfection. Scrambled siRNA was used as non-specific control (NS). A. Total RNA was prepared and converted to cDNA for real-time quantitative PCR analysis of Id1, Id2 and Id3 mRNA expression. Id mRNA levels were represented as fold of NS control. B. Total cell lysates were prepared and Western blotting was applied to detect Id1, Id2 and Id3 protein expression level using specific anti-Id1, anti-Id2 and anti-Id3 antibodies. The lysates of Cos-1 cells transfected with Id1, Id2 and Id3 expression plasmids were used as positive control (PC). C. RIE-1 and RIE-1/Smad3 cells were harvested and stained with Annexin V-FITC and propidium iodide. The percentage of apoptotic cells stained with Annexin V-FITC only was expressed as mean±SEM. The percentage of apoptotic cells stained with Annexin V-FITC only was analyzed in RIE-1 cells treated with and without TGF-β1. *p<0.05 compared to cells with NS transfection.
Id2 repression by TGF-β is Smad3-dependent
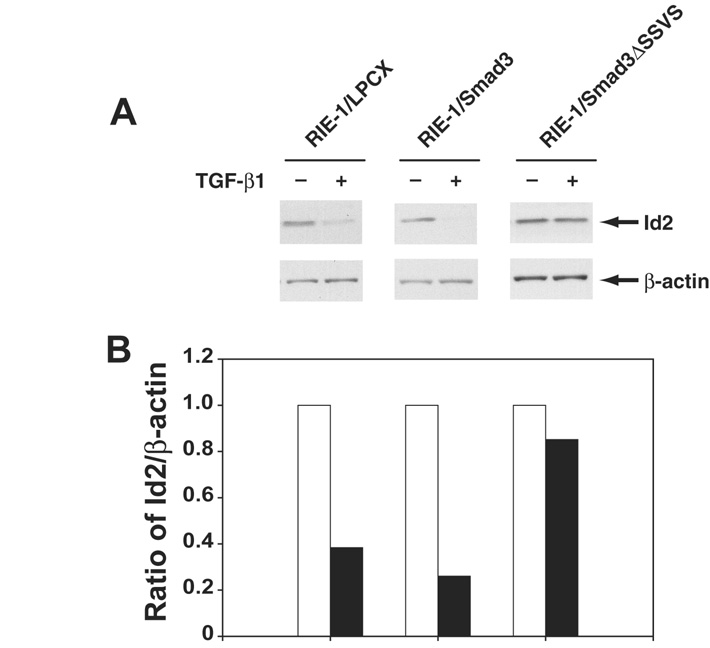
The knockdown assay using Id siRNA revealed differential roles of Id1, Id2, and Id3 in apoptosis induction (Fig. 3) and knockdown of Id2 induced greater apoptosis in both RIE-1 and RIE-1/Smad3 cells. Therefore, we focused our analyses on Id2. Smad proteins are key mediators of TGF-β signaling. If TGF-β is affecting apoptosis through downregulation of Id2, then repression of this gene should depend upon Smad3. We used three cell lines to test this prediction: RIE-1/Smad3 cells, which overexpress Smad3, RIE-1/Smad3ΔSSVS cells, which overexpress dominant negative Smad3, and vector control RIE-1/LPCX cells. TGF-β repressed Id2 protein expression in RIE-1/LPCX and RIE-1/Smad3, confirming our analyses of mRNA abundance. Overexpression of Smad3 enhanced Id2 repression by TGF-β, and dominant negative Smad3 attenuated Id2 repression by TGF-β (Fig. 4). These results indicate that repression of Id2 protein expression by TGF-β is Smad3-dependent.
Figure 4. TGF-β-induced Id repression was Smad3-dependent.
A. RIE-1/LPCX (vector control), RIE-1/Smad3, and RIE-1/Smad3ΔSSVS cells were treated with TGF-β1 (40 pmol/L). The cells were harvested at 24 h. Whole cell lysates were extracted and subjected to Western blot. Id2 protein was measured using specific Id2 antibody, β-actin as a loading control. B. Densitometric analysis was applied for quantification of protein expression from Western blot, and expressed as the percentage of Id2/β-actin.
Over-expression of Id2 inhibits TGF-β-induced apoptosis
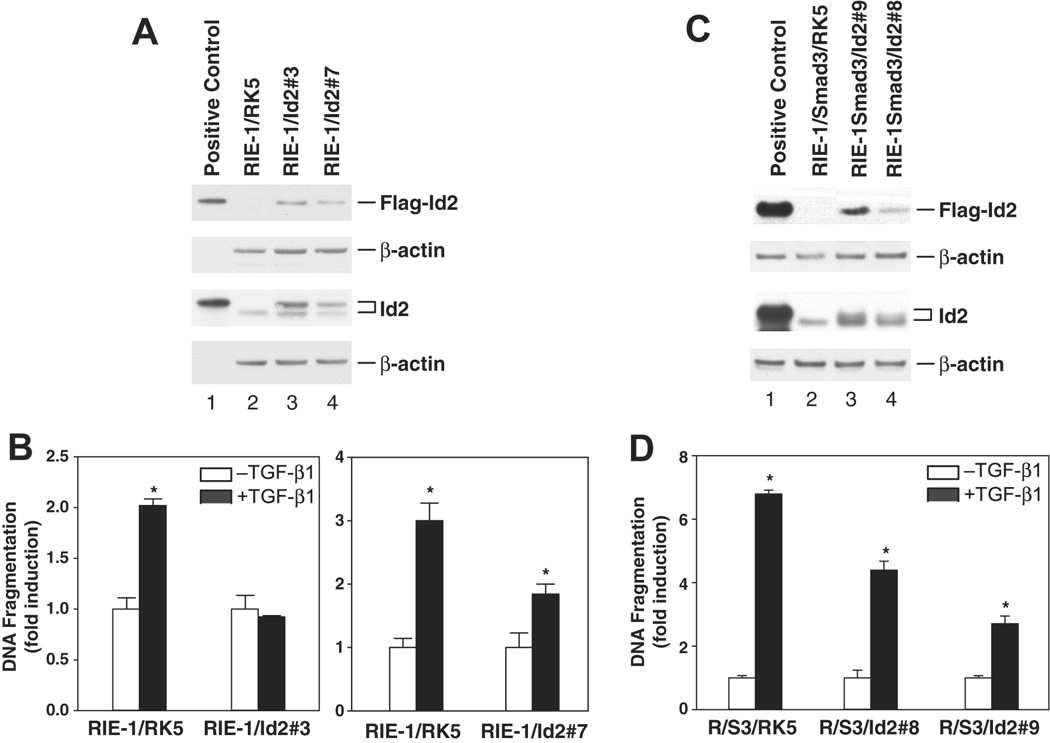
If downregulation of Id2 promotes apoptosis, then overexpression of Id2 should block TGF-β-mediated apoptosis. To test this prediction, we generated stable cell lines that overexpress Id2 in parental cells (RIE-1/Id2) and in RIE-1/Smad3 cells (RIE-1/S3/Id2). Expression of Id2 attenuated TGF-β-induced apoptosis, as shown in two different clones, RIE-1/Id2#3 and RIE-1/Id2#7 (Fig. 5A, 5B). Similar results were obtained with RIE-1/S3/Id2#8 and RIE-1/S3/Id2#9 cells, which over-express both Smad3 and Id2 (Fig. 5C, 5D). These results indicate that downregulation of Id2 is necessary for TGF-β-induced apoptosis in gut epithelial cells.
Figure 5. Overexpression of Id2 inhibited TGF-β-induced apoptosis.
RIE-1 and RIE-1/Smad3 cells were cotranfected with plasmid pRK5-Id2 and pREP4 that carries hygromycin B resistant gene. Stable clones (RIE-1/Id2#3, RIE-1/Id2#7, RIE-1/S3#8, and RIE-1/S3#9) were selected under hygromycin B (200 ug/ml). A, C. Id2 protein level was detected by Western blotting using anti-Id2 and anti-Flag antibodies, RIE-1/RK5 and RIE-1/S3/RK5 as vector control cell lines, β-actin as protein loading control. B, D. Cells were treated with TGF-β1 (40 pM) in DMEM supplemented with 0.5% dFBS for 24 h, apoptosis was analyzed using CDD ELISA kit. DNA fragmentation was represented as fold induction and expressed as mean ± SEM. *p<0.05 compared to the group without TGF-β treatment.
TGF-β activates HIF-1
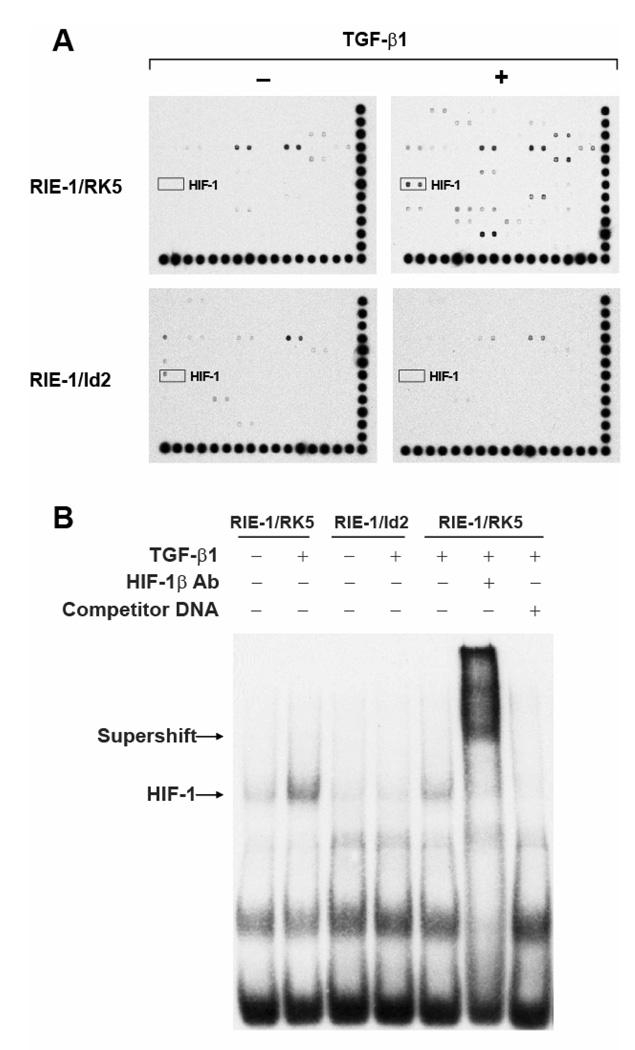
Id2 is a dominant negative inhibitor of bHLH transcription factor, dimerizing with bHLH proteins to prevent DNA binding by such factors. This property of Ids suggests that TGF-β should regulate the activity of bHLH transcription factors. To test this prediction, we undertook a proteomic approach using the TranSignal™ Protein/DNA Array I, II and III to evaluate the effects of TGF-β on DNA binding activity of 244 transcription factors simultaneously within the same sample (54 for Array I, 96 for Array II, and 94 for Array III). Multiple transcription factors were found regulated by TGF-β (Supplemental Table 1). Particularly to our interest, we observed a significant increase in HIF-1 binding (about 3-fold) in nuclear extracts of TGF-β treated RIE-1/RK5 cells, compared to the vehicle control extracts. Moreover, we observed no increase in HIF-1 binding activity when RIE-1/Id2 cells were treated with TGF-β (Fig. 6A). EMSA were performed to validate the result of the transcription factor arrays. An increase in HIF-1 DNA binding was observed in nuclear extracts of RIE-1/RK5 cells treated with TGF-β at 5 h compared to vehicle control (Fig. 6B). However, RIE-1/Id2 cells did not show increased HIF-1 binding activity in the presence of TGF-β. These results were confirmed by DNA precipitation assay (Supplemental Fig. 1). The HIF-1 complexes on EMSA were confirmed by supershift with HIF-1β antibody or competition with cold DNA probe. Our results suggest that TGF-β activates HIF-1 by a mechanism that requires downregulation of Id2.
Figure 6. Id2 over-expression blocked HIF-1 activation by TGF-β.
A. Nuclear extracts of RIE-1/RK5 and RIE-1/Id2 (clone #3) were analyzed using TranSignal™ Protein/DNA Array I, II and III, which include consensus-binding sequences for profiling 54, 96 and 94 transcription factors, respectively. The marked spots were HIF-1. B. Nuclear extracts of RIE-1/RK5 and RIE-1/Id2#3 were analyzed using electrophoretic mobility shift assay (EMSA). The nuclear extracts were incubated with labeled HIF-1 DNA probe, anti-HIF-1β antibody, or an unlabeled competitor DNA as indicated. The mixture was subjected to 6% DNA retardation gel electrophoresis (Invitrogen).
HIF-1 mediates TGF-β-induced apoptosis
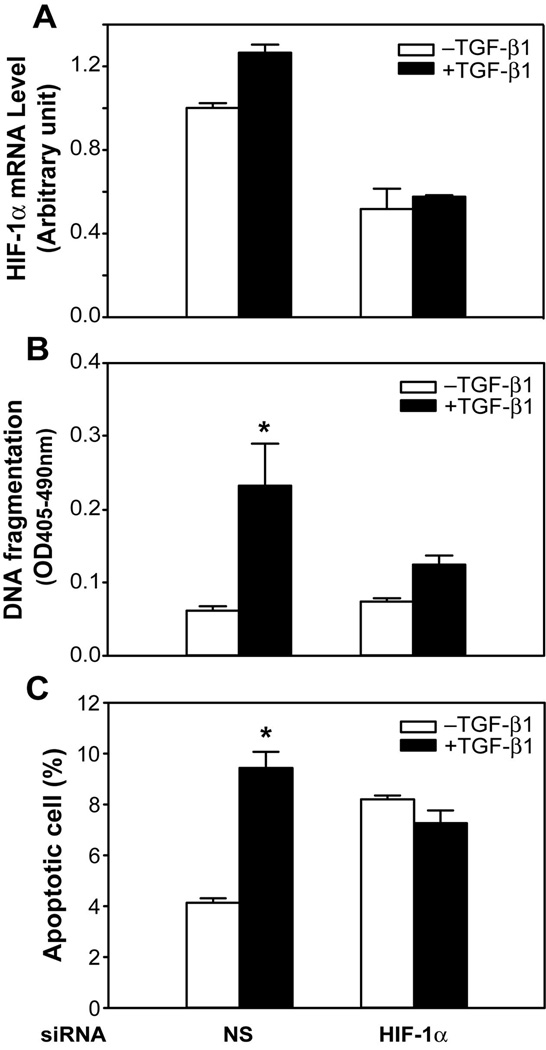
Our data are consistent with the hypothesis that TGF-β downregulates Ids, resulting in activation of HIF-1. HIF-1 is a well-characterized bHLH transcription factor complex that is composed of a heterodimer of two bHLH proteins, the rate-limiting factor HIF-1α and the constitutively expressed HIF-1β. HIF-1 is activated by hypoxia and regulates hypoxia-driven gene expression, and is involved in a broad spectrum of biological functions, including apoptosis. These observations suggest that Id-dependent activation of HIF-1 may play a role in TGF-β-mediated apoptosis. This hypothesis predicts that knockdown of HIF-1 should block TGF-β-mediated apoptosis. To test this prediction we used siRNA knockdown of HIF-1α, the rate-limiting component of the HIF-1 complex. RIE-1/Smad3 were used since these cells have a more robust response to TGF-β and provide a greater dynamic range to evaluate the effects of HIF-1 knockdown on TGF-β-mediated apoptosis. As shown in Fig. 7A, HIF-1α siRNA knocked down HIF-1α mRNA expression in the presence and absence of TGF-β. Furthermore, HIF-1α siRNA blocked TGF-β mediated apoptosis (Fig. 7B and 7C). These observations indicate that HIF-1α is required for TGF-β-induced apoptosis, and are consistent with the hypothesis that TGF-β induces apoptosis by downregulation of Ids, leading to activation of HIF-1, which induces apoptosis.
Figure 7. Knockdown of HIF-1α blocked TGF-β-induced apoptosis in RIE-1/Smad3 cells.
Cells were transfected with rat HIF-1α siRNA (50 nmol/L) using DharmaFECT3 and then replated 24 h after siRNA transfection. Cells were then treated with TGF-β1 (40 pmol/L) in DMEM supplemented with 5% dFBS for 24 h. A. Total RNAs were prepared and cDNAs were converted for real-time PCR analysis of HIF-1α mRNA expression. B. DNA fragmentation was quantified by cell death detection assay. C. The percentage of apoptotic cells stained with Annexin V-FITC only was analyzed by flowcytometry. Results from triplicate wells were expressed as mean of absorbance ± SEM. C. *p<0.05 compared to the group without TGF-β treatment.
DISCUSSION
Previously, we have shown that Smad3 plays an essential role in TGF-β-induced apoptosis (Conery et al., 2004). To gain further insights into TGF-β/Smad3-mediated apoptotic pathway in gut epithelial cells, we utilized a genomic approach to examine potential target genes involved in apoptosis. We found that the inhibitors of differentiation, Id1, Id2 and Id3, were down-regulated by TGF-β. Knockdown of Id1 and Id2 led to apoptosis induction, and over-expression of Id2 inhibited TGF-β-induced apoptosis. Furthermore, using Protein/DNA array, we found the transcription factor HIF-1 was activated by TGF-β, and knockdown of HIF-1 inhibited TGF-β-induced apoptosis. Therefore, we have identified HIF-1 as a novel mediator in TGF-β/Smad3-induced apoptosis.
The profound cytostatic effect that TGF-β exerts on epithelial cells is of particular interest because of its role in tissue homeostasis and the importance that its disruption has in cancer (Gold, 1999; Massague et al., 2000). Genome-wide transcriptional profiling of human epithelial cells using skin keratinocytes, lung epithelial cells, and mammary epithelial cells revealed that repression of Ids is a general feature of the TGF-β cytostatic program (Kang et al., 2003), in which TGF-β enforces homeostasis of epithelia by activating processes such as cell cycle arrest and apoptosis. Current studies mostly focus on the role of Id repression by TGF-β in cell growth suppression, differentiation, and epithelial-mesenchymal transdifferentiation (Kondo et al., 2004; Kowanetz et al., 2004; Siegel et al., 2003). However, the role of Id repression in TGF-β-mediated apoptosis induction has not been addressed previously.
The Id family proteins function as negative regulator of cell differentiation and as positive regulators of G1 cell cycle control (Sikder et al., 2003; Yokota and Mori, 2002; Zebedee and Hara, 2001). Id proteins also drive apoptosis when ectopically over-expressed. However, the pro-apoptotic properties of Id proteins have been mostly reported in cell lines and primary cell models other than epithelial cells, such as fibroblasts (Nakajima et al., 1998; Norton and Atherton, 1998), myeloid progenitors and osteosarcoma cells (Florio et al., 1998), cardiac myocytes (Tanaka et al., 1998), astrocytes (Andres-Barquin et al., 1997), and thymocytes (Kim et al., 1999). There has been a report using constitutively expressed Id1 in mammary epithelial cell cultures showing that Id1 stimulated proliferation in sparse cultures but induced apoptosis in dense cultures (Parrinello et al., 2001). We did not observe apoptosis induction in our study using gut epithelial cell models by over-expression of Id2 protein. These suggest that the pro-apoptotic properties may represent cell-type specific since most of the reports limit to the cells other than epithelial origins. Instead, over-expression of Id2 protein inhibited TGF-β-induced apoptosis, which indicates that repression of Id2 is necessary for apoptosis induction by TGF-β. Furthermore, the observation that Id2 knockdown promotes apoptosis indicates that repression of Ids is sufficient to induce an apoptotic response in RIE cells.
As a member of HLH family lacking basic domain, Id proteins inhibit other bHLH proteins by forming dimmers with bHLH proteins, which are mostly transcription factors. To investigate these transcription factors downstream of Id proteins regulated by TGF-β for the apoptosis induction, we screened 244 transcription factors simultaneously using transcription factor arrays under TGF-β treatment in RIE-1/RK5 cells. Multiple transcription factors were activated by TGF-β (Supplemental Table 1), most notably, a bHLH protein, HIF-1 was significantly activated, which is consistent with the previous report that TGF-β enhanced both AP-1 and HIF-1 DNA binding activities to increase VEGF expression (Shih and Claffey, 2001). Thus, we have identified HIF-1 as a target of TGF-β downstream of Id2. Furthermore, Id2 phyiscially interacts with HIF-1 as shown by immunoprecipitation (IP) assay (Supplement Figure 2) suggesting that Id2 negative regulates HIF-1 activity. Furthermore, to study the role of HIF-1 in TGF-β-induced apoptosis in rat intestinal epithelial cells, we used siRNA to knockdown HIF-1α gene expression, the inducible subunit of HIF-1, and demonstrated that knockdown of HIF-1α inhibited TGF-β-induced apoptosis, suggesting that HIF-1α mediates TGF-β-induced apoptosis at least partially. We speculate that in addition to HIF-1α, other isoforms of HIF may also be involved in TGF-β-induced apoptosis.
Our model is a normal, physiological response by which TGF-β induces apoptosis through repression of Id2 and activation of HIF-1. However, in hypoxia condition, Id1 and Id2 upregulated by HIF-1 could play a significant role in dedifferentiation of hypoxic neuroblastoma cells, which could lead to less mature and more aggressive tumors (Lofstedt et al., 2004; Nemetski and Gardner, 2007). In contrast, in a human breast cancer cell model, Id1 enhances HIF-1α protein stability, nuclear localization, and activity (Kim et al., 2007). These conflicting observations may attribute to different cell models and hypoxia conditions used. Therefore, further studies are needed to determine the molecular mechanisms of the crosstalk between Id and HIF-1 in normoxia and hypoxia conditions, which may play a critical role in physiological and pathological processes.
In conclusion, we have identified Id2 as an important down-stream target of TGF-β/Smad3-induced apoptosis in intestinal epithelial cells. We have shown that downregulation of Id2 is both necessary and sufficient for apoptosis in RIE cells. Furthermore, we have determined that activation of HIF-1 accounts, at least in part, for the Id2-dependent apoptotic effects of TGF-β. The TGF-β/Smad signaling pathway has been shown to interact with HIF-1α to regulate expression of erythropoietin, VEGF, and endoglin (Sanchez-Elsner et al., 2001; Sanchez-Elsner et al., 2002; Sanchez-Elsner et al., 2004). Our study of the role of HIF-1 in TGF-β-induced apoptosis suggests a novel cross-talk between TGF-β and HIF-1 in the regulation of apoptosis to control the turnover of gut epithelial cells and to prevent the development of gut diseases.
MATERIALS AND METHODS
Cell lines
Rat intestinal epithelial cells, RIE-1, were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Mediatech Inc. VA) supplemented with 5% dialyzed fetal bovine serum (dFBS, Invitrogen Co., CA). RIE-1/Smad3 stable cell line was generated by infecting RIE-1 cells with retrovirus pBabe that encodes human Smad3 gene (Conery et al., 2004), and RIE-1/pBabe as a vector control. RIE-1/Smad3ΔSSVS stable cell line was generated by infecting RIE-1 cells with retrovirus pLPCX that encodes a dominant negative human Smad3 gene with C-terminal deletion of SSVS (Choy et al., 2000), and RIE-1/LPCX as a vector control. These cells were maintained in DMEM supplemented with 5% dFBS and 5 µg/ml of puromycin (Calbiochem-Novabiochem Co., CA). RIE-1/Id2 and RIE-1/S3/Id2 (RIE-1/Smad3/Id2) stable cell lines were generated by co-transfecting RK5-rat Id2 plasmid and pREP4 (contains hygromycin B resistant gene), and selected under 200 µg/ml of hygromycin B for RIE-1/Id2 cells, 200 µg/ml of hygromycin B and 5 µg/ml of puromycin for RIE-1/S3/Id2 cells. RIE-1/RK5 and RIE-1/S3/RK5 served as vector controls respectively. Cells were grown in 5% CO2 atmosphere at 37°C with 95% humidity.
Western blotting
Cultured cells were lysed in 1X cell lysis buffer (Cell Signaling Technology, Inc., MA). Protein concentrations of cell lysates were quantified using a protein assay dye (Bio-Rad Laboratories, CA). Western blotting was performed as previously described (Cao et al., 2007; Nguyen et al., 2006). Anti-Id antibodies were from Santa Cruz Biotechnology, Inc., CA, anti-Flag and anti-β-actin antibodies were from Sigma-Aldrich Inc., MO, anti-Smad3 was from Invitrogen Co., and anti-HIF-1α antibody was from R&D Systems. Horseradish peroxidase (HRP) conjugated goat anti-rabbit and anti-mouse antibodies were from Bio-Rad Laboratories.
Apoptosis assays
DNA fragmentation was quantified by cell death detection ELISA assay (Roche Molecular Biochemicals, CA) according to the manufacturer’s instructions as previously described (Cao et al., 2007). Apoptotic cells were stained with Annexin V-FITC and propidium iodide using BD ApoAlert™ Annexin V-FITC Apoptosis kit (BD Bioscience, CA). Cells were analyzed by flow cytometry and cells stained only with Annexin V were identified as apoptotic cells as previously described (Cao et al., 2006).
Microarray analysis
Microarray analysis was performed as previously described (Cao et al., 2007). Briefly, confluent RIE-1 and RIE-1/Smad3 cells were incubated in DMEM supplemented with 5% dFBS for 24 h and then in serum-free DMEM for 48 h. Cells were trypsinized and split into two 100-mm tissue culture plates. In the presence of 2 mmol/L thymidine, cells were maintained in DMEM supplemented with 5% dFBS for 16 h and then treated with or without 120 pmol/L of TGF-β1 (R&D Systems, MN) for 6 h. The University of Texas Medical Branch Genomics Core Facility carried out RNA extraction, labeling, hybridization, and scanning.
Real-time quantitative PCR
Real-time quantitative PCR was described previously (Cao et al., 2007; Nguyen et al., 2006). For detecting rat Id1, Id2, and Id3 mRNA transcripts, the following primers and probes designed from rat cDNA sequences (synthesized by Applied Biosystems, CA) were used: Id1 forward primer 5’-ctacgacatgaacggctgctac-3’, reverse primer 5’-gcggttctgaggcagggt-3’, and probe FAM (6-carboxyfluorescein)-5’-cacgcctcaaggagctggtgcc-3’-TAMRA (6-carboxy-tetramethylrhodamine); Id2 forward primer 5’-ggaccacagcttgggcat-3’, reverse primer 5’-tagagcagactcatcgggtcg-3’, and probe FAM (6-carboxyfluorescein)-5’-tcccggagcaaaaccccggt-3’-TAMRA (6-carboxytetramethylrhodamine); Id3 forward primer 5’-ctcagcttagccaggtggaaa-3’, reverse primer 5’-tctgccagaaccacttgaagg-3’, and probe FAM (6-carboxyfluorescein)-5’-ctgcagcgtgtcatagactacatcctc-3’-TAMRA (6-carboxy-tetramethylrhodamine). For detecting rat β-actin mRNA transcripts, the following primers and probe were used: forward primer 5’-cgtgaaaagatgacccagatca -3’ (locating in exon 3), reverse primer 5’-cacagcctggatggctacgt -3’ (locating in exon 4), and probe 6 FAM-5’-tgagaccttcaacaccccagccatg-3’-TAMRA (spanning exon 3 and 4).
Id and HIF-1α gene silencing
Rat Id1 siRNA (L-080165-01), Id2 siRNA (M-080063-00), Id3 siRNA (L-080095-01), HIF-1α siRNA (L-091718-00, the siGENOME SMARTpool reagents from Dharmacon Inc.) and the transfection reagent DharmaFECT 3 (Dharmacon Inc.) were used according to the manufacturer’s instructions, as previously described (Cao et al., 2007). Scrambled siRNA (D-001206-13, Dharmacon Inc.) was used as a nonspecific siRNA control.
Transcription factor array
TranSignal™ Protein/DNA array I, II and III were obtained from Panomics, Inc., CA. Biotin-labeled DNA binding oligonucleotides (TranSignal probe mix) were incubated with 25 µg of nuclear extracts (prepared using nuclear and cytoplasmic extraction reagents, Pierce Biotechnology Inc., IL) for 30 min to allow the formation of protein/DNA (or transcription factor/DNA) complexes. The protein/DNA complexes were then separated from the free probes by column chromatography. The probes present in the complexes were eluted and hybridized to the TranSignal Array membrane overnight at 42°C. Membranes were incubated with horseradish peroxidase-labeled streptavidin followed by addition of chemiluminescence substrate. Signals were obtained after exposure to x-ray film. Kodak 1D was used for quantification of the signals. Signals were normalized to untreated controls for each cell line, any spots with ≥1.5-fold increase or decrease were considered significant.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed using HIF-1 probe generated from biotinylated oligonucleotides containing HIF-1 binding element synthesized as 5'-CACAGTGCATACGTGGGCTTCCACA-3' and annealed with unbiotinylated complementary oligonucleotides as 5'-TGTGGAAGCCCACGTATGCAC TGTG-3' (Integrated DNA Technologies, Coralville, IA) (Levy et al., 1995). The HIF-1 probe was incubated with 5 µg of nuclear extracts. Excess unlabeled competitor oligonucleotide was added to the reaction mixture when required. The protein/DNA complexes were separated on 6% non-denaturing polyacrylamide DNA retardation gel (Invitrogen). The gel was transferred to a nylon membrane (Ambion) and detected using streptavidin-HRP and a chemiluminescent substrate by exposure to x-ray film. The shifted bands corresponding to the protein/DNA complexes were visualized relative to the unbound DNA. For supershift assay, 2 µg anti-HIF-1β antibody (Novus Biologicals, Littleton, CO) was incubated with nuclear lysate for 45 min at room temperature before the addition of biotinylated HIF-1 probe.
DNA Precipitation Assay
DNA precipitation assay was performed as described (Alliston et al., 2005). Biotinylated 5' oligonucleotides containing HIF-1 binding element were synthesized and annealed with unbiotinylated complementary oligonucleotides to generate wild type probe 5'-AGCTTGCCCTACGTGCTGTCTAGA-3'. A mutant probe 5'-AGCTTGCCCTAAAAGCTGTCTAGA-3' was also synthesized. Briefly, immobilization of biotinylated oligonucleotide probes and absorption of nuclear extracts were carried out using Dynabeads (Dynal) following the manufacturer’s instructions. After extensive washing, DNA-bound proteins were subjected to SDS-PAGE, followed by Western blotting. In parallel, the nuclear lysates were immunoblotted to demonstrate the protein levels.
Immunoprecipitation
Nuclear lysates were prepared from RIE-1/RK5 or RIE-1/Id2 cells using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Pierce, Rockford, IL). FLAG-tagged proteins were immunoprecipitated from nuclear lysates with Anti-FLAG (M2; Sigma-Aldrich) using magnetic Dynabeads Protein G (Invitrogen).
Statistical analysis
Data were expressed as means ± S.E.M. Differences between groups were analyzed by ANOVA with Tukey-Kramer multiple comparisons test, p< 0.05 is considered significant. All experiments were repeated at least twice and similar results were obtained.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank D. Deng, Dr. D. Song, and Z. Chen for technical support; Dr. H. Guo for real-time quantitative PCR assay at Real-Time PCR Core Facility, Sealy Center for Cancer Cell Biology, UTMB; Dr. C. Cox Jr. and F. Jimenez in Department of Pediatric Surgery, UTHSC-Houston, and M. Griffin in Flow Cytometry and Cell Sorting Core Facility, UTMB for flow cytometric analysis. E. Figueroa and S. Schuenke in Department of Surgery, UTMB for manuscript preparation. This study was supported by a grant from Gastrointestinal Research Interdisciplinary Program at the University of Texas Medical Branch (Y.C.), and Public Health Service grants R01 DK060105 (T.C.K.), P01 DK035608 (C.M.T. and T.C.K.) and P50GM038529 (T.C.K.).
REFERENCES
- Alliston T, Ko TC, Cao Y, Liang YY, Feng XH, Chang C, et al. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J Biol Chem. 2005;280:24227–24237. doi: 10.1074/jbc.M414305200. [DOI] [PubMed] [Google Scholar]
- Andres-Barquin PJ, Hernandez MC, Hayes TE, McKay RD, Israel MA. Id genes encoding inhibitors of transcription are expressed during in vitro astrocyte differentiation and in cell lines derived from astrocytic tumors. Cancer Res. 1997;57:215–220. [PubMed] [Google Scholar]
- Babyatsky MW, Podolsky DK. Growth and development in the gastrointestinal tract. In: Yamada T, Alpers DH, Owyang C, Powell DW, editors. Textbook of Gastroenterology. Philadelphia: JB Lippincott Co; 1991. pp. 475–501. [Google Scholar]
- Cao Y, Chen L, Zhang W, Liu Y, Papaconstantinou HT, Bush CR, et al. Identification of apoptotic genes mediating TGF-beta/Smad3-induced cell death in intestinal epithelial cells using a genomic approach. Am J Physiol Gastrointest Liver Physiol. 2007;292:G28–G38. doi: 10.1152/ajpgi.00437.2005. [DOI] [PubMed] [Google Scholar]
- Cao Y, Deng C, Townsend CM, Jr, Ko TC. TGF-beta inhibits Akt-induced transformation in intestinal epithelial cells. Surgery. 2006;140:322–329. doi: 10.1016/j.surg.2006.05.006. [DOI] [PubMed] [Google Scholar]
- Choy L, Skillington J, Derynck R. Roles of autocrine TGF-beta receptor and Smad signaling in adipocyte differentiation. J Cell Biol. 2000;149:667–682. doi: 10.1083/jcb.149.3.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conery AR, Cao Y, Thompson EA, Townsend CM, Jr, Ko TC, Luo K. Akt interacts directly with Smad3 to regulate the sensitivity to TGF-beta induced apoptosis. Nat Cell Biol. 2004;6:366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- Florio M, Hernandez MC, Yang H, Shu HK, Cleveland JL, Israel MA. Id2 promotes apoptosis by a novel mechanism independent of dimerization to basic helix-loop-helix factors. Mol Cell Biol. 1998;18:5435–5444. doi: 10.1128/mcb.18.9.5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold LI. The role for transforming growth factor-beta (TGF-beta) in human cancer. Crit Rev Oncog. 1999;10:303–360. [PubMed] [Google Scholar]
- Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11:335–347. doi: 10.1016/j.ccr.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasskarl J, Munger K. Id proteins--tumor markers or oncogenes? Cancer Biol Ther. 2002;1:91–96. doi: 10.4161/cbt.50. [DOI] [PubMed] [Google Scholar]
- Heldin CH, Miyazono K, ten Dijke P. TGF-beta signalling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massague J. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 2003;11:915–926. doi: 10.1016/s1097-2765(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Kim D, Peng XC, Sun XH. Massive apoptosis of thymocytes in T-cell-deficient Id1 transgenic mice. Mol Cell Biol. 1999;19:8240–8253. doi: 10.1128/mcb.19.12.8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Chung H, Yoo YG, Kim H, Lee JY, Lee MO, et al. Inhibitor of DNA binding 1 activates vascular endothelial growth factor through enhancing the stability and activity of hypoxia-inducible factor-1alpha. Mol Cancer Res. 2007;5:321–329. doi: 10.1158/1541-7786.MCR-06-0218. [DOI] [PubMed] [Google Scholar]
- Ko TC, Bresnahan WA, Thompson EA. Intestinal cell cycle regulation. In: Meijer L, Guidet S, Philippe M, editors. Progress in Cell Cycle Research. New York: Plenum Press; 1997. pp. 43–52. [DOI] [PubMed] [Google Scholar]
- Kondo M, Cubillo E, Tobiume K, Shirakihara T, Fukuda N, Suzuki H, et al. A role for Id in the regulation of TGF-beta-induced epithelial-mesenchymal transdifferentiation. Cell Death Differ. 2004;11:1092–1101. doi: 10.1038/sj.cdd.4401467. [DOI] [PubMed] [Google Scholar]
- Kowanetz M, Valcourt U, Bergstrom R, Heldin CH, Moustakas A. Id2 and Id3 define the potency of cell proliferation and differentiation responses to transforming growth factor beta and bone morphogenetic protein. Mol Cell Biol. 2004;24:4241–4254. doi: 10.1128/MCB.24.10.4241-4254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- Levy AP, Levy NS, Wegner S, Goldberg MA. Transcriptional regulation of the rat vascular endothelial growth factor gene by hypoxia. J Biol Chem. 1995;270:13333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- Ling MT, Wang X, Tsao SW, Wong YC. Down-regulation of Id-1 expression is associated with TGF beta 1-induced growth arrest in prostate epithelial cells. Biochim Biophys Acta. 2002;1570:145–152. doi: 10.1016/s0304-4165(02)00189-7. [DOI] [PubMed] [Google Scholar]
- Lofstedt T, Jogi A, Sigvardsson M, Gradin K, Poellinger L, Pahlman S, et al. Induction of ID2 expression by hypoxia-inducible factor-1: a role in dedifferentiation of hypoxic neuroblastoma cells. J Biol Chem. 2004;279:39223–39231. doi: 10.1074/jbc.M402904200. [DOI] [PubMed] [Google Scholar]
- Massague J. TGF-beta signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Nakajima T, Yageta M, Shiotsu K, Morita K, Suzuki M, Tomooka Y, et al. Suppression of adenovirus E1A-induced apoptosis by mutated p53 is overcome by coexpression with Id proteins. Proc Natl Acad Sci U S A. 1998;95:10590–10595. doi: 10.1073/pnas.95.18.10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemetski SM, Gardner LB. Hypoxic regulation of Id-1 and activation of the unfolded protein response are aberrant in neuroblastoma. J Biol Chem. 2007;282:240–248. doi: 10.1074/jbc.M607275200. [DOI] [PubMed] [Google Scholar]
- Nguyen KA, Cao Y, Chen JR, Townsend CM, Jr, Ko TC. Dietary fiber enhances a tumor suppressor signaling pathway in the gut. Ann Surg. 2006;243:619–625. doi: 10.1097/01.sla.0000216783.85214.c1. discussion 625–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton JD. ID helix-loop-helix proteins in cell growth, differentiation and tumorigenesis. J Cell Sci. 2000;113(Pt 22):3897–3905. doi: 10.1242/jcs.113.22.3897. [DOI] [PubMed] [Google Scholar]
- Norton JD, Atherton GT. Coupling of cell growth control and apoptosis functions of Id proteins. Mol Cell Biol. 1998;18:2371–2381. doi: 10.1128/mcb.18.4.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello S, Lin CQ, Murata K, Itahana Y, Singh J, Krtolica A, et al. Id-1, ITF-2, and Id-2 comprise a network of helix-loop-helix proteins that regulate mammary epithelial cell proliferation, differentiation, and apoptosis. J Biol Chem. 2001;276:39213–39219. doi: 10.1074/jbc.M104473200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Botella LM, Velasco B, Corbi A, Attisano L, Bernabeu C. Synergistic cooperation between hypoxia and transforming growth factor-beta pathways on human vascular endothelial growth factor gene expression. J Biol Chem. 2001;276:38527–38535. doi: 10.1074/jbc.M104536200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Botella LM, Velasco B, Langa C, Bernabeu C. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-beta pathways. J Biol Chem. 2002;277:43799–43808. doi: 10.1074/jbc.M207160200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Ramirez JR, Sanz-Rodriguez F, Varela E, Bernabeu C, Botella LM. A cross-talk between hypoxia and TGF-beta orchestrates erythropoietin gene regulation through SP1 and Smads. J Mol Biol. 2004;336:9–24. doi: 10.1016/j.jmb.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annu Rev Cell Dev Biol. 1999;15:551–578. doi: 10.1146/annurev.cellbio.15.1.551. [DOI] [PubMed] [Google Scholar]
- Semenza GL, Nejfelt MK, Chi SM, Antonarakis SE. Hypoxia-inducible nuclear factors bind to an enhancer element located 3' to the human erythropoietin gene. Proc Natl Acad Sci U S A. 1991;88:5680–5684. doi: 10.1073/pnas.88.13.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih SC, Claffey KP. Role of AP-1 and HIF-1 transcription factors in TGF-beta activation of VEGF expression. Growth Factors. 2001;19:19–34. doi: 10.3109/08977190109001073. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Shu W, Massague J. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J Biol Chem. 2003;278:35444–35450. doi: 10.1074/jbc.M301413200. [DOI] [PubMed] [Google Scholar]
- Sikder HA, Devlin MK, Dunlap S, Ryu B, Alani RM. Id proteins in cell growth and tumorigenesis. Cancer Cell. 2003;3:525–530. doi: 10.1016/s1535-6108(03)00141-7. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Pracyk JB, Takeda K, Yu ZX, Ferrans VJ, Deshpande SS, et al. Expression of Id1 results in apoptosis of cardiac myocytes through a redox-dependent mechanism. J Biol Chem. 1998;273:25922–25928. doi: 10.1074/jbc.273.40.25922. [DOI] [PubMed] [Google Scholar]
- Wang GL, Semenza GL. Purification and characterization of hypoxia-inducible factor 1. J Biol Chem. 1995;270:1230–1237. doi: 10.1074/jbc.270.3.1230. [DOI] [PubMed] [Google Scholar]
- Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. Faseb J. 2002;16:1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- Yokota Y, Mori S. Role of Id family proteins in growth control. J Cell Physiol. 2002;190:21–28. doi: 10.1002/jcp.10042. [DOI] [PubMed] [Google Scholar]
- Zebedee Z, Hara E. Id proteins in cell cycle control and cellular senescence. Oncogene. 2001;20:8317–8325. doi: 10.1038/sj.onc.1205092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.