Summary
COPI-coated vesicles form at the Golgi apparatus from two cytosolic components, ARF G-protein and coatomer, a heptameric complex that can polymerize into a cage to deform the membrane into a bud. Although coatomer shares a common evolutionary origin with COPII and clathrin vesicle coat proteins, the architectural relationship among the three cages is unclear. Strikingly, the αβ′-COP core of coatomer crystallizes as a triskelion in which three copies of a β′-COP β-propeller domain converge through their axial ends. We infer that the trimer constitutes the vertex of the COPI cage. Our model proposes that the COPI cage is intermediate in design between COPII and clathrin: COPI shares with clathrin an arrangement of three curved α-solenoid legs radiating from a common center; and COPI shares with COPII highly similar vertex interactions involving the axial ends of β-propeller domains.
Introduction
Vesicle transport pathways operate through rounds of vesicle budding and fusion reactions, and the underlying reaction mechanisms are conserved from yeast to humans. Budding occurs when cytoplasmic coat protein (COP) complexes assemble on a membrane surface, where they capture cargo proteins and polymerize into spherical cages to deform the membrane into a bud. Eukaryotic cells contain a series of COP complexes: COPI which buds vesicles from the Golgi apparatus, COPII which operates at the endoplasmic reticulum, and clathrin/adaptin which is involved in budding from the plasma membrane, trans-golgi network and endosomal compartments (Bonifacino and Glick, 2004).
COPI-coated vesicles form on the Golgi apparatus by the stepwise recruitment of two cytosolic components: an ARF-family G-protein and coatomer, a 550 kDa cytoplasmic complex of seven COPs—α-, β-, β′, γ-, δ-, ε- and ζ-COP (Waters et al., 1991; Serafini et al., 1991). Budding is initiated by the exchange of GDP for GTP on ARF catalyzed by a Golgi-localized guanine-nucleotide exchange factor (GEF) of the Sec7 family (Peyroche et al., 1996; Chardin et al., 1996). ARF-GTP binds to the membrane by embedding an N-terminal α helix in the bilayer, and in turn recruits coatomer through a direct, GTP-dependent interaction (Antonny et al., 1997; Zhao et al., 1997). Coatomer complexes, attached to the membrane surface through ARF-GTP, can bind cargo molecules and then self-assemble to form spherical cages that yield COPI-coated vesicles (Bremser et al., 1999; Nickel et al., 1998; Orci et al., 1993; Spang et al., 1998).
The architecture and functional organization of clathrin and COPII vesicle coats have been studied extensively by electron microscopy (EM) and X-ray crystallography (reviewed by McMahon and Mills, 2004; see Stagg et al., 2007 for a structural comparison). The clathrin cage is built from triskelion assembly units—trimers of clathrin heavy chain—that are centered on the vertices of the cage, and the long α-solenoid legs curve towards and interdigitate with neighboring legs as they extend to the adjacent vertices (Fotin et al., 2004). The N-terminal β-propeller domain of the clathrin heavy chain projects inwards from the cage to interact with the adaptor protein (AP) complex, a tetramer of two large and two smaller proteins that is responsible for cargo binding and membrane apposition (Collins et al., 2002; Heldwein et al., 2004; ter Haar et al., 2000, and reviewed by Owen et al. [2004]). Like clathrin, the COPII cage is formed from α-solenoid and β-propeller protein domains, reflecting their common evolutionary origin, but the cage architecture is quite distinct. The COPII cage assembly unit—two copies of Sec13/31—is a 28 nm long rod comprising a central α-solenoid dimer capped by two β-propeller domains at each end. The rod forms the edge of a cuboctahedron cage, and four rods converge to form the vertex with no interdigitation of assembly units (Stagg et al., 2006; Fath et al., 2007; Stagg et al., 2008).
Far less is known about the architecture of the COPI coat, as neither EM reconstructions of the cage nor crystal structures of coatomer complexes have been reported. The cargo-binding and cage-forming portions of coatomer remain together in cytosol and bind en bloc to Golgi membranes during COPI coat assembly, unlike the stepwise accretion of clathrin and COPII coats (Hara-Kuge et al., 1994). The functional complexes within heptameric coatomer have been identified through biochemical dissection experiments and by homology with AP subunits (Eugster et al., 2000; Lowe and Kreis, 1995; Pavel et al., 1998; Schledzewski et al., 1999). A tetrameric complex, βδ/γζ-COP, is identified as the cargo-binding portion based on its sequence similarity to the AP complex. The cage-forming trimer, the αβ′ε-COP complex, is composed of α-solenoid and β-propeller domains like clathrin and the Sec13/31 cage protein of COPII.
The realization that α-solenoid and β-propeller domains form the building blocks of COPI, COPII and clathrin cage proteins, as well as the central proteins of the nuclear pore complex, led to the proposal of a common evolutionary origin for these membrane-curving devices (Devos et al., 2004). Recent studies have highlighted some structural relationships among these systems, but unifying principles of lattice construction have yet to emerge (Fath et al., 2007; Hsia et al., 2007; Brohawn and Schwartz, 2009). Indeed, the symmetry and domain arrangements of clathrin and COPII cages are so distinct as to call into question the proposal of an underlying relationship. In this study, we describe the characterization and crystal structures of αβ′-COP and αε-COP subcomplexes of the coatomer cage. Based on this analysis we propose a model for the COPI cage based on a triskelion vertex with striking similarities to both COPII and clathrin.
Results and Discussion
Domain Structure of the αβ′ε-COP Coatomer Complex
Coatomer purified from mammalian cytosol is a heptameric protein that can be disassembled using high salt concentrations into stable βδ/γζ-COP and αβ′ε-COP complexes (Lowe and Kreis, 1995). To study the cage-forming αβ′ε-COP complex in isolation, we prepared the trimeric protein from Bos taurus by coexpression in baculovirus-infected insect cells. The purified αβ′ε-COP protein was soluble despite the absence of βδ/γζ-COP, and it migrated as a large (∼600 kDa) particle on size-exclusion chromatography columns. To define domain boundaries and identify protein interactions in the αβ′ε-COP complex, we digested the protein with subtilisin and analyzed the proteolysis products by chromatography, SDS-PAGE and N-terminal sequencing (see Experimental Procedures). Three major protease-resistant products were obtained (see Figure 1): a subcomplex comprising almost full-length β′-COP and a central α-solenoid domain of α-COP; a C-terminal domain of α-COP bound to essentially full-length ε-COP (this interaction was identified previously by Eugster et al. [2000]); and an N-terminal region of α-COP that includes the β-propeller domain. (This final α-COP product was more difficult to define since a loop within the β-propeller domain is nicked by subtilisin in the vicinity of residue 191).
Figure 1. Domain Organization of the αβ′ε-COP Complex.
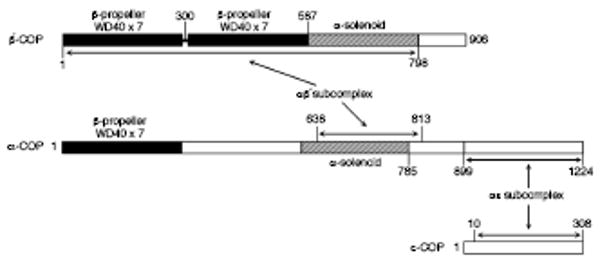
Diagram shows the domain structure of B. taurus αβ′ε-COP, as defined by proteolytic mapping and sequence/structure analysis. The products of limit proteolysis reactions were separated chromatographically to identify discrete subcomplexes and domains (see Experimental Procedures). The two subcomplexes for which crystal structures were determined—αβ′-COP and αε-COP—are indicated with arrows. Note that we determined the crystal structure of the B. taurus αε-COP subcomplex, and the S.cerevisiae αβ′-COP subcomplex comprising β′-COP residues 1-814 (equivalent to B. taurus residues 1-798) and α-COP residues 642-818 (equivalent to B. taurus residues 638-813).
The results of this preliminary analysis suggested that the αβ′-COP and αε-COP subcomplexes are amenable to structural analysis. Indeed, we were able to determine the crystal structures of both subcomplexes, as described below. Together they account for more than two thirds of the mass of the αβ′ε-COP complex.
Crystal Structure Determination
Crystals of the B. taurus αε-COP heterodimer grew in space group I422. The structure was determined by the multi-wavelength anomalous diffraction method using selenium as the anomalous scatterer (Table 1 and Experimental Procedures). The assignment of residues during model building was aided by the selenium atom positions, and the structure was refined with data to 2.6 Å resolution.
Table 1.
Data Collection and Refinement Statistics
| αβ′ε complex: | αβ′ | αβ′ | αβ′ | αβ′ | αβ′ | αε | αε | ||
| Data set: | Native1 | Native2 | Ta6Br121 | Ta6Br122 | Thimerosal | MAD | Native | ||
| PDB accession #: | |||||||||
| Space group: | P3221 | P3221 | P3221 | P3221 | P3221 | I422 | I422 | ||
| Cell parameters a, b, c (Å) | 152.6, 152.6, 294.5 | 153.3, 153.3, 296.0 | 153.3, 153.3, 296.1 | 151.8, 151.8, 294.1 | 152.6, 152.6, 295.6 | 176.9, 176.9, 141.5 | 176.6, 176.6, 141.2 | ||
| Data processing | |||||||||
| Peak | Inflection | Remote | |||||||
| Wavelength (Å) | 0.9790 | 1.0090 | 1.2424 | 1.2424 | 1.0009 | 0.9792 | 0.9794 | 0.9642 | 0.9794 |
| Resolution (Å) | 25-2.5 | 35-3.1 | 35-3.5 | 35-3.4 | 35-3.2 | 35-3.0 | 35-3.0 | 35-3.0 | 50-2.6 |
| Rmerge (%)a | 7.7 (39.9)* | 9.1 (37.9)* | 11.9 (39.2) | 9.0 (32.3) | 8.7 (34.8) | 7.7 (36.0) | 6.9 (45.7) | 8.6 (50.3) | 5.0 (38.3) |
| I/σ | 19.6 (2.8) | 20.7 (4.3) | 8.8 (2.0) | 9.5 (2.4) | 14.8 (3.0) | 13.8 (2.3) | 13.1 (1.9) | 12.1 (1.9) | 28.8 (3.3) |
| Completeness (%0) | 96.3 (92.3) | 96.5 (97.2) | 92.6 (92.3) | 95.3 (97.2) | 93.3 (95.2) | 89.3 (94.1) | 90.0 (94.1) | 85.7 (92.6) | 99.3 (98.8) |
| Redundancy | 3.2 (3.0) | 5.3 (5.3) | 1.9 (1.9) | 1.6 (1.6) | 2.4 (2.4) | 2.1 (2.1) | 2.1 (2.1) | 2.0 (2.0) | 3.9 (3.8) |
| Phasing power | 1.2 | 1.3 | 1.0 | ||||||
| Refinement statistics | |||||||||
| Data range (Å) | 25-2.5 | 30-2.6 | |||||||
| Reflections | 125081 | 32909 | |||||||
| Nonhydrogen atoms | 23463 | 4874 | |||||||
| Water molecules | 270 | 34 | |||||||
| R.m.s. Δ bonds (Å)b | 0.007 | 0.007 | |||||||
| R.m.s. Δ angles (°)b | 1.36 | 1.38 | |||||||
| R-factor (%)c | 22.1 | 22.2 | |||||||
| Rfree (%)c, d | 27.2 | 26.3 | |||||||
Highest resolution shell is shown in parenthesis.
Rmerge = 100 × ΣhΣi | Ii(h) - <I(h)> | / Σh<I(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurement of I(h) for Miller indices h.
Root-mean-squared deviation (r.m.s. Δ) from target geometries.
R-factor = 100 × Σ|FP − FP(calc)|/Σ FP.
Rfree was calculated with 5% of the data.
It proved more difficult to determine the structure of the αβ′-COP subcomplex. Initially, we obtained crystals of αβ′-COP from B. taurus, but they were of low crystallographic quality. Instead, the equivalent αβ′-COP subcomplex from Saccharomyces cerevisiae (residues 1-814 of β′-COP and residues 642-818 of α-COP) crystallized in space group P3221, with three copies of αβ′-COP —a total of 330 kDa—in the asymmetric unit. The structure was determined by the multiple isomorphous replacement method using crystals derivatized with Ta6Br12 and mercury compounds (see Experimental Procedures). The structure of the yeast αβ′-COP subcomplex was refined to 2.5 Å resolution (Table 1).
Structure of the αε-COP Subcomplex
The αβ′-COP subcomplex is the more enlightening of the two crystal structures, so we discuss the αε-COP subcomplex only briefly here.
The molecular models of the two subcomplexes are presented in Figures 2 and 3. The αβ′-COP and αε-COP crystal structures do not share any overlapping elements. Rather the two subcomplexes are connected by a highly acidic ∼80-residue linker region of α-COP (residues 818-900) that may be unstructured according to our proteolysis results (Figure 1). This suggests that the 65 kDa αε-COP subcomplex is flexibly linked to a larger core region of αβ′ε-COP (Figure 3C and D). This is reminiscent of the C-terminal α-solenoid domain of Sec31 in the COPII cage, which is flexibly linked to the assembly unit core and probably projects from the cage in towards the membrane vesicle (Fath et al., 2007).
Figure 2. Structural Analysis of the αβ′-COP Subcomplex.
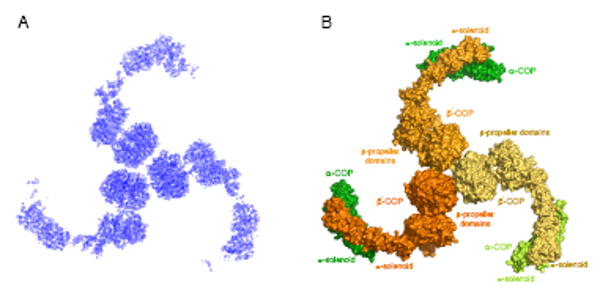
(A) Experimental electron density map (calculated with data to 3.1 Å resolution and contoured at 1.5 σ) of the crystal asymmetric unit of the αβ′-COP subcomplex. This map was calculated with the MIRAS phases following density modification including three-fold non-crystallographic symmetry (NCS) averaging. The map is viewed along the three-fold NCS axis.
(B) Surface representation of the αβ′-COP triskelion. The figure is oriented as in (A), with the three β′-COP subunits colored in three shades of orange, and with the α-COP subunits colored three shades of green. The label “β-propeller domains” denotes to the two β-propeller domains on each copy of β′-COP.
Figure 3. Architecture of the αβ′-COP and αε-COP Subcomplexes.
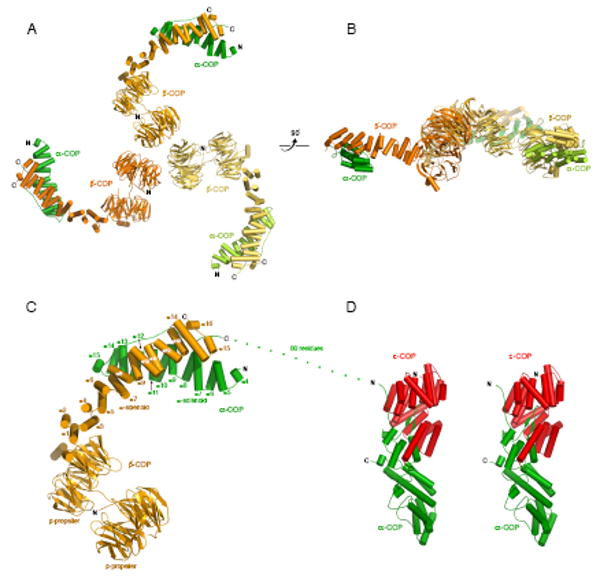
(A) Ribbon diagram of the αβ′-COP triskelion, viewed in the same orientation as Figure 2. Three copies of the αβ′-COP heterodimer associate in the crystal asymmetric unit. We infer that this arrangement corresponds to the vertex of the COPI cage. The crystal structure was determined by MIRAS phasing and refined to 2.5 Å resolution (Table 1). The complex comprises residues 1-814 of β′-COP and residues 642-818 of α-COP (colored as in Figure 2B). β strands are drawn as arrows and α helices as cylinders.
(B) This view of the αβ′-COP triskelion is rotated 90° about a horizontal axis relative to (A).
(C) Close-up view of one copy of the αβ′-COP subcomplex, in the same orientation as the uppermost copy in (A). Starting from its N terminus, the β′-COP subunit (orange) has two β-propeller domains followed by an α-solenoid domain comprising fifteen α helices. In the structure, the α-COP subunit (green) has twelve α helices; these are numbered from helix α4 to highlight the similarity to the solenoid domain of β′-COP (see text for details). See also Figures S1, S3 and S4.
(D) Ribbon diagram of the αε-COP subcomplex drawn as a stereo pair. This structure was determined by MAD phasing and refined to 2.6 Å resolution (Table 1). In the picture α-COP is colored green and ε-COP is red.
The sequence of ε-COP contains tetratricopeptide repeats (TPR), and the αε-COP crystal structure reveals that ε-COP forms a characteristic TPR-protein super-helix (Das, et al., 1998), which coils like a snake around one end of α-COP, to form tight dimer contacts (Figure 3D). Starting from its N-terminus, the TPR domain of ε-COP coils in a right-handed fashion along one end of the 80-Å long rod-shaped α-COP molecule. The TPR domain ends at residue 280, and the C-terminal twenty residues of ε-COP form a final α helix that makes additional contacts with α-COP, so that ε-COP envelops about one-third of the α-COP rod in total. The α-COP C-terminal domain has a mixed tertiary structure of five discrete elements: an N-terminal mostly helical region (residues 915-965); a β-hairpin “finger” (residues 967-983) that is encircled by ε-COP; a three-helix bundle (residues 1003-1074), followed by a short α-solenoid region (residues 1078-1151); and finally a nest of three orthogonally-oriented β-hairpins (residues 1165-1210) toward the C-terminus of the molecule. Overall, the 65 kDa αε-COP subcomplex is a compact rod, 115 Å long and ∼35 Å diameter. Its possible location relative to other elements in the COPI lattice is discussed below.
Architectural Overview of the αβ′-COP Subcomplex
The αβ′-COP subcomplex forms a curved structure composed of the 90 kDa β′-COP molecule and 20 kDa α-COP α-solenoid domain (Figure 1 and 2). β′-COP has the domain arrangement β-propeller–β-propeller–α-solenoid, and three copies of the αβ′-COP subcomplex converge through the N-terminal β-propeller domains to form a triskelion (Figure 2 and 3). The α-solenoid domain of α-COP binds in an anti-parallel manner to the α-solenoid of β′-COP to extend the legs of the triskelion (Figure 3A and C), so that each of the legs is approximately 175 Å along the curved path and 120 Å measured radially (both distances are measured from the triskelion center). The αβ′-COP triskelion does not form around a crystallographic 3-fold axis; rather the three copies of αβ′-COP come together in the asymmetric unit of the crystal. Indeed, the interfacial contacts at the center of the triskelion are not exactly three-fold related, and the long α-solenoid legs deviate incrementally from the three-fold relation the farther they extend from the triskelion center, as might be expected for non-crystallographic symmetry.
Finally, the molecular model we have built for β′-COP most likely includes the entirety of the structured portion of the molecule. A C-terminal element (75-100 residues in yeast and mammalian sequences; see Figure 1) of the β′-COP polypeptide was omitted from the crystallographic study, but we conclude that this region is probably unstructured since it is highly acidic and its sequence is not conserved (indeed, this C-terminal element is absent in β′-COP from Schizosaccharomyces pombe).
Structure and Domain Organization of the αβ′-COP Subcomplex
The arrangement of protein domains in the β′-COP subunit is remarkably similar to that observed in the Sec13/31 complex of the COPII cage (which forms one half of the (Sec13/31)2 assembly unit [Fath et al., 2007]). Sec13/31 adopts a β-propeller–β-propeller–α-solenoid arrangement in which the small Sec13 subunit forms the second β-propeller domain and is thus sandwiched between the N-terminal β-propeller and C-terminal α-solenoid domains of Sec31. The β-propeller domains of β′-COP, like Sec13/31, both comprise seven blades (Figure S1 and 4B). The N-terminal β-propeller (residues 1-301) is characterized by a regular and compact structure involving short connecting loops between the β-strands at both axial ends of the β-propeller. The effect is to create relatively flat axial ends, which seems to be important for interactions at the triskelion center (Figure 3 and 4). The C-terminal β-propeller (residues 304-600) interacts with the N-terminal β-propeller through a relatively small interaction area, whereby loops contributed from blades 1, 2 and 7 of the C-terminal β-propeller interact with residues on loops 5,6 and 7 of the N-terminal β-propeller (see Figure S1 for the numbering scheme). The small interaction area suggests that there might be some flexibility at this site connecting the two β-propellers. Sites of potential flexibility have been identified in the COPII (Sec13/31)2 assembly unit, and these are proposed to be important for lattice adaptability and the formation of different size cages (Lederkremer et al., 2001; Fath et al., 2007; Stagg et al., 2008).
Figure 4. Contact surfaces of the αβ′-COP triskelion.
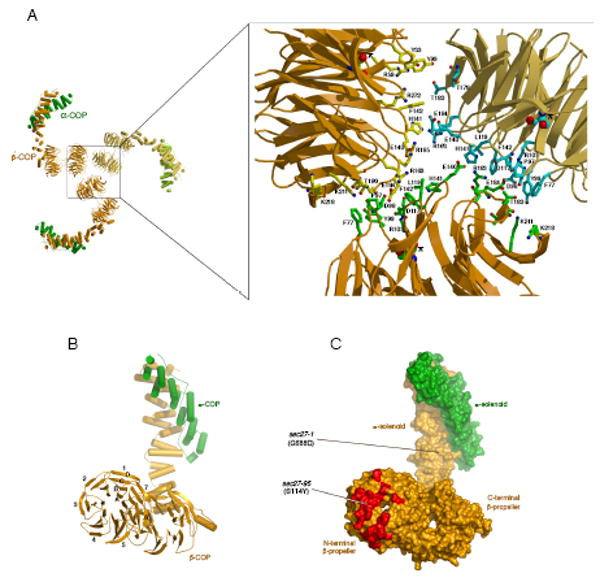
(A) The picture on the left shows the αβ′-COP trimer viewed along the three-fold symmetry axis. The close-up view on the right shows the residues of β′-COP that contribute to the triskelion contact surfaces. The three copies of residue Ser114 are drawn as CPK spheres and their locations indicated by asterisks; this residue is mutated to tyrosine in the sec27-95 mutant (Eugster et al., 2004). Oxygen and nitrogen atoms are colored red and blue, respectively.
(B) In this picture, the top left copy of αβ′-COP from (A) has been rotated 90° about a vertical axis. The view is along the axes of the β-propeller domains of β′-COP. The seven blades of the N-terminal β-propeller domain are labeled 1–7; and the four β strands of blade 1 are labeled A–D (this nomenclature is used in the sequence alignment Figure S1).
(C) Surface representation of the αβ′-COP molecule in (B). Residues on the N-terminal β-propeller involved in interfacial contacts at the triskelion center are colored red. Two yeast mutants that were isolated in genetic screens, sec27-1 and sec27-95, harbor mutations in key regions of the β′-COP molecule: the sec27-1 mutation G688D is located beneath the protein surface near the α-solenoid/α-solenoid interface with α-COP; the sec27-95 mutation S114Y is located just beneath surface residues that form contacts at the triskelion center.
In the COPII system, cage architecture is determined by the geometry of the protein-protein contacts at the dyad vertex center and by the spatial relationships of the β-propeller and α-solenoid domains of the assembly unit (Fath et al., 2007; Stagg et al., 2007; Stagg et al., 2008). In particular, the axes of the β-propeller domains of Sec13 and Sec31 are inclined at a 50° angle and the domains are displaced ∼15 Å from each other. The β-propeller domains of β′-COP are juxtaposed in a slightly different configuration. The axes of the two β-propellers are almost parallel (as can be seen most clearly in Figure 4C, which is viewed along the β-propeller axes and reveals the “pore” of each domain); and the axes are displaced ∼25Å from each other. However, the prevailing observation in this context is that the polarity of the β-propeller domains is conserved in COPI and COPII (the arrows in Figure 5 are meant to convey this relationship). Finally, a characteristic feature of the αβ′-COP subcomplex is the ∼90° angle between the axes of the α-solenoid and the C-terminal β-propeller domain, which helps to create curvature and yields the triskelion form of the trimer (Figure 2 and 3). The similarity of the curved aspect to clathrin heavy chain suggests that this is almost certainly an important feature of COPI cage design (Fotin et al., 2004). The COPII assembly unit is different in this regard, as the axis of the Sec31 α-solenoid domain is roughly parallel with the Sec13 β-propeller axis (Figure 5), the result of which is that the (Sec13/31)2 assembly unit is a relatively straight rod (Figure S2, central panel).
Figure 5. Relationship among COPI, COPII and clathrin cages.
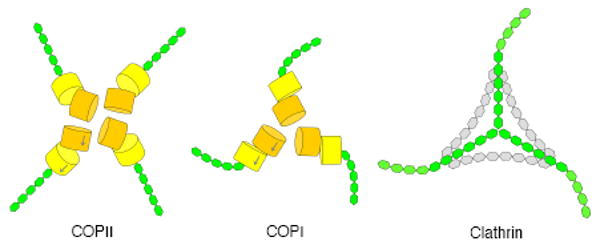
Schematic diagram compares the vertex geometry of COPI, COPII and clathrin cages. The inner β-propeller domains that form vertex contacts are drawn as orange cylinders, the outer β-propeller domains as yellow cylinders, and the α-solenoid domains as linked green hexagons. (The hexagons are purely schematic; their size and number have no meaning). See also Figure S2.
The α-solenoid domain of β′-COP is ∼90 Å long and is composed of sixteen α helices (Figure 3C and S1). The α-solenoid is a relatively straight rod, and the curvature present in the α-solenoid region arises not from β′-COP but from the 40° angled interaction with the α-COP α-solenoid domain (Figure 3A and C). The antiparallel interaction between the α-solenoids of α-COP and β′-COP is highly reminiscent of the homodimer interaction involving the α-solenoid region of Sec31 near the center of the COPII assembly unit (Fath et al., 2007). Indeed, on closer inspection the α-COP and β′-COP α-solenoids are seen to interact around an approximate twofold symmetry axis; the axis runs between the centers of helix 11 of each α-solenoid domain, and is roughly in the plane of the triskelion. To highlight this relationship we numbered the α-COP α-helices to coincide with β′-COP (Figure S3 shows sequence and structural alignments of the α-COP and β′-COP α-solenoid domains). This relationship, together with their similar domain compositions, hints at a common evolutionary origin for the α-COP and β′-COP proteins (Figure 1).
The functional relevance of the interaction we observe between α-COP and β′-COP is supported by the G688D mutation in β′-COP, which is present in the original sec27-1 yeast mutant (Duden et al., 1994; Eugster et al., 2004). The phenotype is a defect in Golgi-to-ER transport (of dilysine cargo) and, strikingly, a destabilization of α-COP in the cell (Eugster et al., 2004). The G688D mutation maps to α helix 7 of β′-COP close to the interface with α-COP (Figure 3C and 4D). The mutation may cause local instability of the α-solenoid structure in this region of β′-COP, and this would weaken the interactions with α-COP in the vicinity of α helices 13–15 (Figure S4).
In summary, through this structural analysis of the αβ′-COP subcomplex, we can recognize differences between the assembly unit cores of COPI and COPII in terms of the juxtaposition of domains in the propeller–propeller–solenoid array. But the overriding conclusion is that the αβ′-COP subcomplex and the (Sec13/31)2 assembly unit of COPII are fundamentally related. The common features—the propeller–propeller–solenoid arrangement, the polarity of the β-propeller domains and their flat axial ends, the anti-parallel interactions of α-solenoid rods—imply an evolutionarily-conserved function for the propeller–propeller–solenoid array. And since these structural features are key to COPII vertex architecture, we propose that the αβ′-COP triskelion constitutes the vertex of the COPI cage (Figure 5).
Model for the COPI Vertex and Common Architectural Principles of COPI, COPII and Clathrin Vertices
The COPI triskelion is intermediate in design between COPII and clathrin: the domain organization and vertex contacts are strikingly similar to COPII, but the triskelion form—curved legs radiating from a three-fold center—closely resembles the clathrin assembly unit (Figure 5).
In the schematic diagram (Figure 5), we have oriented the COPI and clathrin triskelions so that the α-solenoid legs adopt a clockwise curve. This corresponds to a view of the outer face of the triskelion in the clathrin cage (Fotin et al., 2004). The COPII vertex is likewise a view of its outer face (Fath et al., 2007). In the absence of an EM image of the cage, we cannot make this assignment for the COPI triskelion, but the difference (whether clockwise or anticlockwise) is a trivial one with respect to cage design, and the resemblance to clathrin in symmetry and form is evident.
The resemblance of the COPI vertex interactions to COPII is even more striking (Figure 5). The COPI vertex has the simpler arrangement: three αβ′-COP molecules are situated around a threefold rotation axis, so all three N-terminal β-propellers of β′-COP interact in the same way (here we ignore deviations from threefold symmetry in the crystal asymmetric unit). In the COPII vertex, four copies of the assembly unit are situated around a twofold rotation axis, so all four cannot interact in the same way. A proximal pair of Sec31 β-propellers interacts in a somewhat different manner to the distal pair (Fath et al., 2007). Nevertheless, the two types of Sec31 interactions and the β′-COP interaction are all variations on a geometric theme of β-propeller domains interacting via their flat axial ends (Figure 5).
In the absence of an EM image of the COPI cage, we do not have direct evidence that the αβ′-COP triskelion constitutes the vertex. When we tested the αβ′-COP subcomplex for trimer formation in vitro we detected only monomeric αβ′-COP in gel filtration experiments using protein concentrations up to 30 mg/ml protein (data not shown). The same negative result was obtained in previous studies on the COPII vertex: a core Sec13/31 construct of two β-propellers remains monomeric (dimer and tetramer formation is undetectable) using as much as 30 mg/ml protein (Fath et al., 2007). This was despite the fact that the role of Sec31 at the COPII cage vertex has been assigned definitively based on the concordance of the crystal structure and the EM density map (Fath et al., 2007; Stagg et al., 2007; Stagg et al., 2008). The vertex interactions in COPI and COPII cages—both of which lack the α-solenoid interdigitation of the clathrin cage—seem to be exceptionally weak. This fits with the view that, in a protein polyhedron, very weak interactions between assembly units can yield a very stable cage, and even modest-strength interactions may severely compromise the cage disassembly reaction (Zlotnick, 1994).
Indirect evidence for the role of the αβ′-COP triskelion as the vertex comes from the sec27-95 temperature-sensitive mutant of β′-COP, which harbors the mutation S114Y (Eugster et al., 2004; Prinz et al., 2000). Residue Ser114 is located on the N-terminal β-propeller of β′-COP close to the triskelion contact surface (Figure 4A and 4C). The mutation of Ser114 to tyrosine will affect key residues at the triskelion interface, in particular Pro97, Asp98, Tyr99, and possibly also Phe77 and Asp117 (described below). The sec27-95 mutant is defective in retrograde Golgi-to-ER transport (of the dilysine cargo molecule Emp47p) but, unlike the sec27-1 mutant, α-COP is not destabilized in mutant cells grown at the permissive temperature (Eugster et al., 2004). sec27-1 and sec27-95 are the only β′-COP mutants to have been characterized to date, and the two mutations map to strategically important regions of the β′-COP molecule (Figure 4C). This concordance of structure and function lends support to our model for the coatomer vertex, but a direct test for a triskelion COPI vertex geometry probably will require an electron cryomicroscopy analysis of reconstituted cages.
At the triskelion center, the β′-COP subunits associate through pairwise interactions involving a small, circumscribed area of the axial end of one N-terminal β-propeller and the side of the adjacent β-propeller (Figure 4 and 5). (Two of the propeller–propeller interfaces form very similar interactions, whereas the third interface has rotated apart due to crystal packing distortions; hence this description applies to the two similar interfaces). The interface involves residues from loops 2B–2C, 2D–3A, 3B–3C and 3D-4A on the axial end of the N-terminal β-propeller with residues from loops 4B–4C and 4D–5A of the side of the adjacent β-propeller (Figure 4 and S1). Key contacts involve the side chains of residues Phe77, Asp98, Tyr99, Phe142, Arg163 and Glu184 (S. cerevisiae β′-COP numbering). The COPI interface cannot be compared in molecular detail with the corresponding COPII interfaces, since the COPII vertex is based on a model fit into relatively low-resolution EM maps. However, we note that the geometry of the β′-COP–β′-COP interaction seems most similar to the proximal–distal β-propeller contacts (defined as cII and cIII contacts) at the COPII vertex (Fath et al., 2007; Stagg et al., 2008). Finally, the contact interfaces at both the COPI and COPII vertices seem to involve small surface areas, consistent with the weak interactions required to facilitate cage disassembly.
Implications for the Architecture of the COPI Cage
In our schematic representation (Figure 6) we illustrate a possible arrangement of protein components in the COPI cage formed by αβ′ε-COP. The known elements of the cage design are the atomic structures of the αβ′-COP and αε-COP subcomplexes (which account for ∼75% of the structured polypeptide in αβ′ε-COP) and the symmetry and form of the triskelion, which we infer is the vertex of the cage.
Figure 6. Model for the architecture of the COPI lattice.
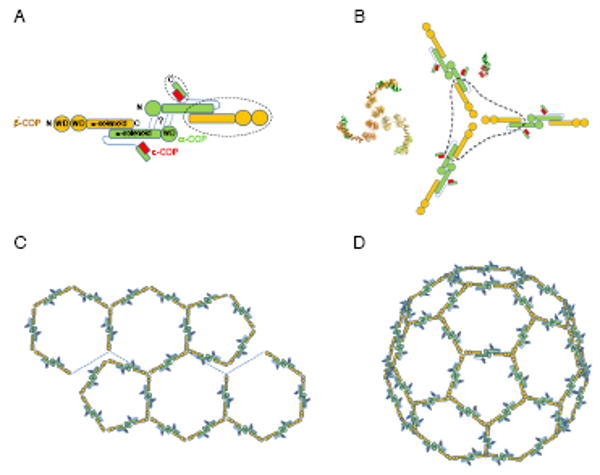
(A) Known and unknown elements of the αβ′ε-COP complex. The αβ′-COP and αε-COP subcomplexes (circled with dotted lines) whose structures we have determined account for more than two thirds of the total mass of the αβ′ε-COP complex. The remaining portion is the N-terminal region of α-COP whose sequence indicates an N-terminal β-propeller domain followed by a ∼300-residue region of unknown structure (possibly a second β-propeller domain). The speculative element of the diagram is the dimer contact (indicated by the question mark) that brings together two copies of αβ′ε-COP. We have drawn this as an α-COP–α-COP interaction as one possibility; alternatively β′-COP might mediate this interaction. Either way, we propose that two copies of αβ′ε-COP would converge to form the assembly unit COPI.
(B) A model for the COPI vertex, and corresponding ribbon diagrams of the αβ′-COP and αε-COP crystal structures.
(C) Based on a three-fold rotation axis at the vertex, the COPI cage symmetry most likely is related to clathrin cage symmetry. Thus the COPI lattice model is composed of hexagons and pentagons.
(D) One possible arrangement of hexagonal and pentagonal units forming an icosahedral COPI cage. This type of structure is formed by clathrin in vitro (Fotin et al., 2004).
The unknown element of the design is the connection that joins adjacent triskelia. In the simplest model, two copies of αβ′ε-COP connect to form a (αβ′ε-COP)2 dimer as the assembly unit of the COPI cage. For simplicity in Figure 6A we have drawn this connection between the N-terminal regions of two copies of α-COP (this is entirely speculative). In this arrangement, the COPI cage assembles in a very similar manner to COPII. Thus, the assembly unit (Figure 6A) is a dimeric molecule—rod-shaped (Sec13/31)2 or S-shaped (αβ′ε-COP)2—with terminal β-propeller domains that interact to drive self-assembly (Figure 6B). In the resultant lattice, Figure 6C, the edge is formed by the assembly unit (αβ′ε-COP)2, but the symmetry is governed by the three-fold centre of the triskelion to yield a clathrin-like array of hexagonal and pentagonal shapes. Finally, the compulsion to maximize the number of stable bonds between assembly units drives the formation of the spherical cage, with the same symmetry as a clathrin cage (Figure 6D).
According to our model, the architectural core of the COPI cage comprises the propeller–propeller–solenoid array of β-COP, the central α-solenoid domain of α-COP, and an additional N-terminal region (possibly the entirety) of α-COP to form the connection at the center of the assembly unit (Figure 1 and 6). We have as yet been unable to express soluble portions of the α-COP N terminus, but future biochemical and structural studies should address the role of this region in cage assembly. We infer that the αε-COP subcomplex is not part of the architectural core of the cage as it is flexibly linked to the αβ′-COP subcomplex via a highly acidic ∼80-residue linker of α-COP (Figure 3C and D). Moreover, the ε-COP protein is not essential for yeast growth, although its absence does compromise the stability of α-COP (Duden, et al., 1998), consistent with the intimate, coiled interaction observed in the crystal structure of the αε-COP subcomplex (Figure 3D). In the case of the COPII cage, the C-terminal α-solenoid domain of Sec31 is flexibly linked to the assembly unit core via a ∼340-residue proline-rich linker; importantly, a 50-residue peptide at the center of this linker interacts with the cargo-binding Sec23/24•Sar1 complex (Bi, et al., 2007). It will be interesting to test the role of the αε-COP subcomplex and acidic linker in the COPI coat, and specifically whether these regions project towards the membrane to interact with the cargo-binding βδ/γζ-COP complex.
In conclusion, this analysis of the αβ′ε-COP complex establishes architectural principles that are common to the three major classes of vesicular cages, and a simple transformation of design—from COPII to COPI to clathrin—is revealed (Figure 5). The findings should provide a foundation for molecular-level studies of the dynamic processes of COPI coat assembly and disassembly.
Experimental Procedures
Protein Production
The αβ′ε-COP complex from B. taurus was prepared by co-expression of the three full-length proteins in insect cells infected with engineered baculoviruses (relative molecular mass of α-COP 135K, β′-COP 100K, ε-COP 34K). Insect cells were harvested 48 hours post-infection, lysed by sonication, and protein was purified by Ni2+-IMAC chromatography, utilizing an N-terminal His6 tag included on the α-COP subunit. The His6 tag was removed using TEV protease (Invitrogen), and the protein was purified further by ion-exchange (Mono Q) and size-exclusion chromatography on a Superdex 200 column.
For crystallographic studies, the S. cerevisiae αβ′-COP subcomplex (comprising residues 642-818 of α-COP and residues 1-814 of β′-COP, as indicated by the domain analysis) was co-expressed in insect cells and purified as before. For the αε-COP subcomplex, B. taurus genes encoding residues 899-1224 of α-COP and residues 10-308 of ε-COP were both cloned into pET28b (Novagen); the COP subunits were expressed separately in E.coli BL21(DE3) cells following induction at 20°C with 0.4 mM IPTG. In both cases, cells were harvested 15 hours post-induction, lysed by sonication, and protein was purified by Ni2+-IMAC chromatography. Histidine tags were removed with thrombin protease (Sigma). At this stage, the α-COP and ε-COP protein solutions were combined, and the subcomplex was purified further by ion-exchange (Mono Q) and size-exclusion chromatography (Superdex 200 column). Selenomethionine-substituted protein was made by expressing the α-COP and ε-COP proteins in B834(DE3) E. coli cells (Novagen) using M9 minimal media plus selenomethionine.
Prior to crystallization experiments, both the αβ′-COP and αε-COP subcomplexes were concentrated to 20 mg/ml and flash-frozen in liquid nitrogen for storage.
Domain Analysis
Limited proteolysis experiments were carried out to define domain boundaries and to identify subcomplexes within the trimeric αβ′ε-COP complex. The full-length complex from B. taurus was digested with subtilisin across a range of protein concentrations and incubation times. The reaction was terminated with 1 mM PMSF and limit products were separated by ion-exchange (Mono Q) and gel filtration chromatography, and then analyzed by SDS-PAGE and N-terminal sequencing. This characterization identified a core αβ′-COP subcomplex and an αε-COP subcomplex (Figure 1).
Protein Crystallization and Structure Determination of αβ′-COP
The yeast αβ′-COP complex was crystallized at 22°C by the hanging-drop method by adding 1 μl of a 20 mg/ml protein solution to 1 μl of well solution comprising 0.8 M K2HPO4 and 0.8 M NaH2PO4,100 mM Bis-Tris propane (pH 8.5) and 1 % (w/v) PEG 4000. The crystals are trigonal, space group P3221 (a = b = 153.2 Å, c = 295.1 Å) and contain three copies of αβ′-COP in the asymmetric unit. For diffraction studies, crystals were transferred to a cryo-protection solution comprising well solution plus 30% glycerol, and flash-frozen in liquid nitrogen.
The crystal structure of the αβ′-COP complex was determined by multiple isomorphous replacement with anomalous scattering (MIRAS). Heavy atom-derivatized crystals were prepared via a 24-hour soak using 1 mM thimerosal and an 18-hour soak using Ta6Br12 (kindly provided by Dimitar Nikolov). Native and derivative X-ray data were collected from frozen crystals at beamline X25 of the National Synchrotron Light Source (NSLS). Data were processed with the program HKL2000 (Otwinowski and Minor, 1997), and MIR phasing analysis was done with the program SOLVE (Terwilliger and Berendzen, 1999) using data between 20 and 3.5 Å resolution (dataset Native-2 in Table 1). SOLVE identified five Ta6Br12 molecules (for both Ta6Br12 derivatives, which gave similar phase information) and ten mercury positions (for the thimerosal derivative) and, following refinement, reported a mean figure of merit (f.o.m.) of 0.37 (f.o.m. of 0.22 for the highest resolution bin). The initial electron density map was improved by density modification with three-fold non-crystallographic symmetry averaging using data between 20 and 3.1 Å resolution. The resulting electron density map was of high quality. Successive rounds of positional refinement using the program CNS (Brünger, 1998) and model building using O (Jones et al., 1991) reduced the R-factor to a final value of 22.1 % (Rfree = 27.2 %) for native data between 25 and 2.5 Å resolution (dataset Native-1 in Table 1). The final model comprises 23,463 protein atoms and 270 water molecules, with just two Ramachandran violations (residues 491 and 679 in one copy of β′-COP). The model contains three copies of β′-COP (residues 1-814) and α-COP (residues 642-818) in the asymmetric unit. Of these, the following residues were not modeled due to weak electron density: β′-COP residues 1, 813 and 814 in the first copy, β′-COP residues 1, 492 and 493 in the second copy, and β′-COP residues 491-493 and 814 and α-COP residues 817 and 818 in the third copy. The X-ray data and refinement statistics are summarized in Table 1.
Crystallization and MAD Structure Determination of αε-COP
For crystallization of the B. taurus αε-COP complex, 1 μl of protein solution (in 20 mM Tris-HCl, 150 mM NaCl, 5 mM DTT, pH 7.5) was equilibrated with 1 μl of well solution comprising 11% PEG-4000, 100 mM sodium citrate (pH 5.5) and 5 mM DTT. The crystals, which appeared after three days, contain one αε-COP complex in the asymmetric unit (space group I422, a = b = 176.9 Å, c = 141.5 Å, 71% solvent). For X-ray diffraction experiments, crystals were transferred to well solution containing an additional 30% glycerol, then flash frozen in liquid nitrogen. Multi-wavelength anomalous diffraction (MAD) data at three wavelengths were collected at beamline 24ID of the Advanced Photon Source (APS) and processed as before (Table 1). Native data to 2.6 Å resolution were collected from a single frozen crystal at beamline X25 of NSLS. The MAD data analysis was done with the program SOLVE (Terwilliger and Berendzen, 1999) using data between 35 and 3.0 Å resolution. SOLVE found 15 of the 16 selenium sites and refined these to give a mean f.o.m. = 0.46 (0.18 for the highest resolution bin). Electron density modification using RESOLVE (Terwilliger and Berendzen, 1999) yielded an initial electron density map of excellent quality. Model building, aided by the position of the selenium atoms, was combined with positional refinement using CNS (Brünger, 1998) to reduce the refinement R-factor to 22.2% (Rfree = 26.3%) for native data between 30 and 2.6 Å resolution (Table 1). The final model comprises 4874 protein atoms and 34 water molecules. There is one outlier in a Ramachandran plot of the final model (Phe906 of α-COP). The following residues have been omitted due to weak electron density: α-COP residues 899-904 and ε-COP residues 10-16 and 308.
Sequence Analysis
For the preparation of Figure S1, β′-COP sequences from thirteen organisms were aligned: H. sapiens (P35606), Mus musculus (O55029), Rattus norvegicus (EDL77461), B. taurus (CAA51285), Xenopus laevis (NP_001080221), Caenorhabditis elegans (NP_501671), Arabidopsis thaliana (NP_001154480), Danio rerio (AAI62672), Drosophila melanogaster (NP_524836), Candida glabrata (XP_447187), Cryptococcus neoformans (XP_777447), S. cerevisiae (EDN61984), and Schizosaccharomyces pombe (XP_001713153).
Supplementary Material
Acknowledgments
We thank staff at beamline X25 of the NSLS and beamline 24ID of the APS for use of and assistance with synchrotron facilities. We thank Felix Wieland and Marianna Breitman for providing full-length cDNA and baculoviruses for expression of COPI subunits. This work was supported by a grant from the Howard Hughes Medical Institute.
Footnotes
Accession Numbers: Atomic coordinates and structure factors for the αβ′-COP and αε-COP structures have been deposited in the Protein Data Bank under PDB codes 3MKQ and 3MKR, respectively.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23•Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bremser M, Nickel W, Schweikert M, Ravazzola M, Amherdt M, Hughes CA, Sollner TH, Rothman JE, Wieland FT. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Brohawn S, Schwartz TU. Molecular architecture of the Nup84-Nup145C-Sec13 edge element in the nuclear pore lattice. Nat Struct Biol. 2009;16:1173–1178. doi: 10.1038/nsmb.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger AT. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Das AK, Cohen PTW, Barford D. The structure of the tetratricopeptide repeats of protein phosphatase 5: implications for TPR-mediated protein-protein interactions. EMBO J. 1998;17:1192–1199. doi: 10.1093/emboj/17.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol. 2004;2:2085–2093. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duden R, Hosobuchi M, Hamamoto S, Winey M, Byers B, Schekman R. Yeast β- and β′-coat proteins (COP): two coatomer subunits essential for ER-to-Golgi traffic. J Biol Chem. 1994;269:24486–24495. [PubMed] [Google Scholar]
- Duden R, Kajikawa L, Wuestehube L, Schekman R. ε-COP is a structural component of coatomer that functions to stabilize α-COP. EMBO J. 1998;17:985–995. doi: 10.1093/emboj/17.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. COPI domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. The α-COP and β′-COP WD40 domains mediate cargo-selective interactions with distinct di-lysine motifs. Mol Biol Cell. 2004;15:1011–1023. doi: 10.1091/mbc.E03-10-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J. structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S, Kuge O, Orci L, Amherdt M, Ravazzola M, Wieland FT, Rothman JE. En bloc incorporation of coatomer subunits during the assembly of COPI-coated vesicles. J Cell Biol. 1994;124:883–892. doi: 10.1083/jcb.124.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A. Architecture of a coat for the nuclear pore membrane. Cell. 2007;131:1313–1326. doi: 10.1016/j.cell.2007.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci U S A. 2001;98:10704–10709. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe M, Kreis TE. In vitro assembly and disassembly of coatomer. J Biol Chem. 1995;270:31364–31371. doi: 10.1074/jbc.270.52.31364. [DOI] [PubMed] [Google Scholar]
- Mancias JD, Goldberg J. Structural basis of cargo membrane protein discrimination by the human COPII coat machinery. EMBO J. 2008;27:2918–2928. doi: 10.1038/emboj.2008.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Op Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, Wieland FT. Uptake by COPI-coated vesicles of both anterograde and retrograde cargo is inhibited by GTPγS in vitro. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- Orci L, Palmer DJ, Amherdt M, Rothman JE. Coated vesicle assembly in the Golgi requires only coatomer and ARF proteins from the cytosol. Nature. 1993;364:732–734. doi: 10.1038/364732a0. [DOI] [PubMed] [Google Scholar]
- Otwinowski W, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR. Adaptors for clathrin coats: structure and function. Annu Rev Cell Dev Biol. 2004;20:153–191. doi: 10.1146/annurev.cellbio.20.010403.104543. [DOI] [PubMed] [Google Scholar]
- Pavel J, Harter C, Wieland FT. Reversible dissociation of coatomer: functional characterization of a beta/delta-coat protein subcomplex. Proc Natl Acad Sci U S A. 1998;95:2140–2145. doi: 10.1073/pnas.95.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–474. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schledzewski K, Brinkmann H, Mendel RR. Phylogenetic analysis of components of the eukaryotic vesicle transport system reveals a common origin of adaptor protein complexes 1, 2, and 3 and the F subcomplex of the coatomer COPI. J Mol Evol. 1999;48:770–778. doi: 10.1007/pl00006521. [DOI] [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Spang A, Matsuoka K, Hamamoto S, Schekman R, Orci L. Coatomer, Arf1p, and nucleotide are required to bud coat protein complex I-coated vesicles from large synthetic liposomes. Proc Natl Acad Sci U S A. 1998;95:11199–11204. doi: 10.1073/pnas.95.19.11199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Balch WE. Structural design of cage and coat scaffolds that direct membrane traffic. Curr Opin Struct Biol. 2007;17:221–228. doi: 10.1016/j.sbi.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Razvi A, Gürkan C, Potter CS, Carragher B, Balch WE. Structural basis for cargo regulation of COPII coat assembly. Cell. 2008;134:474–484. doi: 10.1016/j.cell.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target protein to the β-propeller of clathrin. Proc Natl Acad Sci U S A. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE. ‘Coatomer’: a cytosolic protein complex containing subunits of non- clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
- Zhao L, Helms JB, Brugger B, Harter C, Martoglio B, Graf R, Brunner J, Wieland FT. Direct and GTP-dependent interaction of ADP ribosylation factor 1 with coatomer subunit beta. Proc Natl Acad Sci U S A. 1997;94:4418–4423. doi: 10.1073/pnas.94.9.4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnick A. To build a virus capsid. An equilibrium model of the self assembly of polyhedral protein complexes. J Mol Biol. 1994;241:59–67. doi: 10.1006/jmbi.1994.1473. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


