Abstract
Ion channels form a group of membrane proteins that pass ions through a pore beyond the energy barrier of the lipid bilayer. The structure of the transmembrane segment of membrane proteins is influenced by the charges and the hydrophobicity of the surrounding lipids and the pressure on its surface. A mechanosensitive channel is specifically designed to change its conformation in response to changes in the membrane pressure (tension). However, mechanosensitive channels are not the only group that is sensitive to the physical environment of the membrane: voltage-gated channels are also amenable to the lipid environment. In this article, we review the structure and gating mechanisms of the mechanosensitive channels and voltage-gated channels and discuss how their functions are affected by the physical properties of the lipid bilayer.
Keywords: ion channels, membrane lipid, mechanosensitive channels, mechanosensitive channel of small conductance, mechanosensitive channel of large conductance
1. Introduction
1.1. Structure and conformational change of ion channels
Concentrations of inorganic ions inside cells are kept significantly different from the outside to enable the electrical and chemical signallings by ion channels. High concentrations of potassium ions inside the cells produce negative resting potentials by the leaky potassium channel, which passes potassium ions in the resting state. On the other hand, low intracellular concentrations of sodium and calcium ions generate instantaneous reversal of the membrane potential by allowing sodium or calcium ions to flow into the cells through sodium and calcium channels. These ion concentration gradients are maintained by ion pumps, which consume a significant amount of energy. For example, the activity of K+–Na+–ATPase accounts for 20–45% of the oxygen uptake in resting cells (Whittam 1961). This underscores that the signalling by ion channels is based on a significant cost of energy expenditure.
Ion channels can roughly be grouped into three, according to the signal (stimulus) to activate (gate) the channels. Voltage-gated channels are a group of ion channels whose activity changes with the transmembrane voltage. The opening of ligand-gated channels is regulated by the binding of substances to the ion channel. Mechanosensitive channels comprise a group of channels gated by mechanical force that is generally generated by membrane stretch. Despite the variation in the stimulus type the outcome is the same: channel opening and closing. In this article, we will overview how the channel gating is affected by physical environment. Attention is paid to the influence of the membrane, which is the major physical environment for membrane proteins. In this respect, mechanosensitive channels are of special interest because most of them are activated by the membrane tension and report the physical contact with lipids.
1.2. Membrane proteins and their interaction with membrane lipids
A membrane is not just a sheet where membrane proteins are inserted. The lipid bilayer is a critical environment for membrane proteins and largely affects their structure and function.
Different organisms have different sets of membrane lipids and each lipid varies in its characteristics. The plasma membrane of human cells is composed mainly of phosphatidylcholine (PC) and phosphatidylserine (PS) while minor components such as cholesterol and phosphatidylinositol have important roles in some intracellular signalling. On the other hand, the main component of the inner membrane of Escherichia coli is phosphatidylethanolamine (PE). PC, PS and PE have different charge and spatial occupation depending on the difference in the head group. Besides a negative charge of the phosphate, PC and PE have a positive charge and PS has both positive and negative charge in the head group. These charges of the lipids can interact with the charged residues of the membrane protein and the polar motif can form hydrogen bonds with the protein.
The thickness of the hydrophobic core of the lipid bilayer should match with the thickness of the transmembrane hydrophobic surface of the membrane protein because contact between hydrophobic and hydrophilic moieties is energetically costly. The hydrophobic thickness of the lipid bilayer is generally defined as the distance between the glycerol backbone layers on the two monolayers. When a membrane protein is placed in a bilayer whose hydrophobic core has a thickness different from that of the hydrophobic surface of the protein, the membrane and/or the protein should change the conformation or way of subunit aggregation to adjust the hydrophobic thickness, in other words, to avoid an energetically costly hydrophobic mismatch. For instance, a membrane protein can adjust to a thin membrane by decreasing the hydrophobic thickness of the protein and/or by stretching the hydrophobic core of surrounding lipids close to the protein (figure 1a). The α-helical membrane proteins including mechanosensitive channel of large conductance (MscL), K+ channel from Streptomyces lividans (KcsA) and Ca2+–ATPase presumably adapt to a thin membrane by decreasing the hydrophobic thickness of the transmembrane surface of the protein (Powl et al. 2003 and references therein). The reduction of the hydrophobic thickness is attained by tilting α-helices, as was observed on MscL when reconstituted into a thin lipid bilayer (Perozo et al. 2001). On the other hand, β-barrel proteins like outer membrane porin tend to adapt to the change in the hydrophobic thickness of the membrane by deformation of lipids (O'Keeffe et al. 2000). In this case, the annular lipids (also called boundary lipids), which interact with the transmembrane surface of the protein have a structure and hydrophobic thickness different from the lipids under no influence of the membrane proteins (figure 1a). Thus, residing in the membrane with an optimal hydrophobic thickness is crucial for the membrane proteins because otherwise they should adapt to the non-native thickness by adopting a non-native structure. For example, the activities of melibiose permease and diacylglycerol kinase are maximal at optimal bilayer thickness but reduce either in thinner or thicker membranes (Dumas et al. 2000; Pilot et al. 2001).
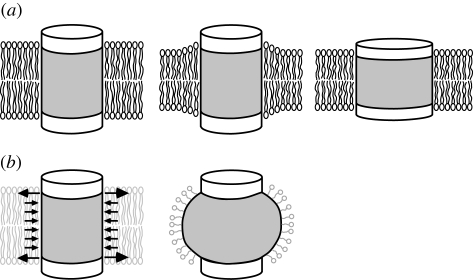
Figure 1.
Membrane protein embedded in lipid bilayer. (a) Cylindrical membrane protein in lipid bilayer when the height of the hydrophobic surface (grey) matches the thickness of hydrophobic core of the lipid bilayer (left). In a thinner bilayer, the hydrophobic mismatch is avoided either by rearrangement of the lipid molecules (middle) or by thinning of the protein (right). (b) The lateral pressure acting on the surface of the membrane protein (left). When the membrane protein is solubilized with detergent, the loss of the lateral pressure is likely to deform the transmembrane segment of the membrane protein (right).
It is not only the hydrophobic thickness that affects the structure of the membrane proteins. The lipid tails are subject to active thermal motion and produce positive pressure in the membrane (i.e. expanding pressure; Cantor 1997). Therefore, the transmembrane surface of the membrane proteins is pushed and compressed by the positive lateral pressure exerted by the lipid tails. To balance the positive pressure in the hydrophobic core of the membrane, a peak of negative lateral pressure (tension) is present at the level of the glycerol backbone (figure 1b). These pressures acting on the surface of the membrane proteins make the mechanical environment that determines the protein structure. It can be easily assumed that the transmembrane domain expands if the positive pressure is absent (figure 1b). Therefore, it stands to reason that a membrane protein adopts a non-native structure when extracted from the lipids for biochemical analyses.
The shape of the lipid molecule affects the profile of the lateral pressure. Insertion of lipids with only one lipid tail to a planer bilayer reduces the positive pressure and thus increases the tension in the inserted monolayer (Yoo & Cui 2009). Lipids with bulky head groups (cone-shaped lipids) also have a similar effect. On the contrary, lipids with smaller head groups (inverted cone-shaped lipids), PE for example, are expected to have the opposite effect. The structures of the membrane proteins change with the pressure profile as is clearly demonstrated by the activation of mechanosensitive channel of small conductance (MscS) upon addition of the single-tailed phospholipid, lysophosphatidylcholine (Perozo et al. 2002b; Machiyama et al. 2009).
1.3. Methods for structure determination of ion channels
The structure of a protein can be deduced at the atomic scale by the X-ray diffraction pattern of the three-dimensional protein crystal. The crystal structures of a number of ion channels have been resolved. The first structure revealed was KcsA, a potassium channel from the Gram-positive bacterium Streptomyces lividans (Doyle et al. 1998). The structure of the selectivity filter accelerated the understanding as to why the channel passes only potassium ions. Although KcsA is devoid of a voltage sensor, following studies disclosed the structures of a range of voltage-dependent K+ channels: voltage-dependent K+ channel from Aeropyrum premix (KvAP), Kv1.2, Kv2.1, Kv3.1 and inward rectifying K+ channel homologue from bacteria (KirBAC; Jiang et al. 2003; Kuo et al. 2003; Long et al. 2005a,b, 2007; Nishida et al. 2007). The crystal structure of the mechanosensitive channels (MscL and MscS) also provides the basis of the structure–function relationship of mechanoreception (Chang et al. 1998; Bass et al. 2002). We now have the structure of a variety of ion channels of the gap junction, NaK, P2X4, aquaporin and pLGIC (Shi et al. 2006; Hilf & Dutzler 2008; Kawate et al. 2009; Maeda et al. 2009).
The three-dimensional crystal is the assembly of the membrane proteins solubilized by detergent. As mentioned above, one of the critical factors that stabilize the membrane proteins is the lipid environment of the membrane, without which membrane proteins may adopt a non-native conformation and lose their functions. In fact, several membrane enzymes are reported to lose their activity on solubilization (Lee 2003). Thus, the caveat for the assessment of the membrane protein in the crystal structure is that the membrane is not there. The voltage sensor paddle spited from the channel core in the KvAP crystal structure and the splayed TM1–TM2 helices in the MscS crystal structure are probably due to the delipidation. Therefore, the crystal structure should always be tested by independent experiments and molecular dynamics simulations.
The structure of the proteins residing in the membrane can be resolved if the protein assembles into two-dimensional crystals in the membrane. This method is advantageous to the three-dimensional crystal in that the structure is devoid of detergent-induced artefacts and is likely to retain the native structure in the membrane. Because the structural data are obtained through the diffraction of an electron beam in an electron microscope, this technique is referred to as electron crystallography. The density maps are typically limited to approximately 3.5 Å, but can be refined up to 1.9 Å (Gonen et al. 2005). Electron crystallography has been developed in a study on bacteriorhodopsin (Henderson & Unwin 1975) and applied to aquaporins (Murata et al. 2000), acetylcholine receptors (Miyazawa et al. 2003), and BK potassium channel (Wang & Sigworth 2009).
The structure of bulky transmembrane proteins that are difficult to crystallize can be viewed by single-particle analysis. The projections of the unstained proteins obtained by cryo-electron microscopy are usually very faint and difficult to detect. Intense digital image analyses and computations enable the detection of the protein particles and their directions, and the reconstitution of the three-dimensional image. The reconstituted image, which typically has a resolution of 15–20 Å, provides a macroscopic view of the protein. The bell-shaped image of sodium channel was the first unveiled by this method (Sato et al. 2001). The structures of the inositol 1,4,5-triphosphate receptor (Sato et al. 2004), canonical transient receptor potential-3 (TRPC3; Mio et al. 2007) and Cav3.1 (Walsh et al. 2009) have been obtained so far.
2. Mechanosensitive channels
2.1. Research history and fundamental issue of mechanogating
The first single channel activities of mechanosensitive channels were recorded on chicken myoblast by applying a negative pressure to a patch membrane caught on a glass pipette (Guharay & Sachs 1984). The negative pressure concaves the membrane towards the inside of the pipette and therefore generates tension in the plane of the membrane according to Laplace's Law. Assessment of the geometry of the membrane showed that the channel activity depends on the tension in the membrane rather than the transmural pressure (Sokabe et al. 1991). Despite an increase in the patch membrane area on suction in the pipette, the thickness of the membrane as monitored by the membrane capacitance does not change, indicating that there is a reservoir of lipids associated with the wall of the glass pipette.
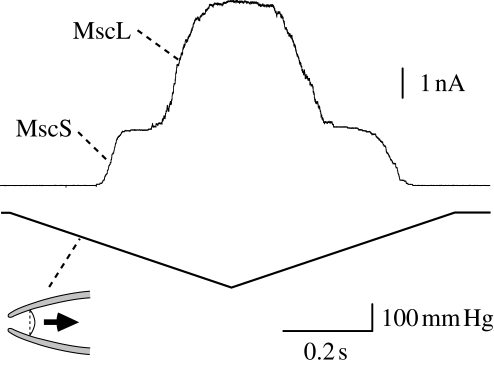
Using this technique, activity of mechanosensitive channels was demonstrated in the spheroplasts of E. coli cells (Martinac et al. 1987). There are at least four types of mechanosensitive channels in the inner membrane (cytoplasmic membrane) of E. coli cells. The channels that are predominantly found on the cell membrane are MscL and MscS (figure 2). MscL gates at tension approximately 1.6 times higher than MscS. Under sustained tension, MscS transits to the desensitized state, in which MscS reopens by supramaximal tension, and then to the inactive state, in which MscS does not open by any tension (Akitake et al. 2007). There are two mechanosensitive channels that are less frequently found on the E. coli patch membrane. MscK, which is a homologue of MscS, differs from MscS in that MscK requires high potassium concentration for activity and does not show inactivation (Li et al. 2002). MscM has a lower conductance than the others but its molecular entity is not fully understood (Berrier et al. 1996).
Figure 2.
Activation of bacterial mechanosensitive channels in a patch clamp experiment. When negative pressure (lower trace) is applied to the membrane in the patch clamp pipette (inset), mechanosensitive channel MscS is activated at low pressure. Further increase in pressure opens MscL. The amplitudes of single MscS and MscL channels are approximately 20 pA and approximately 75 pA, respectively.
Bacterial channels are advantageous over eukaryotic channels because of the availability of genetic information and considerable amount of proteins for biochemical and structural analyses. Thus, MscL and MscS are the first mechanosensitive channels whose genes and structures have been identified (Sukharev et al. 1994; Chang et al. 1998; Levina et al. 1999; Bass et al. 2002).
Because purified MscS and MscL undergo mechanosensitive activation when reconstituted into the lipid bilayer, the activation of either MscL or MscS is brought about solely through tension in the membrane and does not require any supporting structure (Sukharev et al. 1999, 2002). Thus, the contact to the membrane lipid is essential to the activation of MscL and MscS. As was demonstrated in chick muscle cells, the activities of MscL and MscS depend on the membrane tension rather than the curvature or the transmural pressure across the membrane (Sukharev et al. 1999; Sukharev 2002; Moe & Blount 2005). For instance, MscL activates at the same tension when the opening is examined on the patch membrane with considerably different curvatures. It should also be emphasized that MscS and MscL activate when positive pressure is applied to blow up the spheroplast caught on the pipette in whole cell mode (Cui et al. 1995). Because the activity increases with the decrease in the curvature in this mode, the bending of the membrane cannot be the stimulus for gating.
Despite the minor role of the membrane curvature in MscS and MscL opening, incorporation of aliphatic molecules with a significant spontaneous curvature into a single leaflet of bilayer changes the ease of opening of the mechanosensitive channel (Martinac et al. 1990). Trinitrophenol and chlorpromazine, which are crenator and cup-former, respectively, have a similar effect. A more effective amphipath is lysophosphatidylcholine (LPC), which has only one lipid tail and thus is classified as an ‘inverted cone’ type phospholipid. MscS and MscL reconstituted into the lipid bilayer open without applied tension when LPC is applied to the membrane from one side of the membrane (Perozo et al. 2002b; Vásquez et al. 2008a). Although the orientations of the reconstituted channels in the lipid membrane are probably in both directions with respect to the transmembrane axis, the application of LPC to the cytoplasmic side activated MscS indicating that insertion of LPC into the inner leaflet can activate MscS (Vásquez et al. 2008a,b; Machiyama et al. 2009). In spite of the dramatic effect on the channel activity, how LPC activates MscS and MscL is not fully understood. Given that the bending of the membrane does not activate MscS or MscL per se, changes in the surface tension or the pressure profile are probably the factor that gates the channel upon amphipath application. In fact, when the density of the lipid phosphate is monitored by 31P-NMR, the headgroups are compressed in the leaflet where LPC is inserted (Traïkia et al. 2002). On the other hand, the surface tension decreases (i.e. dilated) in the other leaflet. The compression of the inserted leaflet and the dilation in the other leaflet were also observed in coarse-grained simulations on lipid membranes (Yoo & Cui 2009). How these findings relate to the behaviour of MscS and MscL in the patch clamp pipette is to be determined because the outcomes vary with the radius of membrane curvature (size of liposome; Traïkia et al. 2002) or reverses under different simulation conditions (Ollila et al. 2009).
In contrast to the direct activation of MscS and MscL by tension in the lipid bilayer, the functions of some eukaryotic mechanosensitive channels are associated with cytoskeleton and extracellular matrix. For example, the touch receptor of Caenorhabditis elegans requires microtubules and extracellular matrix (Chalfie 2009). The mechanotransduction of hair cells also seems to be correlated with the extracellular links that connect the tips of the stereohair (Grillet et al. 2009). Mutational and morphological experiments favour the idea that the mechanosensitive channels of these cells are activated by stretching the cytoskeleton and extracellular matrix. Recently, direct evidence has been provided that tension in the cytoskeleton (stress fibre) can activate Ca2+-permeable mechanosensitive ion channels expressed in the vicinity of focal adhesion in cultured endothelial cells (Hayakawa et al. 2008). The estimated force required for the activation of single ion channels is as low as 1–2 pN. However, the detailed structure of how these elements, including ion channels, cytoskeleton and focal adhesion, are associated is not known. Therefore, the question remains as to whether the mechanosensitive channels are activated through a direct connection with cytoskeleton or an indirect one via membrane.
2.2. Structure and structural change on gating in MscL
The crystal structures of MscL homologues of Mycobacterium tuberculosis (TbMscL) and Staphylococcus aureus (SaMscL) have been solved although functional studies have been carried out mostly on the MscL of E. coli (EcMscL). TbMscL and SaMscL have an overall sequence identity of 37 per cent and 51 per cent, respectively, with EcMscL. TbMscL has a conductance similar to EcMscL but gated at pressure twice as high as EcMscL when expressed in E. coli cells (Moe et al. 2000).
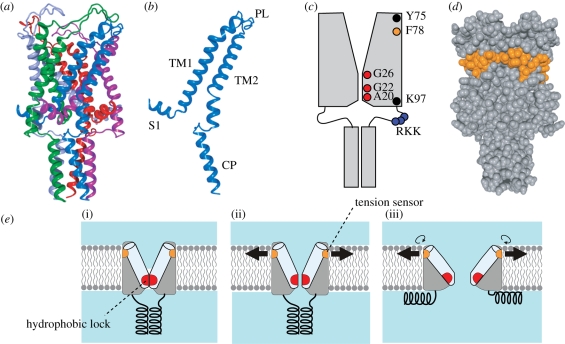
TbMscL is a homopentamer of a subunit that has two transmembrane helices, TM1 and TM2 (Chang et al. 1998; figure 3a,b). The sequence towards the amino-terminus forms the S1 helix and that towards the carboxy-terminus forms a bundle of cytoplasmic helix (CP). The pore is lined with TM1 and narrows towards the cytoplasmic end. Because the pore radius is smallest at Ile-14 and Val-21, the constriction at and between these two residues probably forms the gate. Based on the presence of this constriction, the resolved structure is assumed to represent the closed or nearly closed structure. Study by electron paramagnetic resonance (EPR) spectroscopy on EcMscL shows that this gate region is firmly immobilized, devoid of water and constricted although the region is slightly shifted to the carboxy-terminus (Ala-20 to Gly-26, i.e. Ala-18 to Gly-24 in TbMscL; Perozo et al. 2001). Cysteine and hystidine substitution experiments support the idea that the pore is constricted at Gly-26 in EcMscL (Iscla et al. 2004; Levin & Blount 2004). The peripheral residues facing the membrane comprise the whole length of TM2 and the periplasmic end of TM1.
Figure 3.
Structure and function of the MscL mechanosensitive channel. (a) Crystal structure of M. tuberculosis MscL (PDB accession number: 2OAR). Each subunit is in a different colour. (b) Structure of single subunit of TbMscL. Shown are N-terminal helix (S1), the first and second transmembrane helices (TM1 and TM2), periplasmic loop (PL) and cytoplasmic helix (CP). (c) Positions of important residues in the schematic EcMscL. The pore is constricted between A20 and G26. TM2 is immersed in lipid bilayer between Y75 and K97. Gating threshold changes with the hydrophilicity of G22 and mechanosensitivity is lost on hydrophilic substitution of Y78. Charged residues RKK are involved in pH sensitivity and oligomer assembly. (d) Hydrophobic residues that hamper mechanosensiticity on asparagine substitution. Residues corresponding to EcMscL are indicated in orange in the space fill model of TbMscL. (e) Gating model of MscL. The closed structure is stabilized by the hydrophobic lock of the gate (i). The membrane tension perceived by the tension sensor opens the gate, which results in the exposure of the hydrophobic lock to water (ii). On full opening, the cytoplasmic helices are disassembled and the hydrophobic lock is buried in the protein interior by the rotation of TM1 (iii).
EcMscL has a conductance of 3 nS and passes low molecular substances such as thioredoxin as well as inorganic ions (Ajouz et al. 1998). The diameter of the open pore is estimated to be as large as 3–4 nm while the outer diameter of the TbMscL is only approximately 5 nm (Cruickshank et al. 1997; Chang et al. 1998; Sukharev et al. 1999). This large pore is generated by an iris-like movement of the transmembrane helices: the helices tilt while sliding between neighbouring TM1 helices occurs (Sukharev et al. 2001a,b; Betanzos et al. 2002; Perozo et al. 2002a). Rotation of the transmembrane helices around the helix axis also takes place (Li et al. 2009). The tilting of transmembrane helices leads to the flattening of the transmembrane domain. Consistently, MscL embedded in thinner bilayer gates at low tension in the membrane (Perozo et al. 2002a,b).
The expansion in the transmembrane domain dissociates the CP helices probably through the linker between TM2 and CP (Yoshimura et al. 2008). Thus, it is not likely that CP helices and the TM2–CP linker serve as a prefilter for the open pore. Instead, it is possible that the bundle of the CP helices restrains the transmembrane helices from tilting and stabilizes the closed state because SaMscL with a deletion of CP is in an expanded state (Liu et al. 2009). Because deletion of CP helix and TM2–CP linker results in abnormal oligomerization, it is likely that proper pentamerization occurs through the intersubunit interactions in these regions.
The role of the periplasmic loop is not fully understood although a number of loss-of-function (LOF) mutations are found in this region (Maurer et al. 2000; Li et al. 2004; Yoshimura et al. 2004). The loop can serve as a spring to close the channel because the proteolytic digestion of the loop decreases the gating threshold (Ajouz et al. 2000). In line with this view, when an amino-terminal half including TM1 and a carboxy-terminal half including TM2 are expressed separately and reconstituted into a membrane, they form a functional mechanosensitive channel with increased mechanosensitivity. These observations also indicate that the periplasmic loop is not required for the integrity of MscL.
2.3. Lipid–protein interaction in MscL
The crystal structure of TbMscL suggests that the whole length of TM2 and the periplasmic end of TM1 have contact with membrane lipids although the exact position of the membrane relative to the protein cannot be deduced because of the absence of lipid in the crystal structure. The residues that interact with lipids in EcMscL were identified by labelling each residue in the helices by a spin-labelled probe and assessing the environment of the probe (Perozo et al. 2001). As expected from the helical structure, the residues that are estimated to have contact with membrane lipids appeared periodically. The assessment by Trp, whose fluorescence changes with the depth in the membrane, shows that TbMscL interacts with lipids in a range from Leu-69 to Leu-92, which can be translated from Tyr-75 to Lys-97 in EcMscL (figure 3c; Powl et al. 2005,b).
Here, we will first look over the mutants found through random mutagenesis, because unbiased screening can highlight the residues that cannot be predicted from the existing model. A number of LOF mutations, which resulted in cell lysis on hypo-osmotic shock, were found on the residues in the transmembrane helices as well as in the loop (Maurer & Dougherty 2003). The LOF mutants are frequently found on the hydrophobic residues facing the lipid especially towards the periplasmic end of the transmembrane helices. Independent screen of the mutations that suppress the leaky phenotype of the G22D mutant also shows that mutations at the hydrophobic residues at the periplasmic end of the transmembrane helix and various residues in the loop effectively silence the spontaneous gating (Yoshimura et al. 2004). The mutants identified through the random screen suggest that the function of MscL is more amenable to the disturbance of the hydrophobic interaction in the periplasmic leaflet than that in the cytoplasmic leaflet.
In fact, the outer and inner leaflets of the membrane have different affinity to MscL: the periplasmic residues in the transmembrane domain of MscL are more lipid exposed than the cytoplasmic one (Powl et al. 2005a). When MscL is reconstituted in the lipid bilayer composed of lipid with various tail length, the affinity of MscL to the inner leaflet changes with the thickness of the bilayer, whereas that to the outer leaflet did not change significantly (Powl et al. 2007). This result indicates that the lipid–protein interaction in the outer leaflet is more stable than in the inner leaflet.
Both screens mentioned above identified I41N MscL, which totally lacks the ability to rescue the cells from lysis on hypo-osmotic shock nor open on application of tension under patch clamp although the presence in the membrane is confirmed. Based on the idea that asparagine substitution of the lipid-contacting hydrophobic residue effectively infers the contact necessary for mechanosensitivity, each residue that contacts with lipid is replaced one by one with asparagine (Yoshimura et al. 2004). This asparagine-scanning mutagenesis highlighted seven residues that lead to total loss of the function on substitution. The residues display an impressive alignment along the periplasmic rim of the hydrophobic surface of MscL (figure 3d). Our recent molecular dynamics simulation suggests that one of the seven residues, Phe-78, has strongest interaction with lipid tail among all the lipid-facing amino residues.
Membrane proteins may interact with lipid headgroups through electrostatic and hydrogen bonding interactions. Molecular dynamics simulations showed that extensive hydrogen bonds are formed between MscL and the lipid headgroups, especially the ammonium of PE (Elmore & Dougherty 2001, 2003). However, when PE was supplemented to the PC bilayer, MscL opens at higher tension indicating that the hydrogen bond between PE and MscL does not reinforce the membrane tension to be transmitted to MscL (Moe & Blount 2005). MscL binds to PE, PC, PS and phosphatidic acid (PA) in the outer leaflet with an almost equal affinity indicating that the headgroups of the phospholipids in the outer leaflet do not have specific interactions with MscL (Powl et al. 2005b). On the other hand, anionic phospholipids like PS and PA have higher affinity to the cytoplasmic side of MscL compared with PE and PC. The binding of anionic phospholipid is probably due to the cluster of Arg-98, Lys-99 and Lys-100 (RKK) in TbMscL because neutralization of one of these residues decreases the affinity to anionic phospholipids. Because the high affinity to the anionic lipids is lost in a spontaneously opening mutant, V32K TbMscL, these charged residues presumably do not have access to the lipid in the open state. The role of RKK motif remains still to be explored because the effect of neutralization of all three charged residues in the linker is controversial: the mutant of TbMscL shows a gain-of-function (GOF) phenotype with reduced growth on the induction of expression, whereas that of EcMscL has much higher gating threshold than wild-type (Powl et al. 2005b; Kloda et al. 2006). The interaction between the anionic lipids and MscL is not likely to be essential to the mechanosensitivity because MscL can be activated in pure PC membrane and the mechanosensitivity does not change on addition of PS (Perozo et al. 2002a,b; Moe & Blount 2005).
The discussion above indicates that interaction of the hydrophobic residues at the periplasmic ends of transmembrane helices with the membrane lipid is essential to the tension-dependent gating (figure 3e). The interaction of MscL with the headgroups of lipid is, even if present, not critical for mechanosensitivity.
2.4. Energy barrier for the gating of MscL
In general, a channel in the closed state transits to the open state by overcoming the barrier that separates the energy well of the closed and the open states. In the case of mechanosensitive channels, the energy for activation, i.e. the energy to get over the barrier, derives from membrane tension (Sokabe et al. 1991; Hamill & Martinac 2001). Although the activation energy is used for every conformational change that occurs on gating, it is likely that there is a critical step that dominates the height of the energy barrier.
The mutants with low energetic barrier should be gated at low tension or even in the absence of applied tension. Such GOF mutants have been isolated through a screen for the mutants that show no or slow growth on the induction of expression (Ou et al. 1998). The mutants highlight the pore-lining residues, which decrease the gating threshold on substitution with a more hydrophilic amino acid. Substitution of Gly-22, which is one of the highlighted residues and resides within the gate, with all other 19 amino acids indicates that the hydrophilicity is the key factor that determines the ease of gating (Yoshimura et al. 1999). Reaction of G22C MscL with various methanethiosulphonate reagents confirms this view (Yoshimura et al. 2001). Given that the residues comprising the gate are mostly hydrophobic, it is likely that the exposure of the hydrophobic surface of the gate in TM1 to the aqueous environment is the energy barrier for gating. The gate can be regarded as a ‘hydrophobic lock’ because the MscL is stabilized in the closed conformation by the hydrophobic nature within the gate (figure 3e; Blount & Moe 1999; Yoshimura et al. 1999).
The dwelling in the fully open state, on the other hand, is stabilized by hydrophobic substitution at Gly-22 suggesting that Gly-22 is in a hydrophobic environment in the open state (Yoshimura et al. 1999). It is possible that Gly-22 and the residues comprising the hydrophobic lock are buried in the hydrophobic protein interior in the open state because cysteine scanning mutagenesis and EPR studies indicate that TM1 rotates on gating (figure 3e; Perozo et al. 2002a; Levin & Blount 2004; Li et al. 2009).
Besides the primary gate of the pore constriction, a secondary gate by S1 helices may also exist and open (Sukharev et al. 2001b). In this model, S1 helices associate together to plug the cytoplasmic opening of the pore and an expansion of the transmembrane domain dissociates the S1 helices through the S1–TM1 linker. Kinetic analysis indicates that the opening of the primary gate is associated with most of the in-plane expansion and the free energy change (Anishkin et al. 2005). The opening of the primary gate provides only a fraction of the full conductance; the full conductance is attained by the opening of the S1 gate, which is virtually not tension dependent and energy demanding. While this model is supported by numerous cross-link experiments (note, however, Iscla et al. 2008), the S1 helices in the crystal structure of TbMscL do not form a bundle but spread away from the channel axis. It is possible that the absence of the bundle in the crystal structure is due to the 23 amino acids tag added to the amino-terminus, which may not fit to the space between transmembrane domain and cytoplasmic helices.
2.5. MscS structure and interaction with lipid
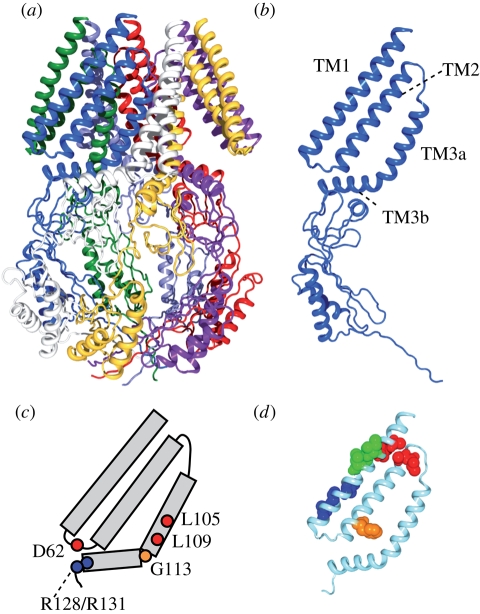
MscS crystal structure of E. coli, in contrast to MscL, demonstrates that MscS is a homoheptamer of a subunit with three transmembrane helices, TM1, TM2 and TM3 (figure 4a,b; Bass et al. 2002). TM1 comprises the outermost layer of the transmembrane domains and faces the lipid bilayer. Some residues of TM2 residues may have access to lipid through the gaps between TM1. TM3 has a characteristic kink at G113, which separates TM3 into amino-terminal helix TM3a and carboxy-terminal helix TM3b. TM3a lines the pore that passes ion on gating. TM3a helices fit each other to form a tightly closed gate by inserting the knobs of alanine residues to the holes of glycine residues (Edwards et al. 2005). The cytoplasmic domain forms a large cage or vestibule, which has a hollow cavity at the centre, seven windows on the side and a small opening at the bottom. TM3b covers the upper surface of the cytoplasmic cage. The 26 amino-terminal residues, which were not resolved in the crystal structure form an α-helix at the membrane interface (Vásquez et al. 2008a,b).
Figure 4.
Structure and function of the MscS mechanosensitive channel. (a) Crystal structure of E. coli MscS (PDB accession number: 2OAU). Each subunit is in a different colour. (b) Structure of single subunit of MscS. Indicated are the first and second transmembrane helices (TM1 and TM2) and the third transmembrane helix, which is separated into two by a kink at G113 (TM3a and TM3b). (c) Important residues for the function of MscS. L105 and L109 form the constriction of the pore. D62 forms a salt bridge with R128 and R131. A kink at G113 occurs in the inactive state. (d) The gating threshold increases on asparagine substitution at the residues indicated by red (A34, I37, A85, L86), blue (I48, A51, L55), and orange (F68). Conversely, the threshold decreases on mutation at I39, V40 and I43 (green). Data from Nomura et al. (2006) and Okada et al. (2002).
The permeation pore is constricted near the cytoplasmic end by the side chains of L105 and L109 (figure 4c). Because the most constricted part of the pore has a diameter of 7 Å in the crystal structure, the authors assigned this structure as an open structure of MscS. However, following molecular dynamics simulation indicated that the pore is devoid of water and ions because of the hydrophobic surface of the pore and thus is not conductive (Anishkin & Sukharev 2004). Although the crystal structure does not seem to be in a conductive state, it does not appear to represent the closed state, either. Note that the pair of TM1 and TM2 helices is splayed away from the core bundle of TM3a helix and there is a gap between TM2 and TM3a in the crystal structure. Assessment of the environment of each residue and computer modelling suggest that TM2 and TM3a are aligned together and all transmembrane helices are packed close to the central axis of the channel (Anishkin et al. 2008a,b; Vásquez et al. 2008a,b). Because preventing the TM3 from kinking at G113 makes the inactivated state unfavourable while the channel normally opens and closes, the G113 kink present in the crystal structure is likely to be the sign that the crystal structure is close to the inactivated state (Akitake et al. 2007; Edwards et al. 2008).
Nearly the whole of the cage is indispensable because the deletion of the 30 carboxy-terminal residues results in failure to be expressed in the membrane (Miller et al. 2003). The cage is not a static mesh but dynamically swells on channel opening (Machiyama et al. 2009). Consistently, inhibition of swelling by adding high molecular substances to the cytoplasmic side accelerated inactivation and cross-linking the carboxy-terminal end prevented the channel opening (Koprowski & Kubalski 2003; Grajkowski et al. 2005). The protein backbone connects the cage to the transmembrane domain only at the hinge between TM3a and TM3b, but there is likely to be additional contact through a salt bridge. The positively charged residues (R128 and R131) at the peripheral end of TM3b interact with the negatively charged residue (D62) in the loop connecting TM1 and TM2 as indicated by molecular dynamics simulation and functional analysis (Sotomayor & Schulten 2004; Nomura et al. 2008). The disruption of the salt bridge elevates the threshold and the rate of inactivation, indicating that the salt bridge is required for proper opening and inactivation. In addition, the loss of the salt bridge decreases the magnitude of the cage expansion suggesting that the conformational change in the transmembrane domain is passed to the cage through the salt bridge at least in part (Machiyama et al. 2009). Fuller understanding of the role of the D62–R128/R131 salt bridge in the dynamics of transmembrane and cytoplasmic domain is expected to provide a global view of MscS gating.
The assessment of the environment using EPR indicates that one side of TM1 is exposed to membrane lipid and the other side is in a proteinous environment (Vásques et al. 2008b). TM2 is mostly buried in the protein. An attempt to disturb the hydrophobic environment by asparagine substitution, which was previously employed on MscL, identified several mutants with increased or decreased gating threshold (Nomura et al. 2006). Remapping the mutants onto the revised MscS crystal structure reveals that they form a cluster (figure 4d). Four hydrophobic residues at the periplasmic ends of TM1 and TM2 increase the threshold on asparagine substitution (indicated by red). The loss of hydrophobic contact between TM1 and TM2 at these ends possibly hampers the transmission of the membrane tension towards the core of the channel. The mutants on one side of TM1 close to the cytoplasmic end also have reduced mechanosensitivity (blue). Because they are at the border of lipidous and proteinous environment, loose packing of TM1 is possible to distort the tension-sensitive conformation. Phe-68 (orange) points towards TM3 and transmits membrane tension to the gate through hydrophobic contact between TM2 and TM3a (Belyy et al. in press). In contrast, three mutants with decreased threshold are found as a group on the lipidic face of TM1 (green). The presence of these GOF mutants and the absence of LOF mutants in the lipid–protein interface contrast with the mutational study on MscL. It is probable that there is no hot spot in the lipid–protein interface for the mechanosensitivity of MscS while contact at the periplasmic rim is significant in MscL. The finding that double mutations on the same side of the membrane are additive in terms of defects while those on both sides are not supports the idea that lipid–protein interaction at both sides should be balanced for the efficient mechanosensitivity of MscS (Nomura et al. 2006).
2.6. Structural change on gating in MscS
Several attempts have been made to solve the MscS structure in different gating states. The crystal structure of A106V MscS has a pore with a diameter of 13 Å, which is wide enough to assume that the protein is in a conductive state (Wang et al. 2008). The open nature of the crystal structure of A106V MscS is possibly due to the absence of membrane because this mutant does not tend to adopt the open state in the membrane when examined by patch clamp; rather, it requires higher tension to gate than wild-type MscS. When compared with the ‘closed’ crystal structure, the pair of TM1 and TM2 is tilted around the cytoplasmic end while the pair also rotates clockwise viewed from the periplasmic side. In the course of pore opening, TM3a is estimated to tilt upward and move apart from the sevenfold axis without axial rotation. This movement is accompanied by the passing of A110 across L115 of the neighbouring subunit. Consistent with the idea that the energy needed for the side chains of these residues to pass each other is the barrier for the interconversion of the closed and the open state, the gating threshold changed with the size of the side chain at these positions.
EPR spectroscopy also resolved the structural change on asymmetric incorporation of LPC (Vásquez et al. 2008a,b). The structure deduced from the resolved information suggests that the pore opening is accompanied by an axial rotation and inclination of TM3a, which were not observed in the structure of A106C MscS. The axial rotation of TM3a exposes A98, A106 and G113 to the pore and the inclination straightens TM3 especially at G113, whose kink is proposed to occur only in the inactive state (Akitake et al. 2007). Rotation of TM1a and TM2 and downward tilting of TM1 also take place.
Yet another model based on computation and electrophysiology suggests a different possibility (Anishkin et al. 2008a,b). On channel opening, pore-constricting L105 and L109 are removed by the rotation of TM3a, as suggested by EPR spectroscopy, but the rotation exposes G101 and G104 to the pore, which is located opposite to A98 and A106 relative to the central axis of TM3a. TM3 is straightened at G113, again consistent with the idea that a kink at G113 is not present in the open state.
The variation of the ‘open’ structures shown above highlights the difficulty in determining the structure that can hardly be reproduced in the process of structure determination. More information on the physiological characteristics of mutants and the structural change monitored in physiological conditions are required for the judgement on which model is plausible.
3. Voltage-gated potassium channel
3.1. Voltage-dependent gating
Voltage-gated potassium channels build a large superfamily of voltage-gated channels together with voltage-gated calcium channels and voltage-gated sodium channels. Typical voltage-gated channels have six transmembrane helices, S1, S2, S3, S4, S5 and S6. The sequence connecting S5 and S6 is referred to as the P-loop because it lines the pore of the channel as will be shown below. S4 has a characteristic array of the positive charges of arginine or lysine at every third position that is involved in voltage sensing. Although a voltage-gated potassium channel is a homotetramer of a subunit that has one set of S1–S6, a voltage-gated calcium or sodium channel is formed by a long polypeptide that has four homologous repetitions of S1–S6. The four segments are believed to be a result of internal duplication of the gene.
The transmembrane voltage tightly regulates the open probability of the voltage-gated channels. The channels can monitor the transmembrane voltage by the translocation of charges or by the tilt of dipoles across the electric field (Hodgkin & Huxley 1952). The movement of the charges within the voltage-gated channels has been experimentally measured as the ‘gating current’ (Armstrong & Bezanilla 1973). The ‘gating charges’ that carry the gating current are attributed to the four arginine residues in the S4 helix and a glutamic acid residue in S2 (Aggarwal & MacKinnon 1996; Seoh et al. 1996).
3.2. Structure of potassium channel
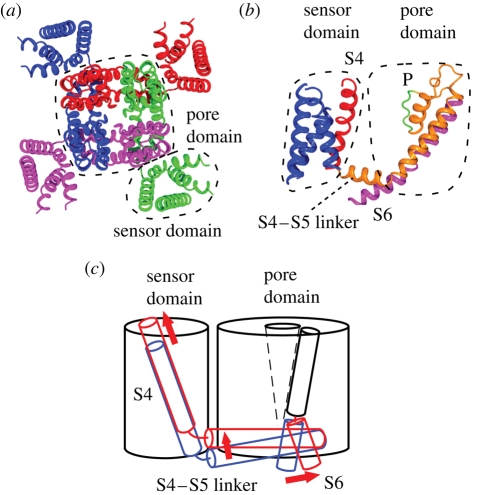
The main functional components of voltage-gated channels are the voltage sensor and the pore, which governs the gate and the selectivity filter. Each of them is also made up of structurally distinct domains: the pore domain and the sensor domain as shown in the crystal structure of Kv1.2 potassium channel (figure 5a,b; Long et al. 2005a). The Kv1.2 potassium channel is a member of the Shaker potassium channel family, whose gene was the first to be identified among the potassium channels and whose electrophysiological characteristics have been studied in detail. The pore domain of the Kv1.2 potassium channel consists of S5, S6 and the P-loop from four subunits. The pore, which is lined by S6 helices, provides the passage of potassium ions at low energetic cost. The gate is at the intracellular end of the pore and opens at the voltage specific to the channel type. On the other hand, the selectivity filter is at the extracellular end of the pore. S1, S2, S3 and S4 helices form the sensor domain, which surrounds the pore domain. The sensor domain loosely connects the pore domain through the S4–S5 linker helix. The crystal structure is likely to be in an open or inactivated state because there is no electric field in the protein crystal.
Figure 5.
Structure and conformational change of the potassium channel. (a) Crystal structure of Kv1.2 potassium channel (PDB accession number: 2A79). Each subunit is in a different colour. The pore domain is surrounded by four sensor domains. (b) Structure of a single subunit showing the position of S4 (red), selectivity filter (P), and gate-forming S6 (magenta). (c) Schematic representation of the conformational change that occurs on gating. Blue and red represent the position in the resting and activated state, respectively.
The selectivity filter comprises a sequence of TVGYGD, which is conserved among potassium channels. It is not the side chains of TVGYGD that line the pore of the selectivity filter (Zhou et al. 2001). Instead, the carbonyl groups of the backbone of these residues surround the selectivity filter as was predicted (Eisenman & Dani 1987). There are four sites where K+ resides in the selectivity filter. Each dehydrated K+ in the selectivity filter is surrounded by eight oxygens of the carbonyl group pointing towards the pore. The oxygens replace the water molecules hydrating K+ and enable high-throughput flow of K+ by reducing the energy requirement for dehydration. Binding of impermeable ions such as Na+ alters the filter into a collapsed non-conducting structure providing K+ selectivity over Na+ (Lockless et al. 2007).
The gate is the hydrophobic constriction of the tight bundling of the S6 helices towards the cytoplasmic end of the pore. The voltage detected by the sensor domain triggers the movement of the S6 helices and opens the gate through which ions flow (Perozo et al. 1999). An outward bend of the cytoplasmic section of the S6 helix associates the gating movement of the S6 helix (figure 5c; Long et al. 2005a). The movement of the voltage sensor can be passed to S6 through the contact with S4–S5 linker (figure 5b).
In the open structure of Kv1.2, the two outer arginines (R1 and R2) among the gating charges in S4 protrude from the transmembrane domain and are exposed to the environment surrounding the channel (Long et al. 2005b, 2007). The outermost arginine (R1) is likely to interact with the phosphate of the membrane lipid. The next three arginines are in the protein interior and interact with acidic residues in S1–S3, forming an electrostatic interaction network. The interior of the sensor domain is penetrated by water through a cleft as has been shown in several accessibility experiments and the crystal structure. Because the water-filled clefts should be isopotential to the environment, the electric field should be focused in the region where water is absent.
3.3. Influence of lipids on voltage-gated potassium channel
While the structural bases of the gating mechanism of the voltage-gated channels have a long history of research, the role of lipids in the function of the voltage-gated channels is a newly evolving issue. The separation of the sensor domains and the pore domain should increase the surface exposed to the membrane lipids compared with a single cylindrical structure. The invasion of lipids into the grooves of the potassium channels was evidenced by the crystallization of the chimaera between Kv1.2 and Kv2.1 in a lipid containing solution (Long et al. 2007). The lipids are found in the gaps between the sensor domain and the pore domain as well as in the grooves between the neighbouring sensor domains. Lipid molecules are also found in the clefts between the transmembrane helices in the sensor domains. The voltage-gated channels are apparently intruded more extensively than the mechanosensitive channels.
Specific interaction between the voltage-gated potassium channel and the lipid is found at the end of the S4 helix of the voltage sensor. The outermost arginine R1 of the isolated sensor domain of KvAP protrudes from the transmembrane domain and is in a position where an interaction with lipid phosphate is possible (Jiang et al. 2003). Consistently, the environment of the R1 of KvAP as assessed by EPR is the water–lipid interface (Cuello et al. 2004). In addition, a molecular dynamics simulation on the isolated S4 of KvAP indicates that the arginines form a hydrogen-bonded network with the lipid phosphates and water (Freites et al. 2005). Intriguingly, KvAP does not open when incorporated into the bilayer of the lipids that lack phosphate, whereas KvAP tolerates changes in the headgroup attached to the phosphate or the length of lipid tail (Schmidt et al. 2006). The negative charge seems to be the essential factor of the phosphate because simple uncharging of the phosphate by ethylation results in the loss of function. Given that the crystal structure is in the open configuration, these results are consistent with the idea that the interaction of the R1 arginine with the lipid phosphate stabilizes the active position of the S4 helix.
The action of the venom of the brown spider Loxosceles reclusa depends also on the interaction between the voltage-gated potassium channels and lipids (Ramu et al. 2006). The venom contains sphingomyelinase D, which removes the positively charged choline group from zwitterionic sphingomyelin yielding a negatively charged lipid ceramide 1-phosphate. The treatment of sphingomyelase D activated the voltage-gated potassium channels, even when the membrane is hyperpolarized, suggesting that negative charge of the headgroups of ceramide 1-phosphate stabilizes the open state. Conversely, removing both the choline group together with the phosphate by sphingomyelinase C stabilized the closed state (Xu et al. 2008). Because the gating current increases and decreases with sphingomyelase D and sphingomyelase C treatment, respectively, it is the movement of the S4 charges that is affected by the cleavage of the headgroup of the lipids. The stabilization of the open state with the lipid with a negative headgroup supports the idea of the interaction between the arginine R1 and the lipid phosphates in the open state.
Another spider toxin, hanatoxin, inihibits the action of the voltage-gated potassium channel by shifting the activation curve to more depolarized voltages (Swartz & MacKinnon 1997a). The binding of hanatoxin is attributed to the residues near the outer edges of S3 while mutations at the S4 arginines also have moderate effect on the binding (Swartz & MacKinnon 1997b). Hanatoxin probably partitions in the lipid bilayer as suggested in a related toxin, VSTX1 (Lee & MacKinnon 2004). The hanatoxin binding site in the Kv1.2 chimaera points outward at the level of the water–lipid interface suggesting that the binding of hanatoxin in the S3–S4 linker hampers the mobility of S4 and demands more positive voltage for the action.
Because the lateral pressure is a general phenomenon occurring on the membrane proteins, it is probable that changes in the lateral pressure alter the gating characteristics of the voltage-gated potassium channels. In fact, the activities of a voltage-gated potassium channel, Shaker-IR, are subject to the membrane tension: they are activated at low tension and depressed at high tension (Gu et al. 2001). Reflecting the general feature of the influence of the lateral pressure on the structure of the membrane proteins, the sensitivity of the ion channel activities to the membrane stretch has been reported for an increasing number of ion channels, including the N-methyl-d-aspartate receptor, transient receptor potential channels and gramicidin (Martinac 2004). One possible scenario of the activation of the voltage-gated potassium channels is that the membrane tension eases the opening of the pore as in the case of bacterial mechanosensitive channels. This idea was tested on a mutant, Shaker ILT, whose rate limiting step is the pore opening (Laitko et al. 2006). The open probability of Shaker ILT is affected by tension but it decreases with tension in contrast to the expectation that pore opening is favoured by the applied tension. This result implies that open probability of the Shaker potassium channel does not change owing to the in-plane expansion of the pore domain but is a result of the modification of some steps preceding the pore opening. The direct action of membrane tension on the gating of MscS and MscL, in turn, highlights the functionally developed and sophisticated structure of the mechanosensitive channels.
4. Conclusion
Membrane proteins are subjected to changes in the physical properties of membrane lipids. Ion channels are no exceptions and their activities are influenced by the length and the number of the lipid tail and the charges, and the size of the head groups. Mechanosensitive channels are particularly amenable to the physical conditions of the membrane and report the membrane tension. The structure–function studies on the bacterial mechanosensitive channels have revealed how dynamically and systematically the protein structure changes with the applied force. Evidence is emerging that the voltage-gated channels operate with the help of membrane lipids. Further understanding of the impacts of lipids on the membrane proteins should unveil the molecular mechanism of the mechanosensitive channels and the environment of the habitat of the membrane proteins, which comprise approximately 25 per cent of the total genes in most organisms.
Acknowledgements
This work was partly supported by grants for Scientific Research on Priority Areas (15086270) and Creative Scientific Research (16GS0308) from MEXT, and a grant for the ICORP/SORST Cell Mechanosensing from JST.
Footnotes
One contribution to a Theme Supplement ‘Mechanobiology’.
References
- Aggarwal S. K., MacKinnon R. 1996. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron 16, 1169–1177. ( 10.1016/S0896-6273(00)80143-9) [DOI] [PubMed] [Google Scholar]
- Ajouz B., Berrier C., Garrigues A., Besnard M., Ghazi A. 1998. Release of thioredoxin via the mechanosensitive channel MscL during osmotic downshock of Escherichia coli cells. J. Biol. Chem. 273, 26 670–26 674. ( 10.1074/jbc.273.41.26670) [DOI] [PubMed] [Google Scholar]
- Ajouz B., Berrier C., Besnard M., Martinac B., Ghazi A. 2000. Contributions of the different extramembranous domains of the mechanosensitive ion channel MscL to its response to membrane tension. J. Biol. Chem. 275, 1015–1022. ( 10.1074/jbc.275.2.1015) [DOI] [PubMed] [Google Scholar]
- Akitake B., Anishkin A., Liu N., Sukharev S. 2007. Straightening and sequential buckling of the pore-lining helices define the gating cycle of MscS. Nat. Struct. Mol. Biol. 14, 1141–1149. ( 10.1038/nsmb1341) [DOI] [PubMed] [Google Scholar]
- Anishkin A., Sukharev S. 2004. Water dynamics and dewetting transitions in the small mechanosensitive channel MscS. Biophys. J. 86, 2883–2895. ( 10.1016/S0006-3495(04)74340-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishkin A., Chiang C. S., Sukharev S. 2005. Gain-of-function mutations reveal expanded intermediate states and a sequential action of two gates in MscL. J. Gen. Physiol. 125, 155–170. ( 10.1085/jgp.200409118) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishkin A., Akitake B., Sukharev S. 2008a Characterization of the resting MscS: modeling and analysis of the closed bacterial mechanosensitive channel of small conductance. Biophys. J. 94, 1252–1266. ( 10.1529/biophysj.107.110171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anishkin A., Kamaraju K., Sukharev S. 2008b Mechanosensitive channel MscS in the open state: modeling of the transition, explicit simulations, and experimental measurements of conductance. J. Gen. Physiol. 132, 67–83. ( 10.1085/jgp.200810000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong C. M., Bezanilla F. 1973. Currents related to movement of the gating particles of the sodium channels. Nature 242, 459–461. ( 10.1038/242459a0) [DOI] [PubMed] [Google Scholar]
- Bass R. B., Strop P., Barclay M., Rees D. C. 2002. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science 298, 1582–1587. ( 10.1126/science.1077945) [DOI] [PubMed] [Google Scholar]
- Belyy V., Anishkin A., Kamaraju K., Liu N., Sukharev S. In press The tension-transmitting ‘clutch’ in a mechanosensitive channel. Nat. Struct. Mol. Biol. ( 10.1038/nsmb.1775) [DOI] [PubMed] [Google Scholar]
- Berrier C., Besnard M., Ajouz B., Coulombe A., Ghazi A. 1996. Multiple mechanosensitive ion channels from Escherichia coli, activated at different thresholds of applied pressure. J. Membr. Biol. 151, 175–187. ( 10.1007/s002329900068) [DOI] [PubMed] [Google Scholar]
- Betanzos M., Chiang C. S., Guy H. R., Sukharev S. 2002. A large iris-like expansion of a mechanosensitive channel protein induced by membrane tension. Nat. Struct. Biol. 9, 704–710. ( 10.1038/nsb828) [DOI] [PubMed] [Google Scholar]
- Blount P., Moe P. C. 1999. Bacterial mechanosensitive channels: integrating physiology, structure and function. Trends Microbiol. 7, 420–424. ( 10.1016/S0966-842X(99)01594-2) [DOI] [PubMed] [Google Scholar]
- Cantor R. S. 1997. The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry 36, 2339–2344. ( 10.1021/bi9627323) [DOI] [PubMed] [Google Scholar]
- Chalfie N. 2009. Neurosensory mechanotransduction. Nat. Rev. Mol. Cell Biol. 10, 44–52. ( 10.1038/nrm2595) [DOI] [PubMed] [Google Scholar]
- Chang G., Spencer R. H., Lee A. T., Barclay M. T., Rees D. C. 1998. Structure of the MscL homolog from Mycobacterium tuberculosis: a gated mechanosensitive ion channel. Science 282, 2220–2226. ( 10.1126/science.282.5397.2220) [DOI] [PubMed] [Google Scholar]
- Cruickshank C. C., Minchin R. F., Le Dain A. C., Martinac B. 1997. Estimation of the pore size of the large-conductance mechanosensitive ion channel of Escherichia coli. Biophys. J. 73, 1925–1931. ( 10.1016/S0006-3495(97)78223-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuello L. G., Cortes D. M., Perozo E. 2004. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science 306, 491–495. ( 10.1126/science.1101373) [DOI] [PubMed] [Google Scholar]
- Cui C., Smith D. O., Adler J. 1995. Characterization of mechanosensitive channels in Escherichia coli cytoplasmic membrane by whole-cell patch clamp recording. J. Membr. Biol. 144, 31–42. ( 10.1007/BF00238414) [DOI] [PubMed] [Google Scholar]
- Doyle D. A., Morais Cabral J., Pfuetzner R. A., Kuo A., Gulbis J. M., Cohen S. L., Chait B. T., MacKinnon R. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280, 69–77. ( 10.1126/science.280.5360.69) [DOI] [PubMed] [Google Scholar]
- Dumas F., Tocanne J. F., Leblanc G., Lebrun M. C. 2000. Consequences of hydrophobic mismatch between lipids and melibiose permease on melibiose transport. Biochemistry 39, 4846–4854. ( 10.1021/bi992634s) [DOI] [PubMed] [Google Scholar]
- Edwards M. D., et al. 2005. Pivotal role of the glycine-rich TM3 helix in gating the MscS mechanosensitive channel. Nat. Struct. Mol. Biol. 12, 113–119. ( 10.1038/nsmb895) [DOI] [PubMed] [Google Scholar]
- Edwards M. D., Bartlett W., Booth I. R. 2008. Pore mutations of the Escherichia coli MscS channel affect desensitization but not ionic preference. Biophys. J. 94, 3003–3013. ( 10.1529/biophysj.107.123448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenman G., Dani J. A. 1987. An introduction to molecular architecture and permeability of ion channels. Annu. Rev. Biophys. Biophys. Chem. 16, 205–226. ( 10.1146/annurev.bb.16.060187.001225) [DOI] [PubMed] [Google Scholar]
- Elmore D. E., Dougherty D. A. 2001. Molecular dynamics simulations of wild-type and mutant forms of the Mycobacterium tuberculosis MscL channel. Biophys. J. 81, 1345–1359. ( 10.1016/S0006-3495(01)75791-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore D. E., Dougherty D. A. 2003. Investigating lipid composition effects on the mechanosensitive channel of large conductance (MscL) using molecular dynamics simulations. Biophys. J. 85, 1512–1524. ( 10.1016/S0006-3495(03)74584-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freites J. A., Tobias D. J., von Heijne G., White S. H. 2005. Interface connections of a transmembrane voltage sensor. Proc. Natl Acad. Sci. USA 102, 15 059–15 064. ( 10.1073/pnas.0507618102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonen T., Cheng Y., Sliz P., Hiroaki Y., Fujiyoshi Y., Harrison S. C., Walz T. 2005. Lipid–protein interactions in double-layered two-dimensional AQP0 crystals. Nature 438, 633–638. ( 10.1038/nature04321) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajkowski W., Kubalski A., Koprowski P. 2005. Surface changes of the mechanosensitive channel MscS upon its activation, inactivation, and closing. Biophys. J. 88, 3050–3059. ( 10.1529/biophysj.104.053546) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillet N., Kazmierczak P., Xiong W., Schwander M., Reynolds A., Sakaguchi H., Tokita J., Kachar B., Müller U. 2009. The mechanotransduction machinery of hair cells. Sci. Signal. 2, pt5. ( 10.1126/scisignal.285pt5) [DOI] [PubMed] [Google Scholar]
- Gu C. X., Juranka P. F., Morris C. E. 2001. Stretch-activation and stretch-inactivation of Shaker-IR, a voltage-gated K+ channel. Biophys J. 80, 2678–2693. ( 10.1016/S0006-3495(01)76237-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guharay F., Sachs F. 1984. Stretch-activated single ion channel currents in tissue-cultured embryonic chick skeletal muscle. J. Physiol. 352, 685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill O. P., Martinac B. 2001. Molecular basis of mechanotransduction in living cells. Physiol. Rev. 81, 685–740. [DOI] [PubMed] [Google Scholar]
- Hayakawa K., Tatsumi H., Sokabe M. 2008. Actin stress fibers transmit and focus force to activate mechanosensitive channels. J. Cell Sci. 121, 496–503. ( 10.1242/jcs.022053) [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. 1975. Three-dimensional model of purple membrane obtained by electron microscopy. Nature 257, 28–32. ( 10.1038/257028a0) [DOI] [PubMed] [Google Scholar]
- Hilf R. J., Dutzler R. 2008. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature 452, 375–379. ( 10.1038/nature06717) [DOI] [PubMed] [Google Scholar]
- Hodgkin A. L., Huxley A. F. 1952. A quantitative description of membrane current and its application to conduction and excitation in nerve. J. Physiol. 117, 500–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscla I., Levin G., Wray R., Reynolds R., Blount P. 2004. Defining the physical gate of a mechanosensitive channel, MscL, by engineering metal-binding sites. Biophys. J. 87, 3172–3180. ( 10.1529/biophysj.104.049833) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iscla I., Wray R., Blount P. 2008. On the structure of the N-terminal domain of the MscL channel: helical bundle or membrane interface. Biophys. J. 95, 2283–2291. ( 10.1529/biophysj.107.127423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Lee A., Chen J., Ruta V., Cadene M., Chait B. T., MacKinnon R. 2003. X-ray structure of a voltage-dependent K+ channel. Nature 423, 33–41. ( 10.1038/nature01580) [DOI] [PubMed] [Google Scholar]
- Kawate T., Michel J. C., Birdsong W. T., Gouaux E. 2009. Crystal structure of the ATP-gated P2X4 ion channel in the closed state. Nature 460, 592–598. ( 10.1038/nature08198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloda A., Ghazi A., Martinac B. 2006. C-terminal charged cluster of MscL, RKKEE, functions as a pH sensor. Biophys. J. 90, 1992–1998. ( 10.1529/biophysj.105.075481) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprowski P., Kubalski A. 2003. C termini of the Escherichia coli mechanosensitive ion channel (MscS) move apart upon the channel opening. J. Biol. Chem. 278, 11237–11245. ( 10.1074/jbc.M212073200) [DOI] [PubMed] [Google Scholar]
- Kuo A., et al. 2003. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300, 1922–1926. ( 10.1126/science.1085028) [DOI] [PubMed] [Google Scholar]
- Laitko U., Juranka P. F., Morris C. E. 2006. Membrane stretch slows the concerted step prior to opening in a Kv channel. J. Gen. Physiol. 127, 687–701. ( 10.1085/jgp.200509394) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A. G. 2003. Lipid–protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta 1612, 1–40. ( 10.1016/S0005-2736(03)00056-7) [DOI] [PubMed] [Google Scholar]
- Lee S. Y., MacKinnon R. 2004. A membrane-access mechanism of ion channel inhibition by voltage sensor toxins from spider venom. Nature 430, 232–235. ( 10.1038/nature02632) [DOI] [PubMed] [Google Scholar]
- Levin G., Blount P. 2004. Cysteine scanning of MscL transmembrane domains reveals residues critical for mechanosensitive channel gating. Biophys. J. 86, 2862–2870. ( 10.1016/S0006-3495(04)74338-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levina N., Tötemeyer S., Stokes N. R., Louis P., Jones M. A., Booth I. R. 1999. Protection of Escherichia coli cells against extreme turgor by activation of MscS and MscL mechanosensitive channels: identification of genes required for MscS activity. EMBO J. 18, 1730–1737. ( 10.1093/emboj/18.7.1730) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Moe P. C., Chandrasekaran S., Booth I. R., Blount P. 2002. Ionic regulation of MscK, a mechanosensitive channel from Escherichia coli. EMBO J. 21, 5323–5330. ( 10.1093/emboj/cdf537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Wray R., Blount P. 2004. Intragenic suppression of gain-of-function mutations in the Escherichia coli mechanosensitive channel, MscL. Mol. Microbiol. 53, 485–495. ( 10.1111/j.1365-2958.2004.04150.x) [DOI] [PubMed] [Google Scholar]
- Li Y., Wray R., Eaton C., Blount P. 2009. An open-pore structure of the mechanosensitive channel MscL derived by determining transmembrane domain interactions upon gating. FASEB J. 23, 2197–2204. ( 10.1096/fj.09-129296) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Gandhi C. S., Rees D. C. 2009. Structure of a tetrameric MscL in an expanded intermediate state. Nature 461, 120–124. ( 10.1038/nature08277) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockless S. W., Zhou M., MacKinnon R. 2007. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5, 1079–1088. ( 10.1371/journal.pbio.0050121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. B., Campbell E. B., Mackinnon R. 2005a Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science 309, 897–903. ( 10.1126/science.1116269) [DOI] [PubMed] [Google Scholar]
- Long S. B., Campbell E. B., Mackinnon R. 2005b Voltage sensor of Kv1.2: structural basis of electromechanical coupling. Science 309, 903–908. ( 10.1126/science.1116270) [DOI] [PubMed] [Google Scholar]
- Long S. B., Tao X., Campbell E. B., MacKinnon R. 2007. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382. ( 10.1038/nature06265) [DOI] [PubMed] [Google Scholar]
- Machiyama H., Tatsumi H., Sokabe M. 2009. Structural changes in the cytoplasmic domain of the mechanosensitive channel MscS during opening. Biophys. J. 97, 1048–1057. ( 10.1016/j.bpj.2009.05.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., Tsukihara T. 2009. Structure of the connexin 26 gap junction channel at 3.5 Å resolution. Nature 458, 597–602. ( 10.1038/nature07869) [DOI] [PubMed] [Google Scholar]
- Martinac B. 2004. Mechanosensitive ion channels: molecules of mechanotransduction. J. Cell Sci. 117, 2449–2460. ( 10.1242/10.1242/jcs.01232) [DOI] [PubMed] [Google Scholar]
- Martinac B., Buechner M., Delcour A. H., Adler J., Kung C. 1987. Pressure-sensitive ion channel in Escherichia coli. Proc. Natl Acad. Sci. USA 84, 2297–2301. ( 10.1073/pnas.84.8.2297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinac B., Adler J., Kung C. 1990. Mechanosensitive ion channels of E. coli activated by amphipaths. Nature 348, 261–263. ( 10.1038/348261a0) [DOI] [PubMed] [Google Scholar]
- Maurer J. A., Dougherty D. A. 2003. Generation and evaluation of a large mutational library from the Escherichia coli mechanosensitive channel of large conductance, MscL: implications for channel gating and evolutionary design. J. Biol. Chem. 278, 21 076–21 082. ( 10.1074/jbc.M302892200) [DOI] [PubMed] [Google Scholar]
- Maurer J. A., Elmore D. E., Lester H. A., Dougherty D. A. 2000. Comparing and contrasting Escherichia coli and Mycobacterium tuberculosis mechanosensitive channels (MscL). New gain of function mutations in the loop region. J. Biol. Chem. 275, 22 238–22 244. ( 10.1074/jbc.M003056200) [DOI] [PubMed] [Google Scholar]
- Miller S., Bartlett W., Chandrasekaran S., Simpson S., Edwards M., Booth I. R. 2003. Domain organization of the MscS mechanosensitive channel of Escherichia coli. EMBO J. 22, 36–46. ( 10.1093/emboj/cdg011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio K., Ogura T., Kiyonaka S., Hiroaki Y., Tanimura Y., Fujiyoshi Y., Mori Y., Sato C. 2007. The TRPC3 channel has a large internal chamber surrounded by signal sensing antennas. J. Mol. Biol. 367, 373–383. ( 10.1016/j.jmb.2006.12.043) [DOI] [PubMed] [Google Scholar]
- Miyazawa A., Fujiyoshi Y., Unwin N. 2003. Structure and gating mechanism of the acetylcholine receptor pore. Nature 423, 949–955. ( 10.1038/nature01748) [DOI] [PubMed] [Google Scholar]
- Moe P., Blount P. 2005. Assessment of potential stimuli for mechano-dependent gating of MscL: effects of pressure, tension, and lipid headgroups. Biochemistry 44, 12 239–12 244. ( 10.1021/bi0509649) [DOI] [PubMed] [Google Scholar]
- Moe P. C., Levin G., Blount P. 2000. Correlating a protein structure with function of a bacterial mechanosensitive channel. J. Biol. Chem. 275, 31 121–31 127. ( 10.1074/jbc.M002971200) [DOI] [PubMed] [Google Scholar]
- Murata K., Mitsuoka K., Hirai T., Walz T., Agre P., Heymann J. B., Engel A., Fujiyoshi Y. 2000. Structural determinants of water permeation through aquaporin-1. Nature 407, 599–605. ( 10.1038/35036519) [DOI] [PubMed] [Google Scholar]
- Nishida M., Cadene M., Chait B. T., Mackinnon R. 2007. Crystal structure of a Kir3.1-prokaryotic Kir channel chimera. EMBO J. 26, 4005–4015. ( 10.1038/sj.emboj.7601828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Sokabe M., Yoshimura K. 2006. Lipid–protein interaction of the MscS mechanosensitive channel examined by scanning mutagenesis. Biophys. J. 91, 2874–2881. ( 10.1529/biophysj.106.084541) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura T., Sokabe M., Yoshimura K. 2008. Interaction between cytoplasmic and transmembrane domains of the mechanosensitive channel MscS. Biophys. J. 94, 1638–1645. ( 10.1529/biophysj.107.114785) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Moe P. C., Blount P. J. 2002. Functional design of bacterial mechanosensitive channels. Comparisons and contrasts illuminated by random mutagenesis. J. Biol. Chem. 277, 27 682–27 688. ( 10.1074/jbc.M202497200) [DOI] [PubMed] [Google Scholar]
- O'Keeffe A. H., East J. M., Lee A. G. 2000. Selectivity in lipid binding to the bacterial outer membrane protein OmpF. Biophys. J. 79, 2066–2074. (doi:10.1016/S0006-3495(00)76454-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollila O. H., Risselada H. J., Louhivuori M., Lindahl E., Vattulainen I., Marrink S. J. 2009. 3D pressure field in lipid membranes and membrane-protein complexes. Phys. Rev. Lett. 102, 078101 ( 10.1103/PhysRevLett.102.078101) [DOI] [PubMed] [Google Scholar]
- Ou X., Blount P., Hoffman R. J., Kung C. 1998. One face of a transmembrane helix is crucial in mechanosensitive channel gating. Proc. Natl Acad. Sci. USA 95, 11 471–11 475. ( 10.1073/pnas.95.19.11471) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., Cortes D. M., Cuello L. G. 1999. Structural rearrangements underlying K+-channel activation gating. Science 285, 73–78. ( 10.1126/science.285.5424.73) [DOI] [PubMed] [Google Scholar]
- Perozo E., Kloda A., Cortes D. M., Martinac B. 2001. Site-directed spin-labeling analysis of reconstituted MscL in the closed state. J. Gen. Physiol. 118, 193–206. ( 10.1085/jgp.118.2.193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perozo E., Cortes D. M., Sompornpisut P., Kloda A., Martinac B. 2002a Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 418, 942–948. ( 10.1038/nature00992) [DOI] [PubMed] [Google Scholar]
- Perozo E., Kloda A., Cortes D. M., Martinac B. 2002b Physical principles underlying the transduction of bilayer deformation forces during mechanosensitive channel gating. Nat. Struct. Biol. 9, 696–703. ( 10.1038/nsb827) [DOI] [PubMed] [Google Scholar]
- Pilot J. D., East J. M., Lee A. G. 2001. Effects of bilayer thickness on the activity of diacylglycerol kinase of Escherichia coli. Biochemistry 40, 8188–8195. ( 10.1021/bi0103258) [DOI] [PubMed] [Google Scholar]
- Powl A. M., East J. M., Lee A. G. 2003. Lipid–protein interactions studied by introduction of a tryptophan residue: the mechanosensitive channel MscL. Biochemistry 42, 14 306–14 317. ( 10.1021/bi034995k) [DOI] [PubMed] [Google Scholar]
- Powl A. M., East J. M., Lee A. G. 2005a Heterogeneity in the binding of lipid molecules to the surface of a membrane protein: hot spots for anionic lipids on the mechanosensitive channel of large conductance MscL and effects on conformation. Biochemistry 44, 5873–5883. ( 10.1021/bi047439e) [DOI] [PubMed] [Google Scholar]
- Powl A. M., Wright J. N., East J. M., Lee A. G. 2005b Identification of the hydrophobic thickness of a membrane protein using fluorescence spectroscopy: studies with the mechanosensitive channel MscL. Biochemistry 44, 5713–5721. ( 10.1021/bi047338g) [DOI] [PubMed] [Google Scholar]
- Powl A. M., East J. M., Lee A. G. 2007. Different effects of lipid chain length on the two sides of a membrane and the lipid annulus of MscL. Biophys. J. 93, 113–122. ( 10.1529/biophysj.107.105130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramu Y., Xu Y., Lu Z. 2006. Enzymatic activation of voltage-gated potassium channels. Nature 442, 696–699. ( 10.1038/nature04880) [DOI] [PubMed] [Google Scholar]
- Sato C., Ueno Y., Asai K., Takahashi K., Sato M., Engel A., Fujiyoshi Y. 2001. The voltage-sensitive sodium channel is a bell-shaped molecule with several cavities. Nature 409, 1047–1051. ( 10.1038/35059098) [DOI] [PubMed] [Google Scholar]
- Sato C., et al. 2004. Inositol 1,4,5-trisphosphate receptor contains multiple cavities and L-shaped ligand-binding domains. J. Mol. Biol. 336, 155–164. ( 10.1016/j.jmb.2003.11.024) [DOI] [PubMed] [Google Scholar]
- Schmidt D., Jiang Q. X., MacKinnon R. 2006. Phospholipids and the origin of cationic gating charges in voltage sensors. Nature 444, 775–779. ( 10.1038/nature05416) [DOI] [PubMed] [Google Scholar]
- Seoh S. A., Sigg D., Papazian D. M., Bezanilla F. 1996. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron 16, 1159–1167. ( 10.1016/S0896-6273(00)80142-7) [DOI] [PubMed] [Google Scholar]
- Shi N., Ye S., Alam A., Chen L., Jiang Y. 2006. Atomic structure of a Na+- and K+-conducting channel. Nature 440, 570–574. ( 10.1038/nature04508) [DOI] [PubMed] [Google Scholar]
- Sokabe M., Sachs F., Jing Z. Q. 1991. Quantitative video microscopy of patch clamped membranes stress, strain, capacitance, and stretch channel activation. Biophys. J. 59, 722–728. ( 10.1016/S0006-3495(91)82285-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M., Schulten K. 2004. Molecular dynamics study of gating in the mechanosensitive channel of small conductance MscS. Biophys. J. 87, 3050–3065. ( 10.1529/biophysj.104.046045) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S. 2002. Purification of the small mechanosensitive channel of Escherichia coli (MscS): the subunit structure, conduction, and gating characteristics in liposomes. Biophys. J. 83, 290–298. ( 10.1016/S0006-3495(02)75169-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S. I., Blount P., Martinac B., Blattner F. R., Kung C. 1994. A large-conductance mechanosensitive channel in E. coli encoded by mscL alone. Nature 368, 265–268. ( 10.1038/368265a0) [DOI] [PubMed] [Google Scholar]
- Sukharev S. I., Sigurdson W. J., Kung C., Sachs F. 1999. Energetic and spatial parameters for gating of the bacterial large conductance mechanosensitive channel, MscL. J. Gen. Physiol. 113, 525–540. ( 10.1085/jgp.113.4.525) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukharev S., Betanzos M., Chiang C. S., Guy H. R. 2001a The gating mechanism of the large mechanosensitive channel MscL. Nature 409, 720–724. ( 10.1038/35055559) [DOI] [PubMed] [Google Scholar]
- Sukharev S., Durell S. R., Guy H. R. 2001b Structural models of the MscL gating mechanism. Biophys. J. 81, 917–936. ( 10.1016/S0006-3495(01)75751-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz K. J., MacKinnon R. 1997a Hanatoxin modifies the gating of a voltage-dependent K+ channel through multiple binding sites. Neuron 18, 665–673. ( 10.1016/S0896-6273(00)80306-2) [DOI] [PubMed] [Google Scholar]
- Swartz K. J., MacKinnon R. 1997b Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron 18, 675–682. ( 10.1016/S0896-6273(00)80307-4) [DOI] [PubMed] [Google Scholar]
- Traïkia M., Warschawski D. E., Lambert O., Rigaud J. L., Devaux P. F. 2002. Asymmetrical membranes and surface tension. Biophys. J. 83, 1443–1454. ( 10.1016/S0006-3495(02)73915-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez V., Sotomayor M., Cordero-Morales J., Schulten K., Perozo E. 2008a A structural mechanism for MscS gating in lipid bilayers. Science 321, 1210–1214. ( 10.1126/science.1159674) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vásquez V., Sotomayor M., Cortes D. M., Roux B., Schulten K., Perozo E. 2008b Three-dimensional architecture of membrane-embedded MscS in the closed conformation. J. Mol. Biol. 378, 55–70. ( 10.1016/j.jmb.2007.10.086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh C. P., Davies A., Butcher A. J., Dolphin A. C., Kitmitto A. 2009. Three-dimensional structure of CaV3.1: comparison with the cardiac L-type voltage-gated calcium channel monomer architecture. J. Biol. Chem. 284, 22310–22321. ( 10.1074/jbc.M109.017152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Sigworth F. J. 2009. Structure of the BK potassium channel in a lipid membrane from electron cryomicroscopy. Nature 461, 292–295. ( 10.1038/nature0829) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Black S. S., Edwards M. D., Miller S., Morrison E. L., Bartlett W., Dong C., Naismith J. H., Booth I. R. 2008. The structure of an open form of an E. coli mechanosensitive channel at 3.45 Å resolution. Science 321, 1179–1183. ( 10.1126/science.1159262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam R. 1961. Active cation transport as a pace-maker of respiration. Nature 191, 603–604. ( 10.1038/191603a0) [DOI] [PubMed] [Google Scholar]
- Xu Y., Ramu Y., Lu Z. 2008. Removal of phospho-head groups of membrane lipids immobilizes voltage sensors of K+ channels. Nature 451, 826–829. ( 10.1038/nature06618) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J., Cui Q. 2009. Curvature generation and pressure profile modulation in membrane by lysolipids: insights from coarse-grained simulations. Biophys. J. 97, 2267–2276. ( 10.1016/j.bpj.2009.07.051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Batiza A., Schroeder M., Blount P., Kung C. 1999. Hydrophilicity of a single residue within MscL correlates with increased channel mechanosensitivity. Biophys. J. 77, 1960–1972. ( 10.1016/S0006-3495(99)77037-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Batiza A., Kung C. 2001. Chemically charging the pore constriction opens the mechanosensitive channel MscL. Biophys. J. 80, 2198–2206. ( 10.1016/S0006-3495(01)76192-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Nomura T., Sokabe M. 2004. Loss-of-function mutations at the rim of the funnel of mechanosensitive channel MscL. Biophys. J. 86, 2113–2120. ( 10.1016/S0006-3495(04)74270-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K., Usukura J., Sokabe M. 2008. Gating associated conformational changes in the mechanosensitive channel MscL. Proc. Natl Acad. Sci. USA 105, 4033–4038. ( 10.1073/pnas.0709436105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Morais-Cabral J. H., Kaufman A., MacKinnon R. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel–Fab complex at 2.0 Å resolution. Nature 414, 43–48. ( 10.1038/35102009) [DOI] [PubMed] [Google Scholar]