Abstract
Human NK cells from the decidua basalis of gravid uteri and from the cycling endometrium of women undergoing hysterectomy were isolated and compared by gene expression profiling using Affymetrix microarrays with probes representing ~47,400 transcripts. Substantial differences indicate that these two types of NK cells represent distinct subsets.
Introduction
The human uterine endometrium harbors various immune cell populations including NK cells, macrophages, T cells, B cells and neutrophils (1, 2). Endometrial NK cells (eNK) increase in number after ovulation becoming the major endometrial immune cell type. With menstruation, shedding of the uterine mucosa carries with it most of the endometrial immune cells. However if pregnancy takes place, NK cells continue to accumulate becoming 70% of the lymphocytes in the human decidua by the end of the first trimester of gestation (3). We refer to these as decidual NK cells (dNK).
Human dNK cells have been fully characterized. Their gene expression profile sets them apart from the two main peripheral blood NK cell (pNK) subsets (4). Like CD56dim, CD16+ pNK cells dNK cells are granular and express killer cell immunoglobulin like receptors (KIR) (5–7). However they lack cytotoxic activity and expression of CD16 (4, 5, 7, 8) as does the minor CD56bright, CD16− pNK subset. dNK cells produce a number of immunosuppressive molecules that may contribute to the establishment of maternal-fetal immune tolerance (4). Their activation also leads to the secretion of proinflammatory, angiogenic and trophoblast migration promoting factors and thus it was proposed that they may play a role in invasion of the endometrium during implantation and placental vascular remodeling during early pregnancy (9–12).
Unlike dNK and pNK cells, characterization of eNK cells is limited. They share many properties with dNK cells such as CD9 expression, the level of CD56 expression, absence of CD16 expression, granularity and lack of cytotoxic activity (2), and therefore they have been thought of as equivalent to dNK cells. More recently an analysis of eNK cells was reported (13). Phenotypic comparison to previously published data on dNK cells revealed some phenotypic differences. Unlike fresh dNK cells, freshly isolated eNK cells were reported not to express the activating receptor NKp30, and failed to secrete the angiogenic cytokine VEGF and placental growth factor (13). However no direct comparison of dNK and eNK cells was provided.
Here we present a genome wide expression profile comparison of the dNK and eNK cell populations. The analysis reveals that dNK and eNK are two distinct NK cell subtypes, as different as CD56bright and CD56dim pNK cells.
Results and discussion
It has been assumed that NK cells from cycling endometrium and NK cells present in the decidua basalis at the maternal-fetal interface during pregnancy are equivalent cells. To compare their gene expression profiles RNA from FACS sorted dNK and eNK cells was hybridized to Affymetrix oligonucleotide microarrays HGU133 Plus 2.0 containing ~54,000 probe-sets representing ~47,400 transcripts. dNK cell were isolated from decidua basalis of first trimester elective terminations as previously described (4), A similar procedure was used to obtain eNK cells from non-affected regions of cycling endometrium of donor women (average age 44 years) undergoing hysterectomy for different reasons.
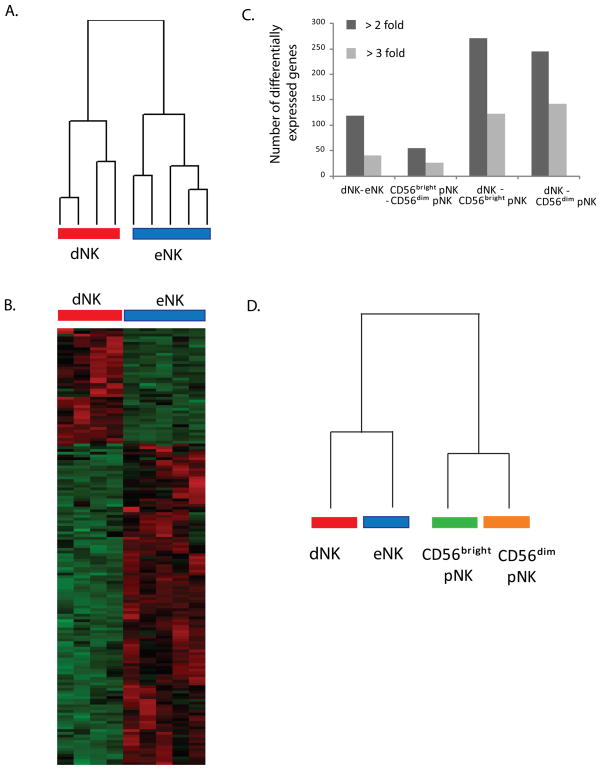
Data obtained from five eNK cell samples and four dNK cell samples were organized based on the expression pattern of genes with variable levels of expression by an unsupervised hierarchical clustering algorithm (14). This resulted in the segregation of dNK and eNK samples into two distinct groups (Figure 1A), thus indicating that the two cell types differ in their gene expression profile.
Figure 1.
dNK and eNK cells present distinct gene expression profiles. A) Unsupervised hierarchical clustering of 4 dNK cell samples and 5 eNK cell samples based on the expression profile of genes with variable expression levels across all samples. 1414 genes were considered in this analysis as they complied with the following criteria: they were expressed by at least 30% of the samples, and σι/μi ratio was >0.5 and <1000 (where σi and μI are the standard deviation and mean of the hybridization intensity values of each individual gene across all samples). The resulting dendogram reflects the similarity in gene expression profiles between each of the samples. B) Relative intensity profiles of genes differentially expressed at p < 0.01 with at least 3 fold change in intensity between dNK and eNK cells. Each row represents the relative hybridization intensities of a gene in the different samples. Each column represents one sample. Color intensities reflect the magnitude of relative expression levels of a particular gene across samples. Red represents higher expression, green represents lower expression, and black represents average intensity across samples. C) The transcription profiles of dNK and eNK cells are as different as the transcription profiles of CD56bright pNK and CD56dim pNK cells. Number of genes differentially expressed at p < 0.01 between dNK and eNK cells and between CD56bright, CD56dim pNK cells and dNK cells. Data correspond to two independent datasets obtained with microarrays HGU133Plus2 for dNK (4 samples) vs eNK cells (5 samples), and with microarrays HGU133A and HGU133B for dNK (4 samples) vs CD56bright pNK cells (3 samples), dNK (4 samples) vs CD56dim pNK cells (3 samples), and CD56bright pNK cells (3 samples) vs. CD56dim pNK cells (3 samples). The average of genes differentially expressed in comparisons involving all possible combinations of three samples of each cell type is shown to correct for unequal sample sizes. T-test comparisons in this analysis were done with Dchip software. The number of genes differentially expressed normalized by the total number of genes present in each array presented a similar trend (not shown). D) Dendogram of the relationship between the four NK cell subsets based on the data of C.
Student’s t-test revealed that 450 genes were differentially expressed between the two subsets with at least a 2 fold change in their transcripts level at p<0.01, and 153 genes were differentially expressed with at least a 3 fold change. About 70% of those genes were over-expressed by eNK cells (Figure 1B).
To evaluate the extent of the differences between dNK and eNK cells in the context of other human NK cell types, the number of genes differentially expressed was compared to the number of genes differentially expressed between CD56bright pNK, CD56dim pNK and dNK cells in an independent dataset using an earlier version Affymetrix HGU133 arrays containing about 45,000 probe-sets representing 39,000 transcripts, a larger number than previously reported (4 and manuscript in preparation). After correcting for uneven sample sizes, the comparisons between dNK and CD56bright or CD56dim pNK cells yielded the largest number of differentially expressed genes at a fixed fold change and p-value. dNK and eNK cells differentially expressed fewer genes but more than those differentially expressed between CD56bright and CD56dim pNK cells in the second dataset, even after normalizing by the number of genes contained in each type of array used in the independent datasets (Figure 1C). Thus, dNK and eNK cells although closely related are at least as dissimilar as CD56bright pNK and CD56dim pNK cells. A dendogram indicating the relationship between the 4 subsets is presented in Figure 1D.
Classification of genes differentially expressed between dNK and eNK cells
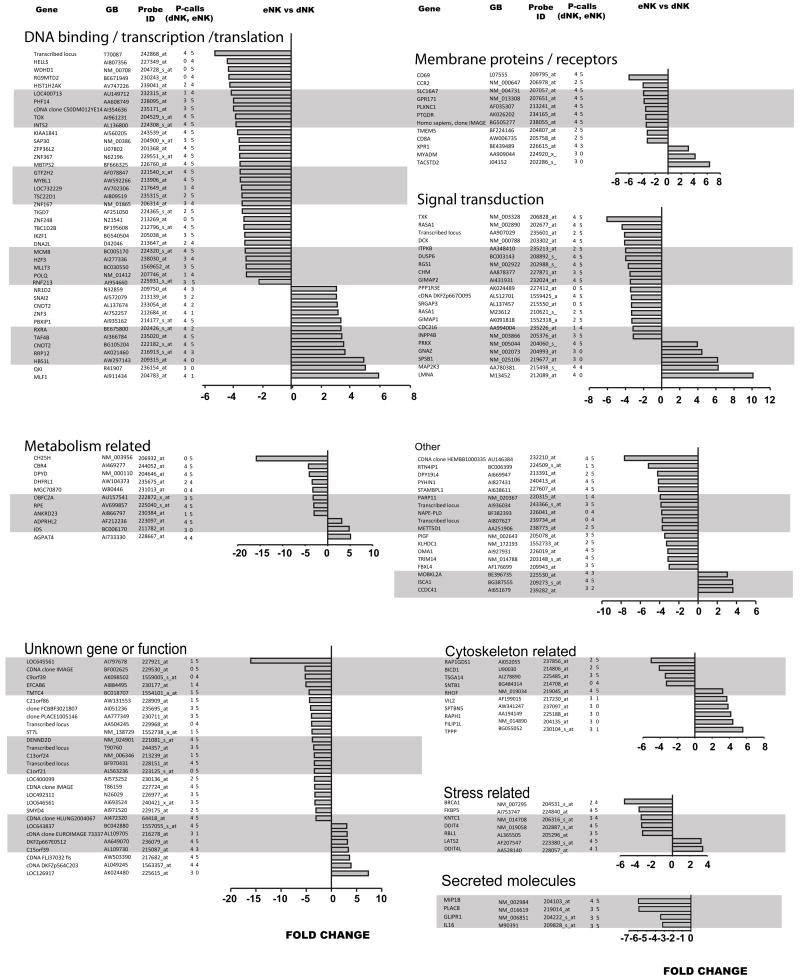
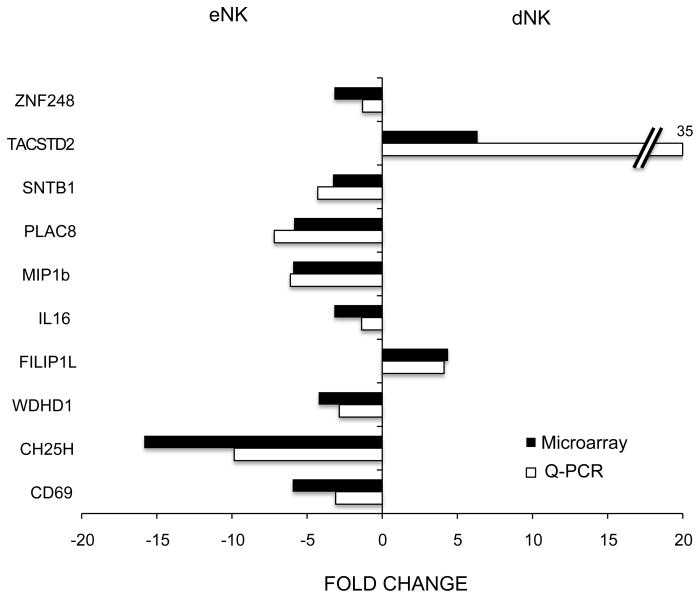
Genes differentially expressed between dNK and eNK cells were classified in functional categories based on gene ontology annotations. The category with the highest number of differentially expressed genes was DNA binding/transcription factors/translation regulators (Figure 2). There were also an important number of differentially expressed genes with unknown function. Two of the genes showed the largest difference among the subsets eNK>dNK differing by about 16 fold. In almost all categories 70% or more of the genes were upregulated in eNK cells with the exception of cytoskeleton related genes that had a few more genes upregulated by dNK cells. The 153 genes differentially expressed with at least a 3 fold change are shown in Figure 2. These differences are substantial. Quantitative RT-PCR was used to validate the gene expression data of 10 differentially expressed genes on one dNK and one eNK sample (Figure 3).
Figure 2.
Genes differentially expressed by dNK and eNK cells. Fold changes of genes that showed greater than or equal to three fold change at P < 0.01 by Student’s t test. 153 genes meeting these criteria were classified into the following categories: membrane proteins/receptors; signal transduction; cytoskeleton related; secreted molecules; metabolism related, DNA binding/transcription/translation; stress related; genes with unknown function; and other genes. Gene, gene name, or gene symbol obtained from Netaffx (Affymetrix, Inc.). GB, GenBank accession number. Probe ID, Affymetrix probeset designation. P-(Presence) calls (dNK, eNK), number of present calls by Affymetrix algorithms in 4 dNK and 5 eNK samples.
Figure 3.
Validation of microarray data by RT-Q-PCR. Fold change in the level of transcripts of 10 genes differentially expressed by dNK and eNK cells as evaluated by microarray gene expression profiling (black bars) and RT-Q-PCR (white bars). Microarray data correspond to the average of 5 eNK and 4 dNK samples as described in figure 2. RT-Q-PCR values are the average of duplicates obtained from one eNK and one dNK sample and were normalized to the expression level of β-actin, that was similar in both samples (not shown), before calculating the fold change in expression level between the two cell types.
It is difficult to figure out the functional meaning of these differences as they are scattered throughout so many different functional categories. For example the 4 most overexpressed genes in the dNK subset (8–16 fold) are present in 4 families metabolism (cholesterol hydroxylation), unknown function, signal transduction and another gene of unknown function. Further exploration of the relationship among these genes may point to important features of the transition from eNK to dNK cells that are important in pregnancy. Particularly interesting could be the study of a number of proteins that may affect the uterine microenvironment and endometrial cellular composition. CH25H (cholesterol 25-hydroxylase) is expressed by eNK but not by dNK cells, based on Affymetrix presence calls. CH25H catalyzes the formation of 25-hydroxycholesterol which acts as a co-repressor of the expression of cholesterol biosynthetic enzymes (15). As cholesterol is a substrate for the synthesis of steroid hormones, changes in the expression of CH25H in pregnant and non pregnant uteri may be relevant to local hormonal changes occurring in pregnancy. Also interesting is the overexpression by eNK cells of a number genes encoding for secreted proteins: Placental specific gene 8 (PLAC8) (16, 17), MIP1β (CCL4) a proinflamatory chemokine, GLI pathogenesis-related protein 1 (GLIPR1) associated with myelomocytic differentiation in macrophages (18), and interleukin 16 (IL16) which stimulates migratory responses of CD4+ lymphocytes, monocytes, and eosinophils (19) In summary these data show that the transcriptional profiles of dNK and eNK cells separate these two NK cell types into distinct subsets that are also different from the two subsets of pNK cells. The number of genes differentially expressed is at least as large as the differences between CD56bright CD16− and CD56dim CD16+ pNK cells. Endometrial NK cells and dNK cells should therefore be thought of as distinct cells that likely reflect differences in the hormonal and cellular environment where they reside. Thus, their potential responses to environmental stress are likely to be different and one should not assume that a response with one would be similar to the other. These cells may reflect a spectrum of NK cell developmental stages which can occur under different microenvironmental conditions.
Methods
Samples
Decidual samples from patients undergoing elective termination in the first trimester between 6 and 12 weeks of gestation were collected and processed as described (4). dNK cells were FACS sorted as CD3- CD56+ CD16- cells. To obtain endometrial NK cells, fertile reproductive tract specimens from women undergoing elective hysterectomy at the Dartmouth Hitchcock Medical Center were used. Samples from 5 patients with an average age of 44 years were employed. The preliminary patient diagnoses included genital prolapse, fibroids, cervical dysplasia, or menorrhagia. Tissue samples used were distal to the pathological changes and processed as described (4, 2). All patients had histology consistent with cycling endometrium and were in the secretory phase of the cycle with exception of sample eNK6 that was from the proliferative phase. The resulting isolated single cell suspensions were used for cell sorting. Endometrial NK cells were isolated by staining and selection for CD45+, CD56+, CD3− cells (20, 21). Cell pellets were washed with PBS and stored at 80 C until used for RNA preparation.
RNA labeling and microarray hybridization
Pellets from flow-sorted cells were washed with PBS and frozen at −80C. Total RNA isolation was done with TRIzol following manufacturer’s instructions, with addition of linear polyacrylamide (Genelute LPA; Sigma Aldrich) to make RNA pellets visible. RNA was cleaned-up (Rneasy, QIAGEN)(4), amplified with Acturus RiboAmp OA kit (Arcturus) and transcribed in vitro for generation of biotin-labeled cRNA target (Affymetrix, CA), all according to kit manufacturers instructions. Biotin-labeled cRNA fragmentation was done in 40 mM Tris-acetate (pH 8.1), 100 mM KOAc, 30 mM MgOAc. This fragment mixture was hybridized to the Human Genome HGU133 Plus 2 chips displaying probes for ~47,400 transcripts, gene transcript levels determined using algorithms in the GCOS software and based on decision matrices, each probe assigned a call of present or absent (all Affymetrix,). In total four dNK and five eNK cell samples were analyzed.
Microarray data analysis
Samples were clustered with an unsupervised hierarchical clustering algorithm (14) using D-chip software (http://www.dchip.org). Intensity values were normalized by the model-based expression analysis method (22). Genes considered for the samples clustering analysis were those presenting variable level of expression across all 9 samples according to the following criteria: genes should be expressed (have presence calls) in at least 30% of the 9 samples, and σi/μi ratio should be >0.5 and <1000, where σi and μi are the standard deviation and mean of the hybridization intensity values of each particular gene across all samples, respectively.
Hybridization intensities of the 9 samples were normalized by setting the mean intensity of each array to be the same with Affymetrix GCOS software. All hybridization intensity values below 35 were set to 35. Samples were grouped according to their biological origin in dNK (four samples) or eNK (five samples). Genes considered in the comparison analysis showed a 2 or 3 fold change at p<0.01 in a Student’s t test and complied with the following criteria: if the gene was overexpressed by dNK cells, it had to be present, according to the Affymetrix algorithm, in at least 3 dNK samples; if it was overexpressed by eNK cells, it had to be present in at least 4 eNK samples. For genes represented by multiple probesets, the result for only one representative probeset is shown.
Genes were arranged in functional categories based on information available from Affymetrix (http://www.affymetrix.com/analysis/index.affx), the Gene Ontology Consortium functional annotations (http://www.geneontology.org), OMIM and PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez) databases.
Raw Microarray Data, the original “.CEL” files for the 9 chip hybridizations, were deposited in gene expression omnibus datasets (GEO, http://www.ncbi.nlm.nih.gov/gds).
Quantitative real-time reverse-transcriptase polymerase chain reaction (qPCR)
cDNA was prepared from 50ng of total RNA using the SuperScript II Kit (Invitrogen). qPCR reactions were prepared in duplicate using iTaq Fast SYBR Green Supermix With ROX (BioRad) and gene specific primers listed in supplemental Table 1. qPCR was performed on a 7500 Fast Real-Time PCR System (Applied Biosystems, Foster City, California). Cyclophyllin A and β-actin were used as internal housekeeping references. Gene expression was quantified using the2−ΔΔCT quantification method of Livak (23).
Supplementary Material
Acknowledgments
We thank B. Houser for helpful discussions and F. Rosetti for help in the preparation of Figure 2. This research was supported by the National Institutes of Health (grants AI053330 to JLS, AI-071761 to CRW, AI-51877 to CRW), and Fogarty (FIC-5-D43-TW006807).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Givan AL, White HD, Stern JE, Colby E, Gosselin EJ, Guyre PM, Wira CR. Flow cytometric analysis of leukocytes in the human female reproductive tract: comparison of fallopian tube, uterus, cervix, and vagina. Am J Reprod Immunol. 1997;38:350–359. doi: 10.1111/j.1600-0897.1997.tb00311.x. [DOI] [PubMed] [Google Scholar]
- 2.Eriksson M, Meadows SK, Wira CR, Sentman CL. Unique phenotype of human uterine NK cells and their regulation by endogenous TGF-{beta} J Leukoc Biol. 2004;76:667–675. doi: 10.1189/jlb.0204090. [DOI] [PubMed] [Google Scholar]
- 3.Loke YW, King A. Human Implantation: Cell Biology and Immunology. Cambridge: Cambridge University Press; 1995. [Google Scholar]
- 4.Koopman LA, Kopcow HD, Rybalov B, Boyson JE, Orange JS, Schatz F, Masch R, Lockwood CJ, Schachter AD, Park PJ, Strominger JL. Human Decidual Natural Killer Cells Are a Unique NK Cell Subset with Immunomodulatory Potential. J Exp Med. 2003;198:1201–1212. doi: 10.1084/jem.20030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.King A, Jokhi PP, Burrows TD, Gardner L, Sharkey AM, Loke YW. Functions of human decidual NK cells. Am J Reprod Immunol. 1996;35:258–260. doi: 10.1111/j.1600-0897.1996.tb00041.x. [DOI] [PubMed] [Google Scholar]
- 6.Hiby SE, King A, Sharkey AM, Loke YW. Human uterine NK cells have a similar repertoire of killer inhibitory and activatory receptors to those found in blood, as demonstrated by RT-PCR and sequencing. Mol Immunol. 1997;34:419–430. doi: 10.1016/s0161-5890(97)00032-1. [DOI] [PubMed] [Google Scholar]
- 7.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 8.Kopcow HD, Allan DS, Chen X, Rybalov B, Andzelm MM, Ge B, Strominger JL. Human decidual NK cells form immature activating synapses and are not cytotoxic. Proc Natl Acad Sci U S A. 2005;102:15563–15568. doi: 10.1073/pnas.0507835102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, Gazit R, Yutkin V, Benharroch D, Porgador A, Keshet E, Yagel S, Mandelboim O. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med. 2006;12:1065–1074. doi: 10.1038/nm1452. [DOI] [PubMed] [Google Scholar]
- 10.Kopcow HD, Karumanchi SA. Angiogenic factors and natural killer (NK) cells in the pathogenesis of preeclampsia. J Reprod Immunol. 2007;76:23–29. doi: 10.1016/j.jri.2007.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der Meer A, Joosten I, Long EO. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4:e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Houser BL, Nicotra ML, Strominger JL. HLA-G homodimer-induced cytokine secretion through HLA-G receptors on human decidual macrophages and natural killer cells. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0901173106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Manaster I, Mizrahi S, Goldman-Wohl D, Sela HY, Stern-Ginossar N, Lankry D, Gruda R, Hurwitz A, Bdolah Y, Haimov-Kochman R, Yagel S, Mandelboim O. Endometrial NK cells are special immature cells that await pregnancy. J Immunol. 2008;181:1869–1876. doi: 10.4049/jimmunol.181.3.1869. [DOI] [PubMed] [Google Scholar]
- 14.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund EG, Kerr TA, Sakai J, Li WP, Russell DW. cDNA cloning of mouse and human cholesterol 25-hydroxylases, polytopic membrane proteins that synthesize a potent oxysterol regulator of lipid metabolism. J Biol Chem. 1998;273:34316–34327. doi: 10.1074/jbc.273.51.34316. [DOI] [PubMed] [Google Scholar]
- 16.Galaviz-Hernandez C, Stagg C, de Ridder G, Tanaka TS, Ko MSH, Schlessinger D, Nagaraja R. Plac8 and Plac9, novel placental-enriched genes identified through microarray analysis. Gene. 2003;309:81–89. doi: 10.1016/s0378-1119(03)00508-0. [DOI] [PubMed] [Google Scholar]
- 17.Rissoan MC, Duhen T, Bridon JM, Bendriss-Vermare N, Peronne C, Saint Vis B, Briere F, Bates EEM. Subtractive hybridization reveals the expression of immunoglobulinlike transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood. 2002;100:3295–3303. doi: 10.1182/blood-2002-02-0638. [DOI] [PubMed] [Google Scholar]
- 18.Gingras MC, Margolin JF. Differential expression of multiple unexpected genes during U937 cell and macrophage differentiation detected by suppressive subtractive hybridization. Exp Hematol. 2000;1:65–76. doi: 10.1016/s0301-472x(99)00126-5. [DOI] [PubMed] [Google Scholar]
- 19.Cruikshank W, Center DM. Modulation of lymphocyte migration by human lymphokines. II. Purification of a lymphotactic factor (LCF) J Immun. 1982;128:2569–2574. [PubMed] [Google Scholar]
- 20.Mselle TF, Meadows SK, Eriksson M, Smith JM, Shen L, Wira CR, Sentman CL. Unique characteristics of NK cells throughout the human female reproductive tract. Clin Immunol. 2007;124:69–76. doi: 10.1016/j.clim.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Sentman CL, Wira CR, Eriksson M. NK cell function in the human female reproductive tract. Am J Reprod Immunol. 2007;57:108–115. doi: 10.1111/j.1600-0897.2006.00448.x. [DOI] [PubMed] [Google Scholar]
- 22.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.