Abstract
The authors of a recent manuscript in “Nature” claim to have discovered “universal trends” of amino acid gain and loss in protein evolution. Here, we show that this universal trend can be simply explained by a bias that is unavoidable with the 3-taxon trees used in the original analysis. We demonstrate that a rigorously reversible equilibrium model, when analyzed with the same methods as the “Nature” manuscript, yields identical (and in this case, clearly erroneous) conclusions. A main source of the bias is the division of the sequence data into “informative” and “noninformative” sites, which favors the observation of certain transitions.
Keywords: amino acid bias, ancestral reconstruction, molecular evolution, parsimony
Introduction
It is obvious to most practitioners of protein sequence analysis that protein evolutionary rates and processes change continually. Over substantial periods of evolutionary time, substitutions can be irreversible and amino acid composition may not be in equilibrium, resulting in a net compositional “flux” from one amino acid type to another. Substantial variation in evolutionary processes also exists among proteins and among sites within the same protein. It is of considerable interest to analyze the nature of these variations and fluxes in order to understand their structural, functional, and genetic causes. Also, as most modeling of molecular evolution ignores such variation and assumes (for mathematical, statistical, and computational convenience) that protein evolution is a stationary, homogeneous, and reversible process, an important additional question is whether these assumptions dramatically impact the resulting inferences.
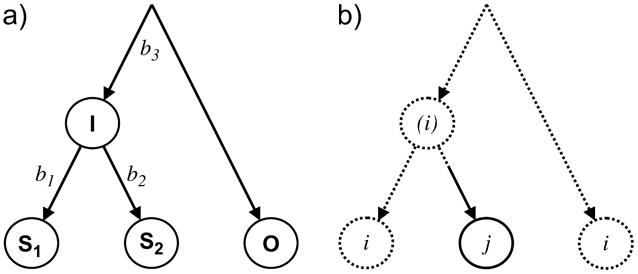
One common approach to analyzing flux is to infer the ancestral state of proteins at internal nodes, or branching points, and to then consider whether there have been changes in state or function or amino acid frequencies between the ancestor and the descendant taxa. An example of this type of flux analysis is the recent work of Jordan and colleagues (2005a), which concluded that the flux in amino acid frequencies was significant and similar in a wide variety of organisms, leading to convergent directional changes. Their study was based on a maximum parsimony evaluation of simple 3-taxon trees (fig. 1a). We will refer to the 2 closer taxa as “sister” taxa (S1 and S2) and the third, more divergent taxon, as the outgroup O. In their study, a location was informative if the amino acid in the outgroup was the same as in one of the sister taxa, yet the amino acids in the 2 sister taxa were different (fig. 1b). For these informative sites, it was assumed that the shared amino acid existed in the common ancestor and remained unchanged on the branch leading to the outgroup, on the branch leading to the internal node I, and on the branch leading from I to one of the sister taxa; thus a single substitution occurred along either branch b1 or b2. Locations with different patterns were deemed noninformative and were excluded from consideration, at least for the primary analysis. The flux between 2 amino acid types, i and j, was thus measured as the difference between the number of informative sites for which i was in the outgroup and one sister taxon and j was in the other (and thus an amino acid substitution from i to j had presumably taken place) minus the number for which the roles of i and j were reversed.
Fig. 1.
A simple 3-taxon tree. In (a), the 2 closest “sister” taxa (S1 and S2) diverge at an internal node (I), which represents the most recent common ancestor of these 2 taxa. At some unknown root point lies the common ancestor of these 2 taxa and the outgroup (O). The distances, or branch lengths, separating I from S1, S2, and O are labeled b1, b2, and b3, respectively. In (b) parsimony reconstruction of an “informative” site is depicted. The amino acid i (dashed lines) is inferred to exist at the root and in the common ancestor of the sister taxa because it is in one of the sisters and in the outgroup. The amino acid in the ancestral node is in parentheses to emphasize that the state here (and therefore the substitution from i to j along the branch leading to S2) is inferred rather than observed as in the 2 sister taxa and the outgroup.
Such an analysis presents significant difficulties. Firstly, it is possible to have asymmetric substitution rates between individual pairs of amino acids and still have static amino acid frequencies (McDonald 2006). More generally, although the analysis is conceptually simple and it is tempting to view the ancestral states as “observations,” they are inferred, not observed; bias in this inference will lead, erroneously, to exactly the sorts of conclusions made by Jordan and colleagues. Thus, findings of disequilibrium and convergence are suspect. Unfortunately, there are a number of known causes of statistical bias:
Under parsimony, the only evolutionary scenarios considered are those that lead to informative sites (fig. 1b). The informative sites included in the analysis might represent a biased set of all locations. Certain substitutions might be preferentially excluded from the analysis due to their higher probability of occurring at noninformative sites, resulting in a sampling bias.
At informative sites, it is assumed that only one amino acid substitution has occurred. At a fraction of such locations, multiple changes will have occurred, resulting in erroneous reconstructions. It is possible for these errors to produce systematic undercounting of some substitutions with respect to others.
All locations are assumed to follow identical substitution patterns, but there is strong evidence that substitution patterns vary widely among locations (Overington et al. 1992; Goldman and Yang 1994; Wako and Blundell 1994; Koshi and Goldstein 1995; Bruno 1996; Kinjo and Nishikawa 2004). Faster evolving locations are more subject to the erroneous reconstruction and sampling bias described above. The effect of this “bias of the biases” may be difficult to predict.
McDonald, for instance, demonstrated that a 2-allele model under nearly neutral selection results in apparent gains of the mildly deleterious allele without any real changes in allele frequency (McDonald 2006).
Correcting for any of these biases with likelihood methods depends on the accuracy of an evolutionary model that may be highly uncertain. In addition, any method such as parsimony (or maximum likelihood) that chooses a single best reconstruction of the evolutionary history at every site may be highly biased. A good example of the effects of such biases was provided by Krishnan et al. (2004), who demonstrated that parsimony and maximum likelihood inference uniformly and incorrectly show consistent convergent trends of nucleotide gain and loss in primate mitochondrial evolution, whereas Bayesian inference implies that changes in nucleotide frequency are not convergent and much reduced (Krishnan et al. 2004). In particular, the incorporation of biologically reasonable variation in processes among sites can plausibly account for all of the apparent derived trends predicted by simpler methods.
Given that Jordan’s group used a method known to lead to systematic biases that could erroneously produce precisely the results they infer, it is important to investigate whether their results can be explained by such biases, and if so, the major sources and causes. Can we truly learn something about the nature of protein evolution and the order of amino acid incorporation from their analysis? Are the results obtained by Jordan et al. consistent with reversibility or static equilibrium frequencies? We approach this question by analyzing evolutionary scenarios where the assumptions of reversibility and static equilibria between the different amino acid types are unquestionable. We then apply the methods of Jordan et al. to this data and explore whether we obtain the same results as they did with their analysis of real sequence data, that is, apparent irreversibility and consistent long-term trends in amino acid gain and loss. If their analysis applied to our simulations yields similar results, leading to (in this case) erroneous conclusions of irreversibility and amino acid flux, this suggests that their conclusions about real proteins may also be erroneous. Our studies suggest that the observation of parallel trends obtained from ancestral reconstruction are problematic and that trends such as convergent evolution that mimic known biases should be treated with suspicion.
Methods
Simple 2-State Model of Evolution
We first consider the simple case where there are only 2 amino acid types at a position or site in a protein sequence, and there are 3 taxa arranged as in figure 1a. In this case, the model of evolution, the instantaneous rate of substitution between these 2 amino acids, can be summarized as a rate matrix (table 1). All such 2-state matrices are “necessarily” reversible, and the expected equilibrium frequencies of the 2 types of amino acids, πA and πB=1 − πA, can be calculated directly from the substitution rates using the relationship πA/πB = αA/αB.
Table 1.
Instantaneous Rate Matrix for the Simple 2 Amino Acid State Model
| A | B | |
|---|---|---|
| A | −αB | αB |
| B | αA | −αA |
Given this model of evolution, and assuming that the substitution rate is constant over time and over positions and that the initial frequencies of the 2 different amino acids are identical to the equilibrium frequencies, one can easily and exactly calculate the probability of any set of residue types being observed at positions S1, S2, and O. Although there is no net flux in this model, we can use the method of Jordan to calculate the “apparent” difference between nA→B, the number of A to B substitutions occurring along branches b1 and b2, and nB→A, the number of B to A substitutions along these same 2 branches. The apparent normalized flux difference for residue A (dA) is defined as the difference in the number of apparent substitutions creating and removing amino acid A divided by the sum of these 2 terms: dA = (nB→A − nA→B)/(nB→A + nA→B).
In the scenario with 2 sister taxa and an outgroup (fig. 1a), it is most straightforward to consider the case where the branches leading from the internal node to the 2 sister taxa are equal (b1 = b2) and the branch leading to the outgroup (b3) is longer. The b3/b1 ratio for the genome data analyzed by Jordan’s group (henceforth, we will refer to this as simply the “genome data”) ranged from approximately 1.5 (Pseudomonas) to over 80 (Staphylococcus). To determine apparent flux levels under the 2-state model for reasonable amino acid frequencies and branch lengths, we performed the analysis for 3 different values of πA, (0.6, 0.7, and 0.8), a variety of branch lengths b1 to the sister taxa, and a b3/b1 ratio of 3.
From proteins that are more densely sampled than the genome data, it is known that there are large differences between the overall rates of substitution in different types of proteins, with fibrinopeptides evolving approximately 3 orders of magnitude faster than histone H4 (Dayhoff et al. 1978). Even within a single protein, it is now commonly understood that many amino acid positions are much less likely to substitute than others due to the greater effect on structure and function these positions have (Pollock et al. 2000). In some proteins, over half the sites may be nearly invariant, and many more may evolve only slowly compared with the few sites (mostly on the surface and distant from the active site) that evolve quickly. Rate variation can be modeled by assuming that a proportion of sites are invariant and that the rest are reasonably well modeled using the flexible gamma distribution (Nei 1976; Yang 1997; Yang 2002). We use a simpler model for rate variation where there are 3 different types of locations: invariant (40% of the sites), slowly varying (40%), and rapidly varying (20%).
There are strong correlations between the rate of substitutions at different locations and the types of amino acids found in these locations (Koshi and Goldstein 1998; Dimmic et al. 2000; Soyer et al. 2003). For instance, hydrophobic residues are more likely to be found in slower changing interior locations, whereas certain residues (i.e., proline, histidine, and cysteine) are especially prevalent at invariant locations. We can again adjust our simple model to include different types of locations, where there is a correlation between the local rates of substitution and the equilibrium frequencies.
More Complex Model of Protein Evolution
Although instructive, this 2-residue model is overly simplistic. What happens with a more realistic model including some of the factors described above? Can we develop a biologically reasonable rigorously reversible model consistent with the results of Jordan and colleagues? If so, it would cast serious doubt on those results because flux would have been detected where, by definition, none exists.
We have developed models for the process of amino acid substitution that include different substitution models for different types of locations (Koshi and Goldstein 1998; Dimmic et al. 2000; Soyer et al. 2003). In these models, different substitution matrices are constructed, each appropriate for different types of site, called a “site class.” These models represent the substitution process in real proteins better (decreased Akaike Information Criterion [Akaike 1978]) than models that neglect such types of site heterogeneity. We created a model with 5 different site classes where each site class was defined by 1) the frequency of that particular site class in the protein set, 2) the frequency of each amino acid in that site class, and 3) an overall rate of substitution for that site class. Given these parameters, a reversible model was constructed based on the JTT symmetric substitution matrix (Jones et al. 1992), and the probability of the various possible amino acids at the 3 terminal taxa were computed (b1 = b2 = 0.05, b3 = 0.15). The various parameters in the model were then adjusted to maximize the similarity between the apparent flux from this model (both with and without correction) and the apparent normalized flux values from the genome data. The overall equilibrium distribution of the different amino acids (averaged over all of the site classes) was constrained to the values described in the JTT model.
Results
Analysis of the Simple 2-State Model of Evolution
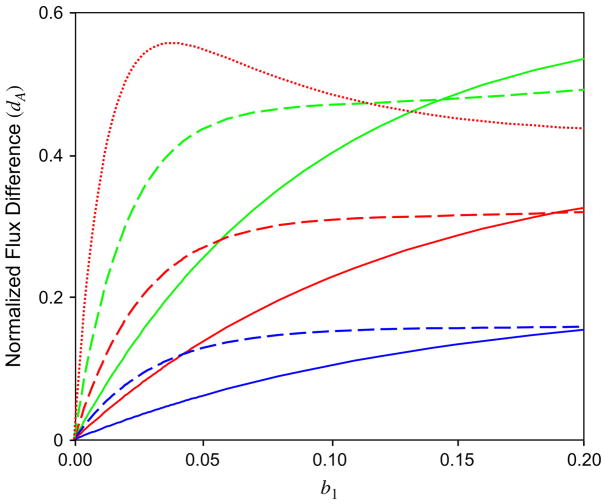
Averaging over the various proteins in the genome data set, Jordan and colleagues observed apparent normalized flux values range from −0.362 for proline to 0.452 for cysteine, with a root mean squared value of 0.165 (Jordan et al. 2005a). As shown in figure 2, values obtained from our evolutionary analyses with the simple 2-state model were comparable, even though the models used in our calculations were strictly reversible with no net gain of either A or B. Notably, these systematic biases occurred for moderate branch lengths.
Fig. 2.
Normalized flux difference versus length of branch to sister taxa for 2 amino acid types. The normalized flux for residue A, dA, was calculated with b1=b2=1=3b3 for 3 different equilibrium frequencies for the majority amino acid: πA = 0.6 (blue), 0.7 (red), and 0.8 (green). Results are shown for homogeneous (solid lines) or variable (dashed lines) rates across sites, for which 40% of the sites are invariant, 40% are slowly varying, and 20% vary 10 times faster than the slowly varying sites. Results are also shown (dotted red line) when πA = 0.6 for the invariant sites, πA = 0.7 for the slowly varying sites, and πA = 0.9 for the rapidly varying sites, for an average equilibrium frequency of πA = 0.7.
With a modest amount of rate variation, large flux imbalances are expected for relatively short branch lengths (fig. 2). More extreme amounts of rate heterogeneity would make these effects more apparent at even shorter branch lengths. This occurs because the faster evolving sites are more likely to have suffered multiple substitutions even when the “overall” number of substitutions is low. Even for our simple model, if equilibrium frequency differences are associated with rate differences, appreciable flux imbalances are found for branch lengths considerably shorter than 0.01 (fig. 2).
Some of the potential causes of biases, and a “correction” to adjust for these effects were previously described (Jordan et al. 2005b). The observation that their results are not affected when this correction applied to the genome data is also claimed as support for their conclusions (Jordan et al. 2005b). We refute this claim in detail in the discussion, but for now, we note that this correction relies on the relative number of examples where 3 different residues exist at the 3 taxa; it is thus inapplicable to the simple 2 amino acid model described above.
Analysis of the More Complex Model of Protein Evolution
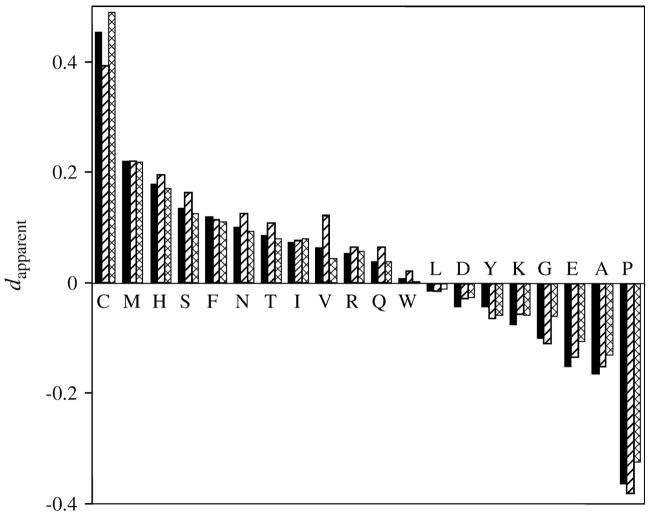
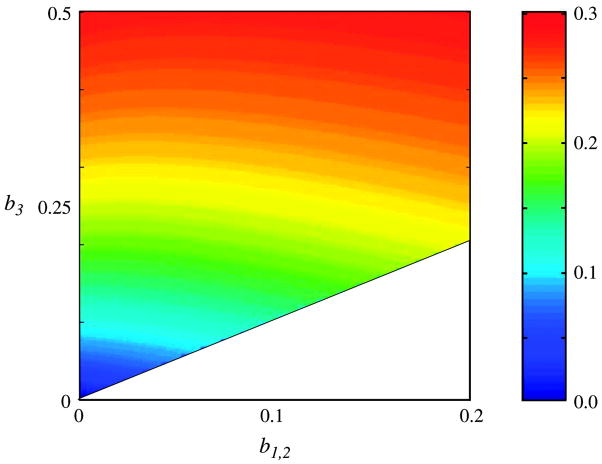
We were able to adjust the parameters of the more complex reversible site-class model to agree with the normalized flux values from the genome data, even when the flux values were corrected as suggested by Jordan et al. (fig. 3). The correlation coefficient is 0.97 for both the corrected and uncorrected flux. A rigorously reversible model with realistic parameters can therefore easily mimic the results obtained by Jordan and colleagues, even when corrections are made that supposedly account for statistical biases. We found that the measured normalized fluxes were high for a wide range of sister and outgroup branch lengths (fig. 4). The results from the genome data are comparable to the results from our simulations, even for moderate branch lengths. The results were also surprisingly independent of the branch lengths to the sister taxa (b1,2).
Fig. 3.
Apparent normalized flux for the various amino acids as reported by Jordan and colleagues (solid) versus those obtained by applying their analysis to synthetic data prepared from a rigorously reversible model, both without (striped) and with (crosshatched) their recommended corrections for correcting statistical bias. The model was based on the symmetric JTT matrix with 5 site classes, each defined by a relative substitution rate, a relative fraction of all locations, and the equilibrium frequencies of the 20 amino acids. Overall equilibrium frequencies were constrained to the JTT values. Branch lengths were b1 = b2 = 0.05, b3 = 0.15, corresponding to 8% sequence difference between sister taxa and 15% between either sister taxon and the outgroup. Seventy-eight percentage of the locations where the 2 sister taxa are different are informative, similar to the values for the real data analyzed by Jordan and colleagues. Published values represent the average over all data sets.
Fig. 4.
Root mean square (rms) flux imbalance for the site-class model as a function of branch lengths. The lengths between both sister taxa and the internal node (b1 and b2) were equal, and the length between the internal node and the outgroup (b3) was constrained to be longer. The flux imbalance was measured as rms values , where the average is over all residue types. Other details of the model were the same as in figure 3. The corresponding quantity for the genome data was 0.165, which corresponds approximately to the turquoise stripe. Because of the different rate of change for different locations, the center point on the plot (b1 = b2 = 0.10, b3 = 0.25) corresponds to sister taxa with a sequence difference of 15%, whereas the outgroup sequence differs from each of the sister taxa by 21%.
The simplicity of our model allows us to examine individual pairs of residues to help understand the source of the flux reconstruction bias. Consider the relative flux between proline (pro), the residue supposedly decreasing in frequency the fastest, and cysteine (cys), the residue supposedly increasing the fastest. The actual number of simulated cys → pro substitutions along the branches from the common ancestor to the sister taxa exactly equals the number of pro → cys substitutions, both in total and within each site class, as expected from a rigorously reversible model. The observed number of cys → pro substitutions is only 36% of the actual number of such substitutions, whereas the observed number of pro → cys substitutions is 92% of the actual number. The result is an apparent flux ratio of 2.6:1 away from pro and toward cys. The main cause of the bias is that the majority (58%) of cys → pro changes occur in noninformative sites, much more often than the 21% of the pro → cys changes that occur in such sites. Essentially all of the cys → pro substitutions (as well as essentially all of the pro → cys substitutions) occurred in the fastest changing locations, at which cys was much less common than pro. Because cys has a lower frequency at these variable locations, it is more probable that it will substitute to some other amino acid along one of the other branches on the tree, making the location uninformative.
The flux correction suggested by Jordan’s group has a relatively small effect, actually “increasing” the apparent flux ratio to 3.4:1. Why is this so? It is difficult to characterize a heterogeneous substitution model with the limited data available from only 3 taxa, and thus a necessary assumption in the correction is that there is a single well-defined frequency for each amino acid. The frequencies of the various amino acids, however, can depend on the type of location in the protein, which may be correlated with the overall rate of substitution. In this case, the highly variable sites have few cys, whereas cys is much more frequent in the more common invariant sites that contribute heavily to the average frequency. The frequency of cys at the highly variable sites is much more relevant than the much higher average frequency for determining the magnitude of the correction, and as a result, the correction is too small to account for most of the missed cys → pro substitutions. Because a higher proportion of the pro are in the fastest changing locations, the correction actually does better in finding the fewer missed pro → cys substitutions, resulting in the mistakenly increased apparent flux ratio.
Discussion
We have demonstrated that the result of simulations of simple, fully reversible models of evolution, when subjected to the flux analysis of Jordan and colleagues (2005a), yield the erroneous conclusion that the evolution must have been irreversible. This means that such analyses are subject to serious and systematic biases. This does not mean that the conclusions of Jordan and colleagues are necessarily wrong but rather that it is not possible to obtain definitive evidence from the 3-taxon case and that their data does not offer meaningful support for their conclusions.
Jordan and colleagues argue that because their results are not sensitive to branch length, their conclusions cannot be due to systematic biases. This actually leads to a serious difficulty for their conclusions: for truly long branches under a simple model, we “expect” differences in reciprocal substitution probabilities to lead to large erroneous flux results (on the order of what is observed). If flux patterns are similar for distantly related organisms and for more closely related organisms (where the biases should be much reduced), then the true flux must, coincidentally, have the same pattern as the bias. This seems improbable, suggesting another mechanism at work, such as that the percentage of amino acids different between 2 genomes is not an accurate measure of the relative number of substitutions at informative sites. For short branch lengths, the fastest evolving locations will be most subject to sampling bias and will also quickly constitute the greatest proportion of informative sites. As branch lengths get longer and divergence continues, these rapidly changing sites will quickly experience multiple substitutions and will become noninformative and thus not included in the analysis. More slowly changing locations, however, will join the informative classes and will have similar forms of sampling bias. The analysis will be biased toward the faster changing locations when the branches are short, and toward the slower changing (but not invariant) locations when the branches are long, buffering the results against changes in branch length. The result is a compensation effect that can preserve the appearance of flux imbalance as divergence continues.
With only 3 taxa in each genome data set, it is difficult to make meaningful estimates of the rate at individual protein positions, but it is possible to evaluate the variation in rate among different groups of proteins. For Bordetella, the most extreme example in these genomes, the average percentage difference between the sister taxa is only 0.3%, but there is a wide range of sequence differences between various proteins: over 45% of the proteins have no sequence divergence between the sister taxa, whereas some have over 10% sequence divergence. The amount that each protein contributed to the overall analysis depended heavily on the degree of sequence divergence: proteins that were identical in the 2 sister taxa contributed no data at all to the analysis, whereas the most diverged 10% of proteins (with 1.8% average difference between the sister taxa or approximately b1 = 0.02) contributed almost 40% of the informative sites. Given typical intraprotein rate variations, it is not difficult to imagine even higher substitution rates in the “regions” of the proteins containing the majority of informative sites. Again, this supports the idea that the calculated branch lengths are relatively meaningless as predictors of the expected strength of the bias in flux.
Further arguments were given as reasons that systematic biases cannot explain the apparent trends in the genomic data (Jordan et al. 2005a). One argument is that the correction for multiple substitutions has little effect, but as is clear from figure 3, this is true for the synthetic data set as well, for which there are no trends. Another argument, that the results are similar to those arrived at by looking at single-nucleotide polymorphism (SNP) databases, is a false comparison. SNPs represent the pattern of population polymorphisms caused by nonlethal (but likely deleterious) SNPs, most of which eventually will be deleted from the genetic pool. This pattern is likely to be quite different from the pattern of long-term accepted amino acid substitutions that results from the action of purifying selective pressure. It is reasonable to conjecture that amino acid SNP variants are more random than accepted substitutions and that those SNPs that are more likely to be selected against in the future are also less frequent in the overall protein; these are exactly the amino acids that will tend to have falsely positive flux due to reconstruction bias. A third argument, that it is the “archaic” amino acids that are gradually disappearing, also appears spurious. The biases in flux analyses generally lead toward the conclusion that there is an overall deletion of the common amino acids. Common amino acids are likely to be simpler—if for no other reason than to avoid extra synthesis work for the cell—and are therefore also most likely to result from experiments that attempt to recreate the conditions for the origin of amino acids. There is a need here to avoid unintended logical circularity: are inferences concerning the order of amino acid acquisition based on much more than the genetic code and their current frequencies?
Conclusion
It is likely that amino acid frequencies in various proteins have changed over evolutionary time and that this has resulted in fluxes. It is difficult to believe, however, that the direction of gain and loss should be convergent in different lineages and that it should be the same in fast- and slow-evolving sites. It is also hard to believe that the rate of gain or loss of amino acids should be constant over billions of years of evolution. Such inferences require convincing evidence and are more easily explained by normal evolutionary processes and statistical biases. Unfortunately, the genome data used by Jordan et al. (2005a) does not contain sufficient information to support their conclusions.
To accurately assess and correct for the statistical error requires an accurate model of the evolutionary process, including differences in equilibrium frequencies and substitution rates at different locations in different proteins. A site-specific model with the required accuracy is impossible to obtain with only 3 taxa, but site-specific models become better defined and more useful for predictive purposes as taxonomic sampling increases(Pollock and Bruno 2000); such model accuracy for a wide variety of proteins will be a great benefit of the rapidly increasing number of complete genomes being obtained by genome sequencing projects. Evolutionary genomics approaches have great potential benefits for understanding organismal function, physiology, and mutation, but there is need to approach the subject carefully. Casual use of these techniques without careful consideration of inherent pitfalls can be treacherous, and claims must be well substantiated from a statistical viewpoint before they are advanced.
Acknowledgments
We would like to thank Drs I. K. Jordan and S. Sunyaev for helpful explanations regarding their paper and Dr B. Blackburne for computational assistance. This work was supported by the Medical Research Council and grants to D.D.P. from the National Institutes of Health (GM065612-01 and GM065580-01), the National Science Foundation through the Louisiana Experimental Program to Stimulate Competitive Research and the Center for Bio-modular Multi-scale Systems, and the State of Louisiana Board of Regents (Research Competitiveness Subprogram and the Louisiana Education Quality Support Fund (2001–04)-RD-A-08 and the Millennium Research Program’s Biological Computation and Visualization Center) and Governor’s Biotechnology Initiative.
Literature Cited
- Akaike H. A Bayesian analysis of the minimum AIC procedure. Ann Inst Stat Math. 1978;30:9–14. [Google Scholar]
- Bruno WJ. Modeling residue usage in aligned protein sequences via Maximum likelihood. Mol Biol Evol. 1996;13:1368–74. doi: 10.1093/oxfordjournals.molbev.a025583. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO, Schwartz RM, Orcutt BC. Atlas of protein sequence and structure. In: Dayhoff MO, editor. A model of evolutionary change in proteins. Washington, DC: National Biomedical Research Foundation; 1978. p. 345. [Google Scholar]
- Dimmic MW, Mindell DP, Goldstein RA. Modeling evolution at the protein level using an adjustable amino acid fitness model. Pac Symp Biocomput. 2000:18–29. doi: 10.1142/9789814447331_0003. [DOI] [PubMed] [Google Scholar]
- Goldman N, Yang Z. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 1994;11:725–36. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S. A universal trend of amino acid gain and loss in protein evolution. Nature. 2005a;433:633–8. doi: 10.1038/nature03306. [DOI] [PubMed] [Google Scholar]
- Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S. A universal trend of amino acid gain and loss in protein evolution. Nature. 2005b;433(Suppl 2):633–8. doi: 10.1038/nature03306. [DOI] [PubMed] [Google Scholar]
- Kinjo AR, Nishikawa K. Eigenvalue analysis of amino acid substitution matrices reveals a sharp transition of the mode of sequence conservation in proteins. Bioinformatics. 2004;20:2504–8. doi: 10.1093/bioinformatics/bth297. [DOI] [PubMed] [Google Scholar]
- Koshi JM, Goldstein RA. Context-dependent optimal substitution matrices derived using Bayesian statistics and phylogenetic trees. Protein Eng. 1995;8:641–5. doi: 10.1093/protein/8.7.641. [DOI] [PubMed] [Google Scholar]
- Koshi JM, Goldstein RA. Mathematical models of natural site mutations including site heterogeneity. Proteins. 1998;32:289–95. [PubMed] [Google Scholar]
- Krishnan NM, Seligmann H, Stewart CB, De Koning AP, Pollock DD. Ancestral sequence reconstruction in primate mitochondrial DNA: compositional bias and effect on functional inference. Mol Biol Evol. 2004;21:1871–83. doi: 10.1093/molbev/msh198. [DOI] [PubMed] [Google Scholar]
- McDonald JH. Apparent trends of amino acid gain and loss in protein evolution due to nearly neutral variation. Mol Biol Evol. 2006;23:240–4. doi: 10.1093/molbev/msj026. [DOI] [PubMed] [Google Scholar]
- Nei M. Mathematical models of speciation and genetic distance. In: Karlin S, Nevo E, editors. Population genetics and ecology. New York: Academic press; 1976. pp. 723–65. [Google Scholar]
- Overington J, Donnelly D, Johnson MS, Sali A, Blundell TL. Environment-specific amino-acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1992;1:216–26. doi: 10.1002/pro.5560010203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock DD, Bruno WJ. Assessing an unknown evolutionary process: effect of increasing site-specific knowledge through taxon addition. Mol Biol Evol. 2000;17:1854–8. doi: 10.1093/oxfordjournals.molbev.a026286. [DOI] [PubMed] [Google Scholar]
- Pollock DD, Eisen JA, Doggett NA, Cummings MP. A case for evolutionary genomics and the comprehensive examination of sequence biodiversity. Mol Biol Evol. 2000;17:1776–88. doi: 10.1093/oxfordjournals.molbev.a026278. [DOI] [PubMed] [Google Scholar]
- Soyer OS, Dimmic MW, Neubig RR, Goldstein RA. Dimerization in aminergic G-protein-coupled receptors: application of a hidden-site class model of evolution. Biochemistry. 2003;42:14522–31. doi: 10.1021/bi035097r. [DOI] [PubMed] [Google Scholar]
- Wako H, Blundell T. Use of amino acid environment-dependent substitution tables and conformational propensities in structure prediction from aligned sequences of homologous proteins. II. Secondary structures. J Mol Biol. 1994;238:693–708. doi: 10.1006/jmbi.1994.1330. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;15:555–6. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yang Z. Phylogenetic analysis by maximum likelihood (PAML) 2002 Available from: http://abacus.gene.ucl.ac.uk/software/paml.html.