Abstract
To understand how cells respond to altered oxygenation, a frequent experimental paradigm is to isolate known components of bona fide oxygen responsive proteins. Recent studies have shown that a protein known as CSN5 or JAB1 interacts with both the HIF-1α oxygen-responsive transcription factor and its oxygen-dependent regulator, the Von Hippel-Lindau (pVHL) tumor suppressor. CSN5 is a component of the COP9 Signalosome (CSN) which is a multi-subunit protein that has high homology to the lid of the 19S lid of 26S proteasome. The exact function of the CSN5 interaction with pVHL and HIF-1α remains to be fully elucidated, but it is clear that the interaction is both oxygen dependent and that CSN5 may play different roles under oxic and hypoxic responses. Further, evidence has also been published indicating that pVHL can be potentially post-translationally modified by CSN5 (de-neddylation) and that CSN5 transcription is regulated by hypoxia as are many of the key pVHL/HIF-1α regulatory genes such as the PHDs and OS-9. This review will give a broad overview of known CSN5 and COP9 Signalosome functions and how these functions impact the pVHL/HIF-1α signaling complex and potentially other oxygen-sensitive response networks.
Keywords: COP9 signalosome, CSN, Hypoxia, VHL, CSN5, Jab1
1. Oxygen sensing and response
Over the last two decades, research from a variety of laboratories has revealed a tremendous amount of information on cellular aspects of oxygen sensing and response (reviewed in [1]). One of the key regulators of this response is the pVHL ubiquitin E3 ligase. This E3 ligase mediates the O2-dependent destruction of proteins required for cellular and tissue-specific responses to hypoxia and anoxia. It has also become apparent that deregulation of this ubiquitin ligase can occur independently of altered pO2 in various physiologic and pathophysiologic states [2]. For example, growth factors and oncogenes that activate the PI(3)K pathway can lead to aerobic stabilization of pVHL targets such as the alpha subunits of the hypoxia-inducible (HIF-α) family of bHLH-PAS transcription factors [3–5]. The exact molecular dynamics controlling pVHL ligase activity are very complex however, and novel control mechanisms continue to be revealed [2,6]. Some of these mechanisms have also revealed a great deal about key regulatory events controlling protein degradation in general [2,7]. As many excellent reviews including Peter Ratcliffe’s review in this edition of Seminars in Cell & Developmental Biology address pVHL function as an E3 ubiquitin ligase in detail, this review will address other aspects of pVHL control, namely the control of ubiquitin E3 ligase activity by the COP9 signalosome (CSN).
Clearly, proteins other than the pVHL pathway play roles in oxygen sensing and response. There are a wide variety of enzymes that directly utilize dioxygen (i.e. cyclooxygenases, dioxygenases, monooxygenases) or utilize O2 as a cofactor in enzymatic reactions (i.e. some heme-binding proteins, metalloproteases). The reductive state of the cell also impacts the tertiary structure and function of many if not all proteins and various lipids. Thus, dioxygen availability impacts on the cellular microenvironment directly as well as through glycolytic metabolites and reactive oxygen species generation. Although hypoxia and anoxia generate such pleiomorphic alterations in cellular functions via these mechanisms, many of these proteins/lipids remain relatively unstudied with regard to oxygenation. This review focuses on how one aspect of the protein degradation pathway can impact the pVHL hypoxia-responsive pathway and reciprocally how altered cellular oxygenation could directly impact 26S-mediated protein degradation.
2. Cullin-based ubiquitylation
Ubiquitylation is a mechanism for targeting specific proteins for degradation by the 26S proteosome (reviewed in [8,9]). This process is mediated by three types of enzymes termed E1, E2 and E3 that facilitate the transfer of ubiquitin (Ub) moieties to target proteins. While this review focuses primarily on the regulation of the E2 and E3 enzymes utilized in the pVHL degradation pathway, other E2–E3 pathways are likely impacted upon by oxygen availability in a similar fashion. E3 enzymes recognize their substrate protein(s) and tether an E2 Ub conjugating enzyme, thereby catalyzing/promoting the transfer of Ub from the E2 to the target. There are two main classes of Ub E3 ligases. The first class contains a HECT domain and the second class possesses a RING finger motif. While HECT E3s utilize a conserved catalytic cysteine residue forming a thiol–ester intermediate with Ub, RING E3s recruit the E2 via the RING domain and promote the transfer of Ub from the E2 to a bound substrate.
Cullins, together with their RING finger partner ROC1/Rbx1 (also named Hrt1) [10–14], are the largest subfamily of RING-based E3s. The interaction between a cullin and ROC1/Rbx1 assembles a core complex, which facilitates the synthesis of polyubiquitin chains [14]. The pVHL complex composed of the SOCS-box protein pVHL, elongins C and B and cullin 2 (CUL2) will be the major subject of this review, but as the cullin-based E3s are thought to be regulated similarly, the mechanisms discussed in the following pages are broadly applicable to other Cullin-based E3s. In the pVHL complex, elongins C and B are referred to as the recognition complex that stabilize and assist in the correct folding of pVHL [15]. CUL2 can then bind to the pVHL–elongin complex but the mechanism facilitating this interaction is currently largely unknown. It is thought that the intact pVHL E3 complex (pVHL/elongin C and B/CUL2) can then recruit the ROC1/Rbx1 RING finger protein via the C-terminal cullin domain. In this manner, CUL2 places pVHL and its target within close proximity to ROC1/Rbx1, which then can recruit the E2 conjugating enzyme Ubc12 [12,13,16]. Consequently, a substrate such as HIF-1α when bound by pVHL via its prolyl-hydroxylated domains is positioned optimally for covalently accepting a Ub moiety in a Ubc12-catalysed transfer reaction.
3. The COP9 signalosome (CSN)
Within the last five years, an evolutionarily conserved protein complex that plays a role in protein degradation in eukaryotes has been identified. This complex has been termed the COnstitutive Photomorphogenesis mutant 9 (COP9) signalosome and is a protein complex composed of eight subunits (CSN1–CSN8 based on mass) and fractionates as a 450–550 kDa complex in gel filtration columns. The CSN was originally identified in Arabidopsis thaliana as a regulator of photomorphogenesis (COP9) [17] and the mammalian complex was isolated as a co-purifying byproduct of the 26S proteasome [18]. The CSN has subsequently been reported to control pleiotropic functions in various eukaryotes from yeast to mammals. For example, the CSN has been implicated in various functions including cytokine and growth factor signaling, the control of invertebrate development, nuclear transport, mating in budding yeast, aspects of DNA repair, and oxygen homeostasis [2,19–30].
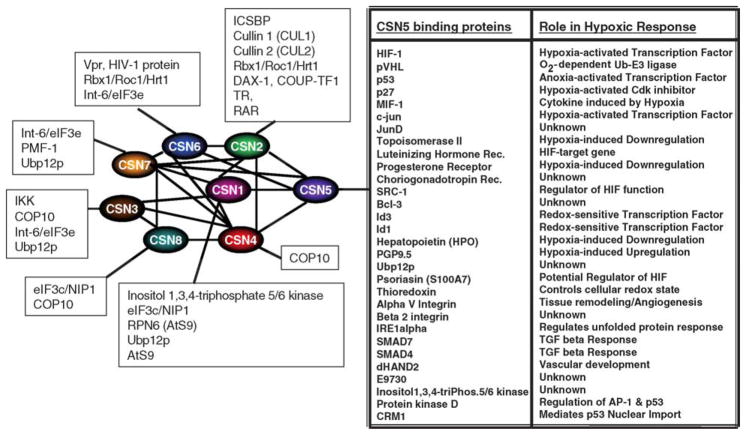
Currently, the exact function(s) of the CSN complex in these various phenotypes remains undefined although it clearly plays a role in protein stability within the 26S proteosomal pathway. Indeed, the CSN has high homology to the 19S lid of the 26S proteosome, and the CSN subunits are considered paralogs of the eight lid subunits of the proteasome complex [31]. Thus, one of the possible functions speculated for the CSN is as an alternative lid for the proteosome. The overall identity and similarity between the CSN and 19S subunits ranges from 34% and 68%, respectively. The most conserved subunits are CSN2 and CSN5, which are over 60% identical between mammals and plants. Of note, the CSN7 subunit is encoded by two similar genes, CSN7a and CSN7b in mammals although the significance of this is unknown [32]. Six of the CSN subunits have a PCI domain (Proteasome, COP9 signalosome, Initiation factor 3) and two subunits have an MPN domain (Mpr1-Pad1-N-terminal). The PCI domain subunits (COP1–4, 7, 8) are believed to act as scaffolds for CSN assembly, while the MPN domain subunits (COP5 and 6) have been shown to encode metalloprotease domains [33,34]. It has also been reported that all cullin (Cul)-based E3s directly associate with all subunits of CSN in human cells [24,27,36,37]. In support of this observation, CSN1 and CSN6 have been shown to directly interact with ROC1/Rbx1 and CSN2 binds CUL1 in yeast two-hybrid assays [36–39]. Additionally, purified CSN was found to bind to the ROC1/Rbx1–CUL1 and the pVHL/elongin C–B/CUL2/Rbx1 complexes in vitro ([39, W.Z., unpublished observation]). Fig. 1 depicts an interaction model of the CSN subunits and proteins found to be associated with individual subunits.
Fig. 1.
Known COP9 signalosome interactions in mammals. Shown are the intercomplex interactions between CSN subunits (in bold) and the interactions of individual CSN subunits with proteins outside of the CSN.
4. Nedd8/Rub1 conjugation (neddylation)
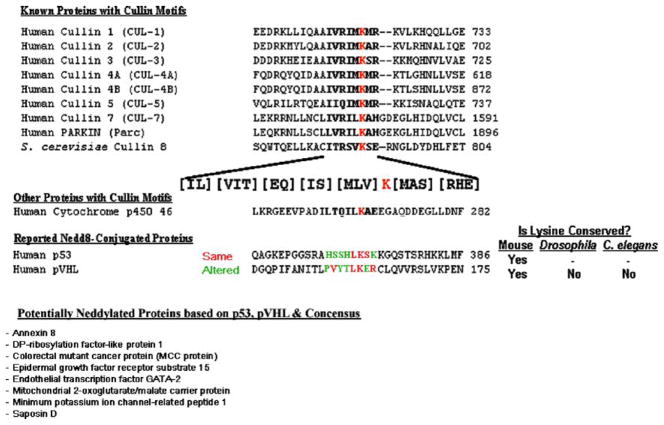
Nedd8 (Neural precursor cell Expressed, Developmentally Downregulated 8) is a small ubiquitin-like protein that was originally found to be conjugated to proteins much like ubiquitin [40]. Neddylation is similar to ubiquitylation or sumolation in that the small protein Nedd8 is covalently linked via an isopeptide bond to a lysine on the target protein by Nedd8-specific E1, E2 and E3 conjugating enzymes. In animals, Nedd8 conjugation is essential for embryonic development and deficiency leads to accumulation of SCF and pVHL substrates such as b-catenin/Armadillo, cyclin E and HIF-1α [30,41,42]. Nedd8 can be directly conjugated to Cdc53, a cullin component of the SCF (Fbox-Skp1-Cdc53/CUL1) E3 Ub ligase complex in Saccharomyces cerevisiae directly linking Nedd8 to ubiquitylation [43,44]. Interestingly, Nedd8 appears to differ from Ub by forming principally mononeddylated species. The cullin neddylation motif contains the consensus sequence: [IL][VIT][RQ][IS][MLV]K[MAS][RHE] (Fig. 2).
Fig. 2.
Cullin neddylation consensus site. The mammalian cullin subunits (exclusive of APC) and a Saccharomyces cullin were used to generate the consensus sequence. Using the web-based Emotif Search Engine [98], the consensus sequence was used to query protein databases. The single conserved mammalian hit is shown. The putative p53 and pVHL sites are also aligned and used to generate a modified consensus sequence that was queried using Emotif.
Interestingly, several proteins that have not been examined for neddylation contain this motif. Cytochrome p450 46 (24S-cholesterol hydroxylase) is an oxygen-regulated enzyme important for the maintenance of brain cholesterol homeostasis. Recent studies have shown elevated plasma concentrations of 24-hydroxycholesterol in patients with Alzheimer’s disease and vascular dementia, suggesting increased brain cholesterol turnover during neurodegeneration. Although speculative, if this enzyme is indeed regulated by neddylation, the implications in 24-hydroxycholesterol turnover by this enzyme and the documented role of oxygen in this process could have significant effects on neurodegeneration.
The X-ray crystal structure of the Skp2/Skp1/CUL1–Rbx1 complex by Zheng et al. revealed that the neddylation site in human CUL1 is K720 [45]. This residue is positioned at the rim of a pocket formed by a WH-B helix and four-helix bundle as well as the ROC1/Rbx1 RING domain. These domains are conserved within the cullin family with K689 being the neddylation site in human CUL2 [46]. The Skp2/Skp1/CUL1–Rbx1 structure suggests a mechanism by which the cullin-linked Nedd8 could assist Rbx1 in positioning the E2 conjugating enzyme juxtaposed to the E3 target for the ubiquitin transfer. Furthermore, the Nedd8 platform may help position an E2-cojugated Ub in a manner that favors the transfer of Ub to the substrate that is sequestered by the recognition component of the E3 (Fig. 3A). Additionally, the Cdc53 C-terminal Domain (CCD) region located just C-terminal to the Nedd8 conjugation site is required for neddylation in vivo and is conserved between all cullin paralogues [43]. Structurally, the CCD starts in the middle of the H31 helix and contains both the S10 and S11 β-sheets with residues V716, R717 and I718 in helix H29 being surface exposed and located in close proximity to ROC1/Rbx1. A deletion of the CCD region in hCUL1, spanning residues 727–776, abolished Nedd8 conjugation in vitro supporting this observation.
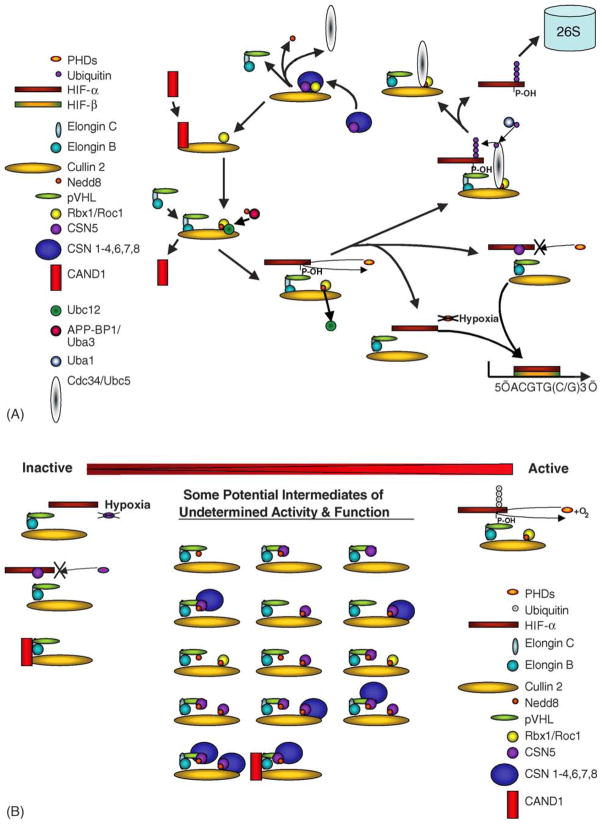
Fig. 3.
(A) pVHL Ub E3 cycle. Shown is a putative model of cycles of pVHL activation and recycling. (B) Potential pVHL protein complexes of unknown activity. Shown are a variety of potential pVHL complexes based on known pVHL interacting proteins and some reported post-translational modifications. This is by no means a comprehensive listing.
Neddylation requires ROC1/Rbx1 in vitro as RING domain mutations prevent Nedd8 conjugation [47,48]. It has also been shown that ROC1/Rbx1 directly interacts with the Nedd8-conjugating enzyme Ubc12 [49,50]. In addition, Nedd8 residues E14 and R25 are required for maintaining Nedd8 activity, suggesting that Nedd8 could facilitate electrostatic interactions to assist ROC1/Rbx1 in the E2-catalysed polyubiquitylation. These observations led to the hypothesis that ROC1/Rbx1 functions as an E3 Nedd8-cullin isopeptide ligase, being capable of binding to both cullins and Ubc12 [47–49]. This supports a model in which neddylation is initiated by cullin-bound ROC1/Rbx1 binding to the Ubc12 E2 conjugating enzyme. The cullin CCD domain may also interact with Ubc12 leading to optimal positioning of the E2 for the subsequent Nedd8 transfer reaction.
To examine the effect of neddylation on cullin-based E3 assembly Read et al. demonstrated that neddylation did not affect the assembly of the SCFb–TrCP complex in vitro, nor did neddylation alter the binding of this E3 to its substrate. It has also been shown that the purified cullin-based SCFb–TrCP E3 was more active in the ubiquitylation of its target than an unneddylatable SCFb–TrCP [49]. Furthermore, Nedd8 conjugation directly increases the binding of SCFb–TrCP to the E2 Ub conjugate. These studies have led to the conclusion that neddylation does not affect E3 assembly, but directly activates the E3 ligase facilitating ubiquitylation of the target protein.
Until last year, the cullin family of E3 ubiquitin ligase scaffolds was thought to be the only target of Nedd8 conjugation, but recent evidence has suggested neddylation of other proteins can occur [51,52]. An important regulator of p53 is Mdm2, an E3 ubiquitin ligase that directly interacts with p53 causing its ubiquitylation and degradation by the 26S proteasome. Xirodimas et al. reported that Mdm2 is neddylated and can promote neddylation of p53 although the sites of Mdm2 neddylation were not found. They further demonstrated that Mdm2-dependent neddylation of p53 inhibits its transcriptional activity [52]. Likewise, the pVHL tumor suppressor has also been shown to be neddylated and is discussed below in Section 10. Interestingly, both p53 and pVHL can associate with CSN5. These observations suggest that many proteins other than cullins could be neddylated and that proteins associated with CSN5 could be neddylation candidates. It should be noted however that the neddylation consensus sequences of pVHL and to a lesser degree, p53 vary significantly from those of the cullins (Fig. 2). Should these finding be substantiated however, several other mammalian proteins including those shown at the bottom of Fig. 2 could be regulated by neddylation. However, if these proteins are substrates for cullin-dependent Ub E3 activity or if neddylation is playing an alternative signaling/targeting roles similar to ubiquitylation is currently undetermined.
5. CSN isopeptidase activity of Nedd8
Increasing evidence indicates that removal of Nedd8 from its cullin targets via CSN-mediated proteolysis plays a significant role in the regulation of cullin-dependant E3 ubiquitylation. This hypothesis is supported by the observation that deletion of a CSN subunit in S. pombe resulted in the accumulation of a predominantly neddylated cullin, thus suggesting that the CSN plays a critical role in Nedd8 deconjugation in vivo. Furthermore, addition of purified CSN to neddylated cullins from yeast extracts resulted in deneddylation of the target cullin. Also, in various genetic models (Drosophila, C. elegans and Arabidopsis), CSN activity is required for the deneddylation of various cullins including CUL1, CUL2 and CUL3 [21,25,26,38,35]. These results paradoxically suggest that contrary to its inhibition of in vitro ubiquitylation, the CSN is required for the degradation of cellular cullin E3 substrates.
Wolf et al. have postulated that the CSN paradox may reflect that the in vivo function of CSN-mediated cullin inhibition is to ‘license’ CSN for its subsequent activation [53]. In this model, the licensing mechanism would be similar to the assembly of pre-replication complexes (pre-RCs) at replication origins. In much the same way that low Cdk activity permits pre-RC assembly, they suggest that CSN-mediated cullin inhibition may create an environment conducive to the assembly of new cullin–ubiquitin ligase complexes that are subsequently activated after release from the CSN. Cullin complexes may then have to return to the CSN for re-assembly (Fig. 3A).
Cope and Deshaies have also postulated a role for CSN in regulating cycles of SCF–ROC1 assembly [54]. In this scenario, an active deneddylated cullin-based E3 ligase–Rbx1 complex that is bound to its degradation target recruits the CSN, which cleaves Nedd8 from the cullin. This allows the binding of p120CAND1 (discussed below) to the cullin, which results in the separation of the Ub E3 ligase recognition complex from the cullin. Subsequent neddylation of the cullin by the Rbx1-dependant Ubc12 E2 conjugating enzyme facilitates the dissociation of p120CAND1, resulting in formation of an active Ub E3. This model suggests that cullin neddylation controls cycles of Ub E3 ligase assembly. Additionally, cullin mutants that are neddylation deficient should exhibit constitutive association with p120CAND1, thus exhibiting a significant deficiency in Ub E3 core–cullin–Rbx1 assembly. The Nedd8 E2 conjugating enzyme Ubc12 has also been shown to bind CUL3 independently of the CSN [26]. This suggests that cullin neddylation occurs following the release of the CSN holocomplex. The regulatory mechanisms facilitating association/dissociation of the CSN from the cullin remain undefined. Also, several critical question such as how CSN-mediated functional inhibition or assembly is regulated and how Ub E3 recognition complexes (i.e. pVHL/elongin C and B) are assembled with the cullin–Rbx1 complex remain unanswered. Thus, given the current models, the CSN would act as a platform that ensures the transient inactivation of cullin complexes by promoting the release of their associated E2s. Therefore, the cycle of neddylation stimulating the association of cullins with Ub E2s would be followed by CSN-mediated deneddylation and Ub E2 dissociation. This would suggest that CSN-mediated cullin deneddylation prevents inappropriate Ub E3 activity. The question of why the CSN can stably associate with both unneddylated and neddylated cullins however, remains a great mystery.
CSN-dependent isopeptidase activity is sensitive to metal ion chelators due to CSN5 containing a conserved, putative metal-binding motif (EXnHS/THX7 SXXD), referred to as the JAMM motif embedded within the larger MPN domain. The JAMM metalloprotease domain within the CSN5 subunit of the CSN is necessary for deneddylase activity, as mutations disrupting this motif accumulate neddylated cullins in S. pombe [33]. Furthermore, the integrity of the putative JAMM catalytic domain is required for proper photoreceptor cell development in Drosophila. These results suggest that the CSN is a metalloprotease whose proteolytic activity is physiologically important. As all fission and budding yeast csn mutants are deficient in cullin deneddylation, an intact CSN complex seems to be required for deneddylation activity [20,26,55,56]. Reciprocally, the CSN5 subunit alone is not able to catalyze the deneddylation reaction, suggesting that the deneddylation function requires CSN5 association with other subunits of the CSN complex. Theoretically, CSN isopeptidase activity would also be regulated by the reduced/oxidized state of the CSN5-metalloprotease as hypoxia can directly and indirectly affect the function of other metalloproteases [57,58].
The CSN has also been associated with cullin-based Ub E3 subcellular localization and stability. For instance, CSN5 mutants in Drosophila accumulate high levels of neddylated CUL1 in the cytoplasm thus suggesting a role for the CSN in influencing the subcellular localization of cullins [25]. Another example is the observation that in the dark, the putative E3 ligase of the plant transcription factor HY5 (COP1) translocates into the nucleus from the cytoplasm, resulting in degradation of HY5 [17]. Importantly, nuclear translocation of COP1 requires the CSN, although exactly how CSN targets COP1 to the nucleus remains undefined. Surprisingly, Gemmill et al. also reported that wt pVHL influenced a notable perinuclear accumulation of CSN5 that was not evident in the original mutant pVHL cell line [59]. It is currently unknown whether the CSN5 observed in this study was a component of the CSN holocomplex or a smaller CSN5-containing complex. LFA-1 engagement also results in nuclear translocation of CSN5 and the subsequent nuclear export of p27 is CSN5-dependent [60]. It has also been shown that CSN1 affects the subcellular distribution of Suc22 (RNR small subunit) [73]. These observations suggest that the CSN can mediate subcellular localization of Ub E3 targets and that the Ub E3 targets could reciprocally influence subcellular localization of CSN subunits.
Importantly, a recent study suggested that neddylation is a regulated cellular process. It was shown that the CSN-free DDB2–DDB1–CUL4A–ROC1/Rbx1 complex became associated with chromatin in response to UV [24]. Concomitantly, a large portion of CUL4A was found to be neddylated. Subsequently, however, the CSN joined the CUL4A ligase–chromatin complex, resulting in the conversion of neddylated CUL4A into its unmodified form. It has also been shown that light and oxygen can also influence CSN assembly with Ub E3 ligases [2,17]. These findings imply that CSN-mediated deneddylase activity and/or association with Ub E3 ligases can be regulated by cellular environmental queues.
Several questions stand out in evaluating the models considered here. For instance, what triggers the release and activation of cullin complexes held in custody by the CSN? How do external signals such as sunlight, hypoxia, auxin, and UV regulate the CSN? What is the role of the CSN in nuclear targeting of various proteins? What is the role of CSN sub-complexes in cullin regulation? Finally, what other CSN-associated enzymatic activities are uncharacterized? For example, some CSN subunits associate with 19S proteasome and eIF3 components and there is almost no information concerning the functions associated with these interactions.
6. CSN ubiquitin isopeptidase activity
Zhou et al. recently reported that the CSN complex is associated with the deubiquitylating enzyme Ubp12, and that this interaction facilitates the suppression of cullin Ub ligase activity in fission yeast [26]. Importantly, the purified CSN holocomplex inhibited the activity of these E3 ligases independent of the deneddylation activity of CSN. More recently, Groisman et al. identified an evolutionarily conserved CSN-associated deubiquitylation activity [24]. This report identified two distinct CSN-containing complexes that associate with either the CSA or the DDB2 two-nucleotide excision repair proteins. Similarly to cullin-based Ub E3 ligases, these complexes, termed ‘CSA.com’ and ‘DDB2.com’, contain a cullin (CUL4A) and a ring finger (Rbx1) in addition to DDB1. DDB1 and DDB2 are the two subunits that comprise the UV-damaged DNA-binding protein (DDB). Both CSA.com and DDB2.com possess polyubiquitylation activity in vitro, but only if purified free of CSN. The isolated CSN was found to possess two different deubiquitylation activities, the first corresponding to CSN5/JAMM-dependent cleavage of monoubiquitin from CUL4A and the second a depolymerization activity toward polyubiquitin chains. The latter activity possibly explains why CSN-associated DDB2.com and CSA.com are catalytically inactive. Importantly, the CSN-associated ubiquitin depolymerization activity does not require the JAMM motif. The CSN was also shown to promote efficient nuclear accumulation of Ubp12, potentially explaining why the inhibitory effect of Ubp12 depends on the CSN. On the basis of these results, it was proposed that the CSN has a dual activity in inhibiting cullin functionality in vitro: the first being mediated by an intrinsic CSN deneddylase and the second depending on the associated Ubp12.
7. Other regulators of neddylation
Recently, several groups reported the discovery of a novel 120 kDa protein termed CAND1 (cullin-associated neddylation dissociated) [61–65]. It appears that CAND1 can specifically interact with an unneddylated cullin–Rbx1 core. Upon cullin neddylation CAND1 was found to dissociate from the complex, allowing the binding of the Ub E3 recognition complex (Fig. 3A). Deneddylation of cullins by the CSN thus leads to dissociation of the Ub E3 recognition complex from the cullin–Rbx1 core followed by the de novo association of CAND1. Therefore, cycles of neddylation and deneddylation could regulate assembly and disassembly of the E3 ubiquitin ligase complex and the subsequent ubiquitylation and degradation of target proteins. These observations also suggest that neddylation can play a role in reversing p120CAND1-mediated inhibition of cullin activity. However, the signal(s) controlling cullin neddylation/deneddylation have currently not been identified.
Three independent groups have recently reported a novel Nedd8-specific protease called DEN1 (deneddylase 1) [66,67] or NEDP1 [68]. Bacterially expressed human DEN1/NEDP1 was shown to bind to Nedd8 selectively and possessed hydrolytic activity toward C-terminal derivatives of Nedd8. These findings suggest a role for DEN1 in processing the C-terminus of Nedd8 precursor, thereby generating the functional form of Nedd8 for conjugation to cullins. Additionally, Wu et al. and Mendoza et al. reported that in vitro, DEN1/NEDP1 contains an isopeptidase activity capable of removing Nedd8 from its cullin targets when present in high concentrations. Co-expression of Nedd8, CUL4A and NEDP1 led to decreased levels of CUL4A–Nedd8 conjugates, suggesting a role for NEDP1 in deconjugating Nedd8 from cullins in vivo. However, Wu et al. reported that the DEN1 deconjugation of Nedd8 from CUL1 was concentration-dependent in vitro. Thus, at low concentrations, DEN1 could process hyperneddylated CUL1 to a mononeddylated form. The authors therefore suggested that the main role for DEN1 as a Nedd8 isopeptidase is to maintain cullins in a mononeddylated form, thereby reversing spurious hyperneddylation that would lead to disruption of normal regulatory interactions for the cullins. Another potential function of DEN1 would be to salvage trapped derivatives of Nedd8 by regenerating Nedd8 from adventitiously formed Nedd8 amides and thiol esters via the C-terminal hydrolytic activity of DEN1. It has also been demonstrated that DEN1 can de-conjugate Nedd8 from Ubc12 in vitro, thus suggesting that this protease may play a role in preventing the autoneddylation of the E2 conjugating enzyme. The elucidation of potential regulatory steps facilitating/inhibiting DEN1–Ubp12 interactions in future studies should be highly illuminating.
8. CSN5 functions independent of the CSN holocomplex
A conserved and distinct feature of CSN5 and some other CSN subunits is the existence of several discrete forms based on relative complex mass as accessed by native gel filtration chromatography. CSN5 masses range from monomeric mass in Arabidopsis, mammalian cells, and Drosophila [69,70,71] to about 200 kDa in fission yeast [55,71] and approximately 450 kD in the CSN holocomplex. Interestingly, in each of the csn mutants of Arabidopsis, CSN5 accumulates only in the free form [72,69]. Similarly, in the csn4 mutant of Drosophila, CSN5 is found only in low molecular mass fractions [70]. This suggests that the association of CSN5 with the CSN holocomplex requires each of the PCI-domain-containing proteins in Arabidopsis and perhaps in Drosophila. Ectopic overexpression of epitope-tagged CSN5 in cultured cells results in limited incorporation of the protein into the CSN complex, with a large fraction found in the free form [73].
Various studies have also shown that CSN1, CSN2, and CSN8 are associated with the holocomplex under normal conditions and are found to be predominantly nuclear-localized [73–77]. CSN-independent CSN5, however, is found both in the nucleus and cytoplasm [69,78,79]. Dependence of CSN5 nuclear accumulation on other subunits has been clearly demonstrated in the budding yeast CSN-like complex [79]. It has further been reported that the mini-CSN complex containing CSN5 and a subset of other CSN subunits is mostly cytosolic [73]. Thus, current evidence suggests that the localization of CSN5 is highly regulated. CSN5 does contain a nuclear export signal, and can bind to CRM1 through its C-terminal region. Point mutations in the nuclear export signal partially impair the ability of CSN5 in nuclear export and downregulation of p27 suggesting that facilitative cellular localization plays a role in at least some of functions reported for CSN5 [73]. In support of this, it has been shown that overexpression of Her-2/neu enhances p27 degradation and stimulates cytoplasmic accumulation of CSN5. This finding also suggests that growth factors as well as environmental factors can regulate CSN5 function although the mechanism underlying these events has yet to be elucidated. Redistribution of CSN5 to the cytoplasm is also reported in response to contact inhibition [80]. Moreover, CSN5 binds to integrin adhesion receptor LFA-1 and co-localizes with it at the cell membrane, again indicating that multiple factors can regulate CSN5 that impact widely diverse functions. Many questions regarding CSN5 function and regulation thus remain unresolved. For instance, are CSN5 interactions with its various binding partners coupled to CSN function in some fashion or does it regulate some of these proteins independently of the CSN?
What is the role of CSN5 in CSN complex assembly? In Drosophila Csn5 null mutants, CSN4 and CSN7 can still form a large protein complex similar to the wild-type CSN, indicating that CSN5 is not critical in complex assembly [70]. Thus, the multiple-pair interaction between CSN5 and other subunits may largely reflect the necessity of those subunits in keeping CSN5 associated with the complex. It also raises the possibility that CSN5 could function as a common recognition component that recruits proteins to the CSN. CSN5 may also affect the assembly or stability of small sub-complexes of the CSN and may also affect CSN8 accumulation in plants [70,37]. In fission yeast, the role of CSN5 in CSN holocomplex formation seems more significant because it is necessary for the association of CSN4 with CSN2 and with the large holocomplex [75]. This suggests evolutionary divergence in some aspects of CSN function.
In the early embryonic stages of mammalian development, CSN5 transcripts are present with low expression levels in all tissues. Preferential expression in selected tissues is detected at embryonic day 11.5, with higher levels in dorsal root ganglia [81]. At later stages of embryonic development, prominent expression of CSN5 transcripts was observed in cranial nerve, spinal and sympathetic ganglia, as well as in the oral and the olfactory epithelium. In the adult brain, additional areas of CSN5 expression were the hippocampus and the Purkinjie layer of the cerebellum. The temporal and spatial expression pattern of Nedd8 was also found to substantially overlap CSN5 expression at all embryonic stages, supporting the model of a functional interaction between CSN5 and Nedd8 during developmental processes. CSN5-null embryos die soon after implantation. As expected, CSN5 null embryonic cells, which lacked other CSN components, expressed higher levels of p27, p53, and cyclin E, resulting in impaired proliferation and accelerated apoptosis [82]. CSN5 heterozygous mice were healthy and fertile but smaller than their wild-type littermates. CSN5+/− mouse embryonic fibroblast cells, in which the amount of CSN5-containing small sub-complex, but not that of CSN, were selectively reduced, proliferated poorly, showed an inefficient downregulation of p27 during G(1), and were delayed in the progression from G(0) to S phase by 3 h compared with the wild-type cells. Most interestingly, in CSN5+/− mouse embryonic fibroblasts, the levels of cyclin E and deneddylated CUL1 were unchanged, and p53 was not induced.
9. CSN5-interacting proteins
A strong word of caution is required concerning CSN5 found in yeast-2-hybrid screens as CSN5 has been shown to interact with the DNA-binding domain of GAL4 alone and must be rejected as a false positive in the GAL4-based two-hybrid system [83]. With that said, CSN5 has been reported to directly interact with a wide variety of proteins with the majority being either targets of ubiquitylation or part of the ubiquitylation/neddylation pathways (Fig. 1). In some cases (i.e. p53, p27, rLHR precursor and Smad4), CSN5 binding is found to stimulate protein degradation, whereas in other cases such as HIF-1α and perhaps c-Jun, CSN5 binding appears to promote stabilization [2,29,84–86]. Yet how the target binding by CSN5 may link to the biochemical activities of the CSN complex or other undetermined cellular processes have yet to emerge.
10. Neddylation and hypoxic function
It is currently unknown whether cellular oxygenation status directly affects neddylation or deneddylation. Because the CSN isopeptidase activity depends on the correct coordination of metallic ions within the JAMM domain of CSN5, it is possible that altered oxygenation could result in a decrease in CSN proteolytic activity due to decreased affinity for the reduced metal ion. This would result in stabilization of proteins targeted by Ub E3 ligases. In this regard, it is interesting to note that some of the binding partners of CSN5 are targets of these Ub E3 ligases (i.e. p27 and p53) and has been shown to accumulate under hypoxic/anoxic conditions [87,88].
Importantly, CSN5 has been shown to interact with both HIF-1α and its E3 ligase pVHL [2,89]. In a report by our group, CSN5 was found to be physically associated with the HIF-1α and to positively regulate HIF function supporting a previous CSN5-HIF-1α interaction found in a yeast-2-hybrid study [89]. However, that study suggested that the mechanism underlying CSN5-induced HIF-1α protein stabilization was related to CSN5 competition with p53 for HIF-1α. As CSN5 stabilizes HIF-1α at higher pO2 concentrations (1–17% O2) than anoxia-stabilized (0.1–0% O2) p53 however, this would suggest that CSN5 mediates aerobic and hypoxic HIF-1α stabilization independent of p53.
The data presented by Bemis et al. demonstrated that CSN5 interacts directly with both the CODD (C-terminal oxygen-dependant degradation domain) of HIF-1α (independent of prolyl-hydroxylation state) as well as the pVHL E3 ligase. Because CSN5, pVHL and the PHDs interact with the same region of HIF-1α, the potential for CSN5 altering pVHL or PHD affinity for HIF-1α seems likely. This poses the possibility that CSN5-associated pVHL and PHD-associated pVHL could have reciprocal and antagonistic association/dissociation mechanisms leading to the dramatic switch-like mechanism seen in vivo. This is supported by the finding that CSN5 expression leads to an inhibition of HIF-1α prolyl-hydroxylation at P564, signifying that CSN5 sterically prevents PHD association with HIF-1α but does not directly interact with or alter PHD enzymatic activity. Why CSN5 interacts directly with the CODD, but not the NODD (N-terminal oxygen-dependant degradation domain) is not clear. It has been suggested that PHD-mediated prolyl-hydroxylation of the CODD could dictate NODD prolyl-hydroxylation, suggesting that inhibition of CODD prolyl-hydroxylation would alter NODD prolyl-hydroxylation as well (Amato Giaccia, personal communication). It is also interesting that CSN5 recognizes both the target of the Ub E3 ligase complex as well as the ligase itself, and it will be informative to determine if CSN5 binds its other targets’ cognate E3 ligases as well.
Thus, increased CSN5 expression is sufficient and necessary for aerobic HIF-1α stabilization. CSN5 expression also affects the kinetics and amplitude of hypoxia-induced HIF-1α stabilization although the mechanism underlying this effect appears distinct from aerobic stabilization. Overexpression of CSN5 has been associated with tumor progression and could reflect the deregulation of aerobic HIF stability, which is thought to be an important transition in tumor progression. Recently, CSN5 mRNA expression has been shown to be downregulated by prolonged and severe hypoxia [90]. Under these same conditions, HIF-1α stability is also decreased, further supporting the functional relationship between the two molecules. Because other known regulators of HIF stability or function (PHD2, ARD1) are also regulated at the message level, CSN5 regulation by hypoxia is consistent with the tight control of HIF expression controlled by feedback mechanisms. It is also interesting that CSN5 associated with HIF-1α independently of the CSN holoenzyme, while pVHL did associate with the CSN holoenzyme. The consequences of this are undetermined and whether CSN5 acts in recruitment of UB E3 targets to their cognate ligases or function to inhibit CSN activity under certain conditions is undetermined. A model of the pVHL E3 ligase activation cycle is presented in Fig. 3A while a portion of the potential intermediates of the pVHL–CSN complex in which the function (or existence) of these intermediates is unknown (Fig. 3B).
Stickle et al. recently reported that pVHL becomes covalently conjugated by Nedd8 [51]. Surprisingly this neddylation was not required for the E3 ligase activity, however, it was needed for binding of pVHL to fibronectin. This would indicate that neddylation indeed plays roles completely unrelated to Ub E3 function and is the first report to suggest such an activity. On the other hand, expression of a cullin neddylation-defective pVHL mutant in RCC cells restored the regulation of HIF-α, however it failed to promote differentiated morphology and was also insufficient to suppress the formation of tumors in SCID mice. The authors concluded that Nedd8 modification of pVHL plays an important role in the proper assembly of fibronectin, and that in the absence of such regulation, an intact HIF pathway is insufficient to prevent tumorigenesis. It is interesting that pVHL (a regulator of hypoxic phenotypes) and p53 (an effector of anoxic phenotypes) can also be modified by neddylation [51,52]. Importantly, these proteins consist of a ubiquitylation target and the recognition component of a ubiquitin E3 ligase suggesting that other proteins targeted for degradation might likewise be neddylated.
Various reports have described a CSN-associated kinase activity associated with the CSN complex. Subsequently, inositol 1,3,4-triphosphate 5/6 kinase, casein kinase 2 (CK2), and protein kinase D (PKD) were found to physically interact with CSN however, their significance in elucidating the various functions of the CSN is currently undetermined [91,92]. It is interesting to note that the kinase activity associated with the CSN was in part characterized by its sensitivity to curcumin. Various reports provide circumstantial evidence that a curcumin-sensitive kinase (possibly the CSN kinase) has distinct effects on hypoxia-mediated cellular functions. Curcumin possesses potent anti-inflammatory activity and can inhibit COX2 induction by inflammatory cytokines or hypoxia [93]. Curcumin also reversed the subcellular redistribution of the inducible Hsp70 protein caused by chronic hypoxia [94]. Further, the induction of triosephosphate isomerase (TPI) gene expression by hypoxia is HIF-1α dependant and was suppressed by curcumin [95]. Curcumin can also mediate the inhibition of c-Jun/AP-1 activation that results in a significant decrease in the activation of GAPDH mRNA and VEGF production by hypoxia [95,96]. Pollman et al. also demonstrated that elevated cellular CSN activity induced by CSN subunit 2 overexpression was sufficient to increase VEGF production in HeLa cells [96]. In a similar study curcumin significantly blocked TGF-β induction of VEGF expression while SP-1 and MKK1 inhibitors did not. It should be noted that TGF-β is a hypoxia-inducible gene and that TGF-β enhanced both AP-1 and HIF-1 DNA binding activities [97]. These studies however are all circumstantial and direct evidence of the CSN kinase(s) role in hypoxic responses remains to be rigorously addressed.
11. Concluding remarks
Although the discovery and study of the CSN is barely a decade old, a tremendous amount of information has been collected that is beginning to elucidate how this protein complex impacts on so many diverse cellular functions. One of the functions regulated by the CSN is oxygen homeostasis via the regulation of HIF-α, the pVHL Ub E3 ligase and potentially other effectors of hypoxic phenotypes. While the details of this regulation are still being defined, the current data supports major roles for the CSN holocomplex, CSN-independent CSN5 functions and protein neddylation in controlling cellular oxygen sensing and responses. As future studies elucidate further functions of this ‘gatekeeper’ of oxygen homeostasis, it will be interesting to see if components of CSN function are deregulated in hematological, vascular, inflammatory pathologies.
Acknowledgments
We would like to thank Dr. Amato Giaccia for his comments on this work and his continuing mentorship. We apologize for any relevant work that was not cited. This research was supported by grants from Pfizer Pharmaceuticals and NCI CA102301 (W.Z.).
References
- 1.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5(5):343–54. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 2.Bemis L, Chan DA, Finkielstein CV, Qi L, Sutphin PD, Chen X, et al. Distinct aerobic and hypoxic mechanisms of HIF-alpha regulation by CSN5. Genes Dev. 2004;18(7):739–44. doi: 10.1101/gad.1180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mazure NM, Chen EY, Laderoute KR, Giaccia AJ. Induction of vascular endothelial growth factor by hypoxia is modulated by a phosphatidylinositol 3-kinase/Akt signaling pathway in Ha-ras-transformed cells through a hypoxia inducible factor-1 transcriptional element. Blood. 1997;90(9):3322–31. [PubMed] [Google Scholar]
- 4.Jiang BH, Agani F, Passaniti A, Semenza GL. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 1997;57(23):5328–35. [PubMed] [Google Scholar]
- 5.Zundel W, Schindler C, Haas-Kogan D, Koong A, Kaper F, Chen E, et al. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 2000;14(4):391–6. [PMC free article] [PubMed] [Google Scholar]
- 6.Brugarolas JB, Vazquez F, Reddy A, Sellers WR, Kaelin WG., Jr TSC2 regulates VEGF through mTOR-dependent and -independent pathways. Cancer Cell. 2003;4(2):147–58. doi: 10.1016/s1535-6108(03)00187-9. [DOI] [PubMed] [Google Scholar]
- 7.Liakopoulos D, Busgen T, Brychzy A, Jentsch S, Pause A. Conjugation of the ubiquitin-like protein Nedd8 to cullin-2 is linked to von Hippel-Lindau tumor suppressor function. Proc Natl Acad Sci USA. 1999;96(10):5510–5. doi: 10.1073/pnas.96.10.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann-Petersen R, Gordon C. Integral UBL domain proteins: a family of proteasome interacting proteins. Semin Cell Dev Biol. 2004;15(2):247–59. doi: 10.1016/j.semcdb.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 9.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5(3):177–87. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 10.Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–61. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- 11.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell. 1999;3(4):535–41. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 12.Seol JH, Feldman RM, Zachariae W, Shevchenko A, Correll CC, Lyapina S, et al. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13(12):1614–26. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skowyra D, Koepp DM, Kamura T, Conrad MN, Conaway RC, Conaway JW, et al. Reconstitution of G1 cyclin ubiquitination with complexes containing SCFGrr1 and Rbx1. Science. 1999;284(5414):662–5. doi: 10.1126/science.284.5414.662. [DOI] [PubMed] [Google Scholar]
- 14.Tan P, Fuchs SY, Chen A, Wu K, Gomez C, Ronai Z, et al. Recruitment of a ROC1–CUL1 ubiquitin ligase by Skp1 and HOS to catalyze the ubiquitination of I kappa B alpha. Mol Cell. 1999;3(4):527–33. doi: 10.1016/s1097-2765(00)80481-5. [DOI] [PubMed] [Google Scholar]
- 15.Feldman DE, Thulasiraman V, Ferreyra RG, Frydman J. Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol Cell. 1999;4(6):1051–61. doi: 10.1016/s1097-2765(00)80233-6. [DOI] [PubMed] [Google Scholar]
- 16.Chen A, Wu K, Fuchs SY, Tan P, Gomez C, Pan ZQ. The conserved RING-H2 finger of ROC1 is required for ubiquitin ligation. J Biol Chem. 2000;275(20):15432–9. doi: 10.1074/jbc.M907300199. [DOI] [PubMed] [Google Scholar]
- 17.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–6. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 18.Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, et al. A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J. 1998;12(6):469–78. [PubMed] [Google Scholar]
- 19.Freilich S, Oron E, Kapp Y, Nevo-Caspi Y, Orgad S, Segal D, et al. The COP9 signalosome is essential for development of Drosophila melanogaster. Curr Biol. 1999;9:1187–90. doi: 10.1016/S0960-9822(00)80023-8. [DOI] [PubMed] [Google Scholar]
- 20.Maytal-Kivity V, Reis N, Hofmann K, Glickman MH. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28–39. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwechheimer C, Deng XW. COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol. 2001;11:420–6. doi: 10.1016/s0962-8924(01)02091-8. [DOI] [PubMed] [Google Scholar]
- 22.Bech-Otschir D, Seeger M, Dubiel W. The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci. 2002;115:467–73. doi: 10.1242/jcs.115.3.467. [DOI] [PubMed] [Google Scholar]
- 23.Seeger M, Gordon C, Dubiel W. Protein stability: the COP9 signalosome gets in on the act. Curr Biol. 2001;11:R643–6. doi: 10.1016/s0960-9822(01)00382-7. [DOI] [PubMed] [Google Scholar]
- 24.Groisman R, Polanowska J, Kuraoka I, Sawada J, Saijo M, Drapkin R, et al. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell. 2003;113(3):357–67. doi: 10.1016/s0092-8674(03)00316-7. [DOI] [PubMed] [Google Scholar]
- 25.Doronkin S, Djagaeva I, Beckendorf SK. The COP9 signalosome promotes degradation of Cyclin E during early Drosophila oogenesis. Dev Cell. 2003;4(5):699–710. doi: 10.1016/s1534-5807(03)00121-7. [DOI] [PubMed] [Google Scholar]
- 26.Zhou C, Wee S, Rhee E, Naumann M, Dubiel W, Wolf DA. Fission yeast COP9/signalosome suppresses cullin activity through recruitment of the deubiquitylating enzyme Ubp12p. Mol Cell. 2003;11(4):927–38. doi: 10.1016/s1097-2765(03)00136-9. [DOI] [PubMed] [Google Scholar]
- 27.Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9-signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint dependent and -independent mechanisms. Genes Dev. 2003;17:1130–40. doi: 10.1101/gad.1090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleemann R, Hausser A, Geiger G, Mischke R, Burger-Kentischer A, Flieger O, et al. Intracellular action of the cytokine MIF to modulate AP-1 activity and the cell cycle through Jab1. Nature. 2000;408(6809):211–6. doi: 10.1038/35041591. [DOI] [PubMed] [Google Scholar]
- 29.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature. 1999;398(6723):160–5. doi: 10.1038/18230. [DOI] [PubMed] [Google Scholar]
- 30.Ohh M, Kim WY, Moslehi JJ, Chen Y, Chau V, Read MA, et al. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO Rep. 2002;3(2):177–82. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu H, Reis N, Lee Y, Glickman MH, Vierstra RD. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO J. 2001;20(24):7096–107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei N, Chamovitz DA, Deng XW. Arabidopsis COP9 is a component of a novel signaling complex mediating light control of development. Cell. 1994;78:117–24. doi: 10.1016/0092-8674(94)90578-9. [DOI] [PubMed] [Google Scholar]
- 33.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, et al. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298(5593):608–11. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 34.Verma R, Aravind L, Oania R, McDonald WH, Yates JR, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–5. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 35.Yang X, Menon S, Lykke-Andersen K, Tsuge T, Di X, Wang X, et al. The COP9 signalosome inhibits p27(kip1) degradation and impedes G1-S phase progression via deneddylation of SCF Cul1. Curr Biol. 2002;12(8):667–72. doi: 10.1016/s0960-9822(02)00791-1. [DOI] [PubMed] [Google Scholar]
- 36.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, et al. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292(5520):1382–5. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 37.Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, et al. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science. 2001;292(5520):1379–82. doi: 10.1126/science.1059776. [DOI] [PubMed] [Google Scholar]
- 38.Pintard L, Kurz T, Glaser S, Willis JH, Peter M, Bowerman B. Neddylation and deneddylation of CUL-3 is required to target MEI-1/Katanin for degradation at the meiosis-to-mitosis transition in C. elegans. Curr Biol. 2003;13(11):911–21. doi: 10.1016/s0960-9822(03)00336-1. [DOI] [PubMed] [Google Scholar]
- 39.Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. Nedd8 on cullin: building an expressway to protein destruction. Oncogene. 2004;23(11):1985–97. doi: 10.1038/sj.onc.1207414. [DOI] [PubMed] [Google Scholar]
- 40.Kamitani T, Kito K, Nguyen HP, Yeh ET. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272(45):28557–62. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 41.Tateishi K, Omata M, Tanaka K, Chiba T. The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol. 2001;155(4):571–9. doi: 10.1083/jcb.200104035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou CY, Lin YF, Chen YJ, Chien CT. Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 2002;16(18):2403–14. doi: 10.1101/gad.1011402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lammer D, Mathias N, Laplaza JM, Jiang W, Liu Y, Callis J, et al. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12(7):914–26. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liakopoulos D, Doenges G, Matuschewski K, Jentsch S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998;17(8):2208–14. doi: 10.1093/emboj/17.8.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, et al. Structure of the Cul1–Rbx1–Skp1–F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416(6882):703–9. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 46.Wada H, Yeh ET, Kamitani T. Identification of NEDD8-conjugation site in human cullin-2. Biochem Biophys Res Commun. 1999;257(1):100–5. doi: 10.1006/bbrc.1999.0339. [DOI] [PubMed] [Google Scholar]
- 47.Kamura T, Conrad MN, Yan Q, Conaway RC, Conaway JW. The Rbx1 subunit of SCF and VHL E3 ubiquitin ligase activates Rub1 modification of cullins Cdc53 and Cul2. Genes Dev. 1999;13(22):2928–33. doi: 10.1101/gad.13.22.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morimoto M, Nishida T, Nagayama Y, Yasuda H. Nedd8-modification of Cul1 is promoted by Roc1 as a Nedd8-E3 ligase and regulates its stability. Biochem Biophys Res Commun. 2003;301(2):392–8. doi: 10.1016/s0006-291x(02)03051-6. [DOI] [PubMed] [Google Scholar]
- 49.Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, et al. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol. 2000;20(7):2326–33. doi: 10.1128/mcb.20.7.2326-2333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawakami T, Chiba T, Suzuki T, Iwai K, Yamanaka K, Minato N, et al. NEDD8 recruits E2-ubiquitin to SCF E3 ligase. EMBO J. 2001;20(15):4003–12. doi: 10.1093/emboj/20.15.4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stickle NH, Chung J, Klco JM, Hill RP, Kaelin WG, Jr, Ohh M. pVHL modification by NEDD8 is required for fibronectin matrix assembly and suppression of tumor development. Mol Cell Biol. 2004;24(8):3251–61. doi: 10.1128/MCB.24.8.3251-3261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP. Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell. 2004;118(1):83–97. doi: 10.1016/j.cell.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 53.Wolf DA, Zhou C, Wee S. The COP9 signalosome: an assembly and maintenance platform for cullin ubiquitin ligases? Nat Cell Biol. 2003;5(12):1029–33. doi: 10.1038/ncb1203-1029. [DOI] [PubMed] [Google Scholar]
- 54.Cope GA, Deshaies RJ. COP9 signalosome: a multifunctional regulator of SCF and other cullin-based ubiquitin ligases. Cell. 2003;114(6):663–71. doi: 10.1016/s0092-8674(03)00722-0. [DOI] [PubMed] [Google Scholar]
- 55.Mundt KE, Liu C, Carr AM. Deletion mutants in COP9/signalosome subunits in fission yeast Schizosaccharomyces pombe display distinct phenotypes. Mol Biol Cell. 2002;13:493–502. doi: 10.1091/mbc.01-10-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wee S, Hetfeld B, Dubiel W, Wolf DA. Conservation of the COP9-signalosome in budding yeast. BMC Genet. 2002;3:15. doi: 10.1186/1471-2156-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nappi AJ, Vass E. Iron, metalloenzymes and cytotoxic reactions. Cell Mol Biol (Noisy-le-grand) 2000;46(3):637–47. [PubMed] [Google Scholar]
- 58.Brenneisen P, Sies H, Scharffetter-Kochanek K. Ultraviolet-B irradiation and matrix metalloproteinases: from induction via signaling to initial events. Ann NY Acad Sci. 2002;973:31–43. doi: 10.1111/j.1749-6632.2002.tb04602.x. [DOI] [PubMed] [Google Scholar]
- 59.Gemmill RM, Bemis LT, Lee JP, Sozen MA, Baron A, Zeng C, et al. The TRC8 hereditary kidney cancer gene suppresses growth and functions with VHL in a common pathway. Oncogene. 2002;21(22):3507–16. doi: 10.1038/sj.onc.1205437. [DOI] [PubMed] [Google Scholar]
- 60.Bianchi E, Denti S, Granata A, Bossi G, Geginat J, Villa A, et al. Integrin LFA-1 interacts with the transcriptional co-activator JAB1 to modulate AP-1 activity. Nature. 2000;404(6778):617–21. doi: 10.1038/35007098. [DOI] [PubMed] [Google Scholar]
- 61.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120Cand1, an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–8. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 62.Oshikawa K, Matsumoto M, Yada M, Kamura T, Hatakeyama S, Nakayama KI. Preferential interaction of TIP120A with Cul1 that is not modified by NEDD8 and not associated with Skp1. Biochem Biophys Res Commun. 2003;303(4):1209–16. doi: 10.1016/s0006-291x(03)00501-1. [DOI] [PubMed] [Google Scholar]
- 63.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, et al. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10(6):1519–26. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 64.Hwang JW, Min KW, Tamura TA, Yoon JB. TIP120A associates with unneddylated cullin 1 and regulates its neddylation. FEBS Lett. 2003;541(1–3):102–8. doi: 10.1016/s0014-5793(03)00321-1. [DOI] [PubMed] [Google Scholar]
- 65.Min KW, Hwang JW, Lee JS, Park Y, Tamura TA, Yoon JB. TIP120A associates with cullins and modulates ubiquitin ligase activity. J Biol Chem. 2003;278(18):15905–10. doi: 10.1074/jbc.M213070200. [DOI] [PubMed] [Google Scholar]
- 66.Wu K, Yamoah K, Dolios G, Gan-Erdene T, Tan P, Chen A, et al. DEN1 is a dual function protease capable of processing the C terminus of Nedd8 and deconjugating hyper-neddylated CUL1. J Biol Chem. 2003;278(31):28882–91. doi: 10.1074/jbc.M302888200. [DOI] [PubMed] [Google Scholar]
- 67.Gan-Erdene T, Nagamalleswari K, Yin L, Wu K, Pan ZQ, Wilkinson KD. Identification and characterization of DEN1, a deneddylase of the ULP family. J Biol Chem. 2003;278(31):28892–900. doi: 10.1074/jbc.M302890200. [DOI] [PubMed] [Google Scholar]
- 68.Mendoza HM, Shen LN, Botting C, Lewis A, Chen J, Ink B, et al. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J Biol Chem. 2003;278(28):25637–43. doi: 10.1074/jbc.M212948200. [DOI] [PubMed] [Google Scholar]
- 69.Kwok SF, Solano R, Tsuge T, Chamovitz DA, Ecker JR, Matsui M, et al. Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell. 1998;10(11):1779–90. doi: 10.1105/tpc.10.11.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oron E, Mannervik M, Rencus S, Harari-Steinberg O, Neuman-Silberberg S, Segal D, et al. COP9 signalosome subunits 4 and 5 regulate multiple pleiotropic pathways in Drosophila melanogaster. Development. 2002;129(19):4399–409. doi: 10.1242/dev.129.19.4399. [DOI] [PubMed] [Google Scholar]
- 71.Zhou C, Seibert V, Geyer R, Rhee E, Lyapina S, Cope G, et al. The fission yeast COP9/signalosome is involved in cullin modification by ubiquitin-related Ned8p. BMC Biochem. 2001;2(1):7. doi: 10.1186/1471-2091-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Kang D, Feng S, Serino G, Schwechheimer C, Wei N. CSN1 N-terminal-dependent activity is required for Arabidopsis development but not for Rub1/Nedd8 deconjugation of cullins: a structure–function study of CSN1 subunit of COP9 signalosome. Mol Biol Cell. 2002;13(2):646–55. doi: 10.1091/mbc.01-08-0427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, et al. The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem. 2002;277(3):2302–10. doi: 10.1074/jbc.M104431200. [DOI] [PubMed] [Google Scholar]
- 74.Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, et al. The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell. 1996;86(1):115–21. doi: 10.1016/s0092-8674(00)80082-3. [DOI] [PubMed] [Google Scholar]
- 75.Mundt KE, Porte J, Murray JM, Brikos C, Christensen PU, Caspari T, et al. The COP9/signalosome complex is conserved in fission yeast and has a role in S phase. Curr Biol. 1999;9(23):1427–30. doi: 10.1016/s0960-9822(00)80091-3. [DOI] [PubMed] [Google Scholar]
- 76.Staub JM, Wei N, Deng XW. Evidence for FUS6 as a component of the nuclear-localized COP9 complex in Arabidopsis. Plant Cell. 1996;8(11):2047–56. doi: 10.1105/tpc.8.11.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wei N, Deng XW. Characterization and purification of the mammalian COP9 complex, a conserved nuclear regulator initially identified as a repressor of photomorphogenesis in higher plants. Photochem Photobiol. 1998;68(2):237–41. doi: 10.1562/0031-8655(1998)068<0237:capotm>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 78.Bounpheng MA, Melnikova IN, Dodds SG, Chen H, Copeland NG, Gilbert DJ, et al. Characterization of the mouse JAB1 cDNA and protein. Gene. 2000;242(1/2):41–50. doi: 10.1016/s0378-1119(99)00525-9. [DOI] [PubMed] [Google Scholar]
- 79.Maytal-Kivity V, Pick E, Piran R, Hofmann K, Glickman MH. The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int J Biochem Cell Biol. 2003;35(5):706–15. doi: 10.1016/s1357-2725(02)00378-3. [DOI] [PubMed] [Google Scholar]
- 80.Caballero OL, Resto V, Patturajan M, Meerzaman D, Guo MZ, Engles J, et al. Interaction and colocalization of PGP9.5 with JAB1 and p27(Kip1) Oncogene. 2002;21(19):3003–10. doi: 10.1038/sj.onc.1205390. [DOI] [PubMed] [Google Scholar]
- 81.Carrabino S, Carminati E, Talarico D, Pardi R, Bianchi E. Expression pattern of the JAB1/CSN5 gene during murine embryogenesis: colocalization with NEDD8. Gene Expr Patterns. 2004;4(4):423–31. doi: 10.1016/j.modgep.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 82.Tomoda K, Yoneda-Kato N, Fukumoto A, Yamanaka S, Kato JY. Multiple functions of Jab1 are required for early embryonic development and growth potential in mice. J Biol Chem. 2004;279(41):43013–8. doi: 10.1074/jbc.M406559200. [DOI] [PubMed] [Google Scholar]
- 83.Nordgard O, Dahle O, Andersen TO, Gabrielsen OS. JAB1/CSN5 interacts with the GAL4 DNA binding domain: a note of caution about two-hybrid interactions. Biochimie. 2001;83(10):969–71. doi: 10.1016/s0300-9084(01)01329-3. [DOI] [PubMed] [Google Scholar]
- 84.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20(7):1630–9. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li S, Liu X, Ascoli M. p38JAB1 binds to the intracellular precursor of the lutropin/choriogonadotropin receptor and promotes its degradation. J Biol Chem. 2000;275(18):13386–93. doi: 10.1074/jbc.275.18.13386. [DOI] [PubMed] [Google Scholar]
- 86.Wan M, Cao X, Wu Y, Bai S, Wu L, Shi X, et al. Jab1 antagonizes TGF-beta signaling by inducing Smad4 degradation. EMBO Rep. 2002;3(2):171–6. doi: 10.1093/embo-reports/kvf024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276(11):7919–26. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 88.Graeber TG, Peterson JF, Tsai M, Monica K, Fornace AJ, Jr, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14(9):6264–77. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bae MK, Ahn MY, Jeong JW, Bae MH, Lee YM, Bae SK, et al. Jab1 interacts directly with HIF-1alpha and regulates its stability. J Biol Chem. 2002;277(1):9–12. doi: 10.1074/jbc.C100442200. [DOI] [PubMed] [Google Scholar]
- 90.Denko NC, Fontana LA, Hudson KM, Sutphin PD, Raychaudhuri S, Altman R, et al. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene. 2003;22(37):5907–14. doi: 10.1038/sj.onc.1206703. [DOI] [PubMed] [Google Scholar]
- 91.Uhle S, Medalia O, Waldron R, Dumdey R, Henklein P, Bech-Otschir D, et al. Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J. 2003;22:1302–12. doi: 10.1093/emboj/cdg127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Y, Wilson MP, Majerus PW. Inositol 1,3,4-trisphosphate 5/6-kinase associates with the COP9 signalosome by binding to CSN1. J Biol Chem. 2002;277:45759–64. doi: 10.1074/jbc.M208709200. [DOI] [PubMed] [Google Scholar]
- 93.Plummer SM, Holloway KA, Manson MM, Munks RJ, Kaptein A, Farrow S, et al. Inhibition of cyclo-oxygenase 2 expression in colon cells by the chemopreventive agent curcumin involves inhibition of NF-kappaB activation via the NIK/IKK signalling complex. Oncogene. 1999;18(44):6013–20. doi: 10.1038/sj.onc.1202980. [DOI] [PubMed] [Google Scholar]
- 94.Rafiee P, Shi Y, Pritchard KA, Jr, Ogawa H, Eis AL, Komorowski RA, et al. Cellular redistribution of inducible Hsp70 protein in the human and rabbit heart in response to the stress of chronic hypoxia: role of protein kinases. J Biol Chem. 2003;278(44):43636–44. doi: 10.1074/jbc.M212993200. [DOI] [PubMed] [Google Scholar]
- 95.Yamaji R, Fujita K, Takahashi S, Yoneda H, Nagao K, Masuda W, et al. Hypoxia up-regulates glyceraldehyde-3-phosphate dehydrogenase in mouse brain capillary endothelial cells: involvement of Na+/Ca2+ exchanger. Biochim Biophys Acta. 2003;1593(2/3):269–76. doi: 10.1016/s0167-4889(02)00397-x. [DOI] [PubMed] [Google Scholar]
- 96.Pollmann C, Huang X, Mall J, Bech-Otschir D, Naumann M, Dubiel W. The constitutive photomorphogenesis 9 signalosome directs vascular endothelial growth factor production in tumor cells. Cancer Res. 2001;61(23):8416–21. [PubMed] [Google Scholar]
- 97.Shih SC, Claffey KP. Role of AP-1 and HIF-1 transcription factors in TGF-beta activation of VEGF expression. Growth Factors. 2001;19(1):19–34. doi: 10.3109/08977190109001073. [DOI] [PubMed] [Google Scholar]
- 98.http://www.dna.stanford.edu/emotif/.