Abstract
Homing endonucleases have long been known as the orchestrators of intron mobility. However, the extent of their influence on the intron and its genetic and cellular environment is still being elucidated. The accompanying paper emphasizes the importance of temporal control of endonuclease expression on splicing, expression of the host gene and cellular metabolism, while it raises questions to guide future inquiry.
Much has been written of the deleterious effects of mobile elements and the forces that have arisen to combat their spread: everything from methylation in mammals, through siRNA-based epigenetic silencing in plants, to ribonucleases in bacteria (Beauregard et al., 2008). The flip side of the coin is the mobile element itself, which must adopt the role of a well-behaved guest in a host cell whose fitness is primary, but must also preserve its invasive properties as an opportunistic parasite, as insurance against host extinction. These mobile elements therefore temper their own promiscuity, until survival of both host and parasite are threatened. Excellent examples of such self-control are provided by mobile self-splicing introns in phage and bacterial systems.
The collaboration that occurs between a successful mobile intron and its host gene involves multiple layers of interdependent molecular interactions: the host gene and its product, the self-spicing intron, and its encoded endonuclease, which cleaves DNA at the homing site, all participate in this social network. This complex relationship is also facilitated by cis regulatory elements within and near the host gene and the intron, and secondary activities that are acquired by the endonuclease (Nielsen and Johansen, 2009). There is not perhaps any other biological system that is more illustrative of the highly choreographed, synergistic co-evolution of a host gene and its invasive element than the thymidylate synthase (TS) host gene of bacteriophage T4, which is interrupted by a mobile group I intron that encodes the I-TevI homing endonuclease (Fig. 1).
Figure 1.
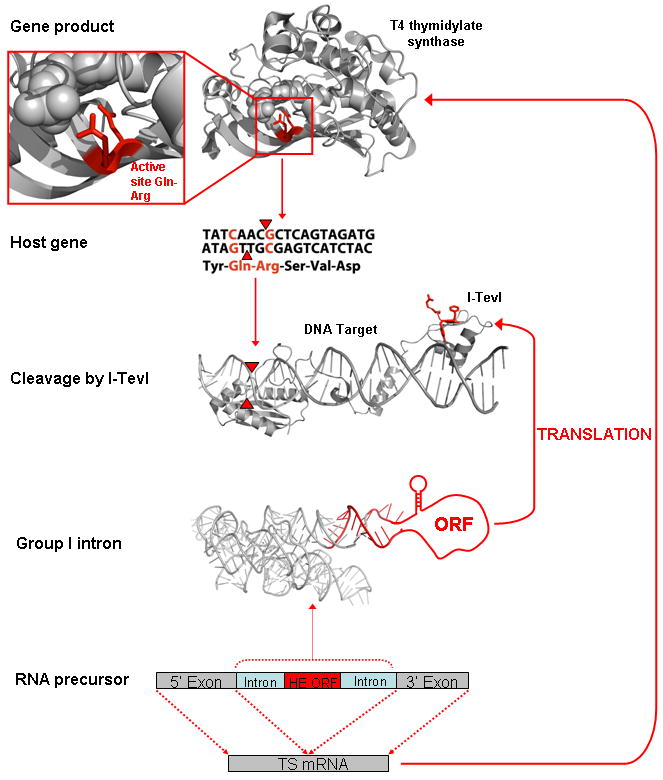
The mobile intron family circle. The RNA precursor of the interrupted T4 thymidylate synthase (TS) gene is depicted at the bottom of the figure, whereas the I-TevI intron-encoded homing endonuclease bound to its DNA target site in the T4 phage TS host gene is shown in the center. The endonuclease is highly specific for the presence of two conserved basepairs (red in host gene) in codons that specify two functionally critical conserved residues in the corresponding TS enzyme (top, Gln-Arg). These side chains contact the dUMP substrate (spheres in TS structure). Cleavage of the DNA target (red triangles) results in invasion of the host gene by the group I intron encoding I-TevI. The I-TevI ORF is inserted at a position corresponding to the P6 stem-loop and extends the final 18 bp at the 3′ end of the endonuclease ORF into the downstream intron core sequence (red bases extending beyond the intron P6 loop into P7). The amino acid residues in the endonuclease that are encoded by these basepairs (red side chains in I-TevI) are therefore constrained by the structure and function required both of the endonuclease and of the group I intron. The regulatory stem-loop, preceding the ORF in the group I intron, represses I-TevI translation, and its removal inhibits both splicing and TS synthesis. The depiction of the structure of full-length I-TevI and its DNA target site is a hypothetical, composite model composed of the crystal structure of its C-terminal domain bound to the 3′ end of its DNA target (pdb ID 1I3J) and a separate crystal structure of the unbound N-terminal catalytic domain (1LN0). The DNA target in the region of the nuclease domain, containing the points of DNA strand cleavage (red triangles) is modeled for illustrative purposes only.
In this issue, Gibb and Edgell describe an elegant series of experiments that clearly illustrate the effect of deregulating the tightly controlled translation of I-TevI on the expression of the host TS gene and cellular metabolism. In the absence of strict temporal controls, the carefully choreographed dance between host gene expression and intron splicing is disrupted by translation of the I-TevI mRNA, because the coding sequence extends into the folded intron core. Translation of the endonuclease reading frame, prior to intron splicing, results in reduced synthesis of the host TS (much in the same the way that a single poorly behaved guest can ruin an otherwise well-planned party).
Tightly regulated control of homing endonuclease translation, and the effects of interfering with this layer of regulation, joins an impressive network of critical interactions and ‘cross-talk’ between the endonuclease, its corresponding self-splicing intron, and the host gene (Gimble, 2000; Gibb and Edgell, 2007). The endonuclease and the intron act jointly to form a highly efficient invasive element. The intron facilitates invasion of coding sequences in host genes without disrupting the underlying expression of that gene and the function of its product. In return, the endonuclease increases the fitness and mobility of the intron, by evolving a specificity profile for DNA cleavage that allows the entire element to persist in its genetic home, while also continuing to search neighboring genomes for new locations to invade.
The I-TevI endonuclease does not gain the cooperation of its cognate intron without making sacrifices of its own. The endonuclease reading frame is inserted at a position in the intron sequence corresponding to the P6a loop in the folded RNA structure, which can accommodate this long additional sequence without significantly interfering with the subsequent splicing activity of the intron (Shub et al., 1988). However, the last 20 nucleotides of the I-TevI coding sequence also correspond to the P6 and P7 structural elements of the folded intron. These elements lie at the heart of the catalytic core of the intron. Thus, these bases are under selective pressure to participate both in folding and splicing of the intron, and also to participate in folding and activity of the endonuclease.
Whereas an ancestor of the I-TevI endonuclease initially succeeded at invading a location in a group I intron sequence while allowing for continued folding and function of that intron, the modern day I-TevI endonuclease ensures the continued success and persistence of the entire mobile element by recognizing a sequence in the T4 phage TS gene that corresponds to the active site of the enzyme (Fig. 1). The DNA target site for I-TevI contains base-pairs that encode functionally critical residues in the TS gene. One of the most critical of these nucleotides is universally conserved in all TS genes (Edgell et al., 2003). This feature of homing endonuclease recognition, namely the concentration of its specificity on the most invariant DNA basepairs in the host gene target site, is a well-documented feature of homing endonucleases in all domains of life. The specificity profiles of these enzymes can be ‘fine-tuned’ to the point where fidelity of recognition at each basepair is well correlated with the underlying degeneracy of the genetic code and the bias in codon use in the host (Scalley-Kim et al., 2007). When a particular host gene sequence offers the appropriate features of strong conservation and a coding sequence that can be readily exploited by an invasive endonuclease, it can become a hotspot that attracts the attention of a myriad of such elements, including both homing endonucleases and transposases (Nomura et al., 2008).
Once safely embedded in its host gene, the mobile endonuclease faces a problem common to many uninvited house guests: its long term residence might become undesirable to the host (Edgell et al., 2000). As a result, mobile introns and their associated homing endonucleases might be periodically removed from the host genome, as part of a life-cycle in which invasion is followed by gradual mutational loss of endonuclease activity, deletion of the intron and possibly reinvasion by a related endonuclease (Burt and Koufopanou, 2004; Goddard and Burt, 1999). However, many homing endonucleases have one further evolutionary trick to avoid eviction from their home: they can acquire a secondary, beneficial activity. This activity is often coupled to the efficient splicing of its cognate intron. This endows the endonuclease with a functional property that is useful or even essential to the host and, therefore, places the endonuclease under positive selection pressure to maintain a well-behaved protein fold. Many homing endonucleases participate directly in intron splicing by interacting physically with the transcribed intron and ‘chaperoning’ the RNA into a functional folded state (Lambowitz and Belfort, 1993). In contrast, I-TevI has acquired an indirect (but no less effective) activity to assist intron splicing: it acts as a repressor of its own transcription and expression (Edgell et al., 2004). This property involves binding (but not cleaving) an operator site that overlaps its late promoter, and is part of the intricate collection of control elements that regulate the timing and level of I-TevI production during the phage lifecycle, thus allowing proper expression of the TS host gene.
Although investigators have long understood that DNA breaks are required for recombination-dependent replication (a mainstay of phage T4 viability), we kept insisting that homing endonucleases have the potential to damage the host genome, and that genomic degradation and resulting toxicity was the reason behind stringent homing endonuclease control. Not until this Gibb and Edgell paper have we understood that I-TevI is down-regulated to preserve TS expression. But now we are forced to ask why it is that the coding sequences all three intron endonucleases in phage T4 extend into RNA structure elements of their host introns (Shub et al., 1988). This overlap might be telling us that the homing endonucleases and nucleotide metabolizing enzymes encoded by the three intron-containing genes are holding hands, or at least talking to each other. But understanding everything that they are saying will require further clever experimentation.
References
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burt A, Koufopanou V. Homing endonuclease genes: the rise and fall and rise again of a selfish element. Curr Opin Gen Develop. 2004;14:609–615. doi: 10.1016/j.gde.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat Struct Mol Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- Edgell DR, Stanger MJ, Belfort M. Importance of a single base pair for discrimination between intron-containing and intronless alleles by endonuclease I-BmoI. Curr Biol. 2003;13:973–978. doi: 10.1016/s0960-9822(03)00340-3. [DOI] [PubMed] [Google Scholar]
- Gibb EA, Edgell DR. Multiple controls regulate the expression of mobE, an HNH homing endonuclease gene embedded within a ribonucleotide reductase gene of phage Aeh1. J Bacteriol. 2007;189:4648–4661. doi: 10.1128/JB.00321-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimble FS. Invasion of a multitude of genetic niches by mobile endonuclease genes. FEMS Microbiol Letters. 2000;185:99–107. doi: 10.1111/j.1574-6968.2000.tb09046.x. [DOI] [PubMed] [Google Scholar]
- Goddard MR, Burt A. Recurrent invasion and extinction of a selfish gene. Proc Natl Acad Sci USA. 1999;96:13880–13885. doi: 10.1073/pnas.96.24.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz AM, Belfort M. Introns as mobile genetic elements. Ann Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Johansen SD. Group I introns: Moving in new directions. RNA Biol. 2009;6:375–383. doi: 10.4161/rna.6.4.9334. [DOI] [PubMed] [Google Scholar]
- Nomura N, Nomura Y, Sussman D, Klein D, Stoddard BL. Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases. Nucleic Acids Res. 2008;36:6988–6998. doi: 10.1093/nar/gkn846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scalley-Kim M, McConnell-Smith A, Stoddard BL. Coevolution of a homing endonuclease and its host target sequence. J Mol Biol. 2007;372:1305–1319. doi: 10.1016/j.jmb.2007.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shub DA, Gott JM, Xu MQ, Lang BF, Michel F, Tomaschewski J, Pedersen-Lane J, Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci USA. 1988;85:1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]


