Abstract
MicroRNAs (miRNAs) are small noncoding RNAs that have recently emerged as critical regulators of gene expression within the immune system. In this study we used mice with conditional deletion of Dicer and Dgcr8 to dissect the roles of miRNAs in NK cell activation, survival, and function during viral infection. We developed a novel system for deletion of either Dicer or Dgcr8 in peripheral NK cells via drug-induced Cre activity. We found that Dicer- and Dgcr8- deficient NK cells were significantly impaired in survival and turnover, and had impaired function of the ITAM-containing activating NK cell receptors. We further demonstrated that both Dicer- and Dgcr8-dependent pathways were indispensable for the expansion of Ly49H+ NK cells during MCMV infection. Our data indicate similar phenotypes for Dicer- and Dgcr8- deficient NK cells, which strongly suggest that these processes are regulated by miRNAs. Thus, our findings indicate a critical role for miRNAs in controlling NK cell homeostasis and effector function, with implications for miRNAs regulating diverse aspects of NK cell biology.
Keywords: NK cells, microRNA, Dicer, Dgcr8, cytomegalovirus, NKG2D
Introduction
NK cells are a key component of the innate immune system, providing early cellular defense against viruses and intracellular pathogens, and contributing to the early detection and destruction of transformed cells (1). NK cells develop from a common lymphoid precursor in the bone marrow and undergo terminal maturation in the periphery, acquiring optimal cytolytic and effector functions (2). Mature NK cells are relatively static in terms of proliferative capacity until transferred into a lymphopenic environment (3, 4) or challenged with pathogens (5).
NK cell activation and function are determined by a balance of signals transmitted by inhibitory and activating NK cell receptors (NKRs) (1). Many of the inhibitory NKRs, including Ly49 receptors in mice and inhibitory Killer cell Immunoglobulin-like Receptors (KIRs) in humans, recognize self-ligands such as MHC class I molecules. Activating NKRs recognize host-encoded molecules induced by transformation or viral infection (e.g., NKG2D ligands) (6), or non-self-ligands, including the mouse cytomegalovirus (MCMV) m157 glycoprotein, which is recognized by Ly49H (7, 8). Some activating receptors associate with adapter proteins that contain immunoreceptor tyrosine-based activation motifs (ITAMs), including DAP12, FcεRIγ, and CD3ζ. Ligation of ITAM-containing receptor complexes results in recruitment and activation of protein tyrosine kinases, which initiate the signaling cascade, resulting in secretion of effector cytokines, such as IFNγ, and target cell cytotoxicity. Lack of NK cells renders both human and mice susceptible to certain infections, particularly the herpesviruses, including human cytomegalovirus (HCMV) and mouse cytomegalovirus (MCMV) (5). Therefore, experimental infection of mice with MCMV provides a useful model for studying factors that are important for NK cell activation in vivo. Although much has been learned about the signaling molecules and transcription factors that control activation of NK cells, less is known about the post-transcriptional mechanisms that regulate NK cell function.
MicroRNAs (miRNAs) are short (~22 nt) noncoding RNAs, expressed from endogenous genes, that act on protein-encoding mRNAs, targeting them for translational repression or degradation (9). The biogenesis of miRNAs involves two processing steps. Primary miRNA (pri-miRNA) transcripts are first cleaved by the nuclear “microprocessor” complex containing the RNAse III enzyme Drosha and its double-stranded RNA (dsRNA)-binding partner Dgcr8, resulting in short hairpin pre-miRNAs (10, 11). These pre-miRNAs are then exported to the cytoplasm, and are further processed by another RNAse III enzyme, Dicer, to generate the mature duplex miRNAs (12). Finally, a single strand of the mature miRNA duplex, loaded into the RNA-induced silencing complex, targets specific sequences within mRNA 3’ untranslated regions (UTRs) resulting in mRNA degradation and/or translational repression.
Genetic studies showed a critical requirement for Dicer in vivo. Ablation of Dicer in the mouse germline produces a lethal phenotype (13), and conditional deletion of Dicer in various hematopoietic lineages has been shown to have detrimental effects, such as impaired cell differentiation, proliferation, and survival (14–21). The function of Dicer, however, is not limited to miRNA biogenesis. Dicer is also required for the processing of small inhibitory RNAs (siRNAs) derived from endogenous dsRNA transcripts or exogenous sources, including viral dsRNAs (22). Endogenous siRNAs were discovered in mammalian oocytes and embryonic stem cells (23–25) and were shown to be essential for oocyte maturation (26). Thus, although the loss of miRNA-dependent regulation has been implicated in the phenotypes caused by Dicer deficiency, at least in some cases they are the consequence of the loss of other small RNA classes. Unlike Dicer, Dgcr8 is essential for the biogenesis of canonical miRNAs, but not in the generation of siRNAs (23, 27). In this study, we developed a novel system for the deletion of either Dicer or Dgcr8 in NK cells via drug-induced Cre activity to dissect the roles of miRNAs in NK cell activation, survival, and function during infection.
Materials and Methods
Mice and infections
The Cre-ERT2 C57BL/6 transgenic mice were kindly provided by Dr. M. Matloubian, University of California San Francisco (28), R26R-YFP knockin C57BL/6 mice were provided by Dr. N. Killeen, University of California San Francisco (29), and Ly49H-deficient (Klra8−/−) mice were from Dr. S. Vidal, McGill University). Mice carrying the conditional floxed alleles of Dicer (DicerF/F) (14) and Dgcr8 (Dgcr8F/F) (30) were backcrossed onto a C57BL/6 background for eight generations. To generate bone marrow chimeras, 6–8 week-old CD45.1-congenic C57BL/6 mice (National Cancer Institute (Frederick, MD)) were lethally irradiated with 1000 rads. Donor bone marrow was harvested from DicerF/+, DicerF/F, Dgcr8F/+, or Dgcr8F/F mice carrying Cre-ERT2 transgene and mixed 1:1 with bone marrow harvested from CD45.1 C57BL/6 mice. Cells were then injected into irradiated recipients by the intraocular vein in the retro-orbital plexus. The chimeras were treated with tamoxifen 7 weeks after reconstitution and infected i.p. with 5×104 PFU of a salivary stock of MCMV (Smith strain) 2 weeks later (31). All experiments were conducted in accordance with UCSF Institutional Animal Care and Use Committee guidelines.
Tamoxifen treatment
Mice were administered tamoxifen (Sigma-Aldrich) dissolved in corn oil via oral gavage for 5 consecutive days. Unless otherwise stated, animals were sacrificed for analysis 10–20 days post-treatment.
Flow cytometry and functional assays
Single cell suspensions were used for flow cytometry. Fc receptors were blocked with anti-CD16 + CD32 mAb (clone 2.4G2) at 10 µg/mL prior to surface staining with the indicated Abs (all purchased from BD Biosciences, eBioscience, or BioLegend). Rae-1 expression was measured using anti-pan-Rae-1 mAb (clone 186107). Splenocytes were stimulated in tissue culture plates treated with N-(1-(2,3-dioleoyloxyl)propyl)-N,N,N-trimethylammonium methylsulfate (Sigma-Aldrich) and coated with anti-NK1.1, anti-NKp46, anti-Ly49H, or control mAb for 4h at 37°C in the presence of GolgiStop (BD Pharmingen), followed by staining for lysosome-associated membrane protein (LAMP)-1 (CD107a, BD Pharmingen) and intracellular IFNγ (BD Pharmingen). For control experiments, splenocytes were stimulated with IL-12 (20 ng/mL) and IL-18 (10 ng/mL). For apoptosis analysis, freshly isolated splenocytes were first stained with anti- NK1.1, anti-TCRβ, and Live/Dead™ fixable near-IR stain (Invitrogen), washed, and then stained with Annexin V-PE (BD Pharmingen), according to manufacture’s protocol. For expansion of NK cells in vitro, splenocytes were stained with anti-CD4, anti-CD8, anti-Ter119, and anti-GR-1 mAbs and then incubated with goat anti-rat IgG-coated and goat anti-mouse IgG-coated magnetic beads (Qiagen) to deplete T cells, B cells and GR-1+ cells, respectively. NK cells were stained with PE-conjugated DX5 mAb and positively selected by using an anti-PE mAb-conjugated magnetic bead system (Miltenyi Biotec). The resulting NK cells were cultured with 1,333 units/ml recombinant human IL-2 (National Institutes of Health Biological Resources Branch Preclinical Repository). NK cells were counted and stained with PE-conjugated anti-NKp46, PerCP-Cy5.5-conjugated -CD3, and annexinV-conjugated A647 after culture for 4 days. Flow cytometry was performed by using a LSRII flow cytometer and analyzed with FlowJo software (Tree Star, Inc).
Cell sorting
NK cells from spleen were enriched with mAbs against CD5, CD4, CD8, Ter119, Gr-1, CD19 (UCSF Antibody Core) and anti-rat IgG-coated magnetic beads (Miltenyi Biotech). NK cells were then stained with anti-NK1.1 and anti-TCRβ mAbs, and NK1.1+ TCRβ− cells were sorted using a FACSAria (Becton Dickinson). For human NK cells, the low-buoyant density cells from the Percoll interface layer were isolated from peripheral blood mononuclear cells (Stanford Blood Center), stained with anti-CD56 and anti-CD3 mAbs, and CD56+ CD3− lymphocytes were sorted. The purity of the recovered NK cells was typically >98%.
In vivo BrdU labeling
Mice were injected i.p. with 200 µg of BrdU (Sigma-Aldrich) every 24 h for 3 consecutive days (Fig. 3 B) or once for 2 h (Fig. 7 A) and then sacrificed. For the detection of incorporated BrdU, cells were first stained for surface antigens, then fixed, permeabilized, treated with DNase I, and stained with APC-conjugated anti-BrdU mAb (BD Pharmingen).
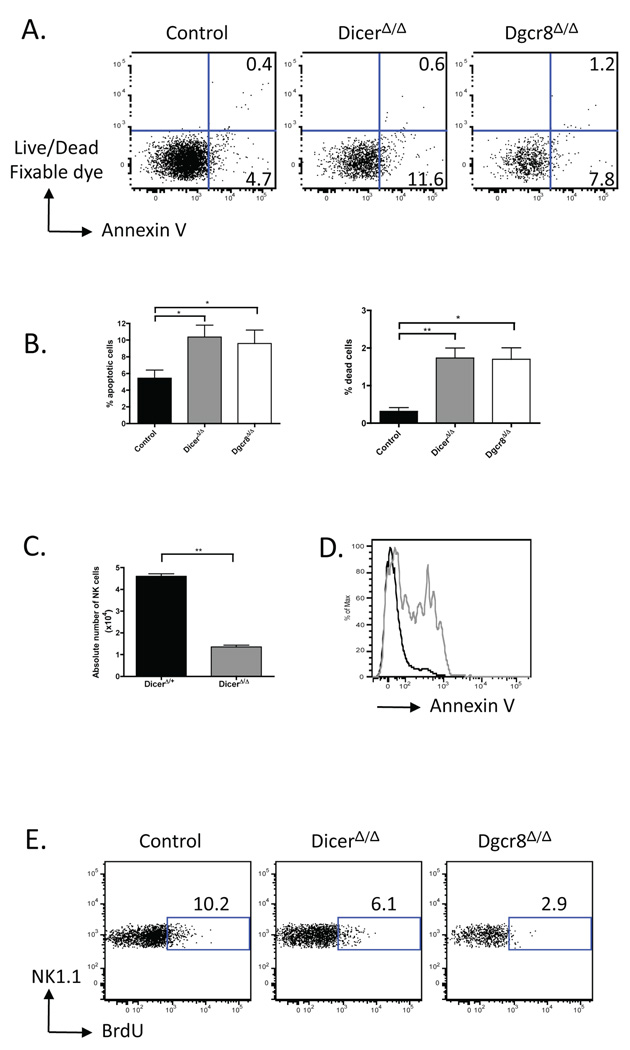
Figure 3. Increased apoptosis and reduced basal turnover of Dicer- and Dgcr8-deficient NK cells.
(A, B) Annexin V staining of freshly isolated splenic NK cells. (A) Percentages of apoptotic (live Annexin V+) and dead cells in control versus DicerΔ/Δ and Dgcr8Δ/Δ mice are indicated within the quadrants. (B) Summary (mean values ± SEM) of data shown in (A) from 3–5 mice per group. (C) Enumeration of NK cells after 4-day culture in medium supplemented with IL-2. Values shown represent the mean ± SEM of NK cell numbers (triplicate wells). (D) Histogram shows Annexin V expression by DicerΔ/+ (black line) and DicerΔ/Δ (grey line) NK cells after 4-day culture in medium supplemented with IL-2. (E) Intracellular staining for the incorporation of BrdU in control, DicerΔ/Δ, and Dgcr8Δ/Δ mice. Percentage of NK cells that incorporated BrdU is shown. (A-E) Gated on YFP+ NK cells. Data are representative of 2–3 independent experiments.
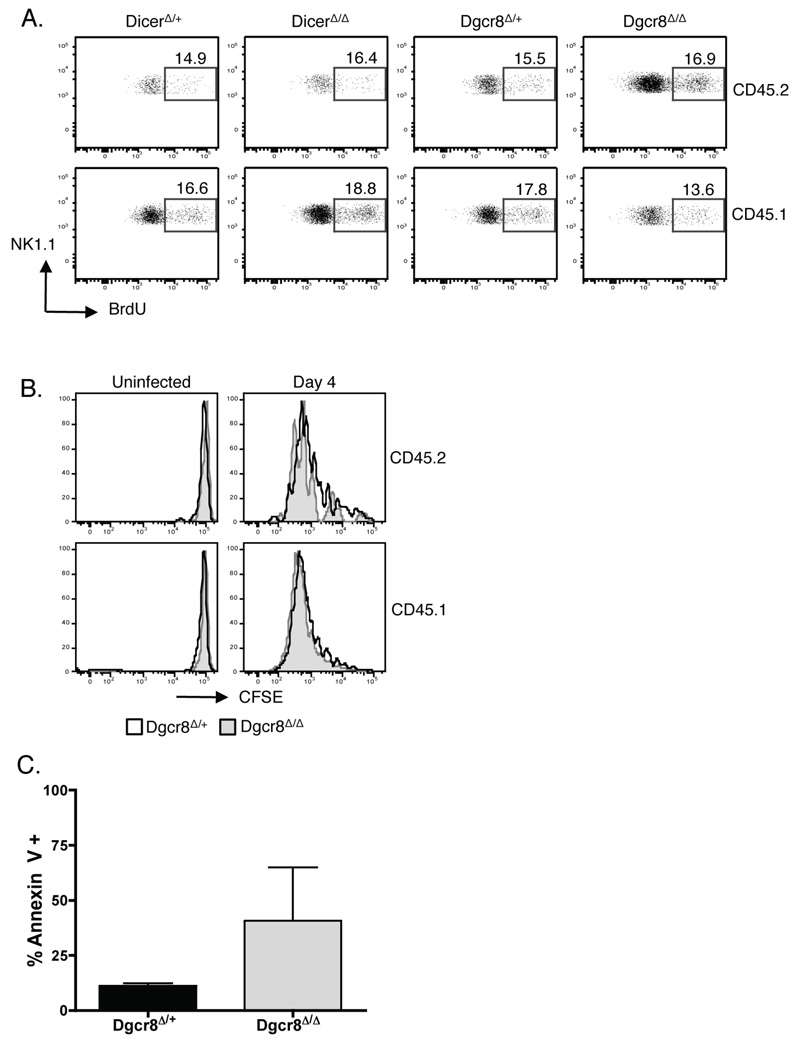
Figure 7. Robust proliferation, but decreased survival, of Dicer- and Dgcr8-deficient NK cells during MCMV infection.
(A) BrdU incorporation (day 5 PI) by NK1.1+ TCRβ− cells from mixed bone marrow chimeric mice (Fig. 6). Percentages of BrdU incorporation by wildtype (CD45.1+) cells (lower panel) and either DicerΔ/+, DicerΔ/Δ, Dgcr8Δ/+, or Dgcr8Δ/Δ (CD45.2+) cells are shown. Data are representative of 2 experiments. (B) CFSE-labeled 1:1 mix of wildtype (CD45.1+) and either Dgcr8Δ/+ or Dgcr8Δ/Δ (CD45.2+) NK cells were transferred into Ly49H-deficient recipients. Ly49H+ NK cells were analyzed at day 4 PI. Data are representative of 3 independent experiments. (C) NK cells from Dgcr8Δ/+ or Dgcr8Δ/Δ mice were transferred into Ly49H-deficient recipients and were stained for Annexin V at day 4 PI. Bar graph indicates the percentage of NK1.1+ TCRβ− cells (mean ± SD) that are live and are staining positive for Annexin V (indicating apoptotic). Data are representative of 3 independent experiments using 2 mice per group.
CFSE labeling and adoptive transfer
A 1:1 splenocyte mixture of CD45.1+ (from wildtype C57BL/6 mice) and CD45.2+ (from either Dgcr8Δ/+ or Dgcr8Δ/Δ C57BL/6 mice) cells was treated with ACK lysis buffer to remove red blood cells, labeled for 8 min with 0.5 µM CFSE, and washed twice in PBS. 1×107 labeled cells were transferred i.v. into Ly49H-deficient C57BL/6 mice. Twenty-four hours later recipient mice were infected with MCMV and sacrificed 4 days later. Adoptively transferred NK cells were analyzed for Ly49H expression and CFSE dilution by flow cytometry.
MicroRNA array profiling
Total RNA was extracted from sorted mouse splenic and human peripheral blood NK cells by using TRIzol (Invitrogen). RNA quality was verified by an Agilent 2100 Bioanalyzer profile. A mixture of equal amounts of RNA from mouse and human NK cells were made as reference. Five microgram of total RNA from the samples (mouse or human NK cells) and a reference pool were labeled with Hy5™ and Hy3™ fluorescent label, respectively, using the miRCURY™ LNA microRNA array labeling kit (Exiqon, Denmark). The Hy5™ – labeled samples and Hy3™ – labeled reference pool RNA samples were mixed pair-wise and hybridized to the LNA array (Gladstone Genomics Core), which contains capture probes targeting human, mouse, and rat miRNAs. The hybridization was performed on GeneTAC hybridization station (Genomic Solutions). The slides were scanned by Axon GenePix 4000B scanner (Molecular Devices). GPR files containing fluorescent ratios (sample/control) were generated using GenePix Pro 6.0 software.
Quantitative RT-PCR
Total RNA was isolated from sorted YFP+ NK cells by using the mirVana miRNA Isolation kit (Ambion). The expression levels of specific miRNAs were examined by using the TaqMan MicroRNA Assay kit (Applied Biosystems) with gene-specific stem-loop PCR primers and TaqMan probes (IDT) to detect mature miRNA transcripts. Quantitative RT-PCR reaction was performed on an Applied Biosystems 7500 Fast Real-Time PCR System. snoRNA202 was used as endogenous control. Relative quantification using the ΔΔct method in tamoxifen-treated DicerΔ/Δ or Dgcr8Δ/Δ versus control mice was carried out and relative changes were calculated for each miRNA.
PCR analysis of DicerF/F and Dgcr8F/F mice
The floxed Dicer allele was genotyped as previously described (18). The same primers were used to quantitate the floxed Dicer alleles by real-time PCR with the SYBR green PCR Master Mix (Applied Biosystems). The PU.1 sequences 5’-CTTCACTGCCCATTCATTGGCTCATCA - 3’ (forward) and 5’-GCTGGGGACAAGGTTTGATAAGGGAA – 3’ (reverse) were used for normalization. The floxed Dgcr8 allele was genotyped using primers 5’-CTGGAGTAGGCATGTTGATTTC - 3’ (forward) and 5’-CCTGATTCACTTACAACACAACC - 3’ (reverse).
Statistical analysis
All data shown are the mean ± SEM unless stated otherwise. Comparisons between samples were performed by using a two-tailed Student t test. Statistics were determined using Prism software (GraphPad Software, Inc.). p values were denoted in the following manner: * p < 0.05, ** p < 0.01, *** p < 0.001.
Results
Inducible deletion of either Dicer or Dgcr8 leads to a global miRNA deficiency in NK cells
We determined the expression profile of miRNAs in mouse and human NK cells. The miRNAs obtained from sorted NK1.1+ TCRβ− mouse NK cells and from CD56+ CD3− human NK cells were profiled with a LNA (locked nucleotide acid)-based microarray (Table S1). The preliminary analysis indicated that 80% of all miRNAs present in human NK cells were also expressed in mouse NK cells, and 59% of mouse miRNAs were present in human NK cells (Suppl. Fig. 1A). Quantitative RT-PCR confirmed expression of the top twenty common miRNAs in mouse NK cells (Suppl. Fig. 1B).
To understand the role of miRNAs in NK cell biology, we induced ablation of the conditional DicerF (14) and Dgcr8F (30) alleles using a drug-inducible Cre recombinase. This inducible system is dependent upon three sets of genes. The first are the genes encoding Dicer and Dgcr8 in which both alleles are flanked by two loxP sites (DicerF/F and Dgcr8F/F, respectively). The second gene is a Cre recombinase-human estrogen receptor (Cre-ERT2) chimeric molecule under the control of the ubiquitin promoter. This fusion protein is sequestered within the cytoplasm in the absence of the estrogen analog, tamoxifen (28, 32). In the presence of tamoxifen, the Cre-ERT2 protein shuttles into the nucleus and gains access to loxP sites. This approach allows for the constitutive expression of a Cre recombinase yet prevents it from acting on loxP sites until tamoxifen is administered. The third component is an enhanced yellow fluorescent protein (YFP) gene inserted into the Rosa-26 locus (29). The YFP cassette is preceded by a loxP-flanked tpA transcriptional stop signal. This construct permits expression of YFP and identification of cells that currently or previously expressed an activated Cre. When tamoxifen is administered to mice, Cre becomes active and deletes the floxed Dicer or Dgcr8 alleles and the R26R stop cassette, resulting in DicerΔ/ΔYFP+ and Dgcr8Δ/ΔYFP+ phenotypes, respectively (Fig. 1A). To serve as a control for these mice, we generated mice in which loxP sites flank only one allele, the other allele is wildtype (DicerF/+ and Dgcr8F/+ mice). Following tamoxifen treatment, these mice would be referred to as DicerΔ/+ and Dgcr8Δ/+, respectively.
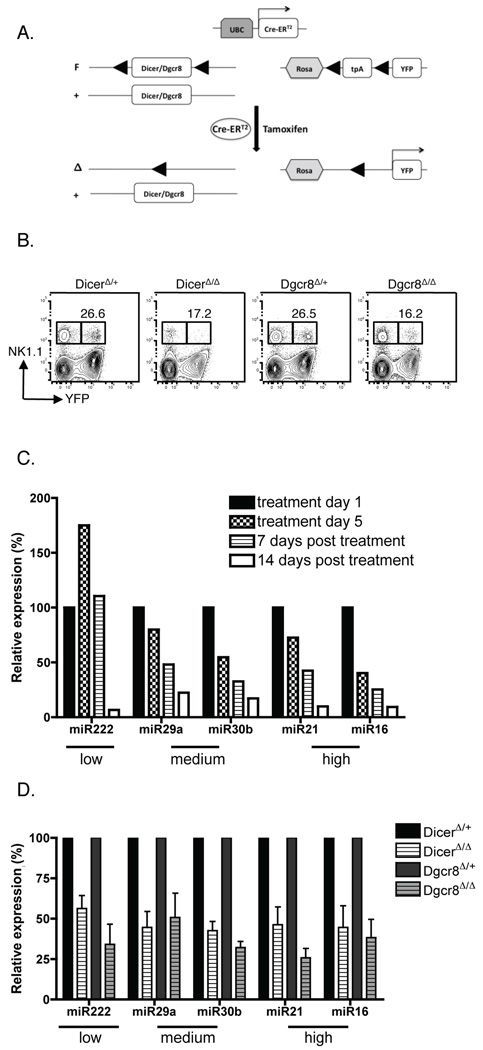
Figure 1. Inducible deletion of Dicer and Dgcr8 leads to miRNA deficiency in NK cells.
DicerF/F or Dgcr8F/F mice were crossed with Cre-ERT2 transgenic and R26R-YFP knockin mice to delete one (DicerΔ/+ or Dgcr8Δ/+) or both (DicerΔ/Δ or Dgcr8Δ/Δ) alleles, expressing YFP upon deletion of a floxed stop cassette inserted into the Rosa-26 locus. (A) Diagram of the floxed loci before and after tamoxifen-induced activation of the Cre-ERT2-fusion protein. (B) NK1.1 and YFP expression in lymphocytes isolated from DicerΔ/+, DicerΔ/Δ, Dgcr8Δ/+, and Dgcr8Δ/Δ mice on day 14 post-treatment. (C, D) Quantification of miRNA levels using qRT-PCR. Levels of mature miRNAs were normalized to snor202 and relative expression of poorly (miR222), moderately (miR29a and miR30b), and highly (miR16 and miR21) expressed miRNAs are shown. (C) RNA was isolated from YFP+ NK1.1+ TCRβ− cells sorted from Dgcr8Δ/Δ mice on treatment day 1 and day 5, and day 7 and day 14 post-treatment. Data are representative of two experiments. (D) RNA was isolated from YFP+ NK1.1+ TCRβ− cells sorted from mice of the indicated genotypes on days 10–20 after treatment. Mean ± SEM is shown for DicerΔ/Δ (n=4) relative to DicerΔ/+ (n=2), and for Dgcr8Δ/Δ (n=4) relative to DicerΔ/+ (n=2).
We determined the kinetics of YFP expression and concomitant reduction of miRNA expression in NK cells from DicerΔ/Δ and Dgcr8Δ/Δ mice after administration of tamoxifen. YFP expression appeared on day 5 after tamoxifen treatment was initiated (data not shown) and was maximal by day 14 post-treatment (Fig. 1B). Only approximately 25% of NK cells in DicerΔ/+ and Dgcr8Δ/+ control mice produced the YFP protein, suggesting that this system under-reports Dicer (and perhaps Dgcr8) deletion, as has been reported for other genes (33). In fact, Dicer was also excised in YFP− NK cells in DicerΔ/Δ mice, albeit not as efficiently as in YFP+ NK cells (Suppl. Fig. 2A). To confirm that deletion occurred in the studied cells, all experiments were performed on cells gated as YFP+.
By day 14 post-treatment, miRNA expression in YFP+ NK cells from Dgcr8Δ/Δ mice decreased to 7–22% of the miRNA expression level in NK cells from untreated Dgcr8F/F mice (Fig. 1C). Because the half-life of naïve NK cells is approximately 17 days (4) (and potential global effects of Dicer and Dgcr8 deficiency on the mouse immune system may exist (18, 21, 34)), we limited our analysis of NK cells from these mice to days 10–20 post-tamoxifen treatment. Approximately 50–75% reduction of select miRNAs was confirmed in YFP+ NK cells from several DicerΔ/Δ and Dgcr8Δ/Δ mice (Fig.1D, Suppl. Fig. 2B). Importantly, miRNAs expressed at high (miR21 and miR16), medium (miR29a and miR30b), and low levels (miR222) were similarly affected by Dicer and Dgcr8 deletion. These studies demonstrate that inducible Cre-ERT2-mediated deletion of either Dicer or Dgcr8 leads to a significant depletion of miRNAs in YFP+ NK cells.
Reduction of the NK cell compartment in DicerΔ/Δ and Dgcr8Δ/Δ mice
To investigate the role for Dicer- and Dgcr8-controlled miRNAs in the regulation of NK cell homeostasis, we first determined the frequency and number of NK cells in peripheral organs of DicerΔ/Δ and Dgcr8Δ/Δ mice. Flow cytometric analyses of YFP+ lymphocytes in the spleen, liver, and blood showed a marked reduction of the overall percentage of NK1.1+ TCRβ− cells (2-fold reduction on average) in DicerΔ/Δ and Dgcr8Δ/Δ mice compared with littermate controls (Fig. 2A, B). Using the same R26R-YFP reporter mice, it was previously demonstrated that in the absence of Dicer, there is a strong deletion of CD4+ and CD8+ T cells that have expressed Cre (16). We also found a smaller proportion of YFP+ cells in the splenic and liver NK populations in DicerΔ/Δ and Dgcr8Δ/Δ mice compared to controls (Fig. 2C). Taken together with the reduced frequency of NK cells within the YFP+ lymphocytes, the absolute numbers of YFP+ NK cells were significantly decreased in the spleen of DicerΔ/Δ and Dgcr8Δ/Δ mice (2.4-fold, P<0.0001 and 3.3-fold, P=0.0024, respectively). Similar reduction in numbers of YFP+ NK cells were seen in liver of DicerΔ/Δ and Dgcr8Δ/Δ mice (6.3-fold, P=0.0066 and 3.3-fold, P=0.033, respectively) (Fig. 2D). This decrease in peripheral NK cell numbers in DicerΔ/Δ and Dgcr8Δ/Δ mice occurs in the context of reduced, but not abolished, miRNA function. As such, it is possible that the reason that we observed only 50–75% reduction in miRNA levels (Fig. 1D) is because these are the only cells that are viable.
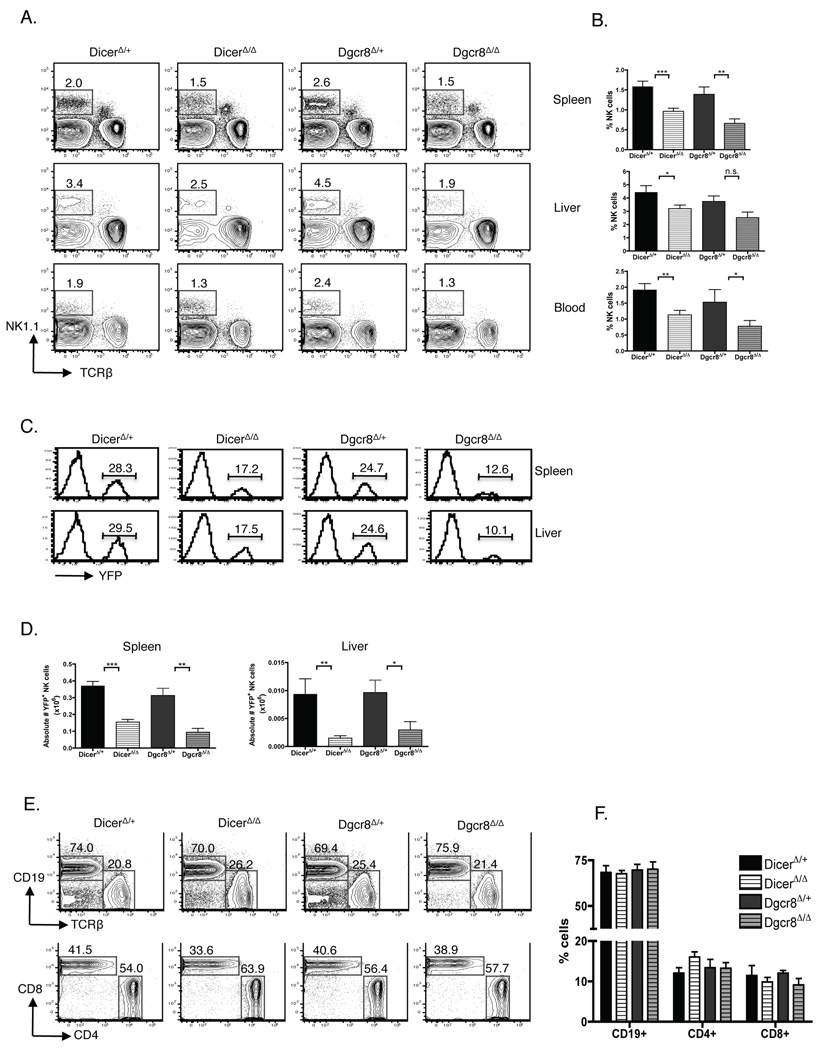
Figure 2. Preferential reduction of NK cells in DicerΔ/Δ and Dgcr8Δ/Δ mice.
Flow cytometric analysis of lymphocytes from DicerΔ/Δ (n=16), Dgcr8Δ/Δ (n=9), and control DicerΔ/+ (n=13) and Dgcr8Δ/+ (n=11) mice. (A) Contour plots depict frequency of NK (NK1.1+ TCRβ−) cells in spleen (top panels), liver (middle panels), and blood (bottom panels). Gated on YFP+ lymphocytes. (B) Summary (mean ± SEM) of data shown in (A). (C) Selective loss of YFP+ DicerΔ/Δ and YFP+ Dgcr8Δ/Δ NK cells. Histograms depict relative cell number versus Cre-induced YFP fluorescence within NK cells in spleen (top row) and liver (bottom row). Gated on NK1.1+ TCRβ− cells and percentages of YFP-expressing cells are indicated. (D) Bar graph shows the number of YFP+ NK cells from spleen (left panel) and liver (right panel) of DicerΔ/Δ, Dgcr8Δ/Δ, and control DicerΔ/+, Dgcr8Δ/+ mice. (E, F) Comparison of B (CD19+), CD4+ T (CD4+ TCRβ+), and CD8+ T (CD8+ TCRβ+) cell frequencies from spleen of DicerΔ/Δ, Dgcr8Δ/Δ and control DicerΔ/+, Dgcr8Δ/+ mice. (E) Flow cytometric analysis of populations in total YFP+ splenocytes (top panel) and YFP+ TCRβ+ splenocytes (bottom panel). Percentages are shown. (F) Summary (mean values ± SEM) of data shown in (E) from 4–6 mice per group. Data are representative of at least 5 experiments.
In contrast to the NK cell compartment, the frequencies of B cells and CD4+ T cells were unchanged in the spleens of DicerΔ/Δ and Dgcr8Δ/Δ mice compared with controls. CD8+ T cells were consistently moderately reduced in DicerΔ/Δ and Dgcr8Δ/Δ mice, although this difference did not reach statistical significance (Fig. 2E, F). Spleen cellularity in Dicer- and Dgcr8-deficient mice, however, was significantly reduced, as compared to controls (67.6x106 ± 8.5x106 (n= 9 DicerΔ/+) vs. 39.8x106 ± 4.9x106 (n=14 DicerΔ/Δ); P=0.006 and 59.4x106 ± 12.8x106 (n=8 Dgcr8/+) vs. 30.0x106 ± 2.8x106 (n=8 Dgcr8Δ/Δ); P=0.04). As a result, the numbers of B cells, CD4+ T cells, and CD8+ T cells, were lower (~2-fold, 2.3 fold, and 5-fold, respectively) in DicerΔ/Δ and Dgcr8Δ/Δ mice compared with that in controls. This is consistent with prior reports showing that Dicer deletion resulted in a dramatic reduction of B cells and T cells (15–17). Although iNKT cell development was recently reported to be severely impaired in the absence of Dicer (19, 20), we detected only minor reduction in the number of NK1.1+ TCRβ+ cells in our DicerΔ/Δ and Dgcr8Δ/Δ mice (~1.3-fold and ~1.2-fold reduction compared to controls, respectively) (data not shown). Taken together these data show that miRNAs are required for the maintenance of normal lymphocyte numbers, particularly those of the NK cell lineage.
MiRNAs regulate survival and turnover of NK cells
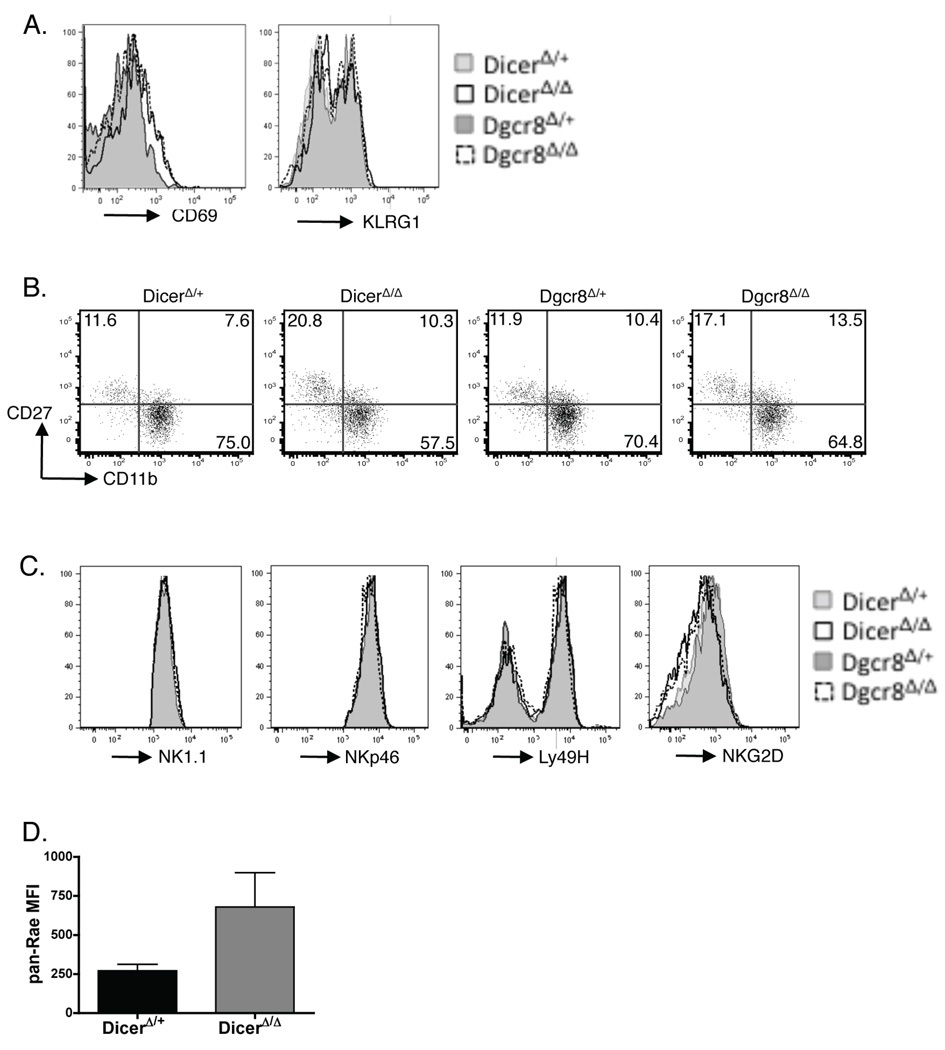
To determine whether the reduction in NK cell numbers in DicerΔ/Δ and Dgcr8Δ/Δ mice was the result of increased cell death, impaired expansion of immature NK cells, or both, we stained freshly isolated cells with Annexin V and a viability dye to assess apoptosis (live, Annexin V+) and cell death (dead, Annexin V+). There was a consistent increase of both apoptotic and dead NK cells from DicerΔ/Δ and Dgcr8Δ/Δ versus control mice (Fig. 3A, B). The rate of apoptosis and death of DicerΔ/Δ NK cells was further increased and significantly higher than that of DicerΔ/+ NK cells if cells were cultured in the presence of IL-2 in vitro (Fig. 3C, D). In addition, we examined NK cell turnover using BrdU incorporation. Approximately 10% of wildtype NK1.1+ TCRβ− NK cells incorporated BrdU over 3 days of labeling, consistent with previous reports (4). In contrast, DicerΔ/Δ and Dgcr8Δ/Δ NK cells incorporated BrdU at a lower rate, 6.3 ± 0.3 % and 5.8 ± 2.0 % of NK cells, respectively (Fig. 3E). Consistent with the decreased turnover rate, DicerΔ/Δ and Dgcr8Δ/Δ NK cells did not exhibit activated phenotypes, shown by the similar expression levels of CD69 and KLRG1 (Fig. 4A). These results indicate that Dicer- and Dgcr8-deficiency affects both the turnover rate, as well as survival, of NK cells.
Figure 4. Dicer- and Dgcr8-deficeint NK cells do not posses an activated phenotype and have normal receptor expression with the exception of NKG2D.
Splenocytes from DicerΔ/Δ, Dgcr8Δ/Δ, and control DicerΔ/+ and Dgcr8Δ/+ mice were stained with mAbs against activation markers CD69 and KLRG1 (A), maturation markers CD11b and CD27 (B), and activating NK receptors NK1.1, NKp46, Ly49H, and NKG2D (C). Gated on YFP+ NK1.1+ TCRβ− cells. Data are representative of at least 5 experiments. (D) Expression levels of Rae-1 on YFP+ NK1.1+ TCRβ+ cells in the spleens of DicerΔ/+ and DicerΔ/Δ mice. Mean fluorescence intensity (MFI) values are shown as mean ± SD using 2 mice per group. Data are representative of 3 experiments.
Phenotype and receptor repertoire of NK cells in DicerΔ/Δ and Dgcr8Δ/Δ mice
Dicer-deficiency has been shown to affect the development of Treg and iNKT lineages, and the differentiation of CD4+ T cell into Th1 and Th2 cells (15, 16, 18–21). To determine whether miRNAs could affect the maturation of peripheral NK cells, we analyzed the expression of maturation markers and surface receptors on freshly isolated YFP+ NK cells. The maturation status of peripheral NK cells is classified based on their expression of CD11b and CD27, with CD27lo CD11bhi NK cells being the most mature population (35). The percentage of this mature NK cell subset in DicerΔ/Δ and Dgcr8Δ/Δ mice was decreased compared to control mice (Fig. 4B). Expression levels of NK1.1, NKp46, Ly49H, Ly49D, CD94, Ly49G2, Ly49C/I, and Ly49A receptors were unaffected in DicerΔ/Δ and Dgcr8Δ/Δ mice (Fig. 4C, Table S2). The only exception was NKG2D whose level was often diminished on NK cells from DicerΔ/Δ and Dgcr8Δ/Δ mice (Fig. 4D). NKG2D recognizes several cellular ligands, which are expressed at low levels in healthy cells, but are often up regulated in stressed, virally infected, or tumor cells (36). We found that the reduced expression of NKG2D correlated with higher expression of Rae-1 ligands on DicerΔ/Δ (Fig. 4D), and Dgcr8Δ/Δ (data not shown) NK cells. Together, our results indicate that Dicer- and Dgcr8-deficiency effects NK cell maturation and elicits up-regulation of NKG2D ligand expression, but does not globally affect the NK cell receptor repertoire.
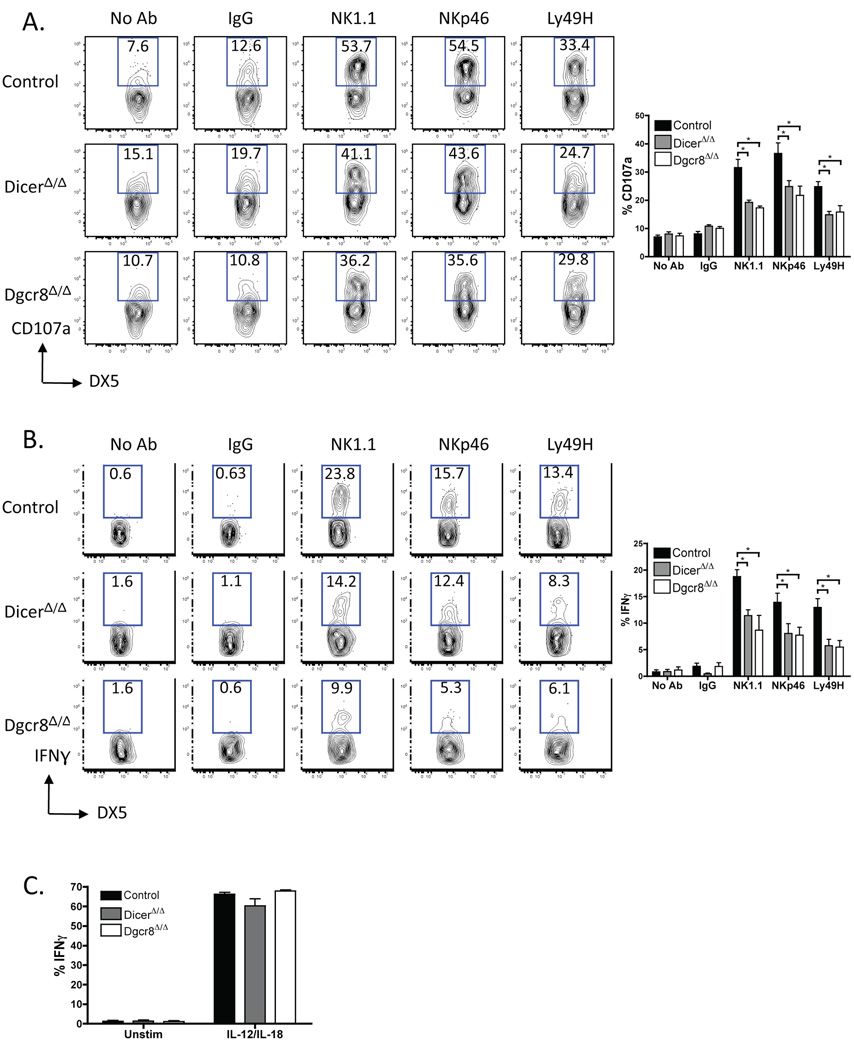
DicerΔ/Δ and Dgcr8Δ/Δ NK cells produce less CD107a and IFNγ when stimulated through ITAM-containing receptors
Many of the activating NK cell receptors, such as NK1.1, NKp46, and Ly49H, associate with the ITAM-containing FcεRIγ or DAP12 adapter proteins (1). Ligation of an ITAM-associated NK cell receptor results in degranulation, cytokine production, and killing of the target cell. Several studies have recently reported that miRNAs regulate activation of signaling pathways downstream of ITAM receptors (37, 38). We asked whether Dicer- or Dgcr8-deficient NK cells show impaired function of their ITAM-containing receptors. Freshly isolated NK cells were activated by plate-bound anti-NK1.1, anti-NKp46, and anti-Ly49H Abs, and surface CD107a (LAMP-1) and intracellular IFNγ were measured. There were fewer CD107a-positive cells among gated YFP+ NK populations from DicerΔ/Δ and Dgcr8Δ/Δ mice compared with controls (Fig. 5A). Furthermore, fewer DicerΔ/Δ and Dgcr8Δ/Δ NK cells produced IFNγ when activated by antibodies to NK1.1, NKp46, or Ly49H (Fig. 5B). To determine whether this is a generalized hypo-responsiveness or an exclusive defect associated with ITAM-containing receptors, we stimulated NK cells with IL-12 and IL-18. Interestingly, DicerΔ/Δ, Dgcr8Δ/Δ, and control NK cells produced comparable amounts of IFNγ in the presence of IL-12 and IL-18 (Fig. 5C). This demonstrates that Dicer- and Dgcr8-deficient NK cells are fully capable of responding through their cytokine receptors; however, function of their ITAM-containing receptors is impaired.
Figure 5. Defective degranulation and IFNγ production after stimulation of the activating NK receptors in Dicer- and Dgcr8-deficient NK cells.
(A) NK cells were incubated in wells either coated with IgG or mAbs NK1.1, NKp46, or Ly49H or with no mAb in the presence of anti-CD107a. Only cells that had degranulated displayed CD107a on the cell surface. Percentages of NK cells (gated on YFP+ DX5+ TCRβ− cells) expressing CD107a. Right, percentages of NK cells expressing CD107a (mean ± SEM of 4–9 mice/group). (B, C) NK cells were stimulated as above or incubated with IL-12 and IL-18 in the presence of brefeldin A. Intracellular staining for IFNγ in YFP+ DX5+ TCRβ− cells is shown. Right, percentages of NK cells producing IFNγ (mean ± SEM of 3–7 mice/group). (C) Data are mean ± SEM of 3 mice/group. Data are representative of at least 5 experiments.
Dicer and Dgcr8 are critical for Ly49H+ NK cell expansion during MCMV infection
To investigate whether miRNAs regulate NK cell function in vivo, we asked whether Dicer and Dgcr8 are necessary for NK cell cytokine production and expansion in response to viral infection. During infection with MCMV, NK cells bearing the activating Ly49H receptor recognize the viral protein m157 (7, 8), and respond to inflammatory cytokines such as IL-12 produced by dendritic cells (39). Signaling primarily via ITAM-containing DAP12, and augmented through DAP10, Ly49H+ NK cells mount a rapid anti-viral response, secrete cytokines including IFNγ, and undergo expansion (40).
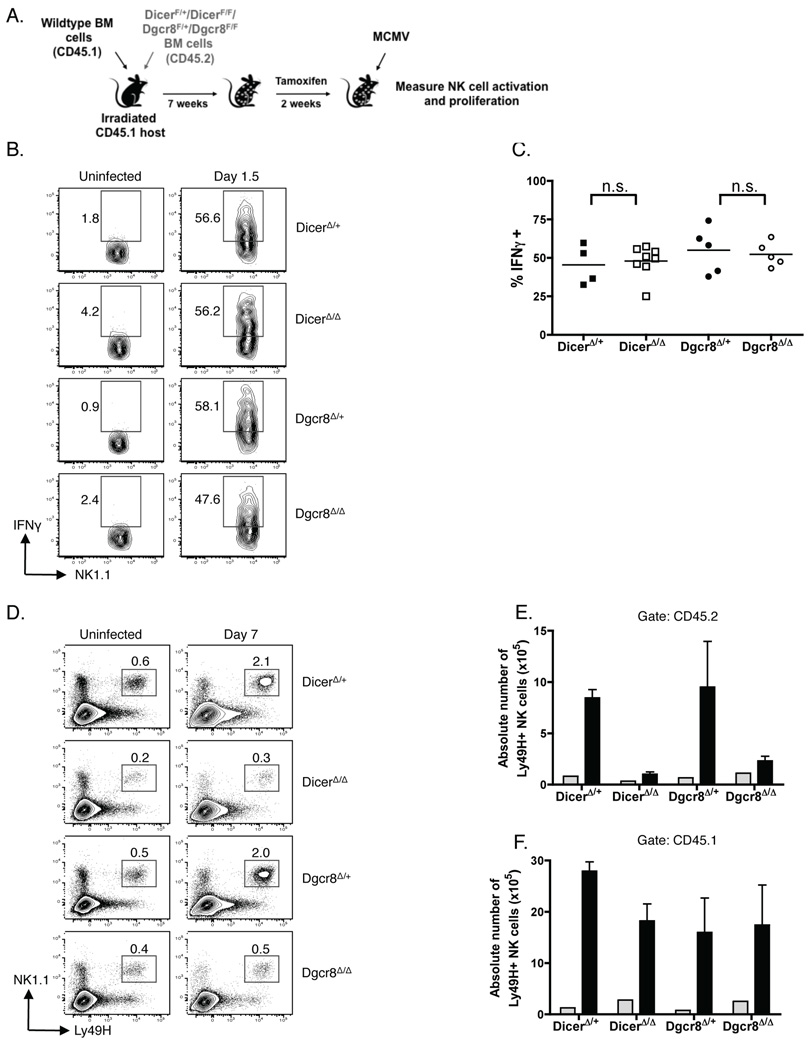
To measure MCMV-specific immune responses by Dicer- and Dgcr8-deficient NK cells, we generated chimeric mice by reconstituting lethally irradiated mice with 1:1 mixed bone marrow from wildtype (CD45.1+) and experimental (DicerF/F, Dgcr8F/F and control DicerF/+, Dgcr8F/+ (CD45.2+)) mice (Fig. 6A). Prior to tamoxifen treatment, wildtype and experimental bone marrow reconstituted the NK cell compartment equally well (Suppl. Fig. 3A). Post-tamoxifen treatment, however, the frequency of NK cells was significantly decreased in the CD45.2+ DicerΔ/Δ and CD45.2+ Dgcr8Δ/Δ compartments of chimeric mice, compared to that in the DicerΔ/+ and Dgcr8Δ/+ chimeric mice, consistent with our earlier findings (Suppl. Fig. 3A, B). Importantly, NK cell frequency in the CD45.1+ compartment was similar in DicerΔ/Δ, Dgcr8Δ/Δ, and DicerΔ/+, Dgcr8Δ/+ chimeric mice, thus providing an internal control as a standard for each sample.
Figure 6. Dicer and Dgcr8 are necessary for Ly4H+ NK cell expansion during MCMV infection.
(A-F) Mixed bone marrow chimeric mice (1:1 mixture of wildtype (CD45.1+) and either DicerF/+, DicerF/F, Dgcr8F/+, or Dgcr8F/F (CD45.2+) cells) were treated with tamoxifen and then given PBS (uninfected) or infected with MCMV. (B, C) Intracellular staining for IFNγ in NK cells immediately ex vivo at day 1.5 PI. (B) Representative scatter plots gated on CD45.2+ NK1.1+ TCRβ− cells from DicerΔ/+, DicerΔ/Δ, Dgcr8Δ/+, and Dgcr8Δ/Δ chimeric mice are shown, and the numbers represent percentages of cells in the indicated quadrants. (C) Cumulative results (n = 4–8 mice/group) are quantified. Percentages (D) and absolute numbers (E) of CD45.2+ Ly49H+ NK cells in DicerΔ/+, DicerΔ/Δ, Dgcr8Δ/+, or Dgcr8Δ/Δ chimeric mice at day 7 PI. (F) Absolute number of wildtype CD45.1+ Ly49H+ NK cells in the same DicerΔ/+, DicerΔ/Δ, Dgcr8Δ/+, or Dgcr8Δ/Δ chimeric mice at day 7 PI. (E, F) The number of Ly49H+ NK cells from uninfected chimeric mice (grey bars) and from MCMV-infected mice (black bars) is shown. Data are mean ± SEM of 3 mice per group and are representative of 3 independent experiments.
Following MCMV infection, we stained for intracellular IFNγ immediately ex vivo on day 1.5 post - infection (PI). 45.5 ± 6.5%, and 47.9 ± 3.6 of CD45.2+ NK cells from DicerΔ/Δ and DicerΔ/+ chimeric mice, respectively, produced IFNγ (Fig. 6B, C). No differences in intracellular IFNγ production were observed between CD45.2+ NK cells from Dgcr8Δ/Δ and Dgcr8Δ/+ chimeric mice (54.9 ± 6.7% and 52.3 ± 3.5%, respectively). Together, these results indicate that IFNγ production by NK cells during MCMV infection proceeds independently of miRNAs.
During the NK cell response against MCMV in wildtype mice, the Ly49H+ NK cells preferentially expand during the first several days of infection (41, 42). When we infected DicerΔ/+ and Dgcr8Δ/+ chimeric mice with MCMV, both mice showed an increase in Ly49H+ NK cell frequency (Fig. 6D), and numbers (Fig. 6E) at day 7 PI. With precursor numbers of ~0.8×105 (DicerΔ/+) and ~0.66×105 (Dgcr8Δ/+) total Ly49H+ NK cells in the spleen, the absolute number of Ly49H+ NK cells at day 7 PI expanded ~10.6- and ~13.8-fold, respectively (Fig. 6E). In contrast, fewer Ly49H+ NK cells were found in both DicerΔ/Δ and Dgcr8Δ/Δ chimeric mice at day 7 PI. The absolute number of Ly49H+ NK cells in DicerΔ/Δ and Dgcr8Δ/Δ chimeric mice expanded only ~3- and ~2.1-fold, respectively. Importantly, wildtype CD45.1+ Ly49H+ NK cells from every set of chimeric mice (DicerΔ/Δ and Dgcr8Δ/Δ, and control DicerΔ/+ and Dgcr8Δ/+) expanded equally well, indicating that all groups of mice were able to mount an effective antiviral response (Fig. 6F). Altogether, these experiments demonstrate that miRNAs are required for robust NK cell expansion during MCMV infection.
MiRNAs regulate survival of Ly49H+ NK cells during MCMV infection
Defective expansion of DicerΔ/Δ and Dgcr8Δ/Δ Ly49H+ NK cells could result from decreased NK cell proliferation and/or impaired survival during MCMV infection. To discriminate between these possibilities, we monitored proliferation in vivo by analyzing BrdU incorporation. Following a 2 h pulse at day 5 PI, similar percentages of DicerΔ/Δ, Dgcr8Δ/Δ and DicerΔ/+, Dgcr8Δ/+ NK cells from mixed bone marrow chimeric mice incorporated BrdU (Fig. 7A). Adoptive transfer of CFSE-labeled NK cells into Ly49H-deficient recipients further confirmed that both Dgcr8Δ/Δ and Dgcr8Δ/+ Ly49H+ cells were dividing to a similar extent, fully diluting their CFSE during the 4 days following infection (Fig. 7B). We also monitored apoptosis by Annexin V staining at the same time point PI. Notably, a higher percentage of apoptotic cells was detected in Dgcr8Δ/Δ mice compared to that in Dgcr8Δ/+ control mice (Fig. 7C). Together these data indicate that miRNAs are not required for the proliferative responsiveness of Ly49H+ NK cells, but regulate the survival of NK cells during MCMV infection.
Discussion
Here we have shown identical phenotypes when either Dicer or Dgcr8 gene was inactivated in NK cells. These results strongly suggest that these two molecules function in the same biological pathway in miRNA biogenesis, and demonstrate that the deficiency in miRNAs is the primary cause underlying the observed phenotypes. Although it is possible that Dicer-deficient cells also exhibit other more subtle phenotypes, such as the derepression of retrotransposons (24, 25), it appears that it is miRNAs, rather than other Dgcr8-independent, Dicer-dependent small RNAs, that are critical for NK cells.
Ablation of the miRNA biogenesis pathway, through deletion of Dicer or Dgcr8, led to increased apoptosis of peripheral NK cells. Similarly, Dicer deletion in developing B cells (17), thymocytes (15, 16), or iNKT cells (19) resulted in increased cell death. These results suggest that the miRNA pathway plays an important role in controlling cell survival. Potential mechanisms include mitotic defects due to centromere dysfunctions (43), defects in heterochromatin maintenance (44), and aberrant over expression of pro-apoptotic protein Bim (17). In addition, our preliminary studies indicate that Bcl-2 mRNA level was consistently decreased in DicerΔ/Δ and Dgcr8Δ/Δ NK cells (Suppl. Fig. 4). Yamanaka et al. recently showed that inhibition of miR21 in the human NKL cell line led to increased apoptosis associated with the up-regulation of pro-apoptotic miR21 targets PTEN, PDCD4, and Bim (38). Although it is very likely that multiple miRNAs take part in the coordinate regulation of NK cell survival, it would be informative to test whether miR21 is a key regulator of NK cell survival.
Maturation of NK cells is characterized by a decrease in the expression of CD27, active proliferation, and a concomitant increase in the levels of CD11b and effector functions (35, 45). Analysis of DicerΔ/Δ and Dgcr8Δ/Δ mice revealed a relative accumulation of more immature CD27hi CD11blo NK cells. This could be due to a selective loss of more mature CD27lo CD11bhi NK cells or a maturation defect. We did not detect any preferential difference in the death or turnover in the CD27hi CD11blo or CD27lo CD11bhi NK cell subsets (unpublished observations). Furthermore, we found a similar accumulation of CD27hi CD11blo NK cells in CD45.2+ subsets of DicerΔ/Δ and Dgcr8Δ/Δ mixed bone marrow chimeras (data not shown). These data indicate that impaired transition from CD27hi CD11blo to CD27lo CD11bhi of Dicer- and Dgcr8-deficient NK cells is hematopoietic cell intrinsic and cell autonomous, suggesting that miRNAs might regulate NK cell maturation. Interestingly, miR150 has been shown to regulate B cell development and this regulation occurs through the inhibition of Myb expression (46, 47). MiR150 is dynamically regulated during NK cell maturation (unpublished observations), and Chiossone et al. reported that Myb is differentially up-regulated in CD27hi CD11blo NK cells (45). Future studies will determine if NK cell development or maturation is regulated by miR150.
In contrast to having no significant effect on many of the activating and inhibitory NK cell receptors analyzed, Dicer and Dgcr8-deficiency had an impact on the level of NKG2D on the surface of NK cells. In addition, we found elevated surface levels of Rae-1 on NK cells from Dicer-deficient mice (Fig. 4D). NKG2D is an activating receptor that binds to a diverse family of ligands that are distant relatives of MHC-class I molecules, including MICA and MICB in humans and Rae-1α-ε, MULT1, and H60 in mice (36). Engagement of NKG2D by its ligands leads to the direct activation of killing and cytokine secretion by NK cells. Because of this potent killing ability, surface expression of NKG2D ligands is tightly regulated so that the immune response is not triggered inappropriately. Our findings suggest that miRNAs may regulate the expression of the mouse NKG2D ligand Rae-1. One possible mechanism would be a specific miRNA targeting Rae-1 mRNA. Several human miRNAs were recently identified to bind the 3’UTR of MICA and MICB and to repress their translation (48, 49). Alternatively, Dicer or Dgcr8 deficiency in NK cells might elicit a DNA damage response, which in turn results in the induction of Rae-1 expression (50). Consistent with this hypothesis, silencing of Dicer in HEK293T and human hepatoma cells led to the upregulation of MICA and MICB (51). In either of these scenarios, the increased levels of Rae-1 would likely lead to down modulation of NKG2D on NK cells, as has previously been demonstrated (36).
We have shown that degranulation and IFNγ production were impaired in Dicer- and Dgcr8-deficient NK cells following stimulation via NK1.1, NKp46, or Ly49H. This impairment in IFNγ production might be caused by the reduction in the CD27lo CD11bhi NK cell subset in DicerΔ/Δ and Dgcr8Δ/Δ mice, because this subset has been demonstrated to play a dominant role in cytokine production (45). To this end we co-stained NK cells with anti-CD27 and anti-CD11b antibodies and measured intracellular IFNγ levels. Both CD27hi CD11blo and CD27lo CD11bhi NK cell subsets from DicerΔ/Δ and Dgcr8Δ/Δ mice produced less IFNγ compared to that in the corresponding NK cell subsets from the control mice (data not shown). Thus, a decline in CD27lo CD11bhi NK cells in DicerΔ/Δ and Dgcr8Δ/Δ mice does not explain the reduction in cytokine generation. Because stimulation with IL-12 and IL-18 induced comparable amounts of IFNγ in DicerΔ/Δ, Dgcr8Δ/Δ, and control NK cells, we conclude that miRNAs are necessary for ITAM-based activation.
Li et al. recently demonstrated that miR181a modulates signal strength of the T cell receptor (TCR) by down-regulating expression of several protein tyrosine phosphatases, including SHP-2, PTPN22, DUSP5, and DUSP6 (37). These phosphatases are expressed in mouse NK cells (52); with the exception of SHP-2, however, it is currently not known whether they regulate NK cell activation. In T cells, miR181a-mediated regulation of these phosphatases leads to enhanced basal activation of the TCR signaling molecules including Src kinase, Lck, and Erk (37). In addition, Yamanaka et al. showed that reducing levels of miR21 in the human NKL cell line resulted in the downregulation of phosphorylated AKTser473 (38). Thus, a potential explanation for the impaired function of ITAM-containing receptors in Dicer- and Dgcr8-deficient NK cells is that specific miRNA(s) might regulate signaling downstream of these ITAM-containing receptors.
NK cells have long been compared to effector and memory CD8+ T cells in phenotype and function (53), and recently several groups have demonstrated that NK cells can become long-lived cells and mount secondary responses against viral antigens (31, 54). In this study, we determined the miRNA expression profile of mouse and human NK cells. Naïve NK cells and CD8+ T cells share a large part of their miRNA profile: miR142-3p, miR142-5p, miR150, miR16, miR23a, miR15b, miR29a, miR29b, miR30b, and miR26a are highly expressed in both naïve NK cells and CD8+ T cells (Suppl. Fig. 1, (55, 56)). Similar to NK cells, the frequency and number of naïve CD8+ T cells were preferentially reduced in Dicer- and Dgcr8-deficient mice. Thus, miRNAs shared by naïve NK cells and CD8+ T cells might be implicated in the regulation of common molecular pathways, such as regulation of homeostasis.
NK cells also share a miRNA profile with effector and memory CD8+ T cells. MiR21, miR221, and miR222 are expressed in both NK and effector CD8+ T cells, whereas miR146a is found in both NK cells and memory CD8+ T cells (Suppl. Fig. 1, (56)). It is intriguing to speculate that the miRNAs shared by NK cells, effector CD8+ T cells, and memory CD8+ T cells might be implicated in the regulation of common pathways that lead to the acquisition of the effector phenotype and survival. Our identification of a requirement of Dicer and Dgcr8 for Ly49H+ NK cell expansion during MCMV infection predicts miRNAs might target factors that are important for maintaining the NK cell population during viral infection. In conclusion, our results present novel evidence for miRNAs regulating diverse aspects of NK cell biology, including basic processes such as turnover and survival, as well as the function of activating NK cell receptors during MCMV infection.
Supplementary Material
Acknowledgement
We thank Drs. Michael McManus and Eric Brown for mice. We thank Ms. Jessica Jarjoura for assistance with cell sorting, Mr. Hernan Consengco for assistance with genotyping, and members of the Lanier laboratory for critical reading of the manuscript.
Abbreviations
- NK
Natural Killer cells
- ITAM
immunoreceptor tyrosine-based activation motif
- MCMV
mouse cytomegalovirus
- miRNA
microRNA
- Dgcr8
DiGeorge syndrome critical region 8
- qRT-PCR
quantitative RT-PCR
- NKL
human natural killer cell line
- Lck
lymphocyte-specific protein-tyrosine kinase
- Erk
extracellular signal-regulated kinase
- MFI
mean fluorescence intensity
- PI
post infection
Footnotes
Disclosures
The authors have no financial conflicts of interest.
N.A.B. is American Cancer Society Postdoctoral Fellow and was supported by NIH training grant T32. D.G.T.H. is a special fellow of the Leukemia and Lymphoma Society. R.H.B. was supported by NIH/NINDS (K08 NS048118 and R01 NS057221). This study was supported by NIH grant AI068129 and L.L.L. is an American Cancer Society Professor.
REFERENCES
- 1.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 3.Prlic M, Blazar BR, Farrar MA, Jameson SC. In vivo survival and homeostatic proliferation of natural killer cells. J Exp Med. 2003;197:967–976. doi: 10.1084/jem.20021847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jamieson AM, Isnard P, Dorfman JR, Coles MC, Raulet DH. Turnover and proliferation of NK cells in steady state and lymphopenic conditions. J Immunol. 2004;172:864–870. doi: 10.4049/jimmunol.172.2.864. [DOI] [PubMed] [Google Scholar]
- 5.Lanier LL. Evolutionary struggles between NK cells and viruses. Nat Rev Immunol. 2008;8:259–268. doi: 10.1038/nri2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 7.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 8.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, Fremont DH, Yokoyama WM. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 10.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 11.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 12.Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 14.Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc Natl Acad Sci U S A. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cobb BS, Hertweck A, Smith J, O'Connor E, Graf D, Cook T, Smale ST, Sakaguchi S, Livesey FJ, Fisher AG, Merkenschlager M. A role for Dicer in immune regulation. J Exp Med. 2006;203:2519–2527. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 18.Zhou X, Jeker LT, Fife BT, Zhu S, Anderson MS, McManus MT, Bluestone JA. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedeli M, Napolitano A, Wong MP, Marcais A, de Lalla C, Colucci F, Merkenschlager M, Dellabona P, Casorati G. Dicer-dependent microRNA pathway controls invariant NKT cell development. J Immunol. 2009;183:2506–2512. doi: 10.4049/jimmunol.0901361. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L, Seo KH, He HZ, Pacholczyk R, Meng DM, Li CG, Xu J, She JX, Dong Z, Mi QS. Tie2cre-induced inactivation of the miRNA-processing enzyme Dicer disrupts invariant NKT cell development. Proc Natl Acad Sci U S A. 2009;106:10266–10271. doi: 10.1073/pnas.0811119106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liston A, Lu LF, O'Carroll D, Tarakhovsky A, Rudensky AY. Dicer-dependent microRNA pathway safeguards regulatory T cell function. J Exp Med. 2008;205:1993–2004. doi: 10.1084/jem.20081062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- 23.Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, Surani MA, Sakaki Y, Sasaki H. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- 26.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J, Blelloch R. MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol. 20:271–277. doi: 10.1016/j.cub.2009.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G, Zediak VP, Velez M, Bhandoola A, Brown EJ. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1:113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao PK, Toyama Y, Chiang HR, Gupta S, Bauer M, Medvid R, Reinhardt F, Liao R, Krieger M, Jaenisch R, Lodish HF, Blelloch R. Loss of cardiac microRNA-mediated regulation leads to dilated cardiomyopathy and heart failure. Circ Res. 2009;105:585–594. doi: 10.1161/CIRCRESAHA.109.200451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun JC, Beilke JN, Lanier LL. Adaptive immune features of natural killer cells. Nature. 2009;457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- 33.Sonnenberg GF, Mangan PR, Bezman NA, Sekiguchi DR, Luning Prak ET, Erikson J, Maltzman JS, Jordan MS, Koretzky GA. Mislocalization of SLP-76 leads to aberrant inflammatory cytokine and autoantibody production. Blood. 2009 doi: 10.1182/blood-2009-08-237438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chong MM, Rasmussen JP, Rudensky AY, Littman DR. The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J Exp Med. 2008;205:2005–2017. doi: 10.1084/jem.20081219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayakawa Y, Smyth MJ. CD27 dissects mature NK cells into two subsets with distinct responsiveness and migratory capacity. J Immunol. 2006;176:1517–1524. doi: 10.4049/jimmunol.176.3.1517. [DOI] [PubMed] [Google Scholar]
- 36.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunological Reviews. 2010 doi: 10.1111/j.0105-2896.2010.00893.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li QJ, Chau J, Ebert PJ, Sylvester G, Min H, Liu G, Braich R, Manoharan M, Soutschek J, Skare P, Klein LO, Davis MM, Chen CZ. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Yamanaka Y, Tagawa H, Takahashi N, Watanabe A, Guo YM, Iwamoto K, Yamashita J, Saitoh H, Kameoka Y, Shimizu N, Ichinohasama R, Sawada K. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood. 2009;114:3265–3275. doi: 10.1182/blood-2009-06-222794. [DOI] [PubMed] [Google Scholar]
- 39.Krug A, French AR, Barchet W, Fischer JA, Dzionek A, Pingel JT, Orihuela MM, Akira S, Yokoyama WM, Colonna M. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 40.Sun JC, Beilke JN, Lanier LL. Immune memory redifined: characterizing the longevity of natural killer cells. Immunological Reviews. 2010 doi: 10.1111/j.1600-065X.2010.00900.x. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dokun AO, Chu DT, Yang L, Bendelac AS, Yokoyama WM. Analysis of in situ NK cell responses during viral infection. J Immunol. 2001;167:5286–5293. doi: 10.4049/jimmunol.167.9.5286. [DOI] [PubMed] [Google Scholar]
- 43.Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- 44.Giles KE, Ghirlando R, Felsenfeld G. Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nat Cell Biol. 12:94–99. doi: 10.1038/ncb2010. sup pp 91-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiossone L, Chaix J, Fuseri N, Roth C, Vivier E, Walzer T. Maturation of mouse NK cells is a 4-stage developmental program. Blood. 2009;113:5488–5496. doi: 10.1182/blood-2008-10-187179. [DOI] [PubMed] [Google Scholar]
- 46.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci U S A. 2007;104:7080–7085. doi: 10.1073/pnas.0702409104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Stern-Ginossar N, Gur C, Biton M, Horwitz E, Elboim M, Stanietsky N, Mandelboim M, Mandelboim O. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–1073. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 49.Yadav D, Ngolab J, Lim RS, Krishnamurthy S, Bui JD. Cutting edge: down-regulation of MHC class I-related chain A on tumor cells by IFN-gamma-induced microRNA. J Immunol. 2009;182:39–43. doi: 10.4049/jimmunol.182.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–1190. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang KF, Ren H, Cao J, Zeng GL, Xie J, Chen M, Wang L, He CX. Decreased Dicer expression elicits DNA damage and up-regulation of MICA and MICB. J Cell Biol. 2008;182:233–239. doi: 10.1083/jcb.200801169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–1094. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 53.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 54.O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol. 2006;7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 55.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foa R, Schliwka J, Fuchs U, Novosel A, Muller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu H, Neilson JR, Kumar P, Manocha M, Shankar P, Sharp PA, Manjunath N. miRNA profiling of naive, effector and memory CD8 T cells. PLoS One. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.