Abstract
Sex-biased demographic events can result in asymmetries in female and male effective population size that can lead to different patterns of genetic variation on chromosome X than are expected based on the patterns on the autosomes. Previous studies point to a period around the time of the dispersal of anatomically modern humans out of Africa when chromosome X experienced a significant reduction in effective population size relative to the autosomes. Here, we explore whether a sex-biased demographic history could explain these observations. We use coalescent simulations to show that a model of primarily male migration during the out-of-Africa dispersal can produce the striking patterns that are observed when comparing patterns of genetic variation on the autosomes and chromosome X. The model involves a history in which after the founder population of non-Africans lost much of its genetic diversity, subsequent mostly male gene flow from an African source brought new diversity into the population. We also explore two additional models, one of sex-biased generation time and one of a substructured population during the dispersal out of Africa with primarily female migration among demes. These latter models cannot account for the magnitude of the observed reduction in chromosome X effective population size, although it is plausible that they played a more minor role in producing the striking chromosome X/autosome patterns.
Keywords: gender-biased demography, chromosome X, autosomes, effective population size, coalescent simulations, human
Introduction
In a population that has been of constant size throughout history and has had equal effective number of reproducing males and females in every generation, the effective population size (Ne) of chromosome X is expected to be ¾ of what it is on the autosomes. However, changes in the historical effective population size over time can lead to a departure from this ratio, even in the face of an equal male and female effective population size, because the smaller effective population size of chromosome X results in more ancient demography affecting chromosome X less than it does the autosomes (Pool and Nielsen 2007; Keinan et al. 2008). Deviations from the expected ratio of ¾ are also predicted by sex-biased demographic events resulting in different Ne of females and males, as well as from forces of selection differentially affecting chromosome X and the autosomes.
In a previous study, we compared patterns of variation on chromosome X and the autosomes in humans on a genomic scale using two independent uniformly collected data sets (Keinan et al. 2008). Independent analyses of different aspects of these data pointed to a period around the time of the out-of-Africa dispersal when the effective population size of chromosome X was transiently much reduced relative to the expectation according to the autosomes. These analyses were based on single nucleotide polymorphism (SNP) allele frequency differentiation between different human populations, on the allele frequency spectrum, and on estimates from resequencing data of the average time to the most recent common ancestor (tMRCA) in different populations.
Our previous results using SNP allele frequencies were based on uniformly discovering SNPs on both the autosomes and the chromosome X and considering their allele frequencies based on genotyping in all HapMap samples (International HapMap Consortium 2005, 2007). We estimated FST in each of these two parts of the genome and translated these estimates to a ratio Q of X-to-autosome effective population size since the time that the two populations have split (Keinan et al. 2008). The results showed that Q is significantly reduced below ¾ when measured between North Europeans and West Africans, as well as between East Asians and West Africans. However, Q is consistent with ¾ when measured between North Europeans and East Asians, as well as between Chinese and Japanese. These results, which were also replicated in a more recent study (Amato et al. 2009), point to a reduction in chromosome X effective population size after the split of West Africans and non-Africans but before the separation of North Europeans and East Asians. In another analysis, we focused on SNP allele frequency spectra within populations and concluded that the spectrum of chromosome X in non-African populations is inconsistent with that of the autosomes in a way that supports a relative reduction in chromosome X effective population size in the history of non-Africans (Keinan et al. 2008).
The previous results of analyzing the tMRCA were based on assembling a data set of over a billion aligned base pairs and estimating sequence diversity in different human populations as the fraction of differences per base pair between two DNA sequences (heterozygosity/nucleotide diversity). The ratio of chromosome X-to-autosomes tMRCA, which we denote as T, is significantly reduced below ¾ in North Europeans and East Asians, although it is consistent with expectation in West Africans. Though known features of non-African history, including the out-of-Africa population bottleneck, predict some reduction below ¾ (Pool and Nielsen 2007), the observed ratios are still significantly reduced below these predictions (Keinan et al. 2008). These T ratio results are thus consistent in pointing to a relative reduction in chromosome X effective population size and show that it occurred in non-African history after the split from Africans. A recent review suggested that the result based on the T ratio might be explained by differences in normalization for background mutation rate between chromosome X and the autosomes (Bustamante and Ramachandran 2009), which can potentially explain discrepancies from a study that has found an increased X-to-autosome effective population size ratio in both African and non-African populations (Hammer et al. 2008). Although estimating the ratio T indeed requires normalizing by an outgroup, our main result of a reduced T ratio in non-African populations relative to Africans is independent of any normalization by an outgroup (comparing estimates of two populations does not require normalizing them because they would have been normalized by the same value) as are the results based on SNP allele frequencies (Keinan et al. 2008).
The Q and T ratios, while both measuring X-to-autosome Ne, capture two types of signals. Q is sensitive to drift in SNP allele frequencies in the recent history after population split. T is based on the history of individual DNA sequences in one population and averages effective population sizes over the longer time period since the most recent common genetic ancestor. Thus, our results based on Q are affected only by the history after the split of African and non-African populations, while T is additionally confounded by the history of the ancestral African population. Another difference is that T is sensitive to historical changes in population size, even if these do not introduce a deviation from an equal female and male effective population size (Pool and Nielsen 2007), but such changes on the non-African lineage do not affect Q. Hence, to explain the differences between X chromosomal and autosomal Ne, it is necessary to account for the observed deviation in Q and for the observed deviation in T from ¾, as well as for the differences between these two ratios of effective population size.
One possible explanation of the relative reduction in chromosome X effective population size is natural selection differently affecting chromosome X. Whereas chromosome X is expected to experience more selection than the autosomes due to the exposure of recessive alleles in hemizygous males, to explain our results the differential selection would have to be restricted to a certain epoch of human history occurring after the ancestors of West Africans split from the ancestors of non-Africans and before the divergence of North Europeans and East Asians. The exposure to new environments when moving out of Africa and the increased competition during that time of human history could have shifted the selective pressures (Storz et al. 2004). For that reason, we searched for features of the data that are predicted by natural selection but to date found no evidence for selection explaining the relative reduction in chromosome X Ne (Keinan et al. 2008). However, these analyses also could not rule out the possibility of selection being the force behind these results, and it remains a plausible explanation.
Another set of potential explanations of the transient reduction in chromosome X Ne has to do with sex-biased demographic history. Sex-biased historical events can account for our results because a reduction in female Ne relative to male translates to a reduction in Ne of chromosome X relative to autosomes. Perhaps the simplest possibility is that females had much greater variability in their reproductive success than males during the out-of-Africa dispersal (Caballero 1995; Charlesworth 2001). However, to even come close to explaining the results, it is required that during many generations throughout that epoch of human history, a tiny fraction of females would have had almost all the offspring (Keinan et al. 2008; Labuda et al. 2010), which is biologically implausible.
Here, we use demographic modeling via coalescent simulations to study possible sex-biased historical events in human history and to examine whether they can account for the empirical observations. The aims of this paper are 3-fold. First, we show that primarily male migration during the human dispersal out of Africa can produce both the T and the Q ratios we observed. Indeed, it is the most parsimonious demographic scenario we identified that is sufficient by itself to explain the magnitude of the relative reduction of chromosome X Ne. Second, we investigate the effect of relevant demographic parameters of such a model of primarily male migration on X-to-autosome effective population size ratios. Third, we explore other sex-biased demographies that can produce a reduction in the effective population size of chromosome X relative to the autosomes.
Materials and Methods
Model of Primarily Male Migration during the Out-of-Africa Dispersal
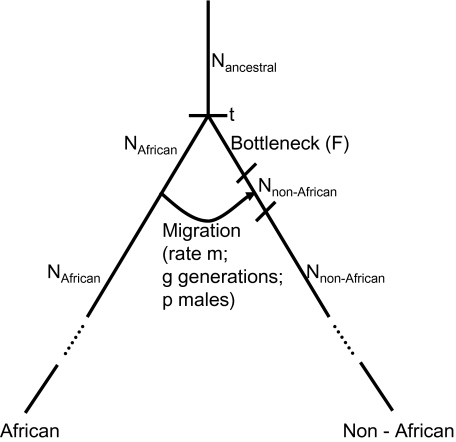
To demonstrate that a model of primarily male migration can produce data that match our observations, we modeled the joint history of West African and non-African populations. We focused on North Europeans as one representative non-African population because all the patterns we observed were shared between Europeans and Asians. The empirical observation of similar patterns in East Asians and in Europeans suggests that if primarily male migration is to explain these patterns, it will have to be into the founder population of non-Africans before the split of European and East Asian populations. The proposed model of primarily male migration is that the founder population of non-Africans was established by an equal dispersal of both males and females. Following the African–non-African population split, the founder population of non-Africans lost much of its genetic diversity due to a population bottleneck. Then, subsequent migrants brought new genetic diversity into the founder population. These migrants, if they have been mostly male, can give rise to X-autosome asymmetries in the same direction as in the empirical observations.
The history and parameters we explored to show that this scenario is sufficient to explain the observations are as follows (and diagrammed in fig. 1):
The out-of-Africa event occurred t generations ago and split a panmictic population of constant diploid effective size of Nancestral into a West African ancestral population of constant size NAfrican and the ancestral population of North Europeans.
Immediately after the population split, the ancestors of North Europeans experienced a population bottleneck. We modeled the population bottleneck as an instantaneous crash in population size, with one parameter, F, capturing bottleneck intensity as the amount of genetic drift during the bottleneck and defined as the number of generations the bottleneck lasted divided by twice the effective population size during the bottleneck (Reich et al. 2001). The effect of a bottleneck on allele frequencies depends primarily on the total amount of genetic drift and, given this ratio, is practically independent of the numerator—the number of generations the bottleneck lasted (Reich et al. 2001; Keinan et al. 2007)—so we kept the latter fixed at 100 generations.
After the population bottleneck and until the present, the effective population size of the ancestors of North Europeans is Nnon-African.
We modeled migration from the West African ancestral population to the ancestral population of North Europeans immediately after the bottleneck. The migration lasted for g generations, with another parameter, m, capturing the migration rate—the fraction of individuals in the North European ancestral population who are migrants in each generation. Last, the parameter allowing for sex-asymmetric migration—the proportion of males among the migrants—is p.
FIG. 1.
Model of sex-biased migration during the dispersal of modern humans from Africa. The figure outlines the primarily male migration model, a detailed description of which is provided in Materials and Methods. Nancestral is the ancestral Ne, NAfrican the African Ne, and Nnon-African the Ne of the ancestral population of North Europeans; t is the split time of the African and non-African populations; F is the severity of the out-of-Africa population bottleneck; g denotes the number of generations migration lasted following the bottleneck; m denotes its rate; and p the proportion of migrants who were male.
Our modeling makes the simplification that male and female effective population size is equal, namely chromosome X effective population size is ¾ that of the autosomes, throughout all epochs of history. The only sex-biased factor in the model is the different proportion of male and female migrants: Chromosome X migration occurs at a rate of m(1 − p) for females and at a rate of mp for males, which amount to
overall because ⅔ of X chromosomes in the founder population are in females. This expression for chromosome X migration rate equals m (for a nonzero m) if and only if there is an equal number of male and female migrants (p = 0.5). Another simplifying assumption in this model is that all individuals have an equal mean and variance of reproductive success, but deviations from this assumption can to some extent be captured by varying model parameters. For example, if migrants are more/less reproductively successful on average than the individuals in the population to which they are immigrating, this results in an effective increase/decrease in the magnitude of the migration rate.
Coalescent Simulations of Primarily Male Migration Model
To generate data that would be expected under the model of sex-biased migration during the dispersal out of Africa, we conducted a series of coalescent simulations for a range of model parameters using the “ms” software (Hudson 2002). Coalescent simulations for assessing allele frequency differentiation (FST) between West Africans and North Europeans and estimating the ratio Q were carried out in a way that exactly matched the sample sizes in our empirical data. Thus, for both populations, 120 chromosomes were sampled on the autosomes and 90 on chromosome X. To emulate the SNP discovery strategy, we simulated two additional West African chromosomes (for a total of 122 for the autosomes and 92 for chromosome X in the African population) and discarded SNPs from analysis if they were not polymorphic between these two discovery chromosomes, which is exactly as we generated our empirical data. Coalescent simulations for assessing sequence diversity and the ratio T were carried out by considering only two North European chromosomes, for which we calculated tMRCA, which exactly matches our methodology in obtaining the empirical results.
We simulated 10,000,000 genealogies for each set of parameters to accurately calculate FST and tMRCA. We estimated FST between West Africans and North Europeans for both the chromosome X and the autosomes and calculated the X chromosome-to-autosomes ratio of effective population size Q from the FST values (Keinan et al. 2008). We also estimated the X-to-autosomes ratio of effective population size T based on estimates of tMRCA in each of the two compartments of the genome with the same set of parameters. We compared the results of the simulations to key summary statistics measured from our data: 1) FST between West Africans and North Europeans on the autosomes, 2) FST between West Africans and North Europeans on chromosome X, 3) X-to-autosomal ratio Q between the two populations, and 4) X-to-autosomal ratio T in North Europeans. (We did not consider the individual sequence diversity estimates for chromosome X and the autosomes because these are sensitive to details of the SNP calling algorithm and the normalization by an outgroup.)
We began by identifying a set of baseline model parameters that we chose to provide a good match to known features of human demographic history, while varying the level of gene flow and bottleneck intensity to match the autosomal FST between West Africans and North Europeans, and at the same time matching other features of the data as closely as possible. This approach is similar to that of a recent study that modeled the processes underlying X-to-autosomal diversity ratios in Drosophila melanogaster (Pool and Nielsen 2008). Due to the simplifying assumptions underlying the model, as well as its complexity with regard to number of parameters and their correlations, we did not attempt to fit model parameters to the data. Rather, after obtaining a baseline set of parameter values, we explored the signature of different historical models on genetic data by varying parameters in our simulations, examining how they affected the predictions of the model, and addressing questions about the domain of viable models.
Model of Sex-Specific Generation Time
To examine the effect that asymmetries in male–female generation length could have on our observations, we considered a scenario of a much shorter female effective generation time. Let x denote average female generation time and c·x denote average male generation time. As ⅔ of X chromosomes are carried in females but only ½ of the autosomes, the X-to-autosome ratio of effective population size would be expected to be decreased by a factor of (3 + 3c)/(2 + 4c) due to such a difference in generation time:
In a rather extreme scenario of a male generation time double that of female (c = 2), this ratio becomes 9/10, that is, the X-to-autosome ratio of effective population size is expected to decrease by 10%. In the absence of other forces resulting in deviations from ¾, this scenario is expected to decrease the ratio from 0.75 to 0.675 for the period in which the male generation time is twice that of the female. In order to explain our observations, the effect has to be restricted to only a small part of human history (after the separation of African and non-African populations but before the separation of North Europeans and East Asians), so the effect that asymmetries in generation time could have on our observations is even smaller. Thus, even this extreme asymmetry in generation time that corresponds, for example, to an average of 20 for the age at which females have offspring and 40 for the average age at which males have offspring is too slight by itself to quantitatively explain our observations.
As shorter female generation time could not by itself account for the observations, we concurrently modeled the effect of primarily male migration and shorter female generation time by considering the above extreme decrease in relative female generation time (c = 2) within the model of primarily male migration during the out-of-Africa dispersal. To have the largest effect, we considered the asymmetry in generation time as spanning the entire period starting from the out-of-Africa event.
Model of Population Substructure during the Out-of-Africa Dispersal with Primarily Females Migrating between Groups
To consider the effect population structure during the dispersal out of Africa could have on our observations, we used coalescent simulations (Hudson 2002) to consider an extreme scenario of sex-biased structure. We simulated the history of West Africans and North Europeans based on best-fit models of demographic history for each of the populations that we have previously fit (Keinan et al. 2007). The model also included a split of West Africans and North Europeans just before the out-of-Africa bottleneck and SNP ascertainment in two West African chromosomes to match the data. For the entire period starting with the split of West Africans and non-Africans and ending in the time of the North European–East Asian split, we simulated the population as composed of two distinct demes, each with half the overall effective population size, with only females migrating between groups. We repeated these simulations for different levels of female migration rate (the fraction of each group made up of female migrants from the other group in each generation) and estimated the X-to-autosome Ne ratio Q based on FST estimates.
Results and Discussion
A Model of Primarily Male Dispersal Out of Africa Is Sufficient to Account for the Observed Reduction in Chromosome X Effective Population Size
Our initial goal was to identify a demographic model that was sufficient to account for the extreme observation of a reduced effective population size of chromosome X in non-African populations (Keinan et al. 2008). We sought to explain two independent results pointing to low Ne of chromosome X: 1) differentiation of SNP allele frequencies shows that chromosome X/autosomal Ne ratio since the split of West African and North European populations is Q = 0.582 (95% confidence interval: 0.524–0.640) and 2) estimates of diversity (heterozygosity) obtained from sequence data show that chromosome X/autosomal Ne ratio in the history of North Europeans is T = 0.635 (0.587–0.683). One possible explanation for our observations is a larger male than female effective population size during the dispersal of anatomically modern humans out of Africa, which could have occurred specifically through primarily male migration. We modeled this possibility by considering a model of North European and West African history, with a population bottleneck in the ancestral population of non-Africans and with primarily male migrants from an African source bringing new genetic diversity to this ancestral population after the bottleneck (fig. 1 and Materials and Methods). For the model to be more realistic, in addition to explaining the two X-to-autosome ratios, we required that it approximately match the observed FST between North Europeans and West Africans, which we observed to be 0.144 (0.138–0.149) for the autosomes and 0.221 (0.203–0.239) for chromosome X.
We began by identifying a baseline model (table 1), which we chose to be consistent with known features of human demographic history, and to match the autosomal FST between West Africans and North Europeans, which has the smallest standard error out of all statistics we sought to match, while at the same time matching other features of the data as closely as possible. We did not attempt to fit model parameters but rather used coalescent simulations with one set of parameters to generate genetic data that satisfy all these criteria. The baseline model we identified has the following parameter values: t = 2,400 (60,000 years assuming 25 years per generation), F = 1.2, g = 10, m = 0.09, p = 1 (only male migrants), Nancestral = NAfrican = Nnon-African = 10,000. This baseline model is intriguing in that is closely matches all the main features of the data that we set out to capture: Autosomal FST is 0.148, chromosome X FST is 0.220, with the derived Q ratio being 0.605, and the sequence diversity ratio T is 0.666, all within the confidence intervals observed in our data (table 1).
Table 1.
Simulation Results of Sex-Asymmetric Migration Model.
| Parameter Valuesa | Autosomal FST | Chromosome X FST | FST-Based Ratio Q | Sequence Diversity–Based Ratio T |
| Estimates from data (95% confidence interval) | 0.144 (0.138–0.149) | 0.221 (0.203–0.239) | 0.582 (0.524–0.640) | 0.635 (0.587–0.683) |
| Baseline model | 0.148 | 0.220 | 0.605 | 0.666 |
| p = ½ | 0.148 | 0.181 | 0.787 | 0.742 |
| p = 0.8 | 0.148 | 0.201 | 0.686 | 0.701 |
| NAfrican = 20,000 | 0.128 | 0.197 | 0.590 | 0.660 |
| t = 1,600 | 0.119 | 0.189 | 0.574 | 0.658 |
| NAfrican = 20,000, t = 1,600 | 0.104 | 0.172 | 0.552 | 0.657 |
| t = 110 | 0.058 | 0.120 | 0.449 | 0.646 |
| m = 0.12 | 0.128 | 0.192 | 0.612 | 0.686 |
| m = 0.06 | 0.186 | 0.264 | 0.621 | 0.641 |
| g = 13 | 0.130 | 0.194 | 0.611 | 0.684 |
| g = 7 | 0.182 | 0.259 | 0.618 | 0.642 |
| g = 20, m = 0.046 | 0.146 | 0.218 | 0.606 | 0.667 |
| g = 400, m = 0.0023 | 0.138 | 0.210 | 0.594 | 0.668 |
| g = 1,000, m = 0.00094 | 0.123 | 0.194 | 0.575 | 0.672 |
| g = 1, m = 0.5 | 0.205 | 0.283 | 0.634 | 0.633 |
| F = 1.5 | 0.154 | 0.228 | 0.603 | 0.659 |
| F = 0.9 | 0.142 | 0.210 | 0.612 | 0.675 |
| Twice male generation time | 0.148 | 0.233 | 0.564 | 0.641 |
Values of parameters that differ from the baseline values of t = 2,400, Nancestral = NAfrican = Nnon-African = 10,000, F = 1.2, m = 0.09, g = 10, and p = 1.
A striking feature of this model is the severity of the bottleneck associated with the out-of-Africa dispersal, with F > 1 associated with it, which is equivalent to Ne < 50 on average during that period of time. However, the extensive gene flow immediately following the bottleneck masks most of the effect of the bottleneck on FST and sequence diversity. The combined effect of the modeled bottleneck and ensuing gene flow, which gives rise to the amount of genetic drift associated with the out-of-Africa dispersal, is consistent with previous estimates (Pluzhnikov et al. 2002; Adams and Hudson 2004; Marth et al. 2004; Voight et al. 2005; Keinan et al. 2007). Indeed, the model accurately captures the moderate level of autosomal FST observed between West African and non-African populations today, as well as all other summary statistics of our data (table 1). Other models that do not assume the bottleneck and migration events to be sequential are not less plausible; for instance, the bottleneck and migration can be part of the same event according to which the founder population receives migrants as it travels away from the ancestral population, perhaps with diminishing migration rate over time and distance.
We carried out a series of simulations with different parameter values to explore how they affect the different ratios of X chromosomal-to-autosomal effective population size, as well as the effective population size of each of these compartments of the genome. The next five sections explore the effect of varying model parameters on the model’s predictions, and the simulations results are summarized in table 1 in the same order as they are described in the text. Specifically, we explored whether sex-asymmetric migration is a mandatory feature for the model to explain the results, how varying the different sex-symmetric demographic parameters varies model predictions, how long the genetic signatures of X-autosome asymmetries are predicted to last, and whether the primarily male gene flow could potentially have occurred in a single migration event. Following these five sections, we turn to explore alternative sex-biased demographic events that can also reduce the effective population size of chromosome X relative to that of the autosomes.
Sex-Asymmetric Migration Is Necessary to Reduce Chromosome X Effective Population Size
To verify that symmetric migration between the genders cannot produce the observed X-to-autosome ratios, we kept all model parameters in the simulations as in the baseline model with the exception of p, which we set to 0.5 to specify an equal number of male and female migrants. This scenario leads to Q = 0.787 and a sequence diversity ratio of T = 0.742 (table 1), close to the prediction of ¾. These results suggest that in the context of this model and the set of parameter values we considered, primarily male migration is a necessary feature in explaining the observed X-autosome asymmetries. Even the slight deviations from ¾ observed when setting p = 0.5 are expected. The sequence diversity ratio is expected to be slightly below ¾ because a bottleneck reduces this ratio (Pool and Nielsen 2007). The allele frequency differentiation ratio Q deviates from ¾ because the theoretical assumptions underlying the estimation of effective population size from FST require an instantaneous population split (Keinan et al. 2008), and this assumption is violated by the gene flow incorporated into the model.
We further explored the effect of including female migrants (p < 1). As shown in table 1 for the case of p = 0.8, we found that the sequence diversity ratio T is higher than in the case of p = 1 and that chromosome X FST is lower, resulting in a higher Q ratio and a poorer fit to our data. For this model of sex-asymmetric migration to explain our data, the migration would have to be heavily male, with female migration weakening both types of signals.
African Population Expansion and a More Recent Out-of-Africa Event
To examine the effect of an expansion in population size on the African lineage, we increased NAfrican from 10,000 to 20,000, while keeping all other model parameters as in the baseline model, including a population size of 10,000 in the ancestral population before population split. The larger African effective population size results in an even more reduced frequency differentiation ratio of Q = 0.590 because a larger proportion of the genetic drift following population split is now being attributed to the non-African branch (table 1). This more closely matches the Q ratio observed in our data, while the sequence diversity ratio does not change much (0.660). However, the values that this model predicts for FST are lower than in the model without expansion and do not match the data. This result by no means rules out the West African population growth reported by several studies (Pluzhnikov et al. 2002; Adams and Hudson 2004; Keinan et al. 2007; Cox et al. 2009) but rather only serves to quantify the effect of a fairly recent and extreme growth event on the model under investigation.
We similarly explored the effect of a more recent out-of-Africa event by changing the population split time in the model from t = 2,400 to t = 1,600 (corresponding to 40,000 years assuming 25 years per generation), which is consistent with some archaeological estimates of the age of the out-of-Africa dispersal. The effect is similar to that of an African population expansion because a larger proportion of the overall genetic drift following population split is now attributed to the out-of-Africa dispersal itself, with Q = 0.574, T = 0.658, and low FST values (table 1). Combining NAfrican = 20,000 with t = 1,600 results in Q = 0.552 and T = 0.657, both of which are still within the ranges observed in our data; however, this model too suffers from too low FST values (table 1).
How Long Are the Signatures of X-Autosome Asymmetries Expected to Last?
To understand how large an effect primarily male migration can have on the X-to-autosome ratios in the context of this model, we kept all model parameters as in the baseline model except for t, which we set to capture sampling the African and non-African populations immediately after the end of the gene flow, thereby reflecting the magnitude of the genetic signature as soon as it is established (t = 110). This model predicts ratios of T = 0.646 and Q = 0.449 (table 1), further highlighting the different nature of these two summary statistics.
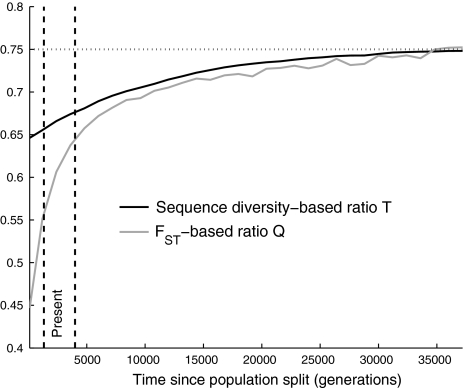
To explore how long the X-to-autosome ratios are expected to be affected by primarily male migration more generally, we simulated the model with varying t corresponding to different times after the out-of-Africa event between immediately after the event as above (t = 110) and about a million years after (t = 40,000 generations). Both the Q and the T signals are expected to be strong for at least a hundred thousand years, and both practically vanish by a million years after the out-of-Africa event (fig. 2). These results also emphasize how the allele frequency differentiation–based effective population size ratio Q is affected by sex-asymmetric migration differently from the sequence diversity–based effective population size ratio T. They show that the difference between the two is larger the more recent the event (fig. 2).
FIG. 2.
Signatures of X-autosome asymmetry decay over tens of thousands of generations after primarily male migration. The figure shows the Q and T ratios as a function of varying the time since the dispersal out of Africa t within the model of primarily male migration. A range of times consistent with the split of Africans and non-African populations is illustrated by the vertical band (40,000–80,000 years ago of 20–30 years per generation; the baseline model discussed in the main text is at the middle of that range). The signals captured by the two different ratios exhibit different behavior with time, but both are predicted to be very strong at present time and to practically vanish by 35,000 generations (∼875,000 years) after the event. (The horizontal dotted line provides the ¾ prediction of a sex-symmetric history of constant Ne.)
Could the Male Migration Have Occurred All in a Single Generation?
We next examined how varying the level of gene flow during the out-of-Africa dispersal affects the prediction of the model. Increasing the amount of gene flow by increasing either the per-generation rate m or the number of gene flow generations g has the expected effect of decreasing both autosomal and chromosome X FST (table 1; m = 0.12, g = 13), and decreasing the amount of gene flow has the opposite effect (table 1; m = 0.06, g = 7). Whereas the sequence diversity ratio T monotonically increases with the amount of gene flow, Q increases with either an increase or a decrease of the amount of gene flow, suggesting that with this specific choice for the values of other parameters, the greatest reduction in Q occurs at a migration level close to that specified by the baseline model.
We found that the effect of parameters m and g is to some extent interchangeable because the probability that a locus at the end of the gene flow epoch has arrived in the population via a migrant is 1 − (1 − m)g. For example, a model with g = 20 and m = 0.046 provides nearly identical predictions to those of the baseline model with g = 10 and m = 0.09 (table 1). We also considered much weaker yet extended gene flow: g = 400 and g = 1,000 (about 10,000 and 25,000 years), while again keeping 1 − (1 − m)g fixed. While these models predict somewhat lower FST than empirically observed on both chromosome X and autosomes, they too provide Q and T ratios that are pretty similar to the baseline model (table 1).
The same predictions as the baseline model are also met by a model with extreme gene flow in a single generation, g = 1 and m = 0.9, which gives rise to the same 1 − (1 − m)g as in the baseline model. However, because only males migrate according to this model, the rate of gene flow in any generation is restricted to m ≤ 0.5, with m = 0.5 being equivalent to replacing 50% of the autosomes and 33% of chromosome X in a single generation. This most extreme possible model of only male migrants replacing all the existing male gene pool in the destination population in a single generation (g = 1, p = 1, m = 0.5) provides a decidedly poor fit to the FST values observed in the data (table 1). We conclude that a single migration event in one generation, however extreme, is unlikely to explain our data in the context of this model. Rather, the modeled primarily male migration must have occurred in multiple waves over generations.
Varying the Intensity of the Out-of-Africa Bottleneck
The role of the very severe out-of-Africa bottleneck in this model is to give rise to an extreme reduction in genetic diversity, some of which is then regained via the migrants. We modeled the bottleneck and migration events as sequential, which results in the intensity of the bottleneck itself being assumed to be very high (F = 1.2). However, as discussed above, the very high level of gene flow masks most of the effect of the bottleneck. As a consequence, varying the bottleneck’s intensity by 25% to either F = 1.5 or F = 0.9 has a minor effect on model predictions (table 1). As expected, we found that increasing F increases both autosomal and X chromosomal FST values and decreases both ratios further below ¾, although not by much, and decreasing F has the opposite effect on all predictions (table 1).
Combining Male Migration with Other Sex-Biased Processes
Although the suggested model fits the data and could in principle account for the phenomena we sought to explain, other parameter values and other models can provide a better fit to our results. Furthermore, there might be other historical and selective processes that when combined with the scenario of primarily male migration can better produce the patterns we observe in real data, and we explore some of these processes in the following. Although we show that the historical processes below are not by themselves sufficient to explain the full magnitude of the signal observed in the data, they might play a role together with primarily male migration in accounting for the results. Similarly, although we did not observe empirical evidence for the possibility of natural selection explaining our results in various tests that we performed (Keinan et al. 2008), it remains possible and even likely that the effects of natural selection have synergized with primarily male migration in reducing the relative effective population size of chromosome X.
One sex-biased demographic force of particular interest to consider is an increase in the male-to-female generation length associated with the dispersal out of Africa (Charlesworth 2001). Because females carry ⅔ of all copies of chromosome X, a relative shorter span between female generations would result in a relative increase in the genetic drift of chromosome X during this period, corresponding to a decrease in its effective population size. Such a transient change in generation time, perhaps due to a shift in social structure, can in principle account for the observed reduction in chromosome X effective population size in that epoch of human history. Although this force by itself cannot explain the magnitude of the results we observed (Materials and Methods), it could compound the effects of primarily male migration. Indeed, a relative increase in male generation time might be expected in a period of primarily male migration because in a population with an excess of males over females, females might elect to start bearing children at a younger age, whereas successful males might be expected to be of higher social status and thus older.
To model this quantitatively, we assumed a rather extreme scenario in which during the dispersal out of Africa, average male generation time was double that of female. This would correspond to, for example, an average of 20 for the age at which females had offspring and an average of 40 for the age at which males had offspring. Our simulations show that under this modified scenario, chromosome X FST changes from 0.220 to 0.233 and Q changes from 0.605 to 0.564. The sequence diversity ratio T decreases, though less considerably, from 0.666 to 0.641 (table 1). These changed predictions of our simulations are still within errors of the observed values and provide even better fit to both ratios, suggesting that a historical scenario of change in generation time is consistent with the model and could contribute to our results. Other sex-biased demographic forces—reducing the effective population size of chromosome X relative to the autosomes—that cannot account for the magnitude of the signal we observed, but can still play some part in explaining it, include effective polyandry leading to higher variability in female than male reproductive success, as well as female out-of-Africa migrants being more closely related to each other than male migrants.
Can Population Substructure with Primarily Females Migrating between Groups Explain the Observations?
An alternative explanation for a reduction of Ne on chromosome X during the dispersal out of Africa is that the founding population of non-Africans was substructured with more females than males migrating between groups (Seielstad et al. 1998; Laporte and Charlesworth 2002). Such a sex-biased population structure would result in higher male Ne and hence lower chromosome X Ne relative to the autosomes. To test the effect such a force would have on X-autosome asymmetries, we considered a model in which the founder population dispersing from Africa consists of two distinct equal-sized groups, with only females migrating between groups, for the entire time after the split of West Africans and non-Africans and before the North European–East Asian split. We simulated this model for different levels of female migration rate between the two groups. Supplementary figure S1 (Supplementary Material online) presents Q as a function of female migration rate, showing that no level of migration rate can account for the magnitude of the signal we observed in our data. The X-to-autosome ratio of effective population size decreases as a function of female migration rate up to a certain point, after which it increases back to its expectation for no migration, because the population becomes practically panmictic (Supplementary figure S1). The ratio never drops below 0.7, while its observed value is 0.582, suggesting that population structure alone cannot explain our empirical observations.
Conclusions
In this study, we demonstrated that a model of primarily male migration during the human dispersal out of Africa can explain the different observations of relatively reduced chromosome X effective population size in non-African populations that we and others have previously made (Keinan et al. 2008; Amato et al. 2009). Although we cannot rule out nondemographic explanations for our observations—natural selection differently affecting chromosome X and the autosomes during the dispersal out of Africa could also explain the observations (Keinan et al. 2008)—we have demonstrated in this study that sex-biased migration can by itself generate a reduction in chromosome X effective population size that is as extreme as observed. Moreover, we showed that the range of alternative demographic scenarios we explored cannot explain the data.
We considered a simplified model of joint African/non-African history, in which the initiation of the dispersal out of Africa, during which the dispersing population maintained relatively little genetic variability, was followed by a period of migration into the dispersing population. If more male than female migrants survived and contributed to the gene pool of the dispersing population (the founder population of non-Africans), there could have been a pronounced reduction of chromosome X effective population size relative to the autosomes. Such male-heavy migration can be envisioned as daughters in the founder population of non-Africans bearing offspring with new immigrant males more often than the converse. We showed that this model with a set of parameters that match known features of human demographic history, together with extensive gene flow, quantitatively predicts the asymmetry in effective population size as measured both via non-African sequence diversity and via allele frequency differentiation between Africans and non-Africans. As our simulation studies did not perform an exhaustive search for parameter values that best fit the data, it is plausible that a wide range of parameter values produces similar predictions and even provides a better fit to our results. It is also important to realize that the model we simulated is only a simple example of a larger family of complex models of primarily male migration, all of which could in principle be consistent with history. This study has pointed to one such model and explored in detail how its predictions vary with variation in parameters’ values.
Primarily male migration is only one possible explanation of a relatively increased autosomal effective population size. We considered a variety of other sex-biased demographic events and also did not rule out natural selection affecting chromosome X differently than the autosomes as an alternative explanation. However, primarily male migration is the most parsimonious explanation for the results that we identified. Furthermore, although it should be treated as a speculative hypothesis pending further lines of evidence, and although other sex-biased historical events can play a part in explaining the observed asymmetry between autosomes and chromosome X when combined with primarily male migration, the general phenomenon of sex-biased migration is well attested to in the literature. According to anthropological studies of hunter-gatherers, female migration dominates at short distances, which is consistent with many results based on contrasting mitochondrial DNA with Y chromosome (Cavalli-Sforza et al. 1994; Seielstad et al. 1998; Kayser et al. 2003). However, male migration in hunter-gatherers dominates at longer distances (Kayser et al. 2003; Marlowe 2005), consistent with our results as well as with archaeological data; for example, in Neolithic archaeological sites across the Levant, arrowheads are uniform in style over a wide geographic range, while domestic tools vary greatly, suggesting more long-range male migration (Bar-Yosef and Belfer-Cohen 1989). Genetic data also provide other striking examples of male-biased migration events at longer distances (Parra et al. 1998; Helgason et al. 2000; Bamshad et al. 2001; Thomas et al. 2006). A modern instance is that of Colombians from Medellin who have about 70% European ancestry despite having essentially no European female ancestors, which is a consequence of multiple waves of male migration (Bedoya et al. 2006), similar to what we propose for the dispersal out of Africa. A priority for future work should be to contextualize these genetic results in light of the archaeological record. Under the hypothesis of primarily male migration, the founding population of non-Africans must have been localized to a constrained geography long enough to have received the migrants. If the population quickly dispersed throughout Eurasia, we would not see the same pattern in North Europeans and East Asians.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (grant number U01 HG004168) and by a Burroughs Wellcome Career Development Award in the Biomedical Sciences to D.R. We thank A. Clark, K. Lohmueller, and S. Musharoff for comments on earlier versions of this manuscript.
References
- Adams AM, Hudson RR. Maximum-likelihood estimation of demographic parameters using the frequency spectrum of unlinked single-nucleotide polymorphisms. Genetics. 2004;168:1699–1712. doi: 10.1534/genetics.104.030171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato R, Pinelli M, Monticelli A, Marino D, Miele G, Cocozza S. Genome-wide scan for signatures of human population differentiation and their relationship with natural selection, functional pathways and diseases. PLoS One. 2009;4:e7927. doi: 10.1371/journal.pone.0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, et al. (18 co-authors) Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.173301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yosef O, Belfer-Cohen A. The Levantine “PPNB” interaction sphere. In: Hershkovitz I, editor. People and culture in change. Oxford: British Archaeological Reports; 1989. pp. 59–72. [Google Scholar]
- Bedoya G, Montoya P, Garcia J, et al. (11 co-authors) Admixture dynamics in Hispanics: a shift in the nuclear genetic ancestry of a South American population isolate. Proc Natl Acad Sci U S A. 2006;103:7234–7239. doi: 10.1073/pnas.0508716103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41:8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero A. On the effective size of populations with separate sexes, with particular reference to sex-linked genes. Genetics. 1995;139:1007–1011. doi: 10.1093/genetics/139.2.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton (NJ): Princeton University Press; 1994. [Google Scholar]
- Charlesworth B. The effect of life-history and mode of inheritance on neutral genetic variability. Genet Res. 2001;77:153–166. doi: 10.1017/s0016672301004979. [DOI] [PubMed] [Google Scholar]
- Cox MP, Morales DA, Woerner AE, Sozanski J, Wall JD, Hammer MF. Autosomal resequence data reveal Late Stone Age signals of population expansion in sub-Saharan African foraging and farming populations. PLoS One. 2009;4:e6366. doi: 10.1371/journal.pone.0006366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Sigureth ardottir S, Nicholson J, Sykes B, Hill EW, Bradley DG, Bosnes V, Gulcher JR, Ward R, Stefansson K. Estimating Scandinavian and Gaelic ancestry in the male settlers of Iceland. Am J Hum Genet. 2000;67:697–717. doi: 10.1086/303046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayser M, Brauer S, Weiss G, Schiefenhovel W, Underhill P, Shen P, Oefner P, Tommaseo-Ponzetta M, Stoneking M. Reduced Y-chromosome, but not mitochondrial DNA, diversity in human populations from West New Guinea. Am J Hum Genet. 2003;72:281–302. doi: 10.1086/346065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat Genet. 2007;39:1251–1255. doi: 10.1038/ng2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2008;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labuda D, Lefebvre JF, Nadeau P, Roy-Gagnon MH. Female-to-male breeding ratio in modern humans-an analysis based on historical recombinations. Am J Hum Genet. 2010;86:353–363. doi: 10.1016/j.ajhg.2010.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte V, Charlesworth B. Effective population size and population subdivision in demographically structured populations. Genetics. 2002;162:501–519. doi: 10.1093/genetics/162.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marlowe FW. Hunter-gatherers and human evolution. Evol Anthropol. 2005;14:54–67. doi: 10.1016/j.jhevol.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra EJ, Marcini A, Akey J, et al. (11 co-authors) Estimating African American admixture proportions by use of population-specific alleles. Am J Hum Genet. 1998;63:1839–1851. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluzhnikov A, Di Rienzo A, Hudson RR. Inferences about human demography based on multilocus analyses of noncoding sequences. Genetics. 2002;161:1209–1218. doi: 10.1093/genetics/161.3.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. Population size changes reshape genomic patterns of diversity. Evolution Int J Org Evolution. 2007;61:3001–3006. doi: 10.1111/j.1558-5646.2007.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. The impact of founder events on chromosomal variability in multiply mating species. Mol Biol Evol. 2008;25:1728–1736. doi: 10.1093/molbev/msn124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, et al. (11 co-authors) Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- Storz JF, Payseur BA, Nachman MW. Genome scans of DNA variability in humans reveal evidence for selective sweeps outside of Africa. Mol Biol Evol. 2004;21:1800–1811. doi: 10.1093/molbev/msh192. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Stumpf MP, Harke H. Evidence for an apartheid-like social structure in early Anglo-Saxon England. Proc Biol Sci. 2006;273:2651–2657. doi: 10.1098/rspb.2006.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight BF, Adams AM, Frisse LA, Qian Y, Hudson RR, Di Rienzo A. Interrogating multiple aspects of variation in a full resequencing data set to infer human population size changes. Proc Natl Acad Sci U S A. 2005;102:18508–18513. doi: 10.1073/pnas.0507325102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.