Abstract
One type of RNA editing converts adenosine residues to inosine in double-stranded regions. Recent transcriptome analysis has revealed that numerous Alu repeats, present within introns and untranslated regions of human transcripts, are subject to this A → I RNA editing. Furthermore, it revealed global transcription of antisense RNAs. Here, we demonstrate that antisense RNAs are also edited extensively but only in their Alu repeat sequences, and editing does not extend to the surrounding sequence. Our findings imply that sense and antisense RNAs form two separate intramolecular double-stranded RNAs consisting of inversely oriented Alu repeats, but rarely form intermolecular duplexes.
Keywords: Antisense RNA, ADARs, Alu repeats, RNA editing, Human transcriptome
1. Introduction
RNA editing refers to a post-transcriptional modification of the base sequence of pre-mRNA, and is recognized as a genetic mechanism for generating RNA and protein diversity [1–4]. Adenosine deaminases that act on RNA (ADARs) are involved in the mechanism of RNA editing that specifically modifies adenosine residues of double-stranded RNAs (dsRNAs) to inosine (A → I RNA editing). Three distinct ADAR genes (ADAR1–3) have been identified in mammals [1–4]. ADARs recognize and edit specific adenosine residues through their interaction with the complete or incomplete dsRNA structure formed between the editing site located within an exon sequence and its complementary sequence usually located in an intron sequence [5–7]. RNA editing of protein-coding sequences could alter codons and functions of target genes [5–7]. Although several cases of A → I editing of protein-coding RNA targets has been identified, bioinformatics analysis of human mRNA and EST (expressed sequence tags) databases revealed a large number of A → I RNA editing sites in non-coding intron and untranslated-region RNA sequences [8–11]. Surprisingly, a majority of editing sites (more than 10000 sites mapped in ~2000 different genes) resides within repetitive sequences, such as Alu and LINE [8–11]. Alu repeats, comprising more than 10% of the human genome, are about 300 nucleotides in length and have relatively high homology among subfamilies [12,13]. An Alu element is likely to form an intramolecular RNA duplex with a nearby inverted Alu repeat sequence, which then could serve as a substrate for A → I RNA editing. In agreement with this prediction, the extent of RNA editing in Alu repeats depends on the distance between two inverted Alu repeats [9,11].
Recent transcriptome analysis indicates that a large fraction ( ≥ P70%) of human transcripts are derived from non-coding antisense strand DNA sequences [14–17]. Among naturally occurring antisense RNA transcripts, cis-encoded antisense RNAs are transcribed from the opposite strand of the same genomic locus and frequently have a long and perfect complementarity to the sense transcript. Notably, sense–antisense RNA pairs may be coexpressed more frequently than the rate expected by pure chance [16], leading to the hypothesis that human genes are regulated by antisense transcripts [14–16]. Although there are a few reported cases of sense–antisense intermolecular duplex formation and consequent regulation of the sense mRNA expression [18], it is not known how frequently coexpression of sense and antisense transcripts and formation of an intermolecular dsRNA occur.
In this study, we demonstrate the extensive editing of cis-encoded antisense transcripts coexpressed with their sense strand RNAs in human lung and NT2-N neurons and, most importantly, that antisense RNA editing occurs intramolecularly in the limited regions containing Alu sequences, as predicted recently by bioinformatics analysis of the human antisense transcriptome [19]. In addition, our in vitro studies confirmed that cis-encoded sense and antisense RNAs remain single-stranded except the region containing Alu repeat sequences. Our results imply that intermolecular pairing of sense and antisense RNAs is a rare occurrence. Formation of intramolecular dsRNA structure made of Alu repeats may function in some cases to prevent formation of the intermolecular RNA duplex between sense and antisense RNAs that could have detrimental effects on processing and expression of sense mRNAs [18].
2. Materials and methods
2.1. Quantification of RNA editing
Maintenance of NT2 cells and their in vitro induction to NT2-N neurons were described previously [20]. Total RNA extracted from human adult lung was obtained from Clontech (Palo Alto, CA). 1 µg of total RNA (human adult lung or NT2-N neurons) was first treated with 1.0 U of DNase I (Invitrogen Corp., Carlsbad, CA) in 7 µl of the DNase I reaction mixture at 37°C for 15 min. DNase-treated RNA was denatured along with 1 µl of 10 µM gene-specific primer (Figs. 1A, 2A and Table 1) and 2 µl of 10 mM dNTP mix at 80°C for 5 min. Denatured RNA was mixed with 4 µl of 5× cDNA Synthesis Buffer, 1 µl of 0.1 M DTT, 40 U of RNaseOUT and 15 U of Thermo-Script (Invitrogen). First-strand cDNA was synthesized at 55°C for 60 min and the reaction was terminated by incubating at 85°C for 5 min, followed by incubation at 37°C for 20 min with 2 U of RNase H.
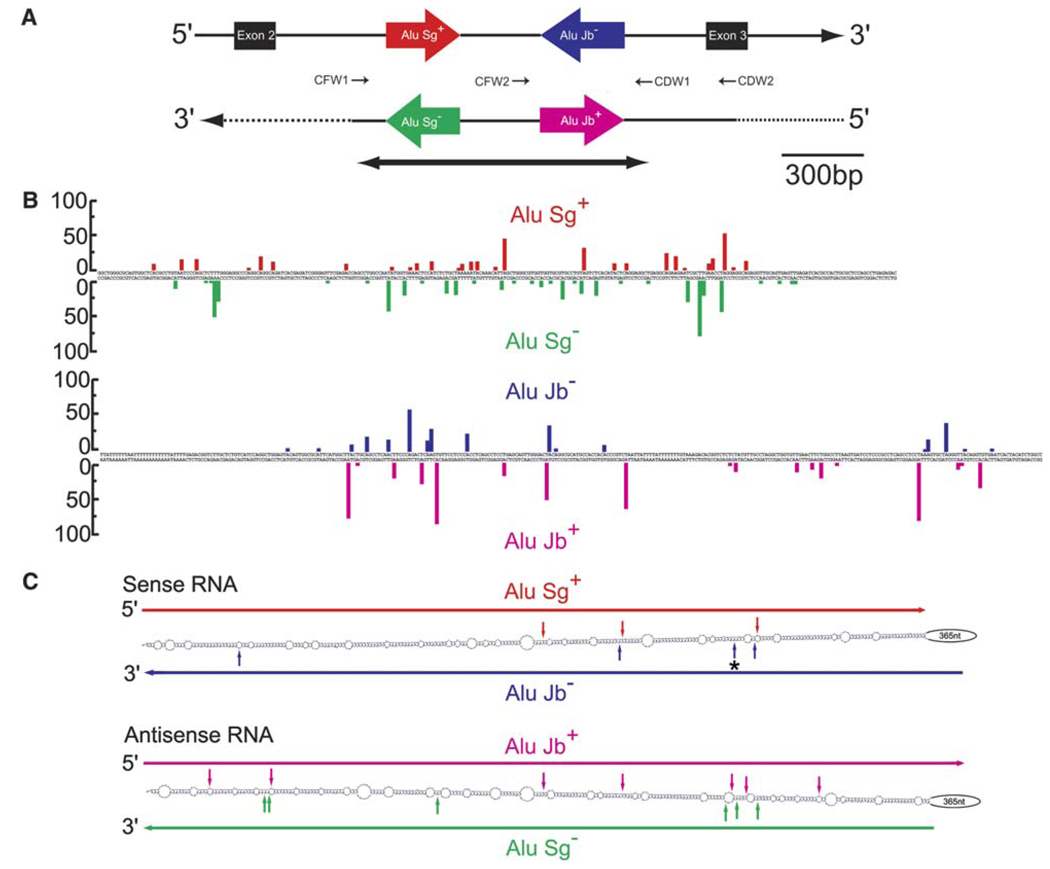
Fig. 1.
Sense and antisense Alu editing in intron 2 of CNNM3. (A) Two inverted Alu repeats present in intron 2 of CNNM3 are shown as red and blue arrows. Antisense RNA corresponding to this region detected in this study is indicated also. The Alu repeats of the antisense RNA are presented as green and pink arrows. The small arrows indicate the RT-PCR primers, their directions, and their relative positions. The bold line with inverted arrows encompasses the region that is transcribed to prepare sense and antisense CNNM3 intron 2 RNAs examined for in vitro RNA editing assay. (B) A → I editing of Alu repeats. The editing frequency (%) at individual sites identified within the entire Alu Sg (262 bp; upper panel) and Alu Jb sequences (309 bp; lower panel) in sense (upper side) and antisense (lower side) RNA is summarized. (C) The secondary structure between two inverted Alu repeats in sense (upper) and antisense (lower) RNA is calculated by MFOLD. Highly edited sites (>30%) are indicated by vertical solid arrows. The blue arrow marked*: the site detected in vivo but not edited in vitro by recombinant ADAR proteins (see Fig. 3).
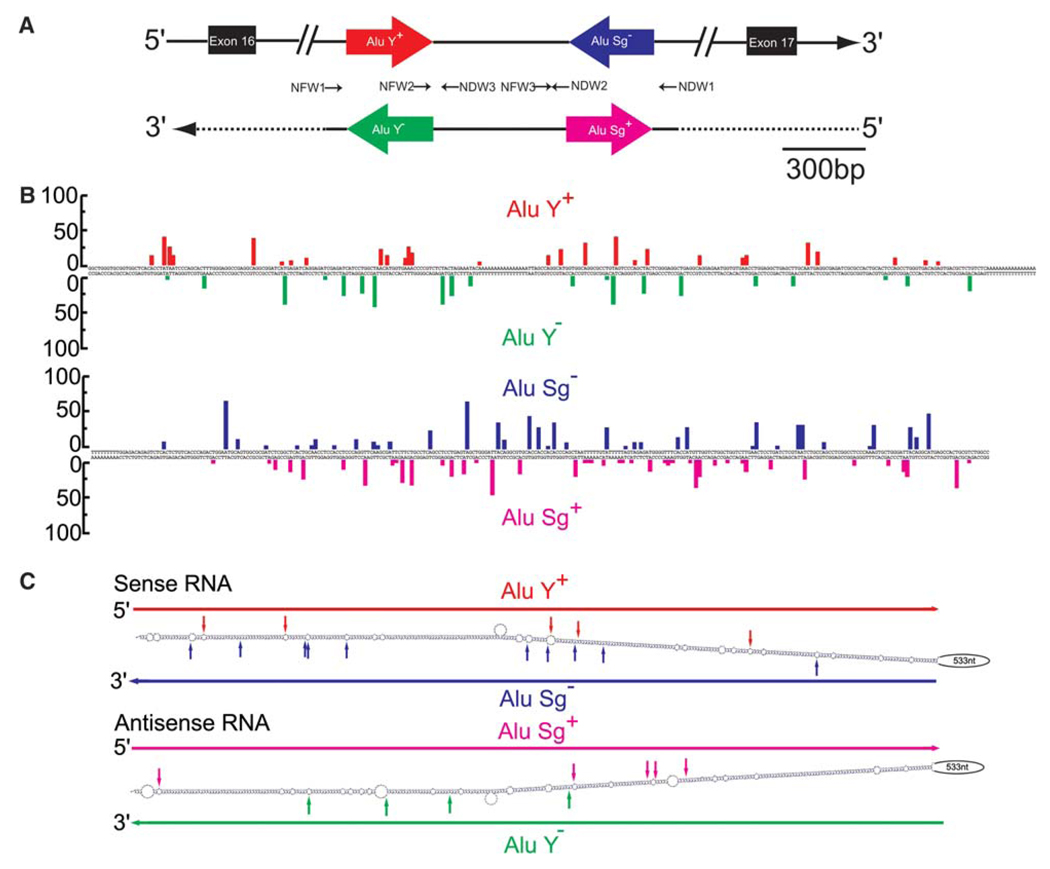
Fig. 2.
Sense and antisense Alu editing in intron 16 of NFκB1. (A) Two inverted Alu repeats present in intron 16 of NFκB1 are shown as red and blue arrows. Antisense RNA corresponding to this region detected in this study is indicated also. The Alu repeats of the antisense RNA are presented as green and pink arrows. The small arrows indicate the RT-PCR primers, their directions, and their relative positions. (B) A → I editing of Alu repeats. The editing frequency (%) at individual sites identified within the entire Alu Y (306 bp; left panel) and Alu Sg sequences (290 bp; right panel) in sense (upper side) and antisense (lower side) RNA is summarized. (C) The secondary structure between two inverted Alu repeats in sense (upper) and antisense (lower) RNA is calculated by MFOLD. Highly edited sites (>30%) are indicated by vertical solid arrows.
Table 1.
Sequences of primers used in this study
| Oligonucleotide sequence | |
|---|---|
| CFW1 | 5′-TCATGGGCCTTTCCCAGTCTT-3′ |
| CFW2 | 5′-TCTTGAGACTGCCATAGGCATG-3′ |
| CDW1 | 5′-GGTGACTATCTGCCTTGGAGCT-3′ |
| CDW2 | 5′-AACTCCTCCTTCCGCTTCAGA-3′ |
| NFW1 | 5′-GGAGAGATGGTTTGCTGGTTCA-3′ |
| NFW2 | 5′-TTGCAATGAGGCGAGATCG-3′ |
| NFW3 | 5′-CAGTCCTCTTTTCAAACCCTGC-3′ |
| NDW1 | 5′-TGCCCATTAATCAGACTGGTGG-3′ |
| NDW2 | 5′-CTGTTGTGCTGTGTCCTTTGC-3′ |
| NDW3 | 5′-GGTCATTTTCCCCTATCAG-3′ |
The resultant cDNA was then amplified by PCR in a reaction mixture of 50 µl containing 200 nM of each primer (Figs. 1A, 2A and Table 1), 1 mM dNTP Mix, 5 µl of 10 × PCR buffer, and 1 µl of Advantage 2 Polymerase mix (Clontech). The PCR amplification began with a 1-min denaturation step at 95°C, followed by 35 cycles of denaturation at 95°C for 10 s, annealing at 64°C for 30 s, and extension at 68°C for 60 s. After gel purification, PCR products were subcloned using the TOPO TA cloning kit (Invitrogen), and then the editing frequency was determined by sequencing from both directions more than 24 individual clones containing appropriately sized inserts. Because the frequency of mismatches other than A → G changes between the genomic sequence and the cDNA sequences cloned were less than 4.0%, only adenosine residues that showed A → G changes with >4.0% frequency were accounted as A → I editing sites.
2.2. In vitro RNA editing assay
Plasmid pBS-CNNM3 contains a 1092-base-pair (bp) fragment of a partial CNNM3 intron 2 including two inverted repeats of Alu sequences (Fig. 1A). The DNA fragment was PCR-amplified using human genomic DNA and PCR primers Bam-CFW3 (5′-CATCCGGATCCCCCTAGGTGCTTTTGTTTTTGC-3′) and Xho-CDW3 (5′-ATAAGAATCTCGAGCTTGGAGCTGGCTGAAGAGAAT-3′). Bam-CFW3 contains a BamHI recognition site (underlined), and Xho-CDW3 contains an XhoI recognition site (underlined). The PCR products were digested with BamHI and XhoI, then inserted into pBlue-script II KS vector (Stratagene, La Jolla, CA) linearized with the same restriction enzymes.
pBS-CNNM3, linearized with XhoI orBamHI, was transcribed with T7 or T3 RNA polymerase (20 U) in the presence of a trace amount of [α-32P] UTP at 37°C for 60 min to synthesize the sense or antisense CNNM3 intron 2 RNA, respectively, as described previously [21].
An equal amount of the sense and antisense CNNM3 intron 2 RNA (2 or 20 fmol each) was mixed in 4 µl of water with or without pre-incubation at 80°C for 5 min followed by rapid chill on ice for 5 min. The sense and antisense RNA mixture (0.02 or 0.2 nM) was then subjected to in vitro editing at 30°C for 60 min with 50 ng of recombinant ADAR1 (p110) or ADAR2 protein as described previously [21]. Quantification of RNA editing frequency was done as already described above.
3. Results and discussion
Recent transcriptome analysis revealed global expression of cis-encoded antisense strand RNAs [14–17]. However, formation of sense–antisense intermolecular RNA duplexes and regulation of sense RNA expression have been a subject of debate [15,18,19]. Any long RNA duplexes such as those formed between sense and antisense RNA pairs, if any, would be extensively edited by ADARs, leading to conversion of >50% of their adenosine residues to inosine along the entire dsRNA region [20,22]. However, editing would be restricted to intramolecular dsRNA regions, e.g., Alu inverted repeats, if they did not form such intermolecular duplexes.
We therefore examined experimentally two randomly selected human intronic sequences for expression of cis-encoded antisense RNAs, if any, and A → I editing patterns of sense and antisense RNAs. We chose cyclin M3 (CNNM3; Fig. 1A) and nuclear factor κB subunit 1 (NFκB1; Fig. 2A) genes. The presence of oppositely oriented Alu repeats in intron 2 of CNNM3 and intron 16 of NFκB1 sense transcripts and their A → I editing was indicated by bioinformatics studies [8,9]. We first conducted reverse transcription (RT) of total RNA extracted from human adult lung with an RT primer corresponding to the sequence complementary to a part of CNNM3 intron 2 (CDW1, Fig. 1A and Table 1). Using a set of PCR primers (CFW1 and CDW1), we followed this by PCR amplification of the intron 2 sequence containing both inverted repeats of Alu sequences. The PCR products containing the Alu repeats were subcloned to quantify the editing frequency by sequencing multiple cDNA clone isolates. Extensive A → I RNA editing (up to 60% at some sites) was detected within the Alu repeats (Fig. 1B), and no editing was detected in the region outside the Alu repeats (data not shown), as predicted by bioinformatics studies [8]. Similarly, we conducted RT for intron 16 of NFκB1 with an RT primer corresponding to the complementary sequence of the region 3′ to the second Alu repeat (NDW1, Fig. 2A and Table 1), followed by PCR amplification of the region containing both Alu repeats with a set of primers (NFW1 and NDW1). However, we could not detect PCR products, possibly due to the stable dsRNA secondary structure formed. Therefore, we divided this region into three partially overlapping sections, and conducted PCR separately using three sets of PCR primer pairs (NFW1 and NDW3, NFW2 and NDW2, or NFW3 and NDW1), which allowed us to determine editing sites and to quantify the editing frequency of sites identified within this region. As predicted previously by bioinformatics studies [9], we once again confirmed that extensive A → I RNA editing was restricted to the Alu sequences of both repeats (Fig. 2B).
We then attempted to detect cis-encoded antisense RNAs corresponding to intron 2 of the CNNM3 gene. PCR products corresponding to antisense strand RNAs encompassing two inverted Alu repeats were detected by using the sense strand primer CFW1 as the RT primer and CFW1 and CDW1 as the PCR primer pairs. We found that both Alu repeats in the antisense transcripts were also extensively edited (up to 87% at hot spots) to an extent similar to those in the sense transcripts (Fig. 1B). Once again, however, A → I RNA editing was limited to the two Alu repeat sequences and did not extend to the surrounding sequences. Similarly, antisense transcripts containing the region corresponding to intron 16 of NFκB1 were detected when RT was carried out with the sense strand primer NFW1 (Fig. 2A). These PCR products must be derived from antisense RNA, because A → G (not T → C) changes of the cDNA sequence were detected in the antisense direction (Fig. 2A and B). These results clearly demonstrate that antisense RNA is also frequently subjected to A → I RNA editing. ADAR modifies almost randomly many adenosine residues to inosine of a long intermolecular dsRNA [20,22]. However, A → I editing of CNNM3 intron 2 or NFκB1 intron 16 was limited to the Alu sequences of both sense and antisense RNAs. Thus, editing must occur intra- but not intermolecularly on the fold-back hairpin structure formed between inverted Alu repeats present in these introns.
The sense and antisense Alu-containing RNAs might be expressed in separate individual cells (cell types) of human lung investigated in the present study. In order to exclude the possibility, we then examined editing of sense and antisense RNAs of CNNM3 in NT2-N neuronal cells. NT2-N cells are postmitotic neurons differentiated fully in vitro from human clonal teratocarcinoma NT2 cells. A → I editing of glutamate receptor subunit mRNAs increases dramatically during terminal differentiation of the clonal neurons [23]. Both sense and antisense Alu-containing RNAs of the CNNM3 intron 2 were detected in NT2-N neurons, and they were highly edited but only in the regions containing Alu repeats (data not shown). It is most likely that both sense and antisense RNAs are expressed simultaneously in individual NT2-N neuronal cells. Our results further support the hypothesis that intermolecular duplex formation of Alu-containing sense and antisense RNAs rarely occurs in vivo.
In order to determine the region transcribed to the antisense CNNM3 RNA, we conducted RT using the sense strand RT primer (specific to the antisense RNA, CFW1) followed by PCR with various primer pairs. Through analysis of editing sites and sequence alterations (A → G changes), we concluded that the PCR product derived from the antisense RNA extends at least from exon 3 to very near the 5′ end of intron 2 of CNNM3 (Fig. 1A). The intramolecular dsRNA structure formed between two inverted Alu repeats, which was predicted by MFOLD, revealed that sites highly edited (>30%) concentrate within several regions (hot spots) (Figs. 1C and 2C). This suggests that the sequence of complementary RNA strand of fold-back hairpin dsRNA structure is also a determinant critical for editing frequency in addition to the neighbor sequence preference previously reported [24].
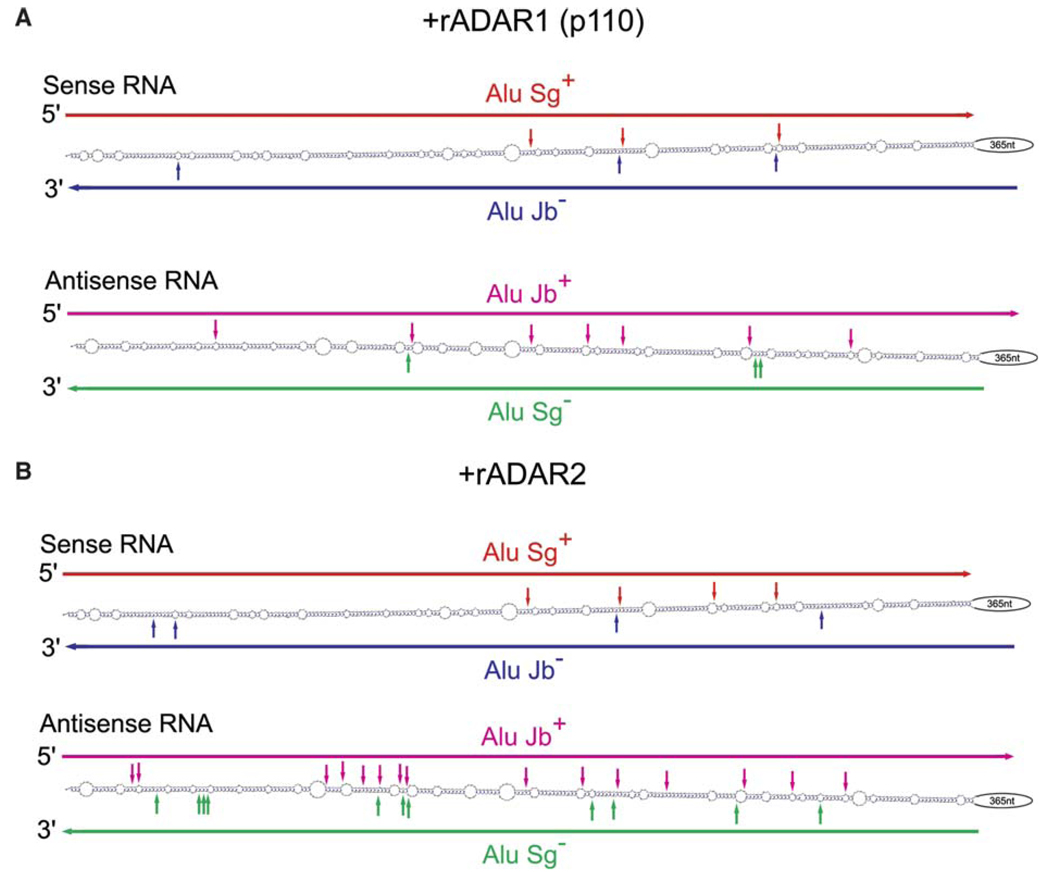
Finally, we investigated in vitro editing of a set of cis-encoded sense and antisense RNAs that contain two inverted Alu repeats (Fig. 3). An equal amount of sense and antisense CNNM3 intron 2 RNAs were mixed together at two concentrations (0.02 or 0.2 nM; some samples were heated and rapidly chilled) prior to in vitro incubation with recombinant ADAR1 (p110) or ADAR2 proteins. Sequence analysis of RT-PCR products derived from in vitro edited CNNM3 intron 2 RNAs revealed that A-to-I editing was once again limited to the Alu sequences. Most importantly, all the hot spots detected in vivo, except for one site in Alu Jb sequences of sense RNA (Fig. 1C), were reproducibly edited also in vitro by either ADAR1 or ADAR2, or both (Fig. 3). These results strongly support our hypothesis that intermolecular duplex formation of Alu-containing sense and antisense RNAs rarely occurs in vivo.
Fig. 3.
In vitro editing of sense and antisense CNNM3 intron 2 RNAs by ADARs. Highly edited sites (>30%) of sense (upper) and antisense (lower) CNNM3 intron 2 RNAs, which were mixed together (0.02 nM) and subjected to in vitro editing with recombinant ADAR1p110 (A) or ADAR2 (B), are indicated by vertical solid arrows. Essentially identical results were obtained at the RNA concentration of 0.2 nM (data not shown).
In the present study, intron 2 of CNNM3 and intron 16 of NFκB1 were chosen arbitrarily, because two inverted Alu repeats of their sense strand transcripts had been predicted to undergo A → I RNA editing [8–11]. In addition to sense strand RNAs, however, we detected antisense strand transcripts derived from these two introns. Human EST clones containing these antisense intronic sequences have not been reported previously, indicating that naturally occurring antisense RNAs containing Alu repeats, such as ones reported in this study, may be transcribed more often than estimated by bioinformatics analysis of human EST databases [15,17]. Furthermore, our present studies demonstrated experimentally that those antisense RNAs rarely form long intermolecular dsRNAs with sense RNAs in human tissues and clonal NT2-N neurons, as predicted by previous bioinformatics studies [15]. Biological functions proposed for antisense RNA at various levels of gene regulation include transcription silencing, alternative splicing, genomic imprinting, and X chromosome inactivation [15]. Since direct duplex formation of antisense transcripts with sense RNAs now appears to be most unlikely, the exact mode and roles of antisense RNAs in these regulatory mechanisms remain to be established.
Acknowledgements
We thank Dr. M.-Y. Lee for NT2-N cells and Dr. J. M. Murray for critical reading of the manuscript. We also thank the Wistar Research Services Department for preparing the manuscript. This work was supported in part by grants from the National Institutes of Health, the March of Dimes, and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
References
- 1.Keegan LP, Gallo A, O’Connell MA. The many roles of an RNA editor. Nat. Rev. Genet. 2001;2:869–878. doi: 10.1038/35098584. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maas S, Rich A, Nishikura K. A-to-I RNA editing: recent news and residual mysteries. J. Biol. Chem. 2003;278:1391–1394. doi: 10.1074/jbc.R200025200. [DOI] [PubMed] [Google Scholar]
- 4.Seeburg PH, Hartner J. Regulation of ion channel/neurotransmitter receptor function by RNA editing. Curr. Opin. Neurobiol. 2003;13:279–283. doi: 10.1016/s0959-4388(03)00062-x. [DOI] [PubMed] [Google Scholar]
- 5.Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 6.Burns CM, Chu H, Rueter SM, Hutchinson LK, Canton H, Sanders-Bush E, Emeson RB. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 7.Aruscavage PJ, Bass BL. A phylogenetic analysis reveals an unusual sequence conservation within introns involved in RNA editing. RNA. 2000;6:257–269. doi: 10.1017/s1355838200991921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 9.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DD, Kim TT, Walsh T, Kobayashi Y, Matise TC, Buyske S, Gabriel A. Widespread RNA editing of embedded alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lander ES, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat. Rev. Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- 14.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 15.Yelin R, et al. Widespread occurrence of antisense transcription in the human genome. Nat. Biotechnol. 2003;21:379–386. doi: 10.1038/nbt808. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Sun M, Hurst LD, Carmichael GG, Rowley JD. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense–antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, et al. Over 20% of human transcripts might form sense–antisense pairs. Nucleic Acids Res. 2004;32:4812–4820. doi: 10.1093/nar/gkh818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okano H, Aruga J, Nakagawa T, Shiota C, Mikoshiba K. Myelin basic protein gene and the function of antisense RNA in its repression in myelin-deficient mutant mouse. J. Neurochem. 1991;56:560–567. doi: 10.1111/j.1471-4159.1991.tb08186.x. [DOI] [PubMed] [Google Scholar]
- 19.Neeman Y, Dahary D, Levanon EY, Sorek R, Eisenberg E. Is there any sense in antisense editing? Trends Genet. 2005;21:544–547. doi: 10.1016/j.tig.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 21.Dabiri GA, Lai F, Drakas RA, Nishikura K. Editing of the GluR-B ion channel RNA in vitro by recombinant double-stranded RNA adenosine deaminase. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl. Acad. Sci. USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai F, Chen CX, Lee VM, Nishikura K. Dramatic increase of the RNA editing for glutamate receptor subunits during terminal differentiation of clonal human neurons. J. Neurochem. 1997;69:43–52. doi: 10.1046/j.1471-4159.1997.69010043.x. [DOI] [PubMed] [Google Scholar]
- 24.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]