Abstract
Maternal care influences hypothalamic-pituitary-adrenal (HPA) function in the rat through epigenetic programming of glucocorticoid receptor expression. In humans, childhood abuse alters HPA stress responses and increases the risk of suicide. We examined epigenetic differences in a neuron-specific glucocorticoid receptor (NR3C1) promoter between postmortem hippocampus obtained from suicide victims with a history of childhood abuse and those from either suicide victims with no childhood abuse or controls. We found decreased levels of glucocorticoid receptor mRNA, as well as mRNA transcripts bearing the glucocorticoid receptor 1F splice variant and increased cytosine methylation of an NR3C1 promoter. Patch-methylated NR3C1 promoter constructs that mimicked the methylation state in samples from abused suicide victims showed decreased NGFI-A transcription factor binding and NGFI-A–inducible gene transcription. These findings translate previous results from rat to humans and suggest a common effect of parental care on the epigenetic regulation of hippocampal glucocorticoid receptor expression.
There are maternal effects on the development of individual differences in behavioral and HPA stress responses in rodents and nonhuman primates1,2. Maternal behavior alters the development of HPA responses to stress in the rat through tissue-specific effects on gene transcription3,4, including forebrain glucocorticoid receptor expression, the activation of which inhibits HPA activity through negative-feedback inhibition5. Thus, selective knockdown of glucocorticoid receptor expression in the corticolimbic system in rodents is associated with increased HPA activity under both basal and stressful conditions6,7. Conversely, glucocorticoid receptor overexpression is associated with a dampened HPA stress response8.
Familial function and childhood adversity are linked to altered HPA stress responses in humans, which are associated with an increased risk for multiple forms of psychopathology9–11. There is evidence for decreased hippocampal glucocorticoid receptor expression in several psychopathological conditions associated with suicide, including schizophrenia and mood disorders12–14. Suicide is also strongly associated with a history of childhood abuse and neglect, and this effect is independent of that associated with psychopathology15,16. Thus, environmental events that associate with decreased hippocampal glucocorticoid receptor expression and increased HPA activity enhance the risk of suicide.
The effects of maternal care on hippocampal glucocorticoid receptor expression, and therefore HPA responses to stress, in the adult rodent are associated with an epigenetic modification of a neuron-specific exon 17 glucocorticoid receptor (Nr3c1) promoter4,17. We attempted to translate these findings to humans. We examined glucocorticoid receptor expression and NR3C1 promoter methylation in hippocampal samples obtained from suicide victims and control subjects who died suddenly of unrelated causes. The focus of our examination was the NR3C1 (also known as GR1F) promoter, the homolog of the exon 17 region in the rat18, which is highly expressed in human hippocampus19. Suicide victims were either positive or negative for history of childhood abuse (sexual contact, severe physical abuse and/or severe neglect), allowing for the separation of the effects associated with childhood abuse from those associated with suicide per se. Our controls were all negative for a history of childhood abuse.
RESULTS
Hippocampal glucocorticoid receptor expression
The human glucocorticoid receptor gene NR3C1 covers a region of more than 80 kb in chromosome 5, containing eight coding exons (exons 2–9) and alternative 5′ noncoding exon 1s19–21. The 5′ untranslated region (UTR) of exon 1 of the NR3C1 gene determines the tissue-specific expression. The 5′ UTR of NR3C1 contains 11 exon 1 splice variants, all of which bear unique splice donor sites and share a common exon 2 splice acceptor site19. Exon 1F of NR3C1 is similar to the rat exon 17, which reveals a maternal effect on cytosine methylation and expression8,18,22. Because individuals with severe forms of major depression show decreased glucocorticoid receptor expression and increased HPA activity, we hypothesized that suicide victims would show decreased expression both of glucocorticoid receptor and glucocorticoid receptor 1F compared with control subjects.
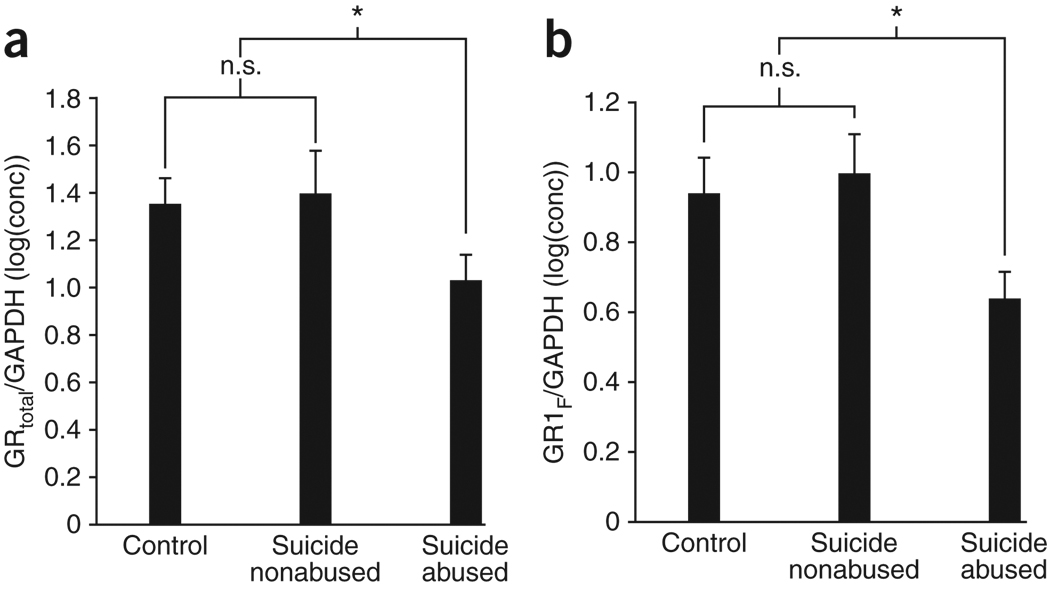
We examined the expression of total glucocorticoid receptor and glucocorticoid receptor 1F using quantitative reverse transcription PCR (qRT-PCR) with RNA extracted from hippocampal tissue of suicide completers with (n = 12) and without (n = 12) a history of childhood abuse and from controls (n = 12). There was a significant effect on glucocorticoid receptor expression (F = 3.17, P = 0.05). Post hoc tests showed that expression of total glucocorticoid receptor mRNA was significantly reduced in suicide victims with a history of childhood abuse relative to nonabused suicide victims or controls (P < 0.05); there was no difference between nonabused suicide victims and controls (P > 0.05; Fig. 1a). There was also a significant effect on the expression of transcripts containing the exon 1F NR3C1 promoter (F = 3.58, P < 0.05). Post hoc tests revealed that glucocorticoid receptor 1F expression was significantly lower in samples from suicide victims with a history of childhood abuse compared with suicide victims without childhood abuse or controls (P < 0.05). Similar to the findings with total glucocorticoid receptor mRNA expression, there was no difference between nonabused suicide victims and controls (P > 0.05; Fig. 1b).
Figure 1.
Hippocampal glucocorticoid receptor expression. (a,b) Mean ± s.e.m. expression levels of total glucocorticoid receptor (GR) mRNA (a) and glucocorticoid receptor 1F (GR1F) in 12 suicide victims with a history of childhood abuse, 12 nonabused suicide victims and 12 control subjects (b). Outliers excluded from analysis included n = 2 control subjects, n = 1 suicide victims with a history of childhood abuse for glucocorticoid receptor 1F and an additional n = 1 suicide victim with a history of childhood abuse, and n = 3 nonabused suicide victims for overall levels of glucocorticoid receptor. * indicates P < 0.05; n.s. indicates not statistically significant.
We examined the relationship between glucocorticoid receptor expression and psychiatric diagnoses (Table 1). Mood disorders and substance abuse disorders are risk factors for suicide and have been linked to alterations of glucocorticoid receptor expression12. There were no significant effects of psychopathology, even after controlling for childhood abuse status, on overall glucocorticoid receptor or glucocorticoid receptor 1F expression (P > 0.05).
Table 1.
Demographic characteristics and psychiatric diagnoses
| Abused suicide | Nonabused suicide | Control | |
|---|---|---|---|
| Male/female | 12/0 | 12/0 | 12/0 |
| Age (years) | 34.2 ± 10 | 33.8 ± 11 | 35.8 ± 12 |
| PMI (h) | 24.6 ± 5.8 | 39.0 ± 25.7 | 23.5 ± 6.0 |
| pH | 6.3 ± 0.24 | 6.5 ± 0.29 | 6.5 ± 0.22 |
| Childhood abuse/neglect | 12/0 (100%) | 0/12 (0%) | 0/12 (0%) |
| Mood disorder | 8/12 (67%) | 8/12 (67%) | 0/12 (0%) |
| Alcohol/drug abuse disorder | 9/12 (75%) | 6/12 (50%) | 5/12 (42%) |
Data are presented as mean ± s.d.
Genotyping and methylation analysis
Because alterations in glucocorticoid receptor 1F activity could be derived from nucleotide sequence variation and/or epigenetic modifications, we sequenced the NR3C1 promoter region from each subject. No sequence variation was seen among subjects and all of the sequences were identical to those published previously19. Moreover, for each subject, the genomic sequences targeted for binding by the primers used for bisulfite mapping were identical to the published sequence19, thus eliminating potential primer bias between subjects in sodium bisulfite mapping.
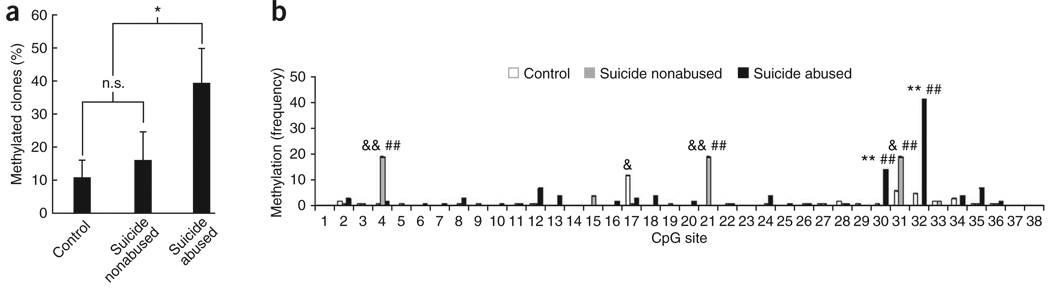
The rat homolog of the exon 1F NR3C1 promoter, the exon 17 region, is differentially methylated as a function of variations in maternal care4,17,22. Cytosine methylation is a highly stable epigenetic mark that regulates gene expression via its effects on transcription factor binding23,24. We used sodium bisulfite mapping25 to examine the methylation status of individual CpG dinucleotides in the NR3C1 promoter sequence extracted from the hippocampal tissue of the same subjects used for glucocorticoid receptor expression analysis. Sodium bisulfite mapping revealed a significant effect on the percentage of methylated clones (that is, the number of clones with at least one methylated CpG site divided by the total number of clones) between groups (F = 3.47, P < 0.05). Post hoc tests revealed a significant difference between suicide victims with a history of childhood abuse compared with nonabused suicide victims (P = 0.05) or controls (P < 0.05). There was no difference in the percentage of methylated clones between suicide victims without childhood abuse and controls (P > 0.05; Fig. 2a). Methylation was limited to specific sites, with no clone showing global methylation (Fig. 2b). There were no significant correlations between levels of exon 1F methylation and age at death (r = 0.15, P > 0.05), brain pH (r = 0.08, P > 0.05) or postmortem interval (PMI, r = 0.24, P > 0.05; Table 1).
Figure 2.
Methylation of the NR3C1 promoter in the hippocampus. Twenty clones were sequenced for each subject for methylation mapping. (a) Mean ± s.e.m. percentage of methylated clones for suicide victims with a history of childhood abuse (n = 12), suicide victims without a history of childhood abuse (n = 12) and controls (n = 12). The methylation percentage was calculated as the number of clones with at least one methylated CpG site divided by the total number of clones (* indicates P ≤ 0.05; n.s. indicates not statistically significant). (b) Methylation of the NR3C1 promoter region, showing the frequency of methylation observed at each CpG site for suicide victims with a history of childhood abuse, suicide victims with no history of childhood abuse and control subjects (*P < 0.05, **P < 0.001, abused suicides versus controls; &P < 0.05, &&P < 0.001, non-abused suicides versus controls; #P < 0.05, ##P < 0.001, abused suicides versus non-abused suicides; Bonferroni post hoc comparisons).
Patch methylation of the NR3C1 promoter
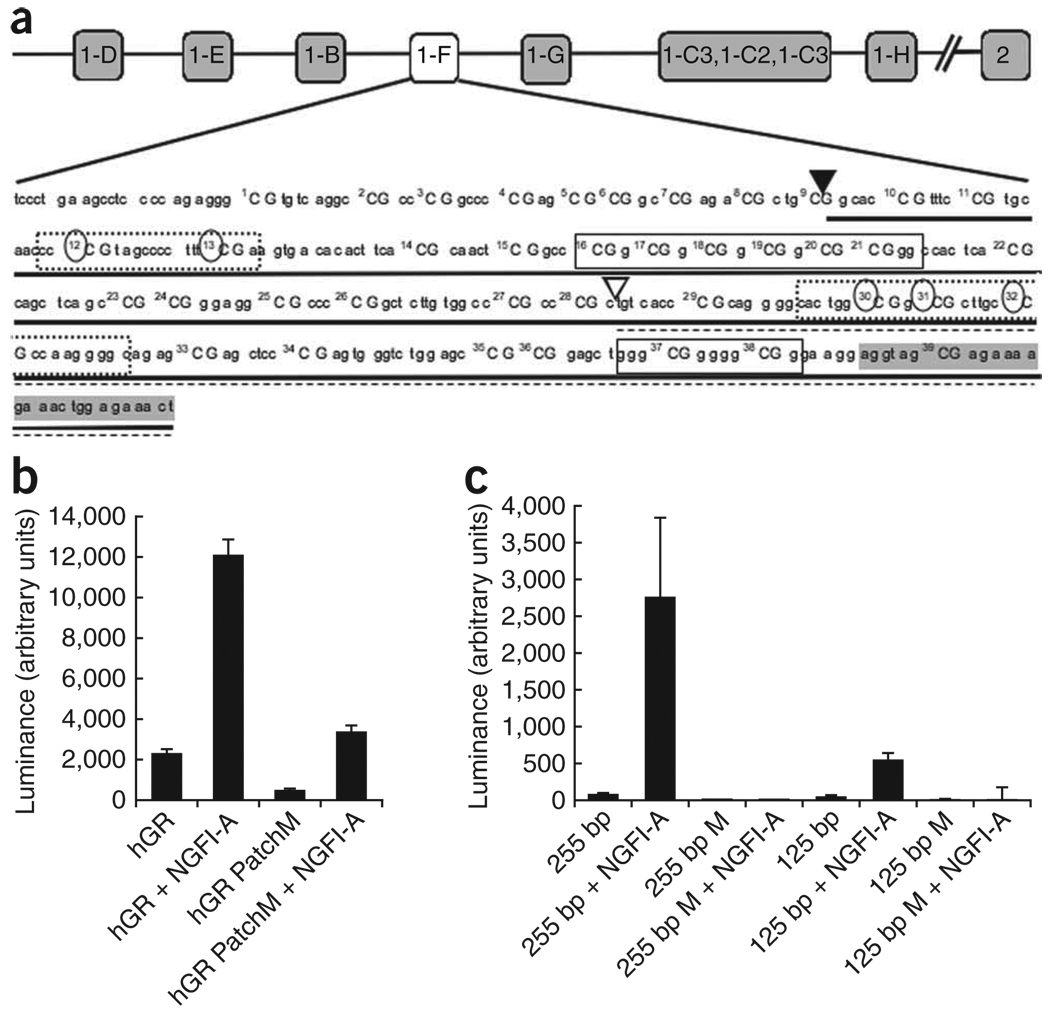
DNA methylation of a limited number of sites in the exon 1F NR3C1 promoter was associated with decreased expression of the glucocorticoid receptor 1F variant and of total glucocorticoid receptor mRNA in suicide victims with a history of childhood abuse. Defining a causal relation between the methylation status and transcriptional efficiency of the NR3C1 promoter is therefore of great importance. We hypothesized that DNA methylation regulates the expression of the NR3C1 promoter through alterations in transcription factor binding. The transcription factor NGFI-A regulates the expression of Nr3c1 promoter in the rat, an effect that is inhibited by DNA methylation17. To our knowledge, the regulation of NGFI-A (also known as Zif268, EGR1, Krox-24 and ZENK) has not been studied in the human hippocampus, although there is evidence that its expression is downregulated in the prefrontal cortex in schizophrenia26. The NR3C1 promoter contains a number of canonical and noncanonical NGFI-A recognition elements (Fig. 3a). We wondered whether, as in the rat17, NGFI-A could regulate gene transcription through the NR3C1 promoter and whether this effect might be influenced by the methylation status of the promoter. We used a transient transfection assay in human HEK293 cells to examine transcriptional activity of a NR3C1 promoter ligated to a promoter-less firefly luciferase expression vector (pGEM-LUC, Promega; Fig. 3a) in the presence or absence of ectopic NGFI-A expression. The use of HEK293 cells allowed us to concurrently transfect a number of expression vectors with high efficiency. The absence of plasmid replication during the transient transfection assay precludes the loss of methylation via passive demethylation27.
Figure 3.
In vitro analysis of NR3C1 promoter methylation. (a) The relative position of the NR3C1 variant and the promoter sequence, showing the location of the CpG dinucleotides. The 255-bp (▼, solid underline) and 125-bp (∇, broken underline) deletion constructs are shown, along with specific CpG dinucleotides that were methylated in each deletion construct, as indicated by circles. Boxes represent known or putative canonical (solid-lined box) and noncanonical (broken-lined box) NGFI-A–binding sites, with the shaded area indicating the beginning of the exon. (b,c) Mean ± s.e.m. levels of luciferase expression in HEK293 cells. Results are shown after the subtraction of expression of the promoter in the antisense orientation. (b) The full NR3C1 promoter was either unmethylated (hGR) or completely patch methylated (hGR PatchM) and transfected in the presence or absence of NGFI-A. (c) The 255-bp and 125-bp NR3C1 deletion construct were either unmethylated (255 bp or 125 bp) or methylated (255 bp M or 125 bp M), as shown in a, and transfected either in the presence or absence of NGFI-A.
Luciferase expression was measured in the presence or absence of NGFI-A from the unmethylated NR3C1 promoter plasmid compared with a methylated version. There was a significant effect of testing condition on the transcriptional activity of the exon 1F NR3C1 promoter (F = 110.6, P < 0.0001; Fig. 3b). Post hoc analysis revealed that the transcriptional activity of the unmethylated NR3C1 promoter was significantly increased in the presence of the NGFI-A expression vector (NR3C1versus NR3C1 + EGR1; P < 0.0001). Furthermore, methylation of the NR3C1 promoter (the entire NR3C1 construct was methylated in vitro and ligated to an unmethylated vector before transfection, NR3C1-M) reduced basal transcriptional activity of the NR3C1 construct (NR3C1 versus NR3C1-M, P < 0.05). Methylation of the NR3C1 construct also blunted NGFI-A induction of transcription (NR3C1 + EGR1 versus NR3C1-M + EGR1, P < 0.0001).
These results indicate that methylation attenuates NGFI-A induction of gene expression through the NR3C1 promoter. However, the decreased glucocorticoid receptor transcription observed in suicide victims with a history of childhood abuse was associated with differences in methylation levels occurring only at specific sites in the exon 1F NR3C1 promoter (Fig. 2b). An ANOVA examining the methylation of CpG dinucleotides across the exon 1F NR3C1 promoter revealed a significant effect of CpG site (F = 13.86, P < 0.0001), a significant effect of group (F = 17.12, P < 0.0001) and a significant interaction between CpG site and group (F = 13.44, P < 0.0001). In NGFI-A recognition elements, methylation was observed at CpG sites 12, 13, 30, 31 and 32 (Fig. 2b). We therefore examined whether such selective, site-specific differences in methylation could alter transcriptional activation through the NR3C1 promoter. Two deletion constructs of the NR3C1 promoter were generated in which selected CpG dinucleotides were patch-methylated (Fig. 3a). CpG sites 12 and 13 were methylated in the 255-bp construct, whereas the 125-bp promoter construct was methylated at CpG sites 30, 31 and 32. Thus, each deletion construct included at least one known or putative NGFI-A binding site28.
We used patch methylation to examine whether selective methylation at specific sites reduces NGFI-A binding to and transactivation through the exon 1F NR3C1 promoter. We found an effect of methylation status on transcription factor–induced gene expression from the NR3C1 promoter (Fig. 3c). For the 125-bp construct, there was a significant effect of methylation status (F = 57.6, P < 0.0001) and NGFI-A treatment (F = 6.3, P < 0.05). As predicted, there was also a significant interaction between methylation status and the NGFI-A expression (F = 48.7, P < 0.0001). Post hoc analysis of the 125-bp NR3C1 promoter construct revealed that the effect of NGFI-A on gene transcription was significantly (P < 0.001) greater in the presence of the unmethylated rather than the patch-methylated NR3C1 promoter construct. The same pattern of results was observed for the 255-bp NR3C1 promoter construct. Thus, for the 255-bp NR3C1 promoter construct, there was a significant effect on transcriptional activity of methylation status (F = 555.4, P < 0.0001) and NGFI-A expression (F = 387.3, P < 0.0001). There was also a significant interaction between methylation status and the presence of NGFI-A (F = 489.1, P < 0.0001). Post hoc analysis revealed a significantly (P < 0.001) greater effect of NGFI-A on gene transcription in the presence of the unmethylated compared with the patch-methylated 255-bp NR3C1 promoter construct.
NGFI-A association with the NR3C1 promoter
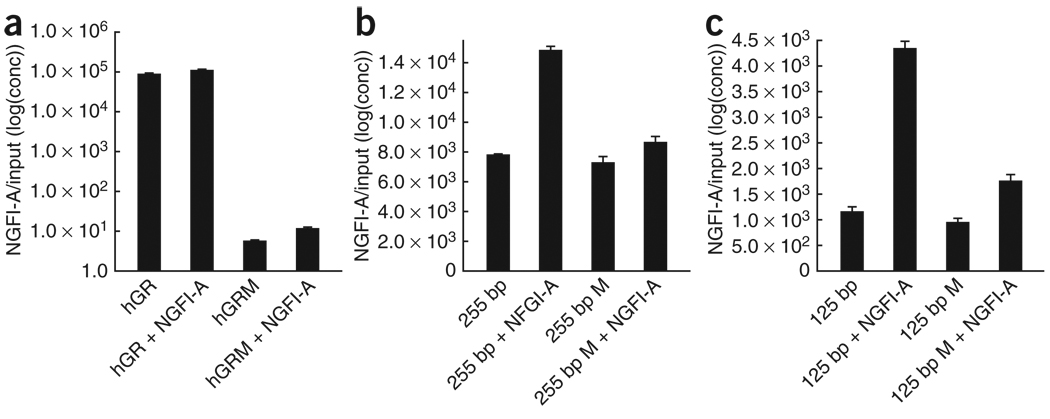
Site-selective differences in methylation of CpG regions in promoters can alter gene transcription by affecting transcription factor binding24. We carried out chromatin immunoprecipitation assays on samples that were obtained using the transfection model described above to examine the association of NGFI-A with the methylated and the nonmethylated NR3C1 promoter constructs. The results revealed a significant effect of methylation status on NGFI-A association with the exon 1F NR3C1 promoter constructs transfected with EGR1 (F = 242.92, P < 0.0001; Fig. 4a). Post hoc tests showed that, in comparison with the nonmethylated construct, patch-methylation inhibited NGFI-A binding to the exon 1F NR3C1 promoter (P < 0.005; Fig. 4a). For the 125-bp promoter construct, there was a significant effect of methylation status (F = 102.28, P < 0.001) and NGFI-A treatment (F = 209.99, P < 0.0005) and a significant interaction between methylation status and the presence of NGFI-A (F = 72.71, P < 0.005). Similarly, for the 255-bp promoter construct, there was a significant effect of methylation status (F = 95.18, P < 0.001) and NGFI-A treatment (F = 152.13, P < 0.0005) and a significant interaction between methylation status and the presence of NGFI-A (F = 67.75, P < 0.005). Post hoc testing revealed that, in comparison with the nonmethylated version, patch-methylation inhibited NGFI-A binding to either the 255-bp or 125-bp promoter constructs (P < 0.01; Fig. 4b,c). These findings suggest that the site-selective methylation of the exon 1F NR3C1 promoter construct, mimicking the differences observed between samples obtained from suicide victims with a history of childhood abuse, reduces NGFI-A binding and transcriptional activation through the exon 1F NR3C1 promoter.
Figure 4.
NGFI-A association with exon 1F NR3C1 promoter constructs. (a–c) Quantification of NR3C1 promoter immunoprecipitated with NGFI-A antibody and normalized to input DNA for the full human NR3C1 promoter (a), the 255-bp deletion construct (b) and the 125-bp deletion construct (c), each of which was either unmethylated or methylated and transfected in the presence or absence of NGFI-A. Data are presented as means ± s.e.m.
DISCUSSION
Our findings indicate that hippocampal NR3C1 gene expression is decreased in samples from suicide victims with a history of childhood abuse compared with controls (victims of sudden, accidental death with no history of abuse). In contrast, we found no differences in glucocorticoid receptor expression between suicide victims without a history of childhood abuse and controls. The pattern of results for hippocampal expression of the glucocorticoid receptor 1F variant was identical to that of total glucocorticoid receptor expression. Our findings suggest that changes in glucocorticoid receptor expression are closely associated with a developmental history of familial adversity, in this case a history of childhood abuse, than with suicide completion. These results are also similar to those of earlier reports in which childhood abuse was associated with an increase in pituitary adrenocorticotropic hormone (ACTH) responses to stress among individuals with or without concurrent major depression11. These findings are particularly relevant, as pituitary ACTH directly reflects central activation of the HPA stress response and hippocampal glucocorticoid receptor activation dampens HPA activity.
Our findings are also consistent with those from studies with rodent and nonhuman primates showing that persistent disruptions of mother-infant interactions are associated with increased hypothalamic corticotrophin-releasing hormone expression and increased HPA responses to stress1,2,29. Variations in maternal care in the rat influence hippocampal glucocorticoid receptor expression, as well as methylation of the rat fetal calf serum Nr3c1 promoter, the homolog of the human exon 1F NR3C1 promoter3,4,17,22. Hippocampal samples from suicide victims showed increased methylation of the exon 1F NR3C1 promoter in comparison with samples from controls, but only in cases with a history of childhood abuse. Neither hippocampal glucocorticoid receptor expression nor the methylation status of the exon 1F NR3C1 promoter was altered in suicide victims with no history of abuse. These findings suggest that variation in the methylation status of the exon 1F NR3C1 promoter, similar to that for glucocorticoid receptor 1F and total glucocorticoid receptor mRNA expression, associates with childhood adversity and not with suicide per se.
A recent study of human cord blood found a correlation between maternal mood and neonatal methylation status of glucocorticoid receptor 1F30. This study reported that increased site-specific methylation of an NGFI-A response element of glucocorticoid receptor 1F is linked to an enhanced cortisol stress response in infants. Maternal mood disorders are associated with decreased maternal sensitivity and impaired mother-infant interactions31, as well as with an increased risk for depression in the offspring32. Decreased hippocampal glucocorticoid receptor expression associates with depression12, and psychotic and severe forms of depression are commonly associated with increased HPA activity13,33. Thus, our findings suggest that the transmission of vulnerability for depression from parent to offspring could occur, in part, through the epigenetic modification of genomic regions that are implicated in the regulation of stress responses.
One limitation of our design is the absence of samples from control subjects with a history of child abuse. Notably, child abuse predicts ACTH responses to stress11. However, the best predictor of HPA responses to stress is the interaction between a developmental history of child abuse and stress in adulthood. One interpretation for such findings is that childhood adversity might alter the development of systems that serve to regulate stress responses, such as hippocampal glucocorticoid receptor expression, and thus enhance the effect of stress in adulthood and vulnerability for mood disorders11. Rodent models provide evidence for a direct effect of variations in forebrain glucocorticoid receptor expression and the behavioral characteristics of depression. Mice bearing a brain-specific glucocorticoid receptor knockdown show behavioral alterations that mimic some of the features of depression6,7,34.
The data reported here are consistent with previous reports of alterations in cytosine methylation associated with psychopathology35–37. We found increased site-specific methylation of the exon 1F NR3C1 promoter in suicide victims with a history of childhood abuse (Fig. 2). Our transfection studies with constructs that replicated the in vivo methylation profiles indicated that there was a relationship between cytosine methylation, transcription factor binding and gene expression. Variations in maternal care in the rat produce differential methylation of the exon 17 Nr3c1 promoter, the rat homolog of the exon 1F NR3C1 promoter, which regulates hippocampal glucocorticoid receptor expression4,17,22 and HPA responses to stress3,4. Increased CpG methylation of the Nr3c1 promoter decreased NGFI-A binding and reduced hippocampal glucocorticoid receptor expression. Manipulations that reversed the differences in exon 17 methylation eliminated the maternal effect on NGFI-A binding, glucocorticoid receptor expression and HPA activity4,22. Likewise, our in vitro data reveal that differential methylation of the human NR3C1 promoter altered NGFI-A binding and NGFI-A–induced gene transcription. These findings suggest that selective differences in methylation status at certain sites affect transcription factor binding and gene expression.
Because cytosine methylation is a highly stable, the differences in CpG methylation are unlikely to be a consequence of events immediately preceding death or during the postmortem period. Therefore, changes in brain pH do not affect DNA methylation38. The PMI did not differ between the groups and was uncorrelated with the methylation of the NR3C1 promoter. The intermittent pattern of methylation and the fact that only a portion of the neuronal population was methylated in each subject is consistent with a model in which alterations in methylation occur at later stages in development, after the completion of embryogenesis and neuronal differentiation. Indeed, the maternal effect on the methylation status of the exon 17 Nr3c1 promoter in the rat occurs during early postnatal life4.
There are precedents for the apparent developmental origins for the observed differences in DNA methylation and glucocorticoid receptor expression. Childhood abuse in humans is associated with altered hippocampal development39, enhanced HPA activity9,11 and an increased risk for psychopathology15,16. Similarly, children exposed to childhood adversity are more likely to engage in suicidal behavior40,41. Variations in the parental care of children are linked with individual differences in HPA and sympathetic and central catecholamine responses to stress1,11,42,43. Interventions that target parental care of high-risk children alter HPA activity44. Thus, it is tempting to speculate that epigenetic processes might mediate the effects of the social environment during childhood on hippocampal gene expression and that stable epigenetic marks such as DNA methylation might then persist into adulthood and influence the vulnerability for psychopathology through effects on intermediate levels of function, such as HPA activity.
In summary, our data reveal increased site-specific methylation in the exon 1F NR3C1 promoter in suicide victims with a history of childhood abuse and suggest that there is a relationship between cytosine methylation, transcription factor binding and gene expression. Our results are consistent with evidence from studies using psychological autopsy methods40 and epidemiological longitudinal designs45, which suggest that suicide has a developmental origin. We acknowledge that such conclusions are based on samples that differ along a wide range of experiential and potentially genetic dimensions. Our data certainly do not exclude alternative mechanisms of vulnerability. Indeed, the challenge for the future is to understand how epigenetic variation overlaying that occurring in nucleotide sequence might explain the developmental origins of vulnerability for chronic illness. Our data are merely consistent with observations from animal studies investigating epigenetic regulation of the Nr3c1 gene and with the hypothesis that early life events can alter the epigenetic state of relevant genomic regions, the expression of which may contribute to individual differences in the risk for psychopathology.
METHODS
Postmortem sample preparation and subject characteristics
We used hippocampal samples from the Quebec Suicide Brain Bank (http://www.douglasrecherche.qc.ca/suicide), which included 12 suicide victims with a history of childhood abuse, 12 suicide victims with a negative history of childhood abuse (matched for psychiatric diagnoses) and 12 controls. All subjects were matched for PMI, gender and age (Table 1). Samples were processed as described previously46 and consisted of hippocampus tissue from male suicides and control subjects of French-Canadian origin, dissected at 4 °C and stored at −80 °C. All subjects died suddenly with no medical or paramedical intervention. Suicides were determined by the Quebec Coroner’s Office and the control subjects were individuals who had died suddenly from causes other than suicide. Homogenates of tissue samples were used for genomic DNA (DNeasy, Qiagen) and RNA (Trizol, Invitrogen) extraction. Possible confounds that were examined included PMI, brain pH and the age of the donor at death. Samples were processed and analyzed blindly with respect to demographic and diagnostic variables. Signed informed consent was obtained from next of kin.
Psychological autopsies
Psychiatric diagnoses were obtained using Structured Clinical Interviews for DSM-III-R I (ref. 47) adapted for psychological autopsies, which is a validated method for reconstructing psychiatric history by means of extensive proxy-based interviews, as described elsewhere48. History of severe childhood sexual and/or physical abuse or severe neglect was determined by means of structured interviews using the Childhood Experience of Care and Abuse49 questionnaire adapted for psychological autopsies45.
Genotyping
Genomic DNA was extracted (DNeasy, Qiagen) according to the manufacturer’s protocol. For PCR, we used a 5′-GGG TTC TGC TTT GCA ACT TC-3′ sense primer and a 5′-CCT TTT TCC TGG GGA GTT G-3′ antisense primer that were directed to the NR3C1 promoter (Genebank accession number AY436590). Primers were selected that covered a 536-bp region that included the region for sodium bisulfite analyses. The resulting PCR products for each subject were sequenced bidirectionally using the forward and the reverse primer on an ABI 3100 genetic analyzer (Applied Biosystems) following the manufacturer’s instructions. Genetic variation was assessed throughout the NR3C1 promoter region used for bisulfite analysis by alignment of genomic DNA with the previously published NR3C1 promoter sequence19 using freely available software (CLC Workbench, CLC bio).
Methylation mapping and expression analyses
Sodium bisulfite mapping was performed as previously described25,50 for 12 suicide victims with a history of childhood abuse, 12 suicide victims with a negative history of childhood abuse and 12 controls. Individual clones were extracted and sequenced (Cequation (8800), Beckman-Coulter). We obtained 20 clones per subject for sequencing from 2–3 independent PCR reactions.
RNA extraction was performed using Trizol (Invitrogen) and was followed by Dnase I treatment, and cDNA conversion was performed using oligo(dT) primers (Invitrogen) according to manufacturers instructions (Roche Molecular Biochemicals). The same subjects used for methylation analysis were studied for expression analyses using quantitative RT-PCR. Outliers with expression values that differed more than 1.5 s.d. from the mean were excluded from analysis (n = 2 control subjects, n = 1 suicide victims with a history of childhood abuse for glucocorticoid receptor 1F and n = 2 control subjects, n = 2 suicide victims with a history of childhood abuse and n = 3 nonabused suicide victims for overall levels of glucocorticoid receptor; see Supplementary Methods online).
HEK293 cell cultures and transient transfection assays
To prepare and transfect (and see ref. 17) the unmethylated and methylated plasmids, we subjected the exon 1F NR3C1 promoter to PCR amplification and cloned the resulting PCR product into a PCR2.1 plasmid (Original TA cloning kit, Invitrogen). The NR3C1 promoter was then ligated into the PCR2.1 plasmid. For patch methylation, the glucocorticoid receptor 1F plasmid was incubated (2 h, 37 °C) with SssI DNA methyltransferase (20 U, New England Biolabs) in a buffer containing S-adenosylmethionine, and this procedure was repeated until full protection from HpaII digestion was observed. Following digestion with HindIII and EcoRV restriction enzymes, each fragment was then ligated into a pGEM-luc vector (Promega) at the HindIII and BamHI or XbaI and BamHI sites in the 5′ to 3′ (sense) or 3′ to 5′ (antisense) orientation, respectively. The concentration of each ligation product was determined by fractionation on a 1.5% agarose gel, by comparing bands of the expected ligation product size against a standard curve of known DNA concentration (ten fivefold serial dilutions of 2 µg/µl−1 micrococal nuclease DNA) to control for possible unequal efficiency of ligation and to ensure that equal amounts of correctly ligated plasmids were used in the transfections. The ligated plasmid was directly transfected into HEK293 cells and was not subcloned to avoid loss of methyl groups from CG dinucleotides during growth in E. coli, which do not express CG DNA methyltransferases. For deletion constructs of the exon 1F NR3C1 promoter plasmids were prepared and ligations verified in the same manner as described above, except that oligonucleotides for NR3C1 promoter amplification were designed that incorporated HindIII and EcoRV restriction sites, obviating the need for ligation into PCR2.1 vector before ligation into the pGEM-luc vector (also see Supplementary Methods).
For the NGFI-A plasmid, we subcloned the EGR1 coding sequence into a TOPO–His expression vector (pEF6/V5-His TOPO TA Expression kit, Invitrogen) 4. In co-transfection studies, human embryonic kidney HEK293 cells were plated at a density of 6 × 104 in six-well dishes and transiently co-transfected with a total amount of 1.5 µg of plasmid DNA (1.0 µg of NR3C1 promoter ligated into the pGEM-luc plasmid and 0.5 µg of NGFI-A expression plasmid or 0.5 µg of control pEF6/V5 plasmid) using the calcium phosphate method. HEK293 cells were maintained as a monolayer in DMEM (Invitrogen) containing 10% fetal calf serum (Colorado Serum Company). The cells were harvested 72 h after transfection and lysed, and luciferase activity was assayed using the Luciferase Assay System (Promega) according to the manufacturer’s protocol.
Chromatin immunoprecipitation assay for NGFI-A
We carried out chromatin immunoprecipitation assays30 by postfixing cells in 37% formaldehyde and then pelleted them before lysis and sonication. We reserved one tenth of the sample as ‘input’ to quantify the amount of DNA before immunoprecipitation. For the remainder of each sample, extracted chomatin was immunoprecipitated using rabbit polyclonal antibody to NGFI-A (antibody) or normal rabbit IgG (nonspecific; both from Santa Cruz Biotechnology). All of the DNA was then uncrosslinked and subjected to qRT-PCR, using primers directed at the luciferase gene immediately downstream of the transfected NR3C1 promoter (sense, 5′-AGA GAT ACG CCC TGG TTC C-3′; antisense, 5′-CCA ACA CCG GCA TAA AGA A-3′; Tm = 54 °C). The signal for each sample was calculated by dividing the value of the antibody by the input. Each resulting value was multiplied by a constant (1 × 106) to plot the values obtained from the experiments on logarithmic axes.
Statistical analyses
Statistical analyses were conducted using Statview or JMP 7 (SAS Institute). Data are presented as mean ± s.e.m. For DNA methylation analysis, the percentage of methylated clones for each subject was tabulated by dividing the number of clones with at least one methylated CpG site by the total number of clones. A factorial ANOVA was used to compare the percentage of methylated clones for each subject as the dependent variable and group (suicide abused, suicide nonabused or control) as the between groups factor. To examine differential methylation across CpG sites methylated in vitro, we compared the total number of methylated CpG sites across the NR3C1 promoter (n = 33) for all clones per group (that is, 12 subjects × 20 clones = 240 clones per group) using ANOVA followed by Bonferroni corrected post hoc comparisons. For RNA expression analysis, ANOVA followed by PLSD post hoc tests were used to examine differences between the suicide victim and control group. Unpaired t tests were used to compare groups of subjects with different clinical diagnoses (for example, subjects with mood disorders versus subjects without mood disorders). The relationships between DNA methylation, expression, age at death, PMI and brain pH were analyzed using linear regression analysis. Factorial ANOVA were used for in vitro analyses of NR3C1 promoter, followed by post hoc analyses. Statistical significance was determined at P ≤ 0.05 and analyses included type 1 error correction for multiple comparisons where applicable.
ACKNOWLEDGMENTS
This research was supported by grants from the US National Institutes of Health (National Institute of Child Health and Human Development; M.J.M. and M.S.), the Canadian Institutes for Health Research (M.J.M., M.S. and G.T.), a Team Grant from the Human Frontiers Science Program (M.J.M. and M.S.) and a Maternal Adversity, Vulnerability and Neurodevelopment Project grant from the Canadian Institutes for Health Research (M.J.M. and M.S.).
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Meaney MJ. Maternal care, gene expression, and the transmission of individual differences in stress reactivity across generations. Annu. Rev. Neurosci. 2001;24:1161–1192. doi: 10.1146/annurev.neuro.24.1.1161. [DOI] [PubMed] [Google Scholar]
- 2.Higley JD, Hasert MF, Suomi SJ, Linnoila M. Nonhuman primate model of alcohol abuse: effects of early experience, personality and stress on alcohol consumption. Proc. Natl. Acad. Sci. USA. 1991;88:7261–7265. doi: 10.1073/pnas.88.16.7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu D, et al. Maternal care, hippocampal glucocorticoid receptors and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–1662. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- 4.Weaver IC, et al. Epigenetic programming by maternal behavior. Nat. Neurosci. 2004;7:847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 5.de Kloet ER, Joels M, Holsboer F. Stress and the brain: from adaptation to disease. Nat. Rev. Neurosci. 2005;6:463–475. doi: 10.1038/nrn1683. [DOI] [PubMed] [Google Scholar]
- 6.Boyle MP, et al. Acquired deficit of forebrain glucocorticoid receptor produces depression-like changes in adrenal axis regulation and behavior. Proc. Natl. Acad. Sci. USA. 2005;102:473–478. doi: 10.1073/pnas.0406458102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ridder S, et al. Mice with genetically altered glucocorticoid receptor expression show altered sensitivity for stress-induced depressive reactions. J. Neurosci. 2005;25:6243–6250. doi: 10.1523/JNEUROSCI.0736-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reichardt HM, Tronche F, Bauer A, Schutz G. Molecular genetic analysis of glucocorticoid signaling using the Cre/loxP system. Biol. Chem. 2000;381:961–964. doi: 10.1515/BC.2000.118. [DOI] [PubMed] [Google Scholar]
- 9.De Bellis MD, et al. Hypothalamic-pituitary-adrenal axis dysregulation in sexually abused girls. J. Clin. Endocrinol. Metab. 1994;78:249–255. doi: 10.1210/jcem.78.2.8106608. [DOI] [PubMed] [Google Scholar]
- 10.Pruessner JC, Champagne F, Meaney MJ, Dagher A. Dopamine release in response to a psychological stress in humans and its relationship to early life maternal care: a positron emission tomography study using [11C]raclopride. J. Neurosci. 2004;24:2825–2831. doi: 10.1523/JNEUROSCI.3422-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heim C, Nemeroff CB. The role of childhood trauma in the neurobiology of mood and anxiety disorders: preclinical and clinical studies. Biol. Psychiatry. 2001;49:1023–1039. doi: 10.1016/s0006-3223(01)01157-x. [DOI] [PubMed] [Google Scholar]
- 12.Webster MJ, Knable MB, O’Grady J, Orthmann J, Weickert CS. Regional specificity of brain glucocorticoid receptor mRNA alterations in subjects with schizophrenia and mood disorders. Mol. Psychiatry. 2002;7:985–994. doi: 10.1038/sj.mp.4001139. [DOI] [PubMed] [Google Scholar]
- 13.Schatzberg AF, Rothschild AJ, Langlais PJ, Bird ED, Cole JO. A corticosteroid/dopamine hypothesis for psychotic depression and related states. J. Psychiatr. Res. 1985;19:57–64. doi: 10.1016/0022-3956(85)90068-8. [DOI] [PubMed] [Google Scholar]
- 14.Isometsa ET, et al. Suicide in major depression. Am. J. Psychiatry. 1994;151:530–536. doi: 10.1176/ajp.151.4.530. [DOI] [PubMed] [Google Scholar]
- 15.Widom CS, DuMont K, Czaja SJ. A prospective investigation of major depressive disorder and comorbidity in abused and neglected children grown up. Arch. Gen. Psychiatry. 2007;64:49–56. doi: 10.1001/archpsyc.64.1.49. [DOI] [PubMed] [Google Scholar]
- 16.Fergusson DM, Horwood LJ, Lynskey MT. Childhood sexual abuse and psychiatric disorder in young adulthood. II. Psychiatric outcomes of childhood sexual abuse. J. Am. Acad. Child Adolesc. Psychiatry. 1996;35:1365–1374. doi: 10.1097/00004583-199610000-00024. [DOI] [PubMed] [Google Scholar]
- 17.Weaver IC, et al. The transcription factor nerve growth factor–inducible protein a mediates epigenetic programming: altering epigenetic marks by immediate-early genes. J. Neurosci. 2007;27:1756–1768. doi: 10.1523/JNEUROSCI.4164-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCormick JA, et al. 5′-heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol. Endocrinol. 2000;14:506–517. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- 19.Turner JD, Muller CP. Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification and tissue distribution of multiple new human exon 1. J. Mol. Endocrinol. 2005;35:283–292. doi: 10.1677/jme.1.01822. [DOI] [PubMed] [Google Scholar]
- 20.Breslin MB, Geng CD, Vedeckis WV. Multiple promoters exist in the human GR gene, one of which is activated by glucocorticoids. Mol. Endocrinol. 2001;15:1381–1395. doi: 10.1210/mend.15.8.0696. [DOI] [PubMed] [Google Scholar]
- 21.Encio IJ, Detera-Wadleigh SD. The genomic structure of the human glucocorticoid receptor. J. Biol. Chem. 1991;266:7182–7188. [PubMed] [Google Scholar]
- 22.Weaver IC, et al. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J. Neurosci. 2005;25:11045–11054. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Razin A. CpG methylation, chromatin structure and gene silencing a three-way connection. EMBO J. 1998;17:4905–4908. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bird A. Molecular biology. Methylation talk between histones and DNA. Science. 2001;294:2113–2115. doi: 10.1126/science.1066726. [DOI] [PubMed] [Google Scholar]
- 25.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamada K, et al. Genetic analysis of the calcineurin pathway identifies members of the EGR gene family, specifically EGR3, as potential susceptibility candidates in schizophrenia. Proc. Natl. Acad. Sci. USA. 2007;104:2815–2820. doi: 10.1073/pnas.0610765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J. Biol. Chem. 2001;276:40778–40787. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- 28.Crosby SD, Puetz JJ, Simburger KS, Fahrner TJ, Milbrandt J. The early response gene NGFI-C encodes a zinc finger transcriptional activator and is a member of the GCGGGGGCG (GSG) element–binding protein family. Mol. Cell. Biol. 1991;11:3835–3841. doi: 10.1128/mcb.11.8.3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plotsky PM, et al. Long-term consequences of neonatal rearing on central corticotropin-releasing factor systems in adult male rat offspring. Neuropsychopharmacology. 2005;30:2192–2204. doi: 10.1038/sj.npp.1300769. [DOI] [PubMed] [Google Scholar]
- 30.Oberlander TF, et al. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 31.Fleming AS, O’Day DH, Kraemer GW. Neurobiology of mother-infant interactions: experience and central nervous system plasticity across development and generations. Neurosci. Biobehav. Rev. 1999;23:673–685. doi: 10.1016/s0149-7634(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 32.Pilowsky DJ, et al. Children of depressed mothers 1 year after the initiation of maternal treatment: findings from the STAR*D child study. Am. J. Psychiatry. 2008;165:1136–1147. doi: 10.1176/appi.ajp.2008.07081286. [DOI] [PubMed] [Google Scholar]
- 33.Holsboer F. The corticosteroid receptor hypothesis of depression. Neuropsychopharmacology. 2000;23:477–501. doi: 10.1016/S0893-133X(00)00159-7. [DOI] [PubMed] [Google Scholar]
- 34.Neigh GN, Nemeroff CB. Reduced glucocorticoid receptors: consequence or cause of depression? Trends Endocrinol. Metab. 2006;17:124–125. doi: 10.1016/j.tem.2006.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abdolmaleky HM, et al. Hypomethylation of MB-COMT promoter is a major risk factor for schizophrenia and bipolar disorder. Hum. Mol. Genet. 2006;15:3132–3145. doi: 10.1093/hmg/ddl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grayson DR, et al. Reelin promoter hypermethylation in schizophrenia. Proc. Natl. Acad. Sci. USA. 2005;102:9341–9346. doi: 10.1073/pnas.0503736102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegmund KD, et al. DNA methylation in the human cerebral cortex is dynamically regulated throughout the life span and involves differentiated neurons. PLoS ONE. 2007;2:e895. doi: 10.1371/journal.pone.0000895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ernst C, et al. The effects of pH on DNA methylation state: in vitro and postmortembrain studies. J. Neurosci. Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 39.Vythilingam M, et al. Childhood trauma associated with smaller hippocampal volume in women with major depression. Am. J. Psychiatry. 2002;159:2072–2080. doi: 10.1176/appi.ajp.159.12.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seguin M, et al. Life trajectories and burden of adversity: mapping the developmental profiles of suicide mortality. Psychol. Med. 2007;37:1575–1583. doi: 10.1017/S0033291707000955. [DOI] [PubMed] [Google Scholar]
- 41.Brezo J, et al. Natural history of suicidal behaviors in a population-based sample of young adults. Psychol. Med. 2007;37:1563–1574. doi: 10.1017/S003329170700058X. [DOI] [PubMed] [Google Scholar]
- 42.Kaufman J, Plotsky PM, Nemeroff CB, Charney DS. Effects of early adverse experiences on brain structure and function: clinical implications. Biol. Psychiatry. 2000;48:778–790. doi: 10.1016/s0006-3223(00)00998-7. [DOI] [PubMed] [Google Scholar]
- 43.Teicher MH, Andersen SL, Polcari A, Anderson CM, Navalta CP. Developmental neurobiology of childhood stress and trauma. Psychiatr. Clin. North Am. 2002;25:397–426. doi: 10.1016/s0193-953x(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 44.Fisher PA, Gunnar MR, Chamberlain P, Reid JB. Preventive intervention for maltreated preschool children: impact on children’s behavior, neuroendocrine activity and foster parent functioning. J. Am. Acad. Child Adolesc. Psychiatry. 2000;39:1356–1364. doi: 10.1097/00004583-200011000-00009. [DOI] [PubMed] [Google Scholar]
- 45.Brezo J, et al. Predicting suicide attempts in young adults with histories of childhood abuse. Br. J. Psychiatry. 2008;193:134–139. doi: 10.1192/bjp.bp.107.037994. [DOI] [PubMed] [Google Scholar]
- 46.Sequeira A, Turecki G. Genome wide gene expression studies in mood disorders. OMICS. 2006;10:444–454. doi: 10.1089/omi.2006.10.444. [DOI] [PubMed] [Google Scholar]
- 47.Spitzer RL, Williams JB, Gibbon M, First MB. The Structured Clinical Interview for DSM-III-R (SCID). I: history, rationale and description. Arch. Gen. Psychiatry. 1992;49:624–629. doi: 10.1001/archpsyc.1992.01820080032005. [DOI] [PubMed] [Google Scholar]
- 48.Dumais A, et al. Risk factors for suicide completion in major depression: a case-control study of impulsive and aggressive behaviors in men. Am. J. Psychiatry. 2005;162:2116–2124. doi: 10.1176/appi.ajp.162.11.2116. [DOI] [PubMed] [Google Scholar]
- 49.Bifulco A, Brown GW, Harris TO. Childhood Experience of Care and Abuse (CECA): a retrospective interview measure. J. Child Psychol. Psychiatry. 1994;35:1419–1435. doi: 10.1111/j.1469-7610.1994.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 50.Frommer M, et al. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]