Summary
MDM2, a negative regulator of p53, is elevated in many cancers that retain wild-type p53. A single nucleotide polymorphism (SNP) in the human MDM2 promoter increases the affinity of Sp1 resulting in elevated MDM2 levels. We generated mice carrying either the MDM2SNP309T or the MDM2SNP309G allele to address the impact of MDM2SNP309G on tumorigenesis. Mdm2SNP309G/G cells exhibit elevated Mdm2 levels, reduced p53 levels, and decreased apoptosis. Importantly, some Mdm2SNP309G/G mice succumbed to tumors before one year of age, suggesting that this allele increases tumor risk. Additionally, the Mdm2SNP309G allele potentiates the tumor phenotype and alters tumor spectrum in mice inheriting a p53 hot-spot mutation. These data provide causal evidence for increased cancer risk in carriers of the Mdm2SNP309G allele.
Introduction
Ablation of the p53 tumor suppressor pathway is a hallmark of tumorigenesis as evidenced by the fact that p53 is the most commonly mutated or inactivated tumor suppressor in neoplastic malignancies. p53 encodes a transcription factor that activates numerous cell cycle arrest, senescence, and apoptotic genes (Vogelstein et al., 2000). Mutations in or deletions of the p53 gene occur in over 50% of human cancers (Soussi and Lozano, 2005). In cancers lacking p53 mutations, other components of the p53 pathway are altered during tumorigenesis, contributing to the functional inactivation of the p53 pathway (Soussi and Lozano, 2005). Most notably, the MDM2 gene is amplified in over 30% of sarcomas and the MDM2 protein is overexpressed in multiple human cancers that retain wild type p53 (Oliner et al., 1992; Valentin-Vega et al., 2007). Mdm2 is a proto-oncogene that encodes an E3 ubiquitin ligase which negatively regulates p53 protein stability and transcriptional activity (Iwakuma and Lozano, 2003). These data underscore the fact that decreased levels of p53, resulting from mutations in the p53 gene or alterations in the stoichiometry of its inhibitor MDM2 play a critical role during tumor development.
A T-to-G single nucleotide polymorphism (SNP) in the second promoter (P2) of the human MDM2 gene (SNP309G) has been identified (Bond et al., 2004). This SNP is present in the heterozygous state (G/T) in approximately 40% and in the homozygous state (G/G) in 15% of healthy individuals, respectively (Bond et al., 2006). MDM2SNP309G enhances the binding of the transcriptional activator Sp1 to the P2 promoter of MDM2 resulting in a constitutive increase in MDM2 transcription. This in turn leads to increased levels of the MDM2 protein and therefore decreased p53 protein levels. Most importantly, the MDM2SNP309G allele has been associated with an increased cancer risk in some human tumors that express wild type p53 (Bond et al., 2006; Dharel et al., 2006; Grochola et al., 2009; Yarden et al., 2008). However, a significant number of reports have failed to corroborate such a notion (Krekac et al., 2008; Sajid et al., 2008). Clinical correlates aimed at supporting the effect of the MDM2SNP309G allele in humans with spontaneous cancers have been controversial, likely as a result of the heterogeneity of the employed datasets and the retrospective nature of these analyses (Bond et al., 2006; Bond and Levine, 2007; Copson et al., 2006; Dharel et al., 2006; Economopoulos and Sergentanis, 2009; Ellis et al., 2008; Grochola et al., 2009; Menin et al., 2006; Schmidt et al., 2007; Yarden et al., 2008)
Additional support of an enhanced cancer risk in MDM2SNP309 carriers is the fact that patients diagnosed with Li-Fraumeni syndrome (LFS) with an inherited germline mutation in p53 and homozygous for the G nucleotide at MDM2SNP309 develop tumors approximately 10 years earlier than LFS patients lacking this polymorphism (Bond et al., 2004; Bougeard et al., 2006; Marcel et al., 2009; Ruijs et al., 2007; Tabori et al., 2007). Additionally, patients with LFS carrying two MDM2SNP309G alleles are more frequently diagnosed with multiple primary tumors compared to LFS patients carrying two MDM2SNP309T alleles. Together, these data suggest an enhanced cancer phenotype in patients carrying germline-inactivating mutations in p53 and MDM2SNP309G. Thus, increased MDM2 levels resulting from the presence of the MDM2SNP309G allele may further down modulate an already muted p53 pathway. These data are however correlative by nature.
Recent attempts to understand the mechanisms that regulate the Mdm2-p53 pathway during tumorigenesis have focused on the generation of mouse models that genetically delete p53, overexpress genes that regulate p53, or produce mutant p53 proteins that mimic human mutations (Donehower and Lozano, 2009). Additionally, though, more subtle changes also affect tumorigenesis. For example, haploinsufficiency at the Mdm2 locus delays tumor onset in mice carrying an Eµ-myc transgene and also renders mice sensitive to DNA damage (Alt et al., 2003; Mendrysa et al., 2003; Terzian et al., 2007). However, little is known about the impact of more subtle genetic modifiers that affect the regulation or expression of proteins involved in tumorigenesis. In this study, we have used the naturally occurring polymorphism in the MDM2 promoter to generate two humanized Mdm2SNP309 murine alleles in order to examine the direct impact of this polymorphism on tumor development.
Results
Generation of Mdm2SNP309 Mice
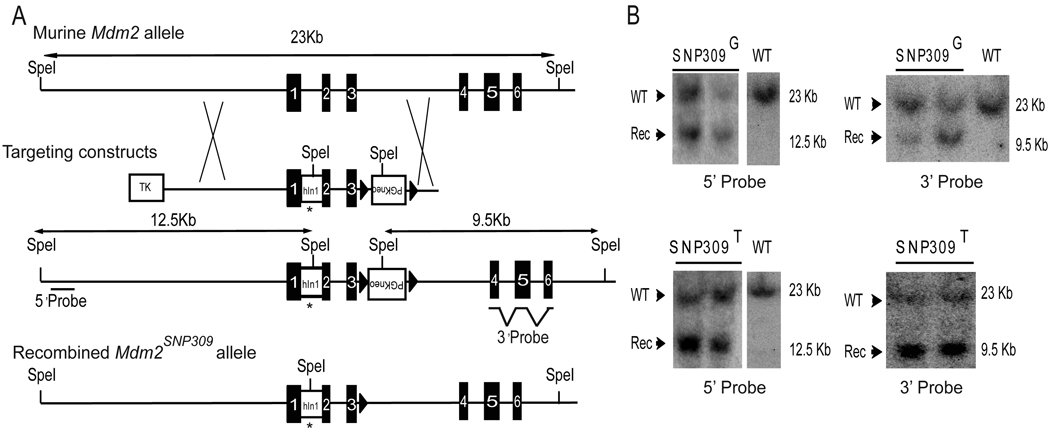
To directly test the significance of Mdm2SNP309 in a prospective manner, we generated humanized Mdm2SNP309 alleles in the mouse. Using a PCR based strategy, we generated Mdm2SNP309G and Mdm2SNP309T targeting constructs by replacing the mouse intron 1 (containing the entire P2 promoter) with the corresponding human intron 1 with either the G or T polymorphism. Both targeting constructs were sequenced in their entirety to rule out the presence of additional changes. MDM2SNP309G and MDM2SNP309T human intron 1 sequences are identical, except for the polymorphism (Figure S1A). The polymorphisms were introduced into the murine Mdm2 locus by homologous recombination in embryonic stem (ES) cells (Figure 1A). Southern blot analysis of the recombination event revealed correct targeting of both constructs at the murine Mdm2 locus (Figure 1B). Additionally, Southern blot analysis using a probe against the neomycin cassette verified single copy integration and indicated the absence of other insertions (Figure S1B and C). Chimeric mice were backcrossed with C57Bl/6 mice for five generations, including one cross with Zp3-Cre mice (also in a C57Bl/6 background) that resulted in Cre-loxP-mediated excision of the PGK-neomycin cassette. A cohort of Mdm2SNP309G/G and Mdm2SNP309T/T mice was established for tumor studies. The background of all mice used in these studies is greater than 98% C57Bl/6.
Figure 1. Generation of mice containing either the humanized Mdm2SNP309G or Mdm2SNP309T allele.
(A) The targeting vectors were designed to incorporate the PGK-neo selectable marker in intron 3 flanked by loxP sites (triangles) containing the entire human intron 1 (hIn1) with a G or T nucleotide at SNP309 (denoted by an asterisk). Black numbered boxes denote Mdm2 exons. Following Cre-mediated recombination, the PGK-neo cassette was deleted. (B) Southern blot analysis of SpeI-digested DNA from ES cell clones electroporated with the Mdm2SNP309G or the Mdm2SNP309T targeting vectors. The 23-kb SpeI fragment represents the wild-type Mdm2 allele while the 12.5-kb SpeI fragment is the expected size for the recombined allele using a 5' probe. Wild type (WT) DNA serves as a control. The 9.5-kb SpeI fragment is the expected size for the recombined alleles using a 3' probe. See also Figure S1.
Mdm2 mRNA Levels are Increased in Tissues from Mdm2SNP309G/G Mice
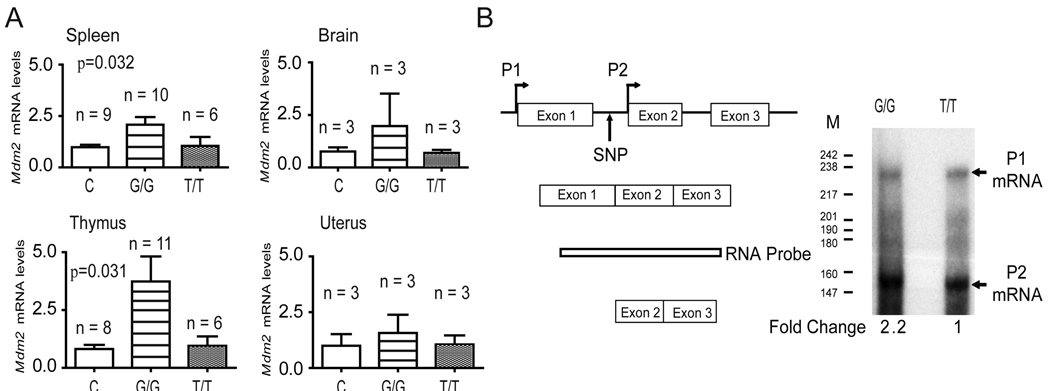
To characterize the polymorphic alleles, we first measured the total level of Mdm2 mRNA in Mdm2SNP309 mice. Real time RT-PCR revealed that the spleens and thymi (two p53 sensitive tissues) of six-week-old Mdm2SNP309G/G mice had significantly higher levels of Mdm2 mRNA (2.08 ± 0.37 and 3.7 ± 1.1, spleens and thymi, respectively) as compared to the control Mdm2SNP309T/T (1.05 ± 0.43 and 0.95 ± 0.41, spleens and thymi, respectively) and wild-type C57Bl/6 (which carry the endogenous murine intron 1) (0.98 ± 0.12 and 0.81 ± 0.18, spleens and thymi, respectively) mice. The levels of Mdm2 in the Mdm2SNP309G/G tissues were significantly higher than in control mice (p=0.032 and p=0.031, spleen and thymus respectively, one-way analysis of variance (ANOVA)) (Figure 2A). Likewise, the brains and uteri of six-week-old Mdm2SNP309G/G mice had higher levels of Mdm2, although not statistically significant, as compared to control mice (Figure 2A). RNAse protection assays were subsequently used to quantitate and distinguish the otherwise identical transcripts arising from the basal (P1) and P2 promoters (the second promoter that has the MDM2SNP309). Data show a 2.2 fold increase in Mdm2 levels from the P2 promoter in spleens taken from Mdm2SNP309G/G mice as compared to those obtained from Mdm2SNP309T/T mice (Figure 2B). The levels of Mdm2 mRNA from the P1 promoter were unchanged in both Mdm2SNP309G/G and Mdm2SNP309T/T mice (Figure 2B). These data were further validated using real time RT-PCR primers specific to the transcript generated from the P1 promoter. No significant difference was noted in the levels of P1 Mdm2 mRNA in C57Bl/6 (0.86 ± 0.12), Mdm2SNP309G/G (1.16 ± 0.28), and Mdm2SNP309T/T (1.16 ± 0.15) mice (ANOVA, p = 0.469, Figure S2). These data indicate that the increased levels of Mdm2 in vivo are specifically due to the presence of Mdm2SNP309G in the P2 promoter.
Figure 2. Mdm2SNP309G/G mice have higher levels of Mdm2 mRNA as compared to Mdm2SNP309T/T and C 57Bl/6 mice.
(A) Real-time RT-PCR analysis for Mdm2 mRNA levels in spleen, brain, thymus, and uterus of C57Bl/6 (C), Mdm2SNP309G/G (G/G), and Mdm2SNP309T/T (T/T) mice. The mean and standard error of the mean (SEM) were determined from triplicate samples after normalization to Rplp0 for the number (n) of mice indicated. Statistical significance in the spleen (p = 0.032) and thymus (p = 0.031) was determined by one-way analysis of variance (ANOVA). (B) RNase protection analysis for Mdm2 mRNA levels in spleen of Mdm2SNP309G/G and Mdm2SNP309T/T mice. Schematic of the Mdm2 locus and RNAse protection probe. P1 (transcript from promoter 1), upper band and P2 (transcript from promoter 2), lower band. Fold change was determined by calculating the ratios of P2/P1 in each sample. See also Figure S2.
Mdm2 Levels in Mdm2SNP309G/G Mouse Embryonic Fibroblasts are Sensitive to the Sp1 Inhibitor Mithramycin A
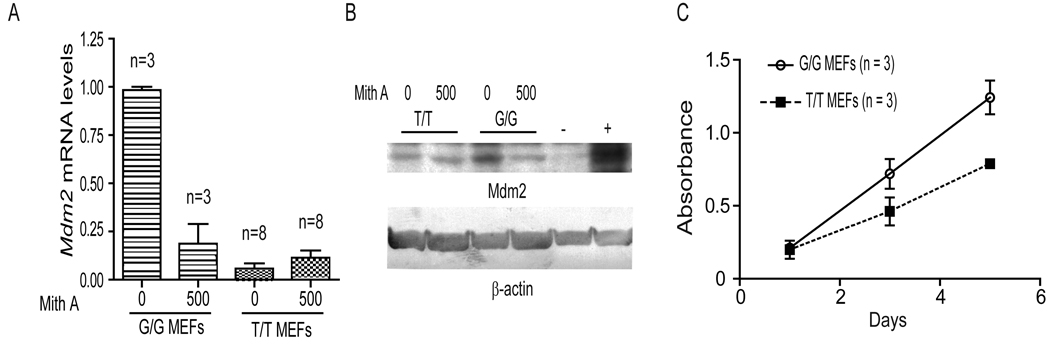
In humans cell lines the MDM2SN309 polymorphism creates a stable binding site for the transcription factor Sp1 (Bond et al., 2004). Mithramycin A disrupts the activity of Sp1, and treatment of MDM2SNP309G/G human cells with Mithramycin A resulted in a decrease in Mdm2 levels (Bond et al., 2004; Chien et al., 2010). We therefore assessed the impact of mithramycin A on Mdm2 mRNA and protein levels. The levels of Mdm2 were significantly reduced in Mdm2SNP309G/G mouse embryo fibroblasts (MEFs) upon mithramycin A treatment (0.98 ± 0.03 and 0.19 ± 0.18, untreated versus treated, respectively, p < 0.0001) (Figure 3A). Western blot analysis revealed that Mdm2SNP309G/G MEFs had lower levels of Mdm2 protein following mithramycin A treatment (Figure 3B). Mithramycin A did not significantly alter the levels of Mdm2 in Mdm2SNP309T/T MEFs, although the Mdm2 levels in the untreated Mdm2SNP309T/T MEFs were much lower than the untreated Mdm2SNP309G/G MEFs (Figure 3A and B). Thus, while the basal levels of Mdm2 were dramatically different in fibroblasts, mithramycin A decreased expression of Mdm2 only in Mdm2SNP309G/G mice.
Figure 3. Mdm2 levels are higher in Mdm2SNP309G/G mouse embryonic fibroblasts as compared to Mdm2SNP309T/T and C 57Bl/6 fibroblasts.
(A) Real time RT-PCR analysis for transcriptional activation of Mdm2 in Mdm2SNP309G/G and Mdm2SNP309T/T mouse embryo fibroblasts (MEFs) isolated from at least 3 separate embryos per genotype. MEFs were either untreated or treated with 500nM mithramycin A for 18 hours. The mean and standard error of the mean (SEM) were determined from triplicate samples after normalization to Rplp0 for the number (n) of different MEF lines indicated. (B) Western blots of untreated or 500nM mithramycin A treated Mdm2SNP309G/G and Mdm2SNP309T/T lysates were performed and blotted with Mdm2 and β-actin antibodies. Lysates from IR treated Mdm2-transgenic (+) and Mdm2−/− p53−/− MEFs (−) serve as positive and negative controls, respectively. (C) Cell proliferation rates of Mdm2SNP309G/G and Mdm2SNP309T/T MEFs. Three separate low passage (P2) MEF cell lines per genotype were plated, grown for 1, 3, or 5 days, and then assayed for cell number. Each data point represents the mean and standard error of the mean (SEM) for three separate MEF lines per genotype.
Given the differences in Mdm2 levels between the Mdm2SNP309G/G and Mdm2SNP309T/T MEFs, we tested whether these differences conferred a growth advantage. We found that the Mdm2SNP309G/G MEFs had an increased rate of proliferation as compared to the Mdm2SNP309T/T MEFs, with a slope of 0.26 ± 0.029 and 0.15 ± 0.022, respectively (Figure 3C). Thus early passage Mdm2SNP309G/G MEFs have a growth advantage as compared to the Mdm2SNP309T/T MEFs.
The p53 Pathway is Partially Attenuated in Mdm2SNP309G/G Mice
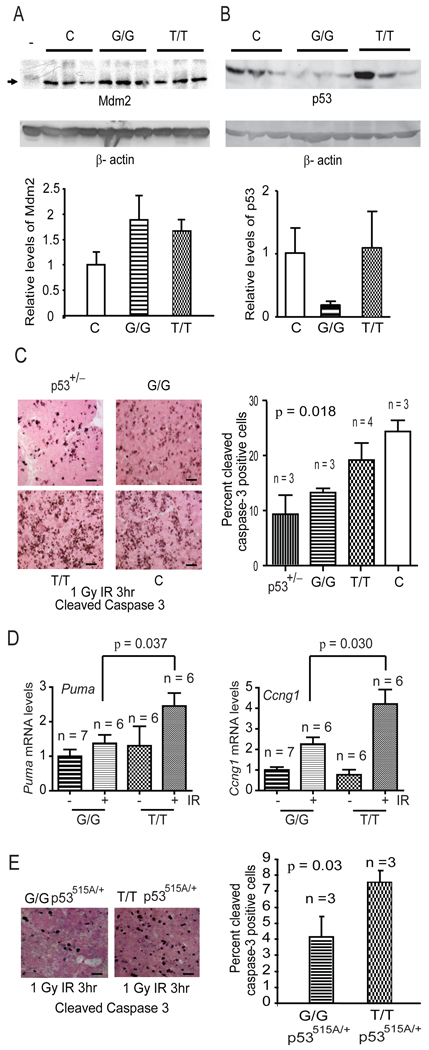
It is widely appreciated that Mdm2 levels tightly control p53 levels in vivo (Iwakuma and Lozano, 2003). In order to determine how small increases in Mdm2 levels impact the p53 pathway, we first examined the levels of Mdm2 and p53 protein in mouse tissues. Spleens from Mdm2SNP309G/G mice had elevated levels of Mdm2 protein and consequently lower levels of p53 as compared to control Mdm2SNP309T/T and wild type C57Bl/6 mice (Figure 4A and B). To test the functional consequences of lower levels of p53 in mice harboring the Mdm2SNP309G allele, Mdm2SNP309G/G, Mdm2SNP309T/T, p53+/−, and C57Bl/6 mice were exposed to low dose (1 Gy) ionizing radiation (IR). This dose of IR triggers a robust p53 response in the thymus (Alvarez et al., 2006). Thymi from Mdm2SNP309G/G mice exhibited a significantly lower apoptotic response following low dose IR (13.3 ± 0.77% cleaved caspase-3 positive cells) as compared to the Mdm2SNP309T/T (19.0%± 3.1% cleaved caspase-3 positive cells) or wild type C57Bl/6 (24.3 ± 2.0% cleaved caspase-3 positive cells) mice (Figure 4C, ANOVA, p = 0.018). In fact, the levels of p53-dependent apoptosis in Mdm2SNP309G/G thymi were comparable to p53 heterozygous mice (p = 0.304). These data suggest that the presence of two Mdm2SNP309G alleles significantly inhibited p53-dependent apoptosis in response to DNA damage. To examine the activation of downstream effectors of p53 function, we quantified activation of apoptotic and cell cycle arrest transcriptional targets of p53 following IR. The pro-apoptotic factor, Puma, and the cell cycle inhibitor, Ccng1 (the gene that codes for Cyclin G1), were significantly lower in thymi from irradiated Mdm2SNP309G/G mice as compared to thymi from irradiated Mdm2SNP309T/T mice, (p = 0.037 and p = 0.030, respectively) (Figure 4D). Two other p53 targets, Pig8 and p21, were decreased in thymi isolated from irradiated Mdm2SNP309G/G mice; however, neither reached statistical significance (Figure S3A). These data suggest that the presence of the Mdm2SNP309G allele attenuates activation of the p53 pathway in the thymus following low dose irradiation.
Figure 4. The Mdm2SNP309G allele results in reduced p53 activity.
(A) Western blots of C57Bl/6 (C), Mdm2SNP309G/G (G/G), and Mdm2SNP309T/T (T/T) splenic lysates were performed and blotted with Mdm2 and β-actin antibodies. Mdm2−/− p53−/− splenic lysates (−) serve as an Mdm2 negative control. The mean and standard error of the mean (SEM) of the three control samples was determined by the ratio between the Mdm2 bands and the corresponding β-actin bands and arbitrarily set to one. The mean and SEM of Mdm2 in splenic lysates from Mdm2SNP309G/G and Mdm2SNP309T/T mice was determined by comparing the ratios of Mdm2/β-actin normalized to the control ratio. (B) Western blots of C57Bl/6 (C), Mdm2SNP309G/G (G/G), and Mdm2SNP309T/T (T/T) lysates were performed and blotted with p53 and β-actin antibodies. The mean and standard error of the mean (SEM) was determined as above. (C) Apoptosis was determined by counting the percentage of cleaved caspase-3 positive cells in the thymi of p53+/− (9.1% ± 3.5%), Mdm2SNP309G/G (G/G) (13.3% ± 0.77%), Mdm2SNP309T/T (T/T) (19.0% ± 3.1%), and C57Bl/6 (C) (24.3% ± 2.0%) mice following 1 Gy ionizing radiation (IR). At least 300 cells were counted from three randomized high magnification fields per animal. The mean and standard error of the mean (SEM) for each genotype were calculated from at least three thymi per genotype. Statistical significance was determined by one-way analysis of variance (ANOVA). n = the number of mice examined for each genotype. The bar scale represents 50 µm. (D) Real-time RT-PCR analysis for mRNA levels of the p53-dependent targets Puma and Ccng1 in the thymus of untreated (−) Mdm2SNP309G/G (G/G) and Mdm2SNP309T/T (T/T) mice or Mdm2SNP309G/G (G/G) and Mdm2SNP309T/T (T/T) mice treated (+) with 1 Gy ionizing radiation. Thymi were harvested from irradiated mice 3 hours post-treatment. The mean and standard error of the mean (SEM) were determined from triplicate samples after normalization to Rplp0 for the number (n) of mice indicated. Statistical significance was determined by unpaired Student t test. (E) Apoptosis was determined by counting the percentage of cleaved caspase-3 positive cells in Mdm2SNP309T/T p53515A/+ and Mdm2SNP309G/G p53515A/+ thymi. At least 300 cells were counted from three high magnification fields per thymus per genotype. The mean and SEM of each genotype were calculated from three separate mice. Statistical significance was determined by unpaired Student t test. n = the number of mice examined for each genotype. The bar scale represents 50 µm. See also Figure S3.
To explore the impact of the Mdm2SNP309G allele on other tissues exposed to low dose IR, we collected epithelial tissues from these mice. Several transcriptional targets of p53 were modestly attenuated in the intestine and breast (female) of Mdm2SNP309G/G mice, specifically Puma in intestine and p21 in breast, as compared to the Mdm2SNP309T/T mice following low dose IR; however, transactivation of no gene reached statistical significance (Figure S3B and C). These results suggest that either epithelial tissues are less sensitive to low dose IR or that transactivation of p53 targets is tissue specific following exposure to IR. Lastly, we measured p53 protein levels and phosphorylation in different tissues after low dose IR. Mdm2SNP309G/G thymus and breast tissues show lower basal levels of p53 (Figure 3D and E), consistent with our observation of low basal p53 levels in the Mdm2SNP309G/G spleens (Figure 4B). No statistically significant differences were observed in p53 levels 3 hours post IR (Figure S3F and G). Together, these findings suggest that following IR the polymorphic Mdm2SNP309 allele has a more prominent impact on p53 activity rather than on p53 stability.
We also examined the impact of Mdm2SNP309G/G on apoptosis in a mutant p53 background by crossing the Mdm2SNP309 mice with mice carrying the p53515A hot-spot mutation that encodes the p53R172H mutant protein (Lang et al., 2004). Thymi from Mdm2SNP309G/G p53515A/+ mice also had a significant reduction in apoptotic cells as compared to the control Mdm2SNP309T/T p53515A/+ mice after low dose IR (p=0.03) (Figure 4E). These data suggest that mice harboring two Mdm2SNP309G alleles have a diminished p53 response following DNA damage both in wild type and heterozygous mutant p53 backgrounds.
Mdm2SNP309G Allele Resulted in Increased Cancer Risk
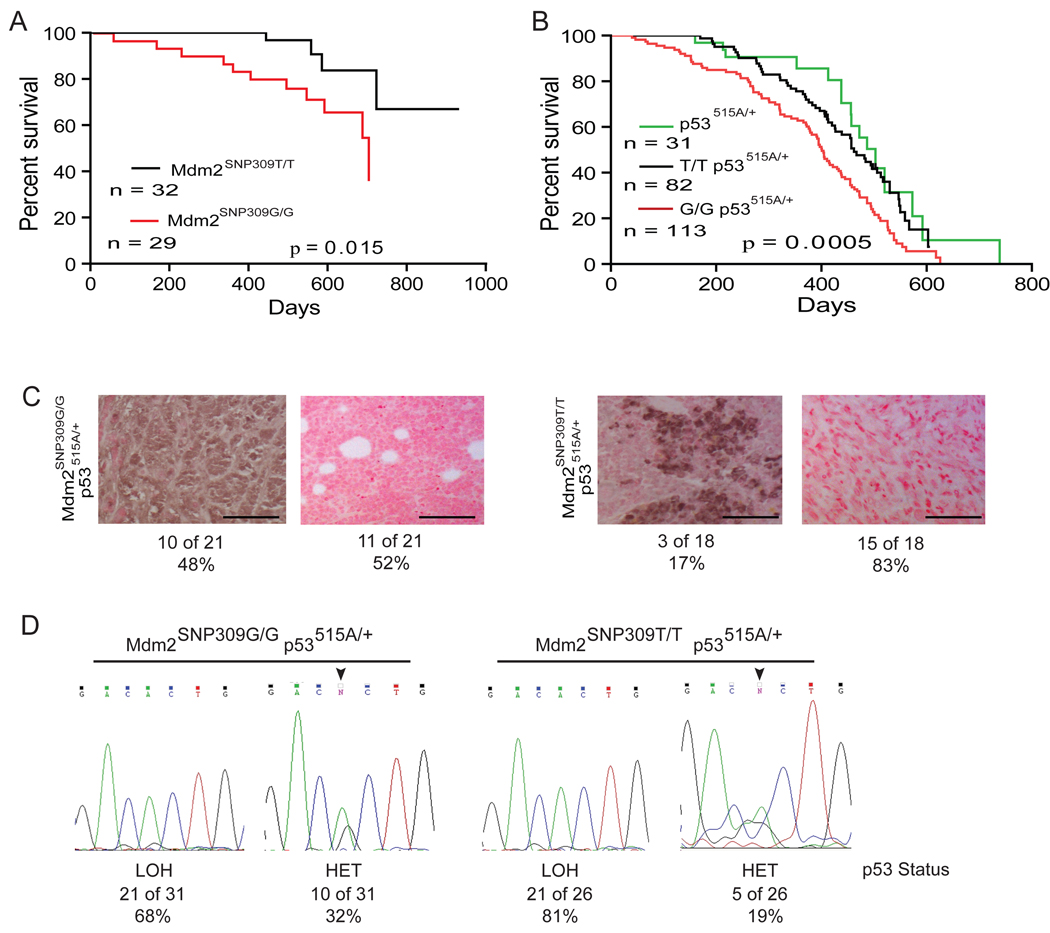
To investigate the impact of Mdm2SNP309G/G on cancer risk in vivo we monitored cohorts of Mdm2SNP309G/G, Mdm2SNP309T/T, Mdm2SNP309G/G p53515A/+, Mdm2SNP309T/T p53515A/+, and p53515A/+ mice for tumor formation. Tumor burdened and moribund mice of each genotype were sacrificed and tumors were harvested for pathological analysis. The overall survival of Mdm2SNP309G/G mice was reduced as compared to the Mdm2SNP309T/T mice (p=0.015) (Figure 5A). Five of ten Mdm2SNP309G/G mice and one of five Mdm2SNP309T/T mice that died had obvious tumor involvement (Table 1). Several Mdm2SNP309G/G and Mdm2SNP309T/T mice died of unknown causes. Mdm2SNP309G/G p53515A/+ mice also succumbed to tumors significantly earlier than the Mdm2SNP309T/T p53515A/+ and p53515A/+ mice (p =0.0005) (Figure 5B). The median overall survival for the Mdm2SNP309G/G p53515A/+ and Mdm2SNP309T/T p53515A/+ cohorts was 401 and 482 days, respectively (Hazard ratio =0.561; Confidence interval = 0.408 to 0.786). These results indicate that the Mdm2SNP309G allele directly increases cancer risk.
Figure 5. Mice harboring two Mdm2SNP309G alleles have decreased survival as compared to mice possessing two Mdm2SNP309T alleles.
(A) Kaplan-Meier curves indicating survival of Mdm2SNP309T/T and Mdm2SNP309G/G mice. Statistical significance was determined by Log-rank test. (B) Kaplan-Meier curves indicating the survival of p53515A/+, Mdm2SNP309T/T p53515A/+, or Mdm2SNP309G/G p53515A/+ mice. Statistical significance was determined by Log-rank test. (C) Mdm2 expression in tumors from Mdm2SNP309G/G p53515A/+ and Mdm2SNP309T/T p53515A/+ mice. Forty-eight percent (10 of 21) of tumors from Mdm2SNP309G/G p53515A/+ mice and 17% (3 of 15) Mdm2SNP309T/T p53515A/+ mice express Mdm2, (p = 0.041). Statistical significance was determined by the χ-squared test. Tumor sections were considered positive if greater than 10% of the total cells were immunoreactive for Mdm2. The bar scale represents 100 µm. (D) Loss of heterozygosity (LOH) varies at the p53 locus in Mdm2SNP309G/G p53515A/+ and Mdm2SNP309T/T p53515A/+ tumors. Exon 5, containing the p53515A mutation, was sequenced in 31 Mdm2SNP309G/G p53515A/+ and 26 Mdm2SNP309T/T p53515A/+ tumors, respectively. LOH was determined by a greater than 50% reduction of the wild type allele, noted by the absence of the G nucleotide at position 515. HET denotes retention of both p53 alleles. See also Figure S4.
Table 1.
Tumor spectrum of Mdm2SNP309G/G,Mdm2SNP309T/T,Mdm2SNP309G/G p53515A/+, and Mdm2SNP309T/T p53515A/+ mice. Parentheses denote number of metastatic tumors.
| Genotype |
|||||
|---|---|---|---|---|---|
| Tumor Type |
Mdm2SNP309G/ G | Mdm2SNP309T/T |
Mdm2SNP309G/G p53515A/+ |
Mdm2SNP309T/T p53515A/+ |
|
| (n = 5) | (n = 1) | (n = 48) | (n = 23) | ||
| Sarcoma | 0 | 1 | 34 | 15 | |
| Fibrosarcoma | 0 | 0 | 10 | 2 | |
| Leiomyosarcoma | 0 | 0 | 1 | 0 | |
| Pleomorphic | 0 | 0 | 2(1) | 0 | |
| Osteosarcoma | 0 | 0 | 13(2) | 6(1) | |
| Histocytic | 0 | 0 | 4 | 5 | |
| Spindle Cell | 0 | 0 | 2(1) | 1 | |
| Histiocytoma | 0 | 0 | 2 | 0 | |
| Ovarian Hemangiosarcoma | 0 | 0 | 1 | 0 | |
| Soft Tissue Sarcoma | 0 | 1 | 1 | 2 | |
| Lipoma | 0 | 0 | 2 | 1 | |
| Chondrosarcoma | 0 | 0 | 1 | 0 | |
| Lymphoma | 4 | 0 | 8 | 1 | |
| Adenoma | |||||
| Lung | 0 | 0 | 1 | 0 | |
| Hepatocellular | 0 | 0 | 1 | 0 | |
| Papilloma | |||||
| Squamous Cell | 0 | 0 | 1 | 0 | |
| Adenocarcinoma | |||||
| Mammary | 1 | 0 | 3 | 0 | |
| Hepatocellular | 0 | 0 | 1 | 2 | |
| Undefined | 0 | 0 | 3 | 3 | |
| Carcinoma | |||||
| Salivary Carcinoma | 0 | 0 | 1(1) | 0 | |
| Squamous Cell | 1 | 0 | 2 | 1 | |
| Undefined | 0 | 0 | 4 | 1 | |
| Total | 6 | 1 | 64 | 24 | |
Mdm2SNP309G Impacts the Mdm2-p53 Axis in Tumors
To explore the molecular events that occur during tumor formation, we investigated the status of Mdm2 and p53. Immunohistochemical analysis of Mdm2 in tumors isolated from the Mdm2SNP309 mice showed 48% (10 of 21) of Mdm2SNP309G/G p53515A/+ tumors expressed Mdm2, while only 17% (3 of 18) Mdm2SNP309T/T p53515A/+ tumors expressed Mdm2 (p = 0.041, Figure 5C). Since cells exert tremendous selective pressure to delete wild type p53 function during tumorigenesis, we also tested whether these tumors underwent loss of heterozygosity (LOH) of the remaining wild type p53 allele. Notably, 32% (10 of 31) of Mdm2SNP309G/G p53515A/+ tumors retained the wild type p53 allele compared to only 19% (5 of 26) of Mdm2SNP309T/T p53515A/+ tumors, although these data were not statistically significant (Figure 5C). Western blot analysis revealed that all but one tumor (12 of 13) that retained the wild type p53 allele expressed p53, regardless of genotype (Figure S4A). Additionally, p53 was phosphorylated in all the tumors that retained both p53 alleles (Figure S4A). In tumors with LOH of the wild type p53 allele, 60% (3 of 5) of the Mdm2SNP309G/G p53515A/+ tumors expressed mutant p53 and 100% (6 of 6) of the Mdm2SNP309T/T p53515A/+ tumors expressed high levels of mutant p53 (Supplemental Figure 6B). Furthermore, 40% (2 of 5) of the Mdm2SNP309G/G p53515A/+ tumors had high levels of phosphorylated mutant p53, while 67% (4 of 6) of the Mdm2SNP309T/T p53515A/+ tumors expressed phosphorylated mutant p53 (Figure S4B).
An examination of p53 targets in tumors revealed that mRNA levels of p53 targets varied greatly in each tumor, likely as a consequence of multiple additional genetic alterations occurring during tumorigenesis. However, one p53-dependent pro-apoptotic target, Perp, was significantly attenuated in the tumors for the Mdm2SNP309G/G p53515A/+ mice as compared to tumors from the Mdm2SNP309T/T p53515A/+ mice (p = 0.023, Figure S4C).
Mdm2SNP309G Alters Tumor Spectrum
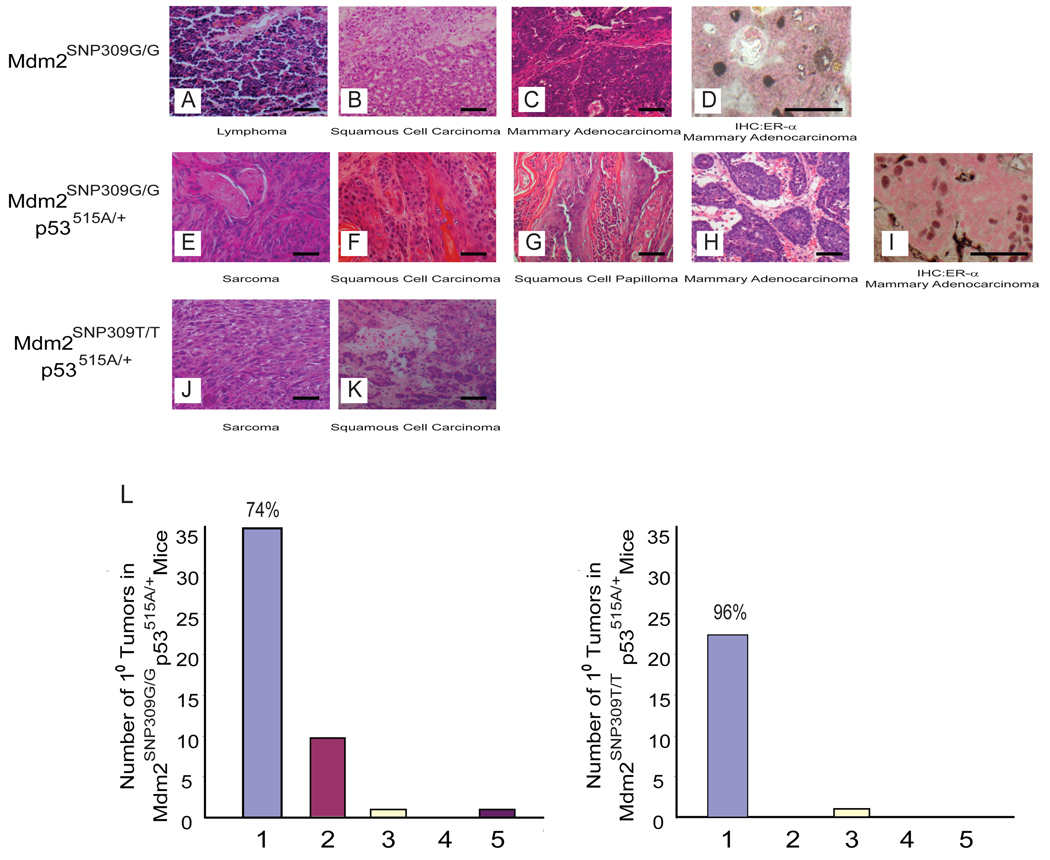
In addition to accelerated tumor onset, Mdm2SNP309G/G and Mdm2SNP309G/G p53515A/+ mice also developed tumor types only anecdotally reported in C57Bl/6 or p53515A mice. One Mdm2SNP309G/G mouse developed a mammary adenocarcinoma in addition to a lymphoma (Figures 6 A–D, and Table 1). Three Mdm2SNP309G/G p53515A/+ mice developed mammary adenocarcinomas (Figures 6 E–I and Table 1), and one developed a squamous cell papilloma (Figure 6G). Since mammary adenocarcinomas have never been detected in our p53 cohorts in a C57Bl/6 background, we confirmed the breast origin of these tumors by demonstrating the expression of the estrogen receptor-a (Figure 6D and I). Additionally and similar to patients with LFS that harbor two Mdm2SNP309G alleles (Bond et al., 2004), Mdm2SNP309G/G p53515A/+ mice also developed multiple primary tumors as compared to Mdm2SNP309T/T p53515A/+ mice (Figure 6L), recapitulating the human phenotype.
Figure 6. Mice harboring two Mdm2SNP309G alleles express high levels of Mdm2 and develop a unique tumor spectrum as compare to mice harboring two Mdm2SNP309T alleles.
(A–C) Hematoxylin and eosin (H & E) staining of paraffin-embedded sections of tumors from Mdm2SNP309G/G mice. (A) Lymphoma. (B) Squamous cell carcinoma. (C) Mammary adenocarcinoma (D) Immunohistochemistry of estrogen receptor-α positive mammary adenocarcinoma from an Mdm2SNP309G/G mouse. (E–H) H & E staining of paraffin-embedded sections of tumors from Mdm2SNP309G/G p53515A/+ mice. (E) Sarcoma. (F) Squamous cell carcinoma. (G) Squamous cell papilloma (H) Mammary adenocarcinoma. (I) Immunohistochemistry of estrogen receptor-α positive mammary adenocarcinoma from an Mdm2SNP309G/G p53515A/+ mouse. (J and K) H & E staining of paraffin-embedded sections of tumors from Mdm2SNP309T/T p53515A/+ mice. (J) Sarcoma. (K) Squamous cell carcinoma. The bar scale represents 50 µm in each figure. (L) The graph depicts the number of primary tumors in Mdm2SNP309G/G p53515A/+ and Mdm2SNP309T/T p53515A/+ mice. The number above the bar represents the percentage of mice that had one primary tumor.
Discussion
The sequencing of the human genome has unveiled the presence of millions of SNPs (Venter et al., 2001). Deciphering the effects of individual SNPs in human cancer has proven extremely challenging given the multitude of SNPs, the number of redundant and overlapping pathways they intersect with, and the heterogeneous genetic make-up of humans. Since the identification of the Mdm2SNP309 allele, multiple studies involving large numbers of patients diagnosed with cancer have suggested that the Mdm2SNP309G allele significantly increases cancer risk, while other studies have failed to show such an association. For less common cancers, such as leukemia, ovarian cancer, lymphoma, or melanoma, the impact of Mdm2SNP309 is even less clear given the retrospective nature of these studies and the small size of the cohorts analyzed.
In order to determine the direct impact of this SNP in tumorigenesis, we generated two genetically engineered mouse models carrying either Mdm2SNP309G or Mdm2SNP309T alleles and followed both cohorts of animals, prospectively. A critical advantage of this mouse model is that we have controlled for the cancer phenotype variability that arises from environmental, stochastic, and modifier gene effects as these mice are greater that 98% C57Bl/6 and, therefore, isogeneic. Mice carrying two Mdm2SNP309G alleles had increased levels of Mdm2 mRNA and protein, decreased p53 levels, and a marked attenuation of the p53 response following ionizing radiation as compared to Mdm2SNP309T/T or wild type C57Bl/6 mice. Our findings suggest that the presence of the Mdm2SNP309G allele contributes to accelerated tumor formation and reduction in overall survival in mice that harbor either wild type or mutant p53. The observation that the Mdm2SNP309G allele alone is sufficient to cause an increased cancer risk in our mouse models supports the retrospective studies associating the MDM2SNP309G allele with enhanced tumorigenesis in humans.
Following the initial description of the MDM2SNP309 allele, the actual impact of this SNP on human cancers has been confounded by conflicting reports vis-à-vis its impact on tumorigenesis in different tissues, ethnicities, or gender. In our study, significant differences in Mdm2 levels were observed in some tissues but not others. Additionally, some of the mice that carry two Mdm2SNP309G alleles developed female cancers such as breast adenocarcinomas regardless of p53 status. This is at variance with the findings observed in Mdm2SNP309T/T p53515A/+ control mice, in which no breast tumors were observed. The development of breast cancer is highly unusual in C57Bl/6 mice, particularly in the absence of an accompanying p53 mutation. These data suggest there may indeed be tissue specific consequences resulting from the presence of two Mdm2SNP309G alleles. Previous studies have shown that forced expression of Mdm2 in murine mammary tissue resulted in increased proliferation and development of breast cancer (Lundgren et al., 1997). The finding that some of these C57Bl/6 mice that carry the Mdm2SNP309G allele also develop breast cancer indicates that even a slight elevation of Mdm2 may result in an increased breast cancer risk.
Our studies emphasize the fact that an ∼2 fold increase in Mdm2 in some tissues significantly increases cancer risk. In comparison, Mdm2 haploinsufficiency (a 2-fold decrease) enhanced survival in several tumor models, further emphasizing the sensitivity of p53 regulation by Mdm2 (Alt et al., 2003; Mendrysa et al., 2006; Terzian et al., 2007). The presence of the Mdm2SNP309G allele alters the homeostasis of the Mdm2/p53 axis resulting in decreased basal steady state levels of p53. The most likely explanation is that Mdm2 has the ability to target p53 for degradation more efficiently than itself. The low steady state levels of p53 observed in these Mdm2SNP309G/G mice may have a significant impact on potential tumor formation, as cells from these tissues may have a greater propensity to give rise to a fully transformed cell in response to even minor genomic insults or mitogenic stimulation. Thus, slight perturbations (up or down) to the p53 pathway may significantly impact tumor formation, and perhaps, response to chemotherapy.
In addition to tumor formation, Mdm2 and p53 also regulate each other in response to DNA damage (Haupt et al., 1997; Kubbutat et al., 1997; Shieh et al., 1997; Siliciano et al., 1997). Loss of one Mdm2 allele renders mice harboring wild-type p53 sensitive to p53-dependent toxicity following IR (Mendrysa et al., 2003; Terzian et al., 2007), further emphasizing the importance of Mdm2 dosage on p53-dependent phenotypes. Our finding that steady state levels of p53 are decreased, coupled with the findings that p53-dependent apoptosis and transactivation of p53 targets are lower in Mdm2SNP309G/G mice exposed to IR suggests that modest increases in Mdm2 levels can disrupt p53 stability and activity. It is noteworthy that p53 levels were similar in both Mdm2SNP309G/G and Mdm2SNP309T/T mice after IR, even though activation of p53 targets was ablated in the Mdm2SNP309G/G mice, suggesting that stabilization of p53 caused by ionizing radiation may overwhelm the destabilizing effects of the Mdm2SNP309G allele on p53. It is therefore tempting to hypothesize that normal individuals harboring two Mdm2SNP309G alleles may be more sensitive to exogenous low level DNA damage caused by environmental stress due to lower steady state p53 levels. A strength of this Mdm2SNP309G mouse model herein reported is that it may aid in understanding the complex and fine-tuned interplay between variants of the p53 pathway (e.g. p53 mutations, Mdm2 SNPs), environmental stresses (e.g. carcinogens), response to chemotherapeutic agents, and tumor-independent variables (e.g. gender) in human cancer.
Experimental Procedures
Generation of Mdm2SNP309G/G and Mdm2SNP309T/T mice
A PCR based strategy was employed to generate a targeting vector containing the deletion of murine intron 1 with replacement by the human intron 1 harboring either the G or T nucleotide at position 309. To this end, the human intron 1 was cloned by PCR amplification of human genomic DNA using chimeric 5’-mouse-human-3’ forward and reverse primers. The forward primer contained 21 nucleotides upstream of the mouse exon 1-intron 1 junction. Three additional C nucleotides were added to the 5’ end of the forward primer in order to create a unique SmaI site. These three C nucleotides were deleted during blunt end digestion and thus resulted in no additions or alterations to the genome. The reverse 5’ murine-human 3’ primer contained 32 nucleotides of the mouse genome downstream of the mouse intron 1-exon 2 junction. Sequence analysis of the PCR product confirmed the presence of the G nucleotide at position 309 in the human MDM2 intron 1 amplicon. The human MDM2 intron 1 containing the T nucleotide was generated by site directed mutagenesis. The resulting human MDM2 intron 1 amplicons, possessing murine exon 1 and exon 2 ends, were used as reverse MEGAprimers along with forward primer 5’-CTAGCGACCATTGCGGTTTCGAG-3’ (∼ 300 base pairs upstream of murine exon 1). This resulted in an amplicon containing murine exon 1, human intron 1, and a portion of murine exon 2. Next, we used PCR to generate a murine amplicon possessing the remainder of exon 2, intron 3, exon 3, a portion of intron 3, a LoxP site, and unique BamHI and SalI sites from a pBSK-based plasmid (B.11) containing genomic murine Mdm2 ∼ 6Kb upstream and 2.5Kb downstream of the first ATG in exon 3. Next, 5.7 kb of genomic DNA, upstream of exon 1, was digested from pBSK-B.11. Four piece ligation was used to generate the 5’ arm in the pGL1-targeting vector containing a PGK-neomycin and a thymidine kinase cassette. The 3’ arm was generated by PCR amplification of intron 3. Of importance, the start of the 3’ arm is precisely at the end of the 5’ arm. This arm contained a unique SalI site (destroyed during cloning), a Lox P site, 1kb of intron 3, and a conserved NheI site. This amplicon was cloned immediately downstream of the neomycin cassette in the targeting vector. The resulting plasmids, containing the G or T alleles, were completely sequenced and contained no mutations. Both targeting constructs were linearized with AscI and electroporated into TC-1 embryonic stem (ES) cells. DNA from G418 and gancyclovir selected ES cells was isolated, digested by SpeI, and subjected to mini-Southern blot analysis using external 5’, 3’, and neomycin probes. Correctly targeted ES cells of the Mdm2SNP309G and Mdm2SNP309T were expanded and Southern blot analysis was repeated to confirm the integrity of the locus. These ES cell lines were used for injection into C57BL/6 blastocysts to generate chimeric mice. Male chimeric mice from each clone were crossed with C57BL/6 female mice and contributed to the germline of offspring mice. The resulting heterozygous mice were backcrossed two generations with C57Bl/6 mice, followed by one cross to C57Bl/6 females containing the Zp3-cre transgene. Female offspring were then further backcrossed two additional times to C57Bl/6 mice, followed by one cross to C57Bl/6 mice harboring the p53515A allele. These mice were then intercrossed to generate a cohort of heterozygous and homozygous mice for tumor studies. The background of all mice used in these studies is greater than 98% C57Bl/6. For mouse embryonic fibroblasts, Mdm2SNP309G/G or Mdm2SNP309T/T mice were crossed and embryos were collected at 13.5 dpc. All animal studies were performed according to M. D. Anderson Cancer Center and IACUC guidelines for animal use, protocol # 079906634.
Genotyping analysis
Tails from mice were genotyped for Mdm2 using forward primers5’-GCATTAGAGAGTGGTCACTGCGAC-3’ and reverse primers 5’-GAACAGTGATAGAACATCATGTCAC-3’ for the Mdm2 alleles and previously described primers for the p53 alleles (Jacks et al., 1994; Liu et al., 2004). In addition to PCR genotyping, tail DNA from Mdm2SNP309G/G and Mdm2SNP309T/T mice were randomly sequenced to confirm the presence of the G or T nucleotide after amplification using forward primer 5’-GGATTTCGGACGGCTCTCG-3’ and reverse primer 5’-CGCGCAGCGTTCACACTAG-3’. The resulting amplicons were sequenced using the forward primer.
Real Time RT-PCR
Tissues and tumors were isolated from mice and RNA was extracted using TRIZOL, followed by DNAaseI treatment. RNA from early passage MEFs (< P2) was extracted as above. 1st strand synthesis was performed per the manufacturer’s protocol (GE Bioscience) on total RNA isolated from each tissue. The resulting 1st strand DNA was amplified in the RT-PCR using primers for Mdm2 (forward 5’-GAAAAGCCTGAGGCTGGTAGAA-3’ and reverse5’-AACATAGGCAACCACCAGGAA-3’), Rplp0 (forward 5’-CCCTGAAGTGCTCGACATCA-3’ and reverse 5’-TGCGGACACCCTCCAGAA-3’), Puma (forward 5’-GCGGCGGAGACAAGAAGA-3’ and reverse 5’-AGTCCCATGAAGAGATTGTACATGAC-3’), Perp (forward 5’- GCTGCAGCCACGCTTTTC and reverse 5’-GGCGAAGAACGAGAGAATGAA-3’), Ccng1 (forward 5’-GCGAAGCATCTTGGGTGTGT-3’ and reverse 5’- TCCTTTCCTCTTCAGTCGCTTT-3), and Pig8 (forward 5’-GGCATCTGTACCATCTCAAAGCT-3’ and reverse 5’- TCGACGCTGTTCCTCTCTCTTC-3’). The RT-PCR primers for p21 have been previously described (Post et al., 2010). Expression of mRNA was normalized to expression of Rplp0 in each reaction.
RNAse protection assay
Total RNA from spleens of different mice were hybridized with a 32P radiolabeled antisense Exon 1–3 Mdm2 probe followed by incubation with RNAse A. The RNA-32P probe complex was resolved on a 5% polyacrymide gel, transferred to Whatman paper, and dried. The gel was then exposed on Bio-Max XRay film.
Cell culture of MEFs
Early passage MEFs were grown in DMEM supplemented with 15% FCS and 1% penicillin/streptomycin. MEFs were either untreated or treated with 500nM mithramycin A (Sigma) for 18 hours and then harvested.
Cell proliferation assays
Three separate Mdm2SNP309G/G and Mdm2SNP309T/T passage 2 MEF cell lines were plated at 15,000 cells per triplicate well in a 24 well plate. MTT assay [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] was performed on days 1, 3, and 5 as previously published (Jackson et al., 1998). Absorbance values were averaged for the triplicate wells for each cell line, and then the three individual cell lines were grouped and averaged with error bars representing standard error of the mean.
Immunoblotting analysis
Spleens, thymi, breast, tumors, and MEFs were collected and frozen in liquid nitrogen. Protein extracts were prepared by homogenizing the tissues or cells in NP-40 lysis buffer supplemented with Complete Protease (Roche) and phosphatase inhibitors. Soluble proteins were boiled in 2X-SDS-sample buffer, separated by SDS-PAGE, and transferred to PVDF membranes (Millipore). Membranes were incubated with rabbit α-p53 (CM5) (Vector Laboratory), mouse α-Mdm2 (2A10) (Calbiochem), rabbit α-phospho-Ser18p53 (Cell Signaling), mouse α-β-actin (Sigma), or mouse α-vinculin (Sigma) antibodies and antigen-antibody complexes were detected by the enhanced chemiluminescence Kit (GE Bioscience) or BCIP/NBT Color Development Substrate (Promega). Determination of band intensity in western blots was performed using ImageQuant software (GE Healthcare).
Gamma irradiation and thymic apoptosis assays
Six week old mice were treated with one Gy IR and sacrificed three hours post treatment. Tissues were harvested and frozen in liquid nitrogen. Western blot analysis was performed as above. For apoptotic assays, thymi were harvested and fixed in 10% phosphate buffered formalin (Sigma) followed by paraffin embedding. Serial sections were either stained with hematoxylin and eosin (H&E) or subjected to immunohistochemistry (IHC). For IHC, sections were deparaffinized and antigens were retrieved using citric acid and steam. IHC was performed using rabbit α-cleaved caspase-3 (Cell Signaling) and visualized by ABC and DAB kits (Vector Laboratories). Slides were counterstained with Nuclear Fast Red.
Pathological analysis of tumors
Moribund mice were sacrificed according to M. D. Anderson Cancer Center and IACUC guidelines. Euthanized mice were necropsied and tissues were harvested and fixed in 10% phosphate buffered formalin followed by paraffin embedding. Tissue sections were stained with hematoxylin and eosin and morphologic analysis was performed by the Department of Veterinary Medicine at MD Anderson Cancer Center. For IHC, sections were deparaffinized and antigens were retrieved using citric acid and steam. IHC was performed using or rabbit α-Mdm2 (R & D Systems) and visualized by ABC and DAB kits. Slides were counterstained with Nuclear Fast Red. Breast tumors were confirmed by immunohistochemistry as above using mouse α-estrogen receptor-α, 1D5 (Dako).
Sequence analysis
Loss of heterozygosity at the p53 locus was determined by PCR amplification of genomic tumor DNA using forward primer 5’ TACTCT CCTCCCCTCAATAAGCTATTC 3’ (exon 5) and reverse primer 5’ AGTCCTAACCCCACAGGCGGTGTT 3’ (intron 5). The PCR amplicons were then sequencing using the exon 5 primer and analyzed using Chromas software.
Statistical analysis
Comparisons of mean values between the groups were analyzed using GraphPad Instat software (GraphPad Software Inc.). Statistical significance of the differences was analyzed by using unpaired Student t test for comparisons of two groups, one-way analysis of variance (ANOVA) for comparisons of more than two groups, or the χ–square test for 2 by 2 comparisons. Survival curves were plotted by the Kaplan-Meier method and compared by the log rank (Mantel-Cox) test using GraphPad Prism. All p values were two-sided and the level of statistical significance was set at less than 0.05.
Significance
Proper regulation of the homeostatic balance between p53 and Mdm2 is critical in tumorigenesis. The identification of the polymorphic MDM2SNP309G allele in humans, which alters this delicate balance, has lead to an intense focus on the impact of this SNP in accelerating tumorigenesis. There have been numerous conflicting reports regarding the importance of this SNP in the clinic. To directly assess the impact of Mdm2SNP309, we generated cohorts of mice that either carry the polymorphic Mdm2SNP309G or Mdm2SNP309T alleles and monitored these mice for tumor development. Mice harboring two Mdm2SNP309G alleles have an attenuated p53 pathway resulting in a shorter latency to tumor formation and decreased survival. These results demonstrate the Mdm2SNP309G allele has a direct impact on cancer risk.
Supplementary Material
Acknowledgments
We thank members of the Lozano lab for helpful discussion and technical advice. DNA sequencing and veterinary core facilities were supported by an NCI Cancer Center Support Grant CA16672. S.M.P. was supported by a Ruth L. Kirschstein NRSA fellowship F32CA119616 and is a recipient of the Dowdy P. Hawn postdoctoral fellowship. J.G.J. was funded as an Odyssey Scholar and supported by the Odyssey Program and The Theodore N. Law Endowment for Scientific Achievement. This study has been supported by NIH Grants CA46392 and CA34936 (to GL), ES015587 and ES011047 (to D.G.J.), and the Cancer Prevention Institute of Texas.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. Embo J. 2003;22:1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez S, Drane P, Meiller A, Bras M, Deguin-Chambon V, Bouvard V, May E. A comprehensive study of p53 transcriptional activity in thymus and spleen of gamma irradiated mouse: high sensitivity of genes involved in the two main apoptotic pathways. Int J Radiat Biol. 2006;82:761–770. doi: 10.1080/09553000600949624. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hirshfield KM, Kirchhoff T, Alexe G, Bond EE, Robins H, Bartel F, Taubert H, Wuerl P, Hait W, et al. MDM2 SNP309 accelerates tumor formation in a gender-specific and hormone-dependent manner. Cancer Res. 2006;66:5104–5110. doi: 10.1158/0008-5472.CAN-06-0180. [DOI] [PubMed] [Google Scholar]
- Bond GL, Hu W, Bond EE, Robins H, Lutzker SG, Arva NC, Bargonetti J, Bartel F, Taubert H, Wuerl P, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Bond GL, Levine AJ. A single nucleotide polymorphism in the p53 pathway interacts with gender, environmental stresses and tumor genetics to influence cancer in humans. Oncogene. 2007;26:1317–1323. doi: 10.1038/sj.onc.1210199. [DOI] [PubMed] [Google Scholar]
- Bougeard G, Baert-Desurmont S, Tournier I, Vasseur S, Martin C, Brugieres L, Chompret A, Bressac-de Paillerets B, Stoppa-Lyonnet D, Bonaiti-Pellie C, Frebourg T. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet. 2006;43:531–533. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien W-P, Wong R-H, Cheng Y-W, Chen C-Y, Lee H. Associations of MDM2 SNP309, Transcriptional Activity, mRNA Expression, and Survival in Stage I Non-Small-Cell Lung Cancer Patients with Wild-Type p53 Tumors. Annals of Surgical Oncology. 2010;4:1194–1202. doi: 10.1245/s10434-009-0853-2. [DOI] [PubMed] [Google Scholar]
- Copson ER, White HE, Blaydes JP, Robinson DO, Johnson PW, Eccles DM. Influence of the MDM2 single nucleotide polymorphism SNP309 on tumour development in BRCA1 mutation carriers. BMC Cancer. 2006;6:80. doi: 10.1186/1471-2407-6-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharel N, Kato N, Muroyama R, Moriyama M, Shao RX, Kawabe T, Omata M. MDM2 promoter SNP309 is associated with the risk of hepatocellular carcinoma in patients with chronic hepatitis C. Clin Cancer Res. 2006;12:4867–4871. doi: 10.1158/1078-0432.CCR-06-0111. [DOI] [PubMed] [Google Scholar]
- Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9:831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- Economopoulos KP, Sergentanis TN. Differential effects of MDM2 SNP309 polymorphism on breast cancer risk along with race: a meta-analysis. Breast Cancer Res Treat. 2009;120:211–216. doi: 10.1007/s10549-009-0467-1. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Huo D, Yildiz O, Worrillow LJ, Banerjee M, Le Beau MM, Larson RA, Allan JM, Onel K. MDM2 SNP309 and TP53 Arg72Pro interact to alter therapy-related acute myeloid leukemia susceptibility. Blood. 2008;112:741–749. doi: 10.1182/blood-2007-11-126508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grochola LF, Muller TH, Bond GL, Taubert H, Udelnow A, Wurl P. MDM2 SNP309 Associates With Accelerated Pancreatic Adenocarcinoma Formation. Pancreas. 2009;39:76–80. doi: 10.1097/MPA.0b013e3181b9f105. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Iwakuma T, Lozano G. MDM2, an introduction. Mol Cancer Res. 2003;1:993–1000. [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Current Biology. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jackson JG, White MF, Yee D. Insulin receptor substrate-1 is the predominant signaling molecule activated by insulin-like growth factor-I, insulin, and interleukin-4 in estrogen receptor-positive human breast cancer cells. J Biol Chem. 1998;273:9994–10003. doi: 10.1074/jbc.273.16.9994. [DOI] [PubMed] [Google Scholar]
- Krekac D, Brozkova K, Knoflickova D, Hrstka R, Muller P, Nenutil R, Vojtesek B. MDM2 SNP309 Does Not Associate with Elevated MDM2 Protein Expression or Breast Cancer Risk. Oncology. 2008;74:84–87. doi: 10.1159/000139135. [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lang GA, Iwakuma T, Suh YA, Liu G, Rao VA, Parant JM, Valentin-Vega YA, Terzian T, Caldwell LC, Strong LC, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–872. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Liu G, Parant JM, Lang G, Chau P, Chavez-Reyes A, El-Naggar AK, Multani A, Chang S, Lozano G. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–68. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
- Lundgren K, Montes de Oca Luna R, McNeill YB, Emerick EP, Spencer B, Barfield CR, Lozano G, Rosenberg MP, Finlay CA. Targeted expression of MDM2 uncouples S phase from mitosis and inhibits mammary gland development independent of p53. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- Marcel V, Palmero EI, Falagan-Lotsch P, Martel-Planche G, Ashton-Prolla P, Olivier M, Brentani RR, Hainaut P, Achatz MI. TP53 PIN3 and MDM2 SNP309 polymorphisms as genetic modifiers in the Li—Fraumeni syndrome: impact on age at first diagnosis. Journal of Medical Genetics. 2009;46:766–772. doi: 10.1136/jmg.2009.066704. [DOI] [PubMed] [Google Scholar]
- Mendrysa SM, McElwee MK, Michalowski J, O'Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol. 2003;23:462–472. doi: 10.1128/MCB.23.2.462-473.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa SM, O'Leary KA, McElwee MK, Michalowski J, Eisenman RN, Powell DA, Perry ME. Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 2006;20:16–21. doi: 10.1101/gad.1378506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menin C, Scaini MC, De Salvo GL, Biscuola M, Quaggio M, Esposito G, Belluco C, Montagna M, Agata S, D'Andrea E, et al. Association between MDM2-SNP309 and age at colorectal cancer diagnosis according to p53 mutation status. J Natl Cancer Inst. 2006;98:285–288. doi: 10.1093/jnci/djj054. [DOI] [PubMed] [Google Scholar]
- Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature. 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- Post SM, Quintas-Cardama A, Terzian T, Smith C, Eischen CM, Lozano G. p53-dependent senescence delays Emu-myc-induced B-cell lymphomagenesis. Oncogene. 2010;29:1260–1269. doi: 10.1038/onc.2009.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruijs MW, Schmidt MK, Nevanlinna H, Tommiska J, Aittomaki K, Pruntel R, Verhoef S, Van't Veer LJ. The single-nucleotide polymorphism 309 in the MDM2 gene contributes to the Li-Fraumeni syndrome and related phenotypes. Eur J Hum Genet. 2007;15:110–114. doi: 10.1038/sj.ejhg.5201715. [DOI] [PubMed] [Google Scholar]
- Sajid AK, Kamran I, Ann F, Zhaoshi Z, Shoshana R, Hanna P, Francis B, Kenneth O, Michael PL, Philip BP. Genetic variants in germline TP53 and MDM2 SNP309 are not associated with early onset colorectal cancer. Journal of Surgical Oncology. 2008;97:621–625. doi: 10.1002/jso.20996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MK, Reincke S, Broeks A, Braaf LM, Hogervorst FB, Tollenaar RA, Johnson N, Fletcher O, Peto J, Tommiska J, et al. Do MDM2 SNP309 and TP53 R72P interact in breast cancer susceptibility? A large pooled series from the breast cancer association consortium. Cancer Res. 2007;67:9584–9590. doi: 10.1158/0008-5472.CAN-07-0738. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–842. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- Tabori U, Nanda S, Druker H, Lees J, Malkin D. Younger age of cancer initiation is associated with shorter telomere length in Li-Fraumeni syndrome. Cancer Res. 2007;67:1415–1418. doi: 10.1158/0008-5472.CAN-06-3682. [DOI] [PubMed] [Google Scholar]
- Terzian T, Wang Y, Van Pelt CS, Box NF, Travis EL, Lozano G. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27:5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentin-Vega YA, Barboza JA, Chau GP, El-Naggar AK, Lozano G. High levels of the p53 inhibitor MDM4 in head and neck squamous carcinomas. Hum Pathol. 2007;38:1553–1562. doi: 10.1016/j.humpath.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The Sequence of the Human Genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Yarden RI, Friedman E, Metsuyanim S, Olender T, Ben-Asher E, Papa MZ. MDM2 SNP309 accelerates breast and ovarian carcinogenesis in BRCA1 and BRCA2 carriers of Jewish-Ashkenazi descent. Breast Cancer Res Treat. 2008;111:497–504. doi: 10.1007/s10549-007-9797-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.