IKK and NF-κB-mediated regulation of Claspin impacts on ATR checkpoint function
Rocha and colleagues identify transcriptional regulation of Claspin (a known ATR activator) by NF-κB. These results molecular intersect the DNA-damage checkpoint with NF-κB-mediated cellular responses, a topic with broad implications as both checkpoint and NF-κB pathways are implicated in cancer development.
Keywords: Chk1, Claspin, IKK, NF-κB
Abstract
In response to replication stress, Claspin mediates the phosphorylation and activation of Chk1 by ATR. Claspin is not only necessary for the propagation of the DNA-damage signal, but its destruction by the ubiquitin–proteosome pathway is required to allow the cell to continue the cell cycle allowing checkpoint recovery. Here, we demonstrate that both the NF-κB family of transcription factors and their upstream kinase IKK can regulate Claspin levels by controlling its mRNA expression. Furthermore, we show that c-Rel directly controls Claspin gene transcription. Disruption of IKK and specific NF-κB members impairs ATR-mediated checkpoint function following DNA damage. Importantly, hyperactivation of IKK results in a failure to inactivate Chk1 and impairs the recovery from the DNA checkpoint. These results uncover a novel function for IKK and NF-κB modulating the DNA-damage checkpoint response, allowing the cell to integrate different signalling pathways with the DNA-damage response.
Introduction
In response to cellular insults, which can result in DNA damage or replication stalling, cells can activate genome surveillance pathways that co-operate to preserve genomic integrity, by preventing inappropriate progression of the cell cycle. In this pathway ATR, a protein kinase, is activated and phosphorylates a number of downstream targets that can co-ordinate cell-cycle progression, with repair of the damaged DNA (Kastan and Bartek, 2004). One of such targets is the Chk1 protein kinase, when phosphorylated by ATR at two critical serine residues at positions 317 and 345 propagates the damage response (Zhao and Piwnica-Worms, 2001). Once activated, Chk1 phosphorylates a number of downstream targets, such as Cdc25, which ultimately results in cyclin-dependent kinase inhibition and cell-cycle delay, allowing the cell time to use the necessary repair mechanisms (Harper and Elledge, 2007). Such a rapid, but reversible, block of cell-cycle progression is essential for the repair of potentially catastrophic perturbations in their genome.
Claspin was originally identified in Xenopus in a screen to identify Chk1-interacting proteins (Kumagai and Dunphy, 2000). It serves as an adaptor protein, binding to both Chk1 and ATR, and is necessary for ATR-dependent phosphorylation of Chk1 in both Xenopus and human systems (Chini and Chen, 2003; Kumagai and Dunphy, 2003; Clarke and Clarke, 2005). Claspin protein levels fluctuate during the cell cycle being low during early G1, accumulating to be high in S-phase and eventually degraded in a manner controlled by the protein kinase, Polo-like kinase-1 (Plk1), at the onset of mitosis (Yoo et al, 2004; Peschiaroli et al, 2006). This cyclical regulation of Claspin has been shown to be involved in the normal progression of the cell cycle, and following checkpoint activation (Peschiaroli et al, 2006). Indeed, inappropriate reduction of Claspin levels by siRNA slows replication fork rates (Petermann et al, 2008) and, following replication stress, promotes premature entry into mitosis before completing DNA replication (Chini and Chen, 2003); conversely, overexpression of a non-degradable form of the protein results in a delay in the inactivation of Chk1 and recovery from the DNA replication checkpoint (Mailand et al, 2006; Mamely et al, 2006; Peschiaroli et al, 2006). Any process involved in the regulation of Claspin levels would therefore be important for maintaining faithful replication of the DNA, and also proper regulation of the DNA replication checkpoint activated by ATR.
NF-κB is the collective name for a family of transcription factors. This family is comprised of five genes, encoding seven proteins: RelA, RelB, c-Rel, p105/p50 (NF-κB1) and p100/p52 (NF-κB2). NF-κB activation pathways can be classified as canonical, non-canonical and atypical regarding the mode and involvement of the IKK complex (Perkins, 2007; Chariot, 2009). Genotoxic agents such as chemotherapeutic drugs and radiotherapy can activate NF-κB through atypical pathways (Perkins, 2007). As such, ATM has been shown to bind and phosphorylate IKKγ/Nemo (NF-κB essential modulator), which results in activation of the IKK complex and hence NF-κB (Wu et al, 2006). NF-κB can have both anti- and pro-apoptotic functions and this is dependent on many factors including genetic context, binding partners and posttranslational modifications of the subunits themselves (Perkins and Gilmore, 2006). Despite the existence of many reports concerning the mode of activation and response of NF-κB following DNA damage, no study thus far as assessed if NF-κB has a function in any of the cellular checkpoints.
In this study, we demonstrate that the IKK complex can regulate Claspin levels by controlling its mRNA expression. This regulation is dependent on the activity of the IKKβ, but independent of the IKKα subunit. Furthermore, we have identified c-Rel as the transcription factor downstream of IKKβ directly controlling Claspin gene transcription. We show that disruption of IKK and c-Rel significantly impairs ATR-mediated checkpoint function following DNA damage. Reduction of IKK or c-Rel results in increased H2Ax foci, in a manner similar to that of Claspin depletion. Importantly, hyperactivation of IKK results in delayed inactivation of Chk1 and recovery from the DNA replication checkpoint, with inappropriate activation of the checkpoint in mitosis.
Results
Depletion of IKKβ results in a severe reduction of Claspin levels
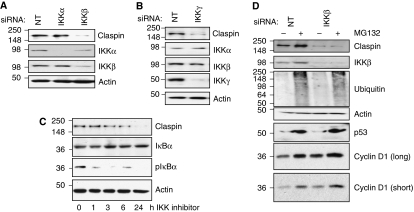
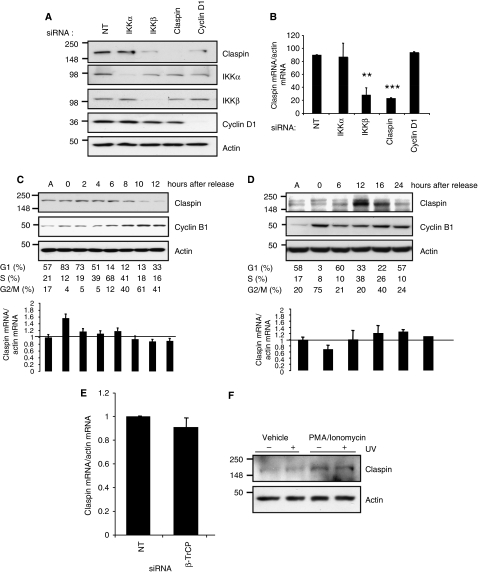
Previous studies have shown that NF-κB is activated following DNA damage and that the IKK complex subunit Nemo interacts with ATM (Wu et al, 2006). In addition, a recent study linked perturbations of IKKβ to genetic instability (Irelan et al, 2007). We noticed that the phospho-degron present in the amino terminus of Claspin is highly similar to that of the IKK substrate IκB-α (Supplementary Figure S1A). As such, we investigated if the IKK complex regulates Claspin. The IKK complex is composed of two catalytic subunits IKKα and IKKβ, and a regulatory subunit IKKγ (Perkins, 2007; Chariot, 2009). Although highly homologous IKKα and IKKβ have largely distinct functions, because of their different substrate specificities and modes of regulation (Hacker and Karin, 2006). Although we could not detect IKK-mediated phosphorylation of a bacterially expressed fragment of Claspin containing the phospho-degron motif, in vitro (Supplementary Figure S1B), we investigated if endogenous IKK could regulate Claspin in cells. RNA interference (RNAi) was used to reduce the expression of the catalytic subunits IKKα or IKKβ. Relative to non-targeting controls, IKKα and IKKβ protein and mRNA levels were specifically decreased in U2OS cells by the respective RNAi (Figure 1A; Supplementary Figure S2A). Although specific knockdown of IKKα had no effect on Claspin levels, reduction of IKKβ unexpectedly correlated with a visible reduction of Claspin protein levels (Figure 1A). Additional siRNA oligonucleotides targeting IKKβ, produced a similar reduction of Claspin protein levels (Supplementary Figure S2B). Furthermore, Claspin protein levels, in cells depleted of endogenous IKKβ, could be partially rescued by the expression of an exogenous IKKβ construct (Supplementary Figure S2C). IKKβ depletion also reduced Claspin levels in MDA-MB-231 and HEK293 cells (Supplementary Figure S2D), demonstrating that this effect is not limited to U2OS cells.
Figure 1.
IKKβ is required for maintaining Claspin protein levels. (A) U2OS cells were depleted of IKKα or IKKβ using siRNA. Whole-cell lysates (WCLs) were subjected to immunoblot analysis for the levels of the indicated proteins. (B) U2OS cells were depleted or IKKγ using siRNA. WCLs were subjected to immunoblot to analyse the levels of the indicated proteins. (C) U2OS cells were treated with the IKK inhibitor, Bay 11-7082, for the indicated times. WCLs were prepared and subjected to immunoblot analysis using the indicated antibodies. (D) U2OS were depleted of IKKβ using siRNA as in (A), but MG132 was added 3 h prior to WCL preparation, and immunoblot analysis was performed using the indicated antibodies.
The IKK kinase complex requires the regulatory subunit IKKγ for its activity (Perkins, 2007; Chariot, 2009); we therefore tested if Claspin levels were sensitive to IKKγ levels. RNAi specifically reduced IKKγ levels, but did not alter levels of IKKα or IKKβ (Figure 1B). Selective reduction of IKKγ significantly diminished levels of Claspin, relative to control-transfected cells (Figure 1B).
The RNAi results indicate that the presence of IKKβ and IKKγ are required for maintaining Claspin protein levels. Given that the IKK complex has kinase activity, we determined if inhibiting this would lower Claspin levels. Inhibition of the IKK complex using an IKK inhibitor (Bay 11-7082), confirmed by a reduction in the basal level of phosho-IκBα (Figure 1C), resulted in a reduction of Claspin levels by 6 h and almost complete loss of Claspin after 24 h of treatment (Figure 1C). IKKβ and IKKγ depletion in unstimulated cells also results in lower NF-κB activity, assessed using an NF-κB luciferase reporter assay (Supplementary Figure S2E), further indicating that IKK and NF-κB have basal activity. In addition, IKK inhibition (Supplementary Figure S3A) or siRNA-mediated depletion of IKKβ and IKKγ (Supplementary Figure S3B and C), in mouse embryo fibroblasts also resulted in reduced Claspin levels, indicating that IKK regulates Claspin in mouse cells. Taken together, these results demonstrate that Claspin levels are sensitive to both IKK levels and activity.
Previous studies demonstrated that Claspin levels are controlled in the cell by changing protein stability, through a mechanism dependent on phosphorylation and degradation by the proteasome (Mailand et al, 2006; Mamely et al, 2006; Peschiaroli et al, 2006). We therefore tested if depletion of IKKβ was altering Claspin protein stability. In cells transfected with non-targeting siRNA, there is a partial elevation of Claspin levels in cells treated with the proteasome inhibitor MG132 (Figure 1D), consistent with previously published observations (Bennett and Clarke, 2006). However, in cells transfected with IKKβ siRNA, no recovery of Claspin levels in the presence of proteasome inhibition could be observed (Figure 1D). Immunoblotting using an anti-ubiquitin antibody revealed that accumulation of poly-ubiquitinated proteins was similar in cells treated with either non-targeting RNAi or IKKβ RNAi, demonstrating that failure to recover Claspin levels in IKKβ-depleted cells, was not due to ineffective proteasome inhibition (Figure 1D, lower panel). Furthermore, MG132 treatment increased the levels of p53 and Cyclin D1 (proteins that are regulated by the proteasome) regardless of IKKβ levels. These results indicate that IKKβ is not altering Claspin levels by regulating its stability.
IKKβ controls the levels of Claspin mRNA through modulation of NF-κB
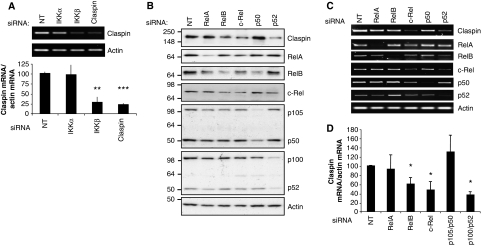
As IKK inhibition does not alter Claspin levels by affecting its protein stability, we investigated if depletion of IKK could alter levels of Claspin mRNA. We performed quantitative RT–PCR (qRT–PCR) using RNA from cells depleted of IKKα, IKKβ or Claspin. Reducing IKKα levels had no significant effect on Claspin mRNA levels, consistent with the observations of the Claspin protein (Figure 2A). Specific knockdown of IKKβ results in a dramatic reduction of Claspin mRNA levels, to ∼20% of the levels observed in the cells transfected with the non-targeting control RNA (Figure 2A). Similar results were also observed using alternative siRNAs targeting IKKβ, and in other cells such as MDA-MB-231 (Supplementary Figure S4).
Figure 2.
Claspin mRNA is regulated by IKKβ and the NF-κB family of transcription factors. (A) Semi-quantitative and quantitative RT–PCR analysis of Claspin mRNA prepared from U2OS cells depleted of IKKα, IKKβ or Claspin by siRNA. (B) U2OS cells were depleted of the individual NF-κB family members; RelA, RelB, c-Rel, p105/p50 or p100/p52, using siRNA and analysed by immunoblot using the indicated antibodies. (C) Semi-quantitative RT–PCR analysis of Claspin, RelA, RelB, c-Rel, p105, p100 and actin mRNA from cells treated as in (A). (D) Quantitative RT–PCR analysis of Claspin mRNA in cells depleted of the individual NF-κB subunits as in (A). ANOVA t-tests were performed on the means, and the P-values were calculated. *P⩽0.05, **P⩽0.01, ***P⩽0.001.
As the best-known function of IKKβ is to activate the NF-κB family of transcription factors (Perkins, 2007), we next investigated if any of the NF-κB transcription factors could regulate the expression of Claspin mRNA. Using previously validated siRNA sequences (Anderson and Perkins, 2003; Schumm et al, 2006; van Uden et al, 2008), we reduced the levels of RelA, RelB, c-Rel, p100/p52 and p105/p50 at both the protein and mRNA levels, as determined by western blot and RT–PCR, respectively (Figure 2B–D; Supplementary Figure S5). NF-κB subunits control each other expression by the presence of κB sites in their promoters, as such depletion of a single subunit will alter the expression levels of additional ones (Figure 2B). As activation of IKKβ usually results in the induction of RelA/p50 heterodimer activity, it was surprising to observe that siRNA oligonucleotides directed against RelA or p105/50, although significantly reducing the levels of their respective targets, did not alter either the levels of the Claspin mRNA or protein (Figure 2B–D). However, reduction of RelB, c-Rel and p100/p52 resulted in a specific decrease in the levels of the Claspin protein (Figure 2B) and mRNA (Figure 2C and D). Taken together, these results demonstrate for the first time that IKKβ and specific NF-κB subunits regulate Claspin levels, through modulation of its mRNA.
c-Rel binds directly to the Claspin promoter
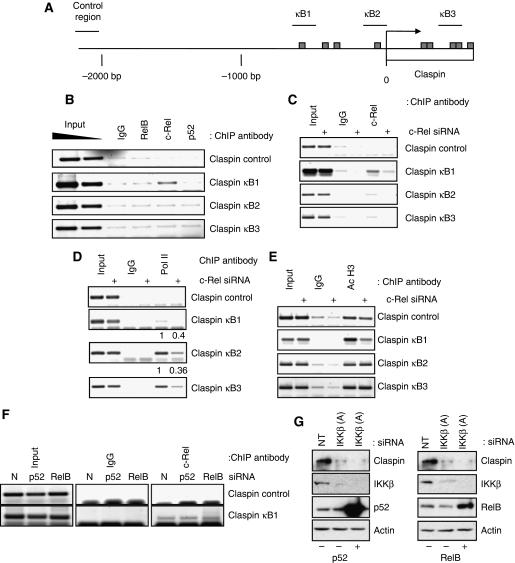
Given the results obtained when analysing the levels of Claspin mRNA, we next determined the function of specific NF-κB subunits in the control of the Claspin gene. Analysis of the Claspin promoter, revealed the presence of putative NF-κB-binding site sequences both upstream and downstream of the transcription start site (http://www.genomatix.de/products/MatInspector) (Figure 3A). As depletion of RelB, c-Rel or p52 altered Claspin levels at both the protein and mRNA levels (Figure 2), we sought to investigate if this effect was direct. To determine if RelB, c-Rel or p52 are directly bound to the promoter of the Claspin gene in vivo, we performed chromatin immunoprecipitation (ChIP) assays in U2OS cells, using specific antibodies against the respective proteins. Primer sets were designed to encompass the NF-κB-binding site upstream of the transcription start site (κB1), at the transcription start site (κB2), downstream of the transcription start site (κB3), and a control region 2000 bp upstream (Figure 3A). Promoter occupancy was assayed by PCR using these sets of primer pairs. We detected enrichment of c-Rel using the κB1 primer pair, indicating binding upstream of the transcription start site (Figure 3B). The specificity of this binding was evident by the lack of signal from the c-Rel ChIP in either the control region or the putative κB consensus sites downstream or at the start site of transcription (Figure 3B). However, little or no binding of RelB or p52 was observed on any of the putative-binding sites (Figure 3B). These results indicate that c-Rel can directly bind to the Claspin promoter.
Figure 3.
The Claspin gene promoter is directly regulated by c-Rel. (A) Schematic of the human Claspin gene promoter with putative NF-κB sites represented as bars, and ChIP PCR primer sets represented as lines. (B) ChIP assays using Control IgG, RelB, c-Rel or p52 antibodies were performed on cell extracts from U2OS cells. Immunoprecipitated DNA was PCR amplified using the indicated primer pairs alongside 2% input genomic DNA. (C) ChIP assays using Control IgG and c-Rel antibodies were performed on cell extract from control or cells depleted of c-Rel. Immunoprecipitated DNA was amplified by PCR using the indicated primer pairs. (D) As in (C) but ChIP was performed using antibodies directed against RNA polymerase II. (E) As in (C) but ChIP was performed using antibodies directed against acetylated histone H3. Levels of RNA polymerase II recruitment to the κB sites of the Claspin gene were quantified, normalized to inputs, and numbers indicate the difference between control and c-Rel-depleted samples. (F) ChIP assays using Control IgG and c-Rel antibodies were performed on cell extract from control or cells depleted of p52 or RelB. Immunoprecipitated DNA was amplified by PCR using the indicated primer pairs. (G) U2OS cells were co-transfected with a non-targeting siRNA or a siRNA to deplete IKKβ, along with either a vector control or a p52 or RelB expression constructs, as indicated. WCLs were prepared and analysed by western blot using the indicated antibodies.
To determine if changes in c-Rel levels directly altered the levels of polymerase recruitment or any hallmark of active transcription at the Claspin promoter, we depleted cells of c-Rel using siRNA and performed ChIPs. When compared with control cells, c-Rel depletion resulted, as expected, in less c-Rel present at the κB1 site of the Claspin promoter (Figure 3C), demonstrating the specificity of the signal obtained in Figure 3B. Importantly, depletion of c-Rel reduced polymerase occupancy at the start site and within the coding region of the Claspin promoter (Figure 3D). Furthermore, levels of acetylated histone H3 were also reduced but only at the promoter region, suggesting that c-Rel is necessary for maintaining high levels of acetylated histones surrounding the Claspin promoter (Figure 3E).
Given that we have found that p52 and RelB also control Claspin levels, we determined if depletion of these subunits would alter c-Rel binding to the Claspin promoter. We found that c-Rel binding to the Claspin promoter remained unaltered in the absence of either p52 or RelB (Figure 3F). Given these results, we sought to determine the contribution of these NF-κB subunits in the regulation of Claspin levels. For this purpose, we overexpressed either p52 or RelB in the absence of IKKβ and analysed Claspin protein levels (Figure 3G). Our results demonstrate that these subunits cannot rescue IKKβ depletion and thus suggest p52 and RelB have indirect functions in the control of Claspin.
IKKβ controls the recruitment of c-Rel to the Claspin promoter and is required for active transcription of the Claspin gene
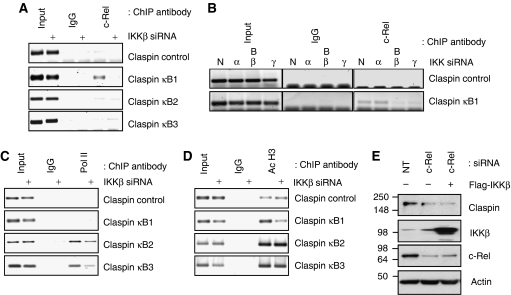
As the IKK complex is immediately upstream of the NF-κB family of transcription factors, including c-Rel, we next sought to determine if Claspin promoter occupancy by c-Rel was dependent on IKKβ. For this purpose, we created U2OS stable cell lines in which IKKβ was selectively depleted using shRNA, but other members of the IKK complex remained unchanged (Supplementary Figure S6). Claspin protein levels were reduced in this cell line, consistent with the observed results with the transient transfections (Supplementary Figure S6). Using control cells carrying a non-targeting shRNA and IKKβ shRNA cells, ChIP assays were performed for the Claspin promoter. c-Rel binding to the upstream κB site could be observed in the control cells (Figure 4A). In contrast, when IKKβ is not present, c-Rel promoter occupancy was significantly decreased (Figure 4A). We also investigated the function of IKKα and IKKγ, as well IKKβ, in the recruitment of c-Rel to the Claspin promoter (Figure 4B). As expected and in accordance with our mRNA and protein analysis, depletion of IKKα did not alter c-Rel levels present at the Claspin promoter (Figure 4B). However, depletion of either IKKβ or IKKγ visibly reduced c-Rel recruitment to this promoter (Figure 4B). These results indicate that IKKβ and IKKγ control c-Rel recruitment to the Claspin promoter.
Figure 4.
The Claspin gene promoter is directly regulated by c-Rel in an IKKβ-dependent manner. (A) ChIP assays using Control IgG or c-Rel antibodies were performed on cell extracts from cells stably expressing a control shRNA plasmid and cells expressing shRNA against IKKβ. Immunoprecipitated DNA was PCR amplified using the indicated primer pairs alongside 2% input genomic DNA. (B) ChIP assays using Control IgG and c-Rel antibodies were performed on cell extracts from control or cells depleted of IKKα (α), IKKβ (β-sequence B) or IKKγ (γ). Immunoprecipitated DNA was amplified by PCR using the indicated primer pairs. (C) As in (A) but ChIP was performed using antibodies directed against RNA polymerase II. (D) As in (A) but ChIP was performed using antibodies directed against acetylated histone H3. (E) U2OS cells were co-transfected with a non-targeting siRNA or a siRNA to deplete c-Rel, along with either a vector control or a Flag-tagged version of IKKβ, as indicated. WCLs were prepared and analysed by western blot using the indicated antibodies.
As IKKβ depletion resulted in lower Claspin mRNA (Figures 1 and 2A), we investigated if this was also evident in RNA polymerase recruitment. When IKKβ was depleted, we observed a reduction in RNA polymerase II recruitment to the start site of transcription and a reduction of polymerase loading within the coding region, consistent with these cells having lower Claspin expression (Figure 4C). We also compared localized levels of acetylated histone H3, a mark of active transcription, surrounding the Claspin promoter. Acetylated H3 levels at the Claspin promoter, surrounding the c-Rel-binding site, were reduced by IKKβ depletion, but remained unaltered at the transcription start site and within the gene in the presence or absence of IKKβ (Figure 4D). To elucidate whether c-Rel and IKKβ exert their effects on Claspin through alternative pathways, we examined if expression of exogenous IKKβ, could rescue Claspin levels in cells depleted of c-Rel. U2OS cells transfected with an siRNA against c-Rel have lower levels of Claspin than those transfected with a control non-targeting siRNA. Overexpression of IKKβ was unable to rescue the Claspin defect in c-Rel knockdown cells, consistent with IKK being upstream of c-Rel in this signalling pathway (Figure 4E). These results indicate that IKKβ and c-Rel regulate polymerase recruitment and activation of transcription of the Claspin gene (Figures 3 and 4).
Regulation of Claspin mRNA is cell-cycle independent but it is responsive to changes in IKK–NF-κB activity
It is well established that the Claspin protein is regulated through the cell cycle, but no study has assessed if Claspin mRNA is also regulated in this manner. Claspin protein levels are low during the G1 phase of the cell cycle. As such, we determined if the reduction of Claspin mRNA we had observed in the absence of IKKβ was merely a consequence of a cell-cycle block. U2OS cells were arrested in the G1 phase of the cell cycle by depletion of the G1-specific cyclin, Cyclin D1 (Figure 5A and B; Supplementary Figure S7A). Cells depleted of IKKβ, Claspin and Cyclin D1 show reduced levels of Claspin protein, compared with the cells depleted of IKKα, or treated with the non-targeting control RNAi (Figure 5A). However, knockdown of Cyclin D1 was not sufficient to reduce the levels of the Claspin mRNA, providing a correlative indication that regulation of Claspin mRNA, unlike the protein, is independent of the cell cycle (Figure 5B; Supplementary Figure S7A).
Figure 5.
Claspin mRNA is regulated independently of the cell cycle but Claspin is responsive to NF-κB activation. (A) U2OS cells were depleted of IKKα, IKKβ, Claspin or Cyclin D1 using siRNA. WCLs were subjected to western blot analysis for the levels of the indicated proteins. (B) Quantitative RT–PCR analysis of Claspin mRNA prepared from U2OS cells depleted of IKKα, IKKβ, Claspin or Cyclin D1 by siRNA. ANOVA t-tests were performed on the means, and the P-values were calculated. **P⩽0.01 and ***P⩽0.001. (C) U2OS cells were synchronized at G1/S using a double-thymidine block. Thymidine was subsequently washed and cells were released for the indicated times. Cells were analysed by flow cytometry and percentage of cells in each stage of the cell cycle is indicated. WCLs and RNA were prepared from these cells and analysed by western blot and quantitative RT–PCR, respectively. Specific antibodies and primer sets are indicated. (D) U2OS cells were synchronized at prometaphase using nocodazole. Nocodazole was subsequently washed and cells were analysed by flow cytometry and percentage of cells in each stage of the cell cycle is indicated. WCLs and RNA were prepared and analysed by western blot and quantitative RT–PCR, respectively. Specific antibodies and primer sets are indicated. (E) Quantitative RT–PCR analysis of Claspin mRNA prepared from U2OS cells depleted of β-TrCP by siRNA. (F) U2OS cells were treated with PMA (100 ng/ml)/Ionomycin (0.5 μM) for 4 h. One hour prior to harvesting, cells were treated with UV (40 J/m2) as indicated. WCLs were prepared and Claspin levels were analysed by western blot.
To test directly if the Claspin transcript is regulated through the cell cycle, U2OS cells were synchronized, either at the G1-S boundary using a double-thymidine block, or in mitosis using nocodazole, then released and followed through the cell cycle by FACS (Figure 5C and D). Using both synchronization approaches, it was possible to observe that Claspin protein levels were maintained through S-phase of the cell cycle before being rapidly degraded during mitosis, consistent with published observations (Figure 5C and D) (Yoo et al, 2004; Peschiaroli et al, 2006). Our results show that the levels of Claspin mRNA do not change significantly throughout the cell cycle (Figure 5C and D). We did see slight fluctuations in the levels of Claspin mRNA in cells which were still subject to chemical block, but these were not observed in samples corresponding to similar stages of the cell cycle in unperturbed conditions, and can therefore be attributed to the chemical intervention (Figure 5C and D and unpublished data). Consistent with Claspin mRNA not fluctuating during the cell cycle, we do not observe any changes in RelB, c-Rel or p100/p52 levels, or IKK levels/activity during the cell cycle using both synchronization approaches (Supplementary Figure S7B and C). These results indicate that Claspin mRNA is not regulated in a cell-cycle manner.
To further separate the cell-cycle-dependent regulation of Claspin by the ubiquitin/proteasome pathway and the IKK-dependent regulation of Claspin, we measured Claspin mRNA levels in the absence of β-TrCP, the ubiquitin ligase required for its degradation. We reduced β-TrCP levels in U2OS cells using previously validated siRNA sequences (Supplementary Figure S8; Mailand et al, 2006; Mamely et al, 2006; Peschiaroli et al, 2006). Depletion of β-TRCP did not change the levels of phosphorylated and total IKK or IκB-α in unstimulated cells (Supplementary Figure S8B). In addition, the levels of nuclear c-Rel were also unaffected by depletion of β-TRCP (Supplementary Figure S8C). Importantly, in the absence of β-TrCP, there is no significant change in Claspin mRNA levels, compared with cells treated with a non-targeting control, indicating that the regulation of Claspin mRNA is independent of the protein degradation pathway (Figure 5E).
We next investigated if Claspin mRNA could be regulated by modulation of IKK–NF-κB activity. IKK responds to a variety of different stimuli such as cytokines, stress signals (Perkins, 2007; Chariot, 2009). We initially tested if Claspin was responsive to TNF-α, but did not detect any significant changes (data not shown); however, when we tested another known NF-κB inducer, PMA/Ionomycin, we observed a modest, but significant induction of Claspin (Figure 5F; Supplementary Figure S9A). Interestingly, UV treatment did not result in higher levels of Claspin. UV is known to activate RelA/p50 (Campbell et al, 2004), but UV-mediated activation of c-Rel has not been reported. Importantly, PMA/Ionomycin-dependent induction of Claspin was lost in cells depleted of IKKβ, demonstrating that Claspin induction is through an IKK-dependent signalling cascade (Supplementary Figure S9B). These results indicate that Claspin is responsive to the levels of IKK–NF-κB activity.
Loss of IKKβ disrupts Chk1 phosphorylation by ATR following DNA damage
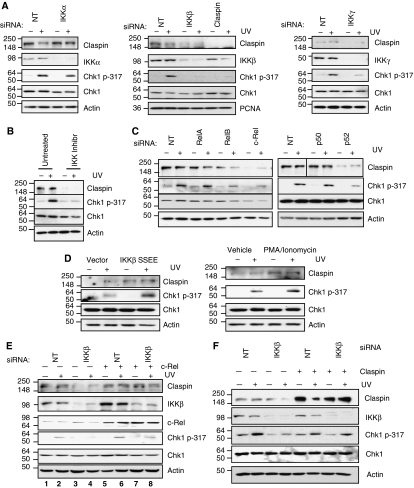
One of the major functions of Claspin is to act as an adaptor between ATR and Chk1. As such, Claspin is critical for the ATR-dependent phosphorylation of Chk1 (Lee et al, 2005). Given our discovery that IKKβ can control Claspin levels through regulation of its mRNA, we wanted to determine what were the functional consequences of IKKβ depletion on the DNA-damage checkpoint and ultimately on cellular fate. To investigate if loss of IKK function could compromise Chk1 phosphorylation in response to DNA damage, we depleted IKKα, IKKβ or IKKγ and then analysed the levels of Chk1 phosphorylation after exposure to UV light (a known stimulus of the ATR-Chk1 pathway). As expected, cells transfected with a non-targeting siRNA displayed a robust level of Chk1 Ser317 phosphorylation when exposed to UV (Figure 6A). Significantly, reduction of IKKβ, but not IKKα, by siRNA attenuated the phosphorylation of Chk1 on the Ser317 residue, but had no great effect on Chk1 protein levels (Figure 6A). Indeed, knockdown of IKKβ produced a similar reduction on Chk1 Ser317 phosphorylation as depletion of Claspin itself (Figure 6A). Importantly, this result is not dependent on method of checkpoint induction or the cell line as similar defects in Chk1 phosphorylation are seen in response to Hydroxyurea in IKKβ-depleted U2OS cells (Supplementary Figure S10A), and UV-induced damage in IKKβ-depleted MDA-MB-231 cells (Supplementary Figure S10B). We had previously shown that IKKγ is required for maintaining Claspin levels (Figure 1B). As predicted from our results, U2OS cells depleted of IKKγ displayed lower levels of Chk1 Ser317 phosphorylation in response to UV-induced DNA damage (Figure 6A). In addition, the IKK inhibitor Bay 11-8072 almost completely abolished UV-induced Chk1 Ser317 phosphorylation when compared with control cells (Figure 6B).
Figure 6.
Loss or inhibition of IKKβ disrupts Chk1 phosphorylation in response to UV. (A) U2OS cells were depleted of IKKα, IKKβ, IKKγ or Claspin with siRNA. Cells were treated with UV (40 J/m2) as indicated, and harvested 4 h later. WCLs were prepared and analysed by western blot using the indicated antibodies. (B) U2OS cell were pretreated with the IKK inhibitor, Bay 11-7082, for 24 h, as indicated. Cells were treated with UV (40 J/m2) as indicated, and harvested 4 h later. WCLs were prepared and analysed by western blot using the indicated antibodies. (C) U2OS cells were depleted of the NF-κB transcription factors RelA, RelB, c-Rel, p50 or p52 with siRNA. Cells were treated with UV as in (A) and WCLs prepared analysed by western blot using the indicated antibodies. (D) U2OS cells were transfected with control or constitutive active IKKβ (left panel) or treated with PMA (100 ng/ml)/Ionomycin (0.5 μM) for 4 h (right panel) prior to treatment with UV (40 J/m2) as indicated. WCLs were prepared and analysed by western blot using the indicated antibodies. (E) U2OS cells were transfected with a non-targeting siRNA or a siRNA to deplete IKKβ. c-Rel was then re-introduced into these cells by transient transfection, and cells were treated with UV (40 J/m2) as indicated, and left to culture for a further 4 h. WCLs were prepared and analysed by western blot using the indicated antibodies. (F) U2OS cells were co-transfected with a non-targeting siRNA or a siRNA to deplete IKKβ, along with a DNA construct to re-introduce Claspin. Cells were treated with UV (40 J/m2) as indicated, and left to culture for a further 4 h. WCLs were prepared and analysed by western blot using the indicated antibodies.
Checkpoint activation by DNA damaging agent such as UV light initiates the action of several signal pathways, which act in concert to repair the damage. To determine how specific the effect of IKKβ depletion on the checkpoint response was, we examined phosphorylation of SMC1 following UV damage, an ATM dependent but Chk1 independent, target of the human S-phase checkpoint (Yazdi et al, 2002). Depletion of IKKβ had no effect on SMC1 Ser966 phosphorylation in response to UV (Supplementary Figure S10C). These results suggest that not all checkpoint activities are lost in the absence of IKKβ, indicating once again a specific effect on Claspin.
We also evaluated the effects of depleting specific NF-κB subunits on the levels of Chk1 phosphorylation following UV (Figure 6C). As predicted from the effects on Claspin mRNA and protein (Figure 2), depletion of RelA and p105/p50 had no significant effect on UV-induced Chk1 Ser317 phosphorylation. However, depletion of RelB, p100/p52 and c-Rel, visibly reduced this phosphorylation event (Figure 6C). Importantly, modulation of endogenous Claspin levels by changing IKK–NF-κB activity results in changes in the levels of checkpoint activation as assessed by the levels of phosphorylated Chk1 (Figure 6D). Similar results were also observed in HEK 293 cells (Supplementary Figure S10D).
Expression of exogenous c-Rel in IKKβ-depleted cells restores checkpoint activation
We have shown that c-Rel can directly regulate the Claspin promoter and this is dependent on IKKβ (Figure 3). In addition, we have found that lack of IKKβ compromises ATR-mediated phosphorylation of Chk1 (Figure 6A and B). To firmly establish the function of c-Rel in this ATR-mediated event, we determined if expression of exogenous c-Rel could rescue Chk1 activation in cells lacking IKKβ. For this purpose, we transiently expressed c-Rel in IKKβ knockdown cells and activated the checkpoint by treating cells with UV. A significant increase in c-Rel levels could be seen in cells transfected with c-Rel expression constructs (Figure 6E). Expression of exogenous c-Rel elevated levels of Claspin in IKKβ-depleted cells to approximately the levels of the control cells (Figure 6E). Importantly, expression of c-Rel almost completely overcame the defect in Chk1 Ser317 phosphorylation in response to UV-induced damage in IKKβ-depleted cells (Figure 6E, compare lane 2 with lane 8). These results demonstrate the importance of IKKβ and c-Rel in the UV-induced ATR/Claspin-mediated Chk1 activation.
Re-introduction of Claspin into IKKβ-depleted cells restores checkpoint activation
We have demonstrated the dependence of Claspin mRNA on IKK signalling, and that depleting IKKβ, results in impaired Chk1 phosphorylation in response to a UV insult. To determine conclusively that the checkpoint impairment in IKKβ cells was due to loss of Claspin, we sought to restore the checkpoint in IKKβ-depleted cells through transfection of a Claspin construct. Expression of exogenous Claspin elevated levels of Claspin in IKKβ depleted to a level higher than that seen in the control cells (Figure 6F). Importantly, expression of Claspin almost completely overcame the defect in Chk1 Ser317 phosphorylation in response to UV-induced damage in IKKβ-depleted cells (Figure 6F, compare lane 2 with lane 4 and lane 6 with lane 8). These results demonstrate that expression of Claspin is sufficient to, at least in part, rescue ATR/Claspin-mediated Chk1 activation in IKKβ compromised cells.
IKKβ-deficient cells are hypersensitive to UV-induced DNA damage
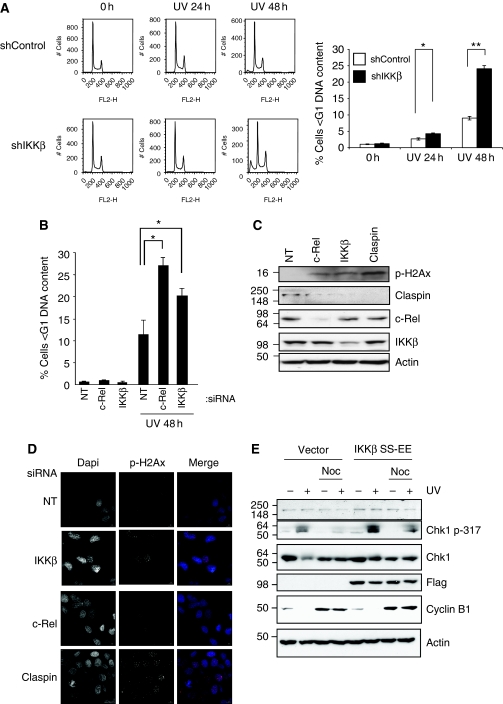
Cell-cycle checkpoints are vital control mechanisms that ensure the fidelity of cell division in response to cellular insults. As we have discovered that depletion of IKKβ specifically compromised Chk1 activation through downregulation of Claspin mRNA, we sought to examine if knockdown of IKKβ renders cells sensitive to cellular insults. Using the U2OS cells line stably depleted of IKKβ, we measured the proportion of cells with a sub-G1 DNA content following UV treatment. As shown in Figure 7A, following UV treatment, control cells showed an increased proportion of cells containing a sub-G1 DNA content of 3% after 24 h treatment raising to 9% after 48 h. Importantly, cells with reduction in IKKβ displayed significantly higher levels of cells with a sub-G1 DNA content, 4.3 and 24%, after 24 and 48 h respectively. This result suggests that IKKβ impaired cells have an increased sensitivity to cell death following UV treatment. To rule out any effects of the stable depletion of IKKβ, we repeated this analysis using transient siRNA transfection. Furthermore, we also investigated the function of c-Rel in this response (Figure 7B). As observed with the stable IKKβ-depleted cells, transient reduction of IKKβ resulted in enhanced sensitivity to UV treatment. Consistent with our previous results, c-Rel depletion also sensitized cells to UV-induced death (Figure 7B).
Figure 7.
Modulation of IKKβ and c-Rel activity regulates Claspin cellular functions. (A) U2OS cells stably depleted of IKKβ were treated with UV (40 J/m2) for 24 and 48 h prior to fixation and flow cytometry analysis. Results are depicted as histograms (left panels) and quantification of sub-G1 content of cells. ANOVA t-tests were performed on the means, and the P-values were calculated. *P<0.05 and **P<0.01. (B) U2OS cells were transiently depleted of c-Rel and IKKβ prior to treatment with UV (40 J/m2) for 48 h. Cells were fixed and analysed by FACS. Graph depicts the mean plus s.d. of the smaller than G1-DNA content of a minimum of three independent experiments. ANOVA t-tests were performed on the means, and the P-values were calculated. *P<0.05 and **P<0.01. (C) U2OS cells were transiently depleted of c-Rel, IKKβ and Claspin for 48 h prior to lysis. WCLs were analysed for the levels of phosphorylated H2Ax by western blot. (D) U2OS were treated as in (A) but cells were fixed and stained for immunofluorescence using the indicated antibodies. Images were acquired using a DeltaVision microscope, deconvolved and processes using OMERO client (opensource software). (E) U2OS cells were transfected with control or constitutive active IKKβ prior to synchronization with nocodazole for 12 h. Cells were treated with UV (40 J/m2) for 2 h prior to lysis. WCLs were analysed by western blot using the indicated antibodies.
Modulation of IKK activity regulates Claspin functions in the cell
Claspin is required for normal replication forks progression (Chini and Chen, 2003; Petermann et al, 2008) and controls genomic stability (Freire et al, 2006; Focarelli et al, 2009). It has been shown that Claspin depletion results in H2Ax foci in cells (Liu et al, 2006). As IKK and NF-κB regulate Claspin levels, we assessed if knockdown of IKK or c-Rel also resulted in the increase of H2Ax phosphorylation and foci. We depleted cells of control, c-Rel, IKKβ and Claspin and analysed by western blot the levels of phosphorylated H2Ax (Figure 7C). Our analysis confirmed the previously published results that Claspin depletion results in high levels of phosphorylated H2Ax. Reduction of c-Rel and IKKβ also resulted in increased levels of this marker when compared with control cells. We confirmed these results using immunofluorescence microscopy (Figure 7D; Supplementary Figure S11A).
Claspin needs to be degraded to allow for checkpoint recovery (Mailand et al, 2006; Mamely et al, 2006; Peschiaroli et al, 2006). Indeed, when degradation of Claspin is inhibited following DNA damage, activation of Chk1 persists and cells fail to recover from the checkpoint. We thus investigated if hyperactivation of the NF-κB signalling pathway resulted in a similar phenotype. For this, we overexpressed a constitutively active form of IKKβ in cells. We synchronized them in mitosis using nocodazole and then treated them with UV for 2 h prior to lysis. Cells with a normal control of Claspin should have no Chk1 phosphorylation present in mitosis. This is evident in control-transfected cells, where no Chk1 phosphorylation could be detected (Figure 7E). However, in the presence of active IKKβ, phosphorylation of Chk1 was still present (Figure 7E), indicating that cells have failed to inactivate the checkpoint in mitosis. Expression levels of Cyclin B1 indicate that cells were synchronized in mitosis.
Claspin levels correlate with c-Rel in a number of cancer cell lines
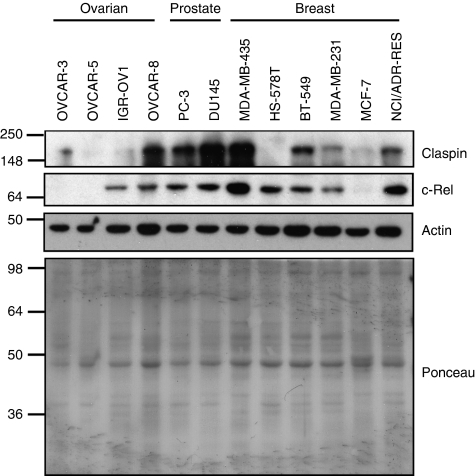
Our analysis demonstrates that IKK and c-Rel control the levels of Claspin in the cells by an alternative mechanism to cell cycle. Given the importance of Claspin for genomic stability and checkpoint responses, we analysed if in cancer cells, there was a correlation between Claspin and c-Rel levels (Figure 8). For this purpose, we obtained cell lysates from 12 different cell lines from the National Cancer Institute (NCI) collection. Our analysis revealed a good correlation between Claspin levels and c-Rel in the majority of the cell lines analysed with two exceptions, an ovarian cancer cell line IGR-OV1 and a breast cancer cell line HS-578T. However, in the rest of the cell lines tested, Claspin levels were high when c-Rel levels were also elevated.
Figure 8.
Claspin and c-Rel levels correlate in a variety of different cancer cell lines. WCLs of different cancer cell lines obtained from the NCI cell line collection were analysed for the levels of Claspin and c-Rel by immunoblot. Actin and ponceau staining were used to demonstrate equal loading.
Our results demonstrate for the first time that interference with IKKβ and c-Rel has an impact on the DNA-damage checkpoints through modulation of the Claspin gene.
Discussion
The DNA-damage checkpoint is crucial for the maintenance of cellular integrity and proper function of the cell. Here, we demonstrate a novel function of IKKβ as being necessary for the proper activation of this checkpoint, in vivo, through regulation of Claspin.
This regulation is dependent on the activity of the IKK complex, and is sensitive to IKK activity modulation with inhibition and activation controlling Claspin function (Figures 6 and 7). Claspin levels are sensitive to disruption of IKKβ or IKKγ. RNAi depletion of IKKβ, or the regulatory subunit IKKγ, but not IKKα, resulted in reduced levels of Claspin protein (Figures 1 and 2). Claspin levels were also reduced when a small molecule inhibitor of the IKK complex was used, indicating that IKK catalytic activity is required to regulate Claspin.
Previous studies have focused on the regulation of Claspin at the level of protein stability (Yoo et al, 2004; Peschiaroli et al, 2006). These studies have shown that Claspin protein levels fluctuate through the cell cycle and closely match the activity profile of Plk1 (Peschiaroli et al, 2006). Plk1 phosphorylates Claspin in its phospho-degron, which results in the recruitment of the SCF-β-TRCP complex. This targets Claspin protein for degradation by the proteasome. Here, we have demonstrated that IKK regulation of Claspin is independent of the proteasome (Figures 1D and 5E; Supplementary Figure S8), but instead depletion of IKKβ results in reduced levels of Claspin mRNA (Figure 2A). Analysis of the regulation of Claspin mRNA levels through the cell cycle also demonstrated that these do not change significantly in our cells (Figure 5). This revealed that Claspin regulation can be achieved by two independent mechanisms, one mediated by protein stability changes and cell-cycle dependent and one mediated by changes in the transcription of the gene itself, which is cell-cycle independent. Changes of Claspin gene expression have not been studied extensively before. However, one research group has suggested that both Chk1 and Claspin are E2F1 targets (Verlinden et al, 2007). These results were based on luciferase assays, but a direct regulation of the Claspin promoter by E2F1 using ChIP has not been demonstrated so far. Furthermore, we did not observe any changes in Claspin mRNA following E2F1 siRNA treatment or either of the synchronization approaches we have used, indicating that in our model systems Claspin mRNA is stable throughout the cell cycle (Figure 5; Supplementary Figure S12).
IKK is the upstream kinase responsible for the activation of the NF-κB family of transcription factors and our data suggest that depletion of RelB, c-Rel or p52 results in reduction of Claspin both at the protein and RNA level (Figure 2). Taken together with the results obtained with the IKK depletion, this indicates that IKK is acting upstream of NF-κB to control Claspin mRNA. Indeed, we could see the NF-κB family member c-Rel bound to the Claspin promoter in an IKKβ-dependent manner (Figures 3 and 4). We could not detect either RelB or p52 binding at the κB sites we have identified in the Claspin promoter (Figure 3). This may be due to RelB/p52 binding at distal sites altering Claspin expression or because the epitopes recognized by these antibodies are occluded in the context of this promoter. However, RelB/p52 may alter Claspin levels indirectly, by altering an unknown factor that controls Claspin. The exact mechanism by which c-Rel transactivates the Claspin gene is not known, but our results indicate that when c-Rel is lost from a region upstream of the transcription start site there is also a localized drop in the levels of histone H3 acetylation (Figure 3E). This suggests that c-Rel may recruit histone acetyl transferases (HATs) to the Claspin promoter. Although we have not investigated the nature of the HAT activity involved in transactivating the Claspin gene, it is interesting to note that c-Rel can act synergistically with p300 to activate target gene promoters in HEK293 cells (Sun et al, 2004). Furthermore, loss of c-Rel also correlated with reduced levels of RNA Polymerase II at the transcription start sites and coding region of the Claspin gene (Figure 3D), indicating an active function for c-Rel on this gene. Claspin protein has been reported to be elevated in cancer cells (Tsimaratou et al, 2007; Verlinden et al, 2007) and an additional study has reported that Claspin mRNA is high breast cancer (Verlinden et al, 2007). Of note, c-Rel (Belguise and Sonenshein, 2007), RelB (Mineva et al, 2009) and p100/p52 (Cogswell et al, 2000) have all been reported to be abnormally active in breast cancer. Given our results, it would be interesting to determine if c-Rel, RelB and p100/p52 were responsible for the high levels of Claspin observed. Our own analysis revealed a good correlation between Claspin and c-Rel levels in a variety of different cancer cell lines (Figure 8).
Functionally, depleting IKKβ results in reduced phosphorylation of Chk1 in response to UV and Hydroxyurea (Figure 6; Supplementary Figure S10). Importantly, c-Rel overexpression can rescue Claspin levels and the impairment in Chk1 phosphorylation when IKKβ is depleted (Figure 6E), further confirming the importance of c-Rel in the control of Claspin. Significantly, expression of exogenous Claspin can functionally rescue the checkpoint defect in IKKβ-depleted cells, demonstrating the function of Claspin in this pathway (Figure 6F). The consequences of the reduction in the checkpoint is evident with the differences between IKKβ-depleted and control cells in response to long-term stresses. IKKβ-depleted cells do not arrest efficiently at the G2/M boundary in response to UV, instead these cells undergo cell death (Figure 7A). Importantly, c-Rel reduction also sensitizes cells to UV-induced cell death (Figure 7B). Depletion of Claspin or Chk1 has been shown to result in similar phenotypes (Chini and Chen, 2003), with cells going through premature mitosis before efficient DNA replication resulting in mitotic catastrophe.
The implications of our results are numerous. They demonstrate that cells depleted of IKKβ are more sensitive to UV damage. This makes IKKβ an attractive anti-cancer agent in combination with DNA damaging agents inhibition of IKK would not only inhibit Chk1 activation but would also prevent other pro-survival genes from being activated by NF-κB. Similarly, Chk1 inhibitors are currently being assessed for their properties as anti-cancer agents in phase I/II trials (Bucher and Britten, 2008). A reverse side to both IKK and Chk1 inhibition would be the increased genetic instability. Our own results indicate that inhibition of IKK and c-Rel leads to increased H2Ax foci, a marker of DNA damage (Figure 7C and D; Supplementary Figure S11A). However, as the majority of the cells in the human body are quiescent, inhibition of Chk1 activity (either by IKK or Chk1 inhibition directly) might only affect cycling cells such as cancer cells.
Conversely, hyperactivation of IKK results in a failure to recover from the DNA-damage checkpoint, with Chk1 phosphorylation persisting in mitosis (Figure 7E). This is exactly how cells behave in the presence of a non-degradable form of Claspin (Mailand et al, 2006). These results suggest that increased activation of IKK might lead to cell-cycle delays because of failure to switch off the checkpoint.
The findings of this study point to the important function of IKKβ and c-Rel in the DNA-damage response pathway. Instead of a single level of control over Claspin through the activity of Plk1, we have uncovered a second and parallel regulatory mechanism. As such, IKKβ and c-Rel are required for proper checkpoint function and recovery and thus important for signalling following DNA-damage checkpoint activation. Furthermore, by controlling the IKK–NF-κB axis and hence Claspin, the cell can integrate signals from various cellular pathways with the DNA-damage response, allowing for different levels of checkpoint activation and hence diverse cellular outcomes.
Materials and methods
Cells
U2OS osteosarcoma, MDA-MB-231 breast carcinoma, mouse embryonic fibroblasts and HEK 293 cell lines were grown in Dulbecco's modified eagle medium (Lonza) supplemented with 10% fetal bovine serum (Gibco), 50 units/ml penicillin (Lonza) and 50 μg/ml streptomycin (Lonza) for no more than 30 passages. U2OS cells containing stably transfected IKKβ or non-targeting shRNAs were created using the pSilencer vectors as described previously (Schumm et al, 2006). shRNA sequences can be found in Supplementary data. Cell lysates presented in Figure 8 were a kind gift from Dr G Sapkota and D Bruce (Dundee) and obtained from the NCI collection.
DNA constructs
Expression plasmid for c-Rel was a kind gift from Professor Neil Perkins (Newcastle, UK). Flag-IKKβ was a kind gift from Professor Ron Hay (Dundee, UK). pcDNA3-IKKβ SS-EE was obtained by site-directed mutagenesis and constructed by the College of Life Sciences University of Dundee cloning service. Claspin expression construct was a kind gift from Professor Paul Clarke (Dundee, UK). Exogenous c-Rel and IKKβ expression plasmids were transfected into cells using GeneJuice (Invitrogen) as per the manufacturer's instructions.
Chemicals inhibitors
MG132 (Merck Biosciences) was dissolved in DMSO and used at the final concentration of 50 μM for 3 h prior to harvest. Bay 11-7082 (Merck Biosciences) was dissolved in DMSO and used at the final concentration of 20 μM. PMA (Sigma) and Ionomycin (Merck Biosciences) were dissolved in DMSO.
siRNA transfection
siRNA duplex oligonucleotides were synthesized by MWG and transfected using Interferin (Polyplus) per the manufacturer's instructions. siRNA sequences can be found in Supplementary data.
siRNA and DNA co-transfection
siRNA duplex oligonucleotides and DNA constructs were co-transfected using JetPrime (Polyplus) as per the manufacturer's instructions.
RT–PCR, Antibodies, ChIP, Microscopy and other methods can be found in Supplementary data.
Statistical analysis
ANOVA and Student's t-tests were performed on the means, and P-values were calculated. *P⩽0.050, **P⩽0.010 and ***P⩽0.001.
Supplementary Material
Acknowledgments
The authors thank Professor Neil Perkins, Professor Julian Blow, Professor Robert White, Professor Paul Clarke, Professor Ron Hay and Dr John Rouse for helpfully comments, suggestions and reagents. The authors are grateful to Adel Ibrahim from the cloning service for providing the constitutively active IKKβ construct and Dr G Sapkota and D Bruce for providing the cancer cell line lysates. NK is funded by an AICR project grant, SR is funded by an RCUK fellowship and the University of Dundee and SM is funded by MRC NIRG.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anderson LA, Perkins ND (2003) Regulation of RelA (p65) function by the large subunit of replication factor C. Mol Cell Biol 23: 721–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguise K, Sonenshein GE (2007) PKCtheta promotes c-Rel-driven mammary tumorigenesis in mice and humans by repressing estrogen receptor alpha synthesis. J Clin Invest 117: 4009–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett LN, Clarke PR (2006) Regulation of Claspin degradation by the ubiquitin-proteosome pathway during the cell cycle and in response to ATR-dependent checkpoint activation. FEBS Lett 580: 4176–4181 [DOI] [PubMed] [Google Scholar]
- Bucher N, Britten CD (2008) G2 checkpoint abrogation and checkpoint kinase-1 targeting in the treatment of cancer. Br J Cancer 98: 523–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell KJ, Rocha S, Perkins ND (2004) Active repression of antiapoptotic gene expression by RelA(p65) NF-kappa B. Mol Cell 13: 853–865 [DOI] [PubMed] [Google Scholar]
- Chariot A (2009) The NF-kappaB-independent functions of IKK subunits in immunity and cancer. Trends Cell Biol 19: 404–413 [DOI] [PubMed] [Google Scholar]
- Chini CC, Chen J (2003) Human claspin is required for replication checkpoint control. J Biol Chem 278: 30057–30062 [DOI] [PubMed] [Google Scholar]
- Clarke CA, Clarke PR (2005) DNA-dependent phosphorylation of Chk1 and Claspin in a human cell-free system. Biochem J 388 (Pt 2): 705–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogswell PC, Guttridge DC, Funkhouser WK, Baldwin AS Jr (2000) Selective activation of NF-kappa B subunits in human breast cancer: potential roles for NF-kappa B2/p52 and for Bcl-3. Oncogene 19: 1123–1131 [DOI] [PubMed] [Google Scholar]
- Focarelli ML, Soza S, Mannini L, Paulis M, Montecucco A, Musio A (2009) Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer 48: 1083–1090 [DOI] [PubMed] [Google Scholar]
- Freire R, van Vugt MA, Mamely I, Medema RH (2006) Claspin: timing the cell cycle arrest when the genome is damaged. Cell Cycle 5: 2831–2834 [DOI] [PubMed] [Google Scholar]
- Hacker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE 2006: re13. [DOI] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Irelan JT, Murphy TJ, DeJesus PD, Teo H, Xu D, Gomez-Ferreria MA, Zhou Y, Miraglia LJ, Rines DR, Verma IM, Sharp DJ, Tergaonkar V, Chanda SK (2007) A role for IkappaB kinase 2 in bipolar spindle assembly. Proc Natl Acad Sci USA 104: 16940–16945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan MB, Bartek J (2004) Cell-cycle checkpoints and cancer. Nature 432: 316–323 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (2000) Claspin, a novel protein required for the activation of Chk1 during a DNA replication checkpoint response in Xenopus egg extracts. Mol Cell 6: 839–849 [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG (2003) Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat Cell Biol 5: 161–165 [DOI] [PubMed] [Google Scholar]
- Lee J, Gold DA, Shevchenko A, Dunphy WG (2005) Roles of replication fork-interacting and Chk1-activating domains from Claspin in a DNA replication checkpoint response. Mol Biol Cell 16: 5269–5282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J (2006) Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26: 6056–6064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Bekker-Jensen S, Bartek J, Lukas J (2006) Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol Cell 23: 307–318 [DOI] [PubMed] [Google Scholar]
- Mamely I, van Vugt MA, Smits VA, Semple JI, Lemmens B, Perrakis A, Medema RH, Freire R (2006) Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr Biol 16: 1950–1955 [DOI] [PubMed] [Google Scholar]
- Mineva ND, Wang X, Yang S, Ying H, Xiao ZX, Holick MF, Sonenshein GE (2009) Inhibition of RelB by 1,25-dihydroxyvitamin D3 promotes sensitivity of breast cancer cells to radiation. J Cell Physiol 220: 593–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins ND (2007) Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8: 49–62 [DOI] [PubMed] [Google Scholar]
- Perkins ND, Gilmore TD (2006) Good cop, bad cop: the different faces of NF-kappaB. Cell Death Differ 13: 759–772 [DOI] [PubMed] [Google Scholar]
- Peschiaroli A, Dorrello NV, Guardavaccaro D, Venere M, Halazonetis T, Sherman NE, Pagano M (2006) SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol Cell 23: 319–329 [DOI] [PubMed] [Google Scholar]
- Petermann E, Helleday T, Caldecott KW (2008) Claspin promotes normal replication fork rates in human cells. Mol Biol Cell 19: 2373–2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumm K, Rocha S, Caamano J, Perkins ND (2006) Regulation of p53 tumour suppressor target gene expression by the p52 NF-kappaB subunit. EMBO J 25: 4820–4832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Lu J, Wei L, Wang X, Xu X, Dong M, Huang B (2004) Histone acetyltransferase activity of p300 enhances the activation of IL-12 p40 promoter. Mol Immunol 41: 1241–1246 [DOI] [PubMed] [Google Scholar]
- Tsimaratou K, Kletsas D, Kastrinakis NG, Tsantoulis PK, Evangelou K, Sideridou M, Liontos M, Poulias I, Venere M, Salmas M, Kittas C, Halazonetis TD, Gorgoulis VG (2007) Evaluation of claspin as a proliferation marker in human cancer and normal tissues. J Pathol 211: 331–339 [DOI] [PubMed] [Google Scholar]
- van Uden P, Kenneth NS, Rocha S (2008) Regulation of hypoxia-inducible factor-1alpha by NF-kappaB. Biochem J 412: 477–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verlinden L, Vanden Bempt I, Eelen G, Drijkoningen M, Verlinden I, Marchal K, De Wolf-Peeters C, Christiaens MR, Michiels L, Bouillon R, Verstuyf A (2007) The E2F-regulated gene Chk1 is highly expressed in triple-negative estrogen receptor/progesterone receptor/HER-2 breast carcinomas. Cancer Res 67: 6574–6581 [DOI] [PubMed] [Google Scholar]
- Wu ZH, Shi Y, Tibbetts RS, Miyamoto S (2006) Molecular linkage between the kinase ATM and NF-kappaB signaling in response to genotoxic stimuli. Science 311: 1141–1146 [DOI] [PubMed] [Google Scholar]
- Yazdi PT, Wang Y, Zhao S, Patel N, Lee EY, Qin J (2002) SMC1 is a downstream effector in the ATM/NBS1 branch of the human S-phase checkpoint. Genes Dev 16: 571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo HY, Kumagai A, Shevchenko A, Shevchenko A, Dunphy WG (2004) Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117: 575–588 [DOI] [PubMed] [Google Scholar]
- Zhao H, Piwnica-Worms H (2001) ATR-mediated checkpoint pathways regulate phosphorylation and activation of human Chk1. Mol Cell Biol 21: 4129–4139 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.