S6K1 is a multifaceted regulator of Mdm2 that connects nutrient status and DNA damage response
In this study, Lai et al implicate p38-Akt-mTOR-S6K1 as a novel regulatory pathway for p53 in response to DNA damage. A model is proposed that downstream of genotoxic stress S6K1-mediated Mdm2 phosphorylation promotes its cytoplasmic retention and p53 stabilization.
Keywords: Mdm2, nuclearcytoplasmic shuttling, p53, S6K1
Abstract
p53 mediates DNA damage-induced cell-cycle arrest, apoptosis, or senescence, and it is controlled by Mdm2, which mainly ubiquitinates p53 in the nucleus and promotes p53 nuclear export and degradation. By searching for the kinases responsible for Mdm2 S163 phosphorylation under genotoxic stress, we identified S6K1 as a multifaceted regulator of Mdm2. DNA damage activates mTOR-S6K1 through p38α MAPK. The activated S6K1 forms a tighter complex with Mdm2, inhibits Mdm2-mediated p53 ubiquitination, and promotes p53 induction, in addition to phosphorylating Mdm2 on S163. Deactivation of mTOR-S6K1 signalling leads to Mdm2 nuclear translocation, which is facilitated by S163 phosphorylation, a reduction in p53 induction, and an alteration in p53-dependent cell death. These findings thus establish mTOR-S6K1 as a novel regulator of p53 in DNA damage response and likely in tumorigenesis. S6K1–Mdm2 interaction presents a route for cells to incorporate the metabolic/energy cues into DNA damage response and links the aging-controlling Mdm2–p53 and mTOR-S6K pathways.
Introduction
The genome is constantly attacked by exogenous factors such as irradiation and endogenous factors such as reactive oxygen species, which are byproducts of mitochondrial energy production and are generated in proportion to the metabolic rates of the cells. Cells have evolved a network of signalling events to sense the liaisons, to transmit signals to alter gene expression, and to halt cell proliferation or induce apoptosis and/or senescence. Although activation of DNA damage response might accelerate the aging process, it can help to maintain genome integrity and prevent tumorigenesis (Serrano and Blasco, 2007). In the early phase of response, PI-3 kinase-like kinases, such as Atm and Atr, are activated, which in turn elicit several pathways, including the Atm/Atr-p38-MK2, the Atm/Atr-Chk2/1, and the Atm/Atr-p53 pathways (Vousden and Lu, 2002; Reinhardt et al, 2007). As ‘the guardian of the genome', transcription modulator p53 has a prominent function in genotoxic stress-induced cell-cycle arrest, apoptosis, and senescence/aging (Vogelstein et al, 2000). A major regulator of p53 is Mdm2, a E3 ligase that is believed to ubiquitinate p53 mainly in the nucleus, which promotes p53 nuclear export and degradation (Kruse and Gu, 2009). There are also studies suggesting that Mdm2 could mediate p53 ubiquitination in the cytoplasm (Xirodimas et al, 2001). Moreover, Mdm2 is a direct target gene of p53. Thus, Mdm2 and p53 form a regulatory loop that determines the protein levels of p53. As such, even a modest change in Mdm2, for example as a result of Mdm2 polymorphism, perturbs the protein levels of p53 and eventually alters the tumorigenesis process (Whibley et al, 2009). In response to genotoxic stress, Mdm2 shows a decrease and Atr-mediated S395 phosphorylation, which is reported to negatively regulates p53 nuclear export and thus facilitate p53 stabilization (Shinozaki et al, 2003). However, how Mdm2 shuttles between the cytoplasm and the nucleus and how Mdm2 activity is regulated are not well understood (Meek and Knippschild, 2003).
Another Atm/Atr-related protein kinase is mTOR, which acts as a sensor for growth factors, nutrients, cellular energy levels, and redox status, and it transmits signals to control cell proliferation, metabolism, and growth (Ma and Blenis, 2009). mTOR can be regulated by PI-3K-Akt1, Ras-MAPKs, AMPK, GSK3β, hVps34, or Redd1 (Woods et al, 2008). The major mTOR downstream signalling molecules are S6K1 and S6K2 (Pende et al, 2004), which in turn phosphorylate a few substrates including rpS6, IRS-1, and eIF4B. Phosphorylation of eIF4B and rpS6 by S6K1, as well as phosphorylation of 4E-BP1 by mTOR, promotes global translation and cell growth (Averous and Proud, 2006; Mamane et al, 2006). Indeed, constitutive activation of mTOR by deficiency of either of the two upstream negative regulators, TSC1 and TSC2, leads to tuberous sclerosis (TSC, benign tumours in several organs) in human (Dann et al, 2007; Guertin and Sabatini, 2007), indicating that mTOR pathway has pro-proliferation activity. Phosphorylation of IRS-1 by S6K1 leads to deactivation and/or degradation of IRS-1, which underlies the alterations in glucose metabolism observed in S6K1-deficient mice (Shah et al, 2004; Um et al, 2006; Aguilar et al, 2007). More interestingly, treatment with rapamycin, a potent inhibitor of mTOR, as well as S6K1 deficiency, has been recently reported to extend the lifespan of mouse (Harrison et al, 2009; Selman et al, 2009), suggesting a function for mTOR-S6K1 in promoting aging. However, how the mTOR-S6K pathway regulates cell proliferation and aging, two seemingly opposite events, is not well defined.
Cells usually exist in a dynamic microenvironment that is constituted of various growth factors, cytokines, and nutrients, which collectively elicit a network of signalling events. Although the activation of the Atm-p53 pathway per se in DNA damage response has been extensively studied, little is known about whether this activation is regulated by the growth conditions and energy status of the cells, which are sensed by pathways such as mTOR-S6K signalling. This study, by investigating Mdm2 phosphorylation on S163, identifies S6K1 as a multifaceted regulator of Mdm2 and reveals a function for the mTOR-S6K1 pathway in regulating p53-mediated DNA damage response. S6K1 physically interacts with Mdm2 and this complex formation not only presents a mechanism by which cells adjust DNA damage response according to their growth conditions, but also links two of the major pathways that control the aging process.
Results
Identification of S6K1/2 as kinases for Mdm2 S163 phosphorylation under genotoxic stress
Mdm2 has an important function in controlling p53 stability in response to genotoxic stress. Recent studies have shown that Mdm2 can be phosphorylated on S163/183 (S166/186 in Hdm2), residues located near the NLS and NES of Mdm2, by Akt, MAPKs, MK2, Pim1/2, and other kinases (Meek and Knippschild, 2003). The phosphorylation is found to regulate Mdm2 nuclearcytoplasmic shuttling under certain conditions. Here, we used primary MEFs to study Mdm2 S163 phosphorylation in response to DNA damage caused by Doxorubicin (Dox), a chemotherapeutic drug that causes double- and single-stranded DNA breaks, or hydroxyurea (HU), a chemotherapeutic drug that causes single-stranded DNA breaks, hoping to identify new regulators of Mdm2. It was found that Mdm2 was phosphorylated on S163 under normal growth conditions and this phosphorylation was augmented by genotoxic stress, even though the protein levels of Mdm2 were transiently downregulated (Figure 1A; Supplementary Figure S1). However, S183 phosphorylation was difficult to detect in these settings (data not shown), likely because of the suboptimal sensitivity of the antibodies, as later experiments showed that overexpressed Mdm2 could be equally phosphorylated at S163 and S183. The co-existence of downregulation of Mdm2 and upregulation of S163 phosphorylation suggests that the S163 phosphorylation might, at least transiently, prevent Mdm2 from degradation. Dox-induced downregulation of Mdm2 was accompanied with a decrease in Mdm2 mRNA levels (Supplementary Figure S2A). Proteosome inhibitor MG132 treatment could increase the protein levels of Mdm2, yet it failed to rescue Dox-induced Mdm2 downregulation (Supplementary Figure S2B), suggesting that Mdm2 is also regulated at the mRNA levels in response to Dox. Genotoxic stress-induced Mdm2 S163 phosphorylation was also observed in primary osteoblasts and mouse embryonic stem cells (data not shown), suggesting that it is a common cellular response.
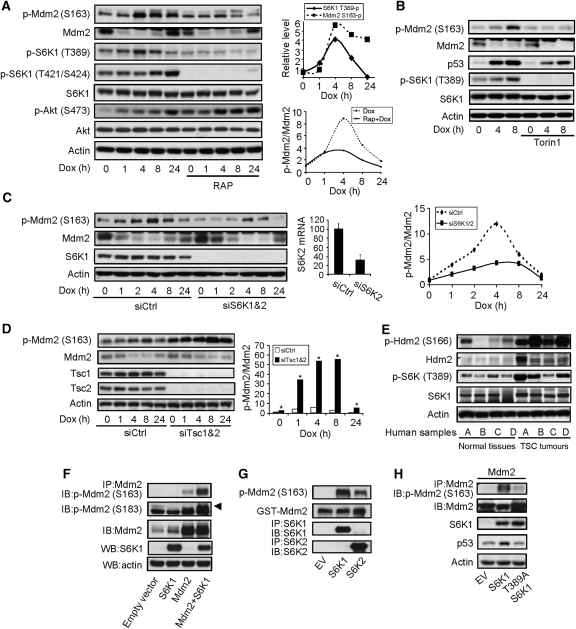
Figure 1.
Genotoxic stress induced Mdm2 S163 phosphorylation through mTOR-S6K. (A) Dox treatment led to Mdm2 S163 phosphorylation in primary MEFs, which was blocked by rapamycin pretreatment. MEFs were pretreated with or without 1 nM of rapamycin for 1 h before adding Dox to a final concentration of 1 μM for different periods of time. Phosphorylation and protein levels of Mdm2, S6K1, and Akt were analysed by western blot. (Right upper panel) Quantitation data of S6K1 T389 phosphorylation and Mdm2 S163 phosphorylation. (Right bottom panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in the absence of RAP was set at 1.0. (B) Dox-induced Mdm2 S163 phosphorylation was blocked by Torin1. The experiments were carried out as in Figure 1A except that 250 nM of Torin1 was used to replace Rapamycin. The value of p-Mdm2 S163 at time 0 in the absence of Torin1 was set at 1.0. (C) Knockdown of S6K1 and 2 led to hypophosphorylation of Mdm2. S6K1 and S6K2 were knocked down with siRNA in primary MEFs for 48 h before addition of Dox. Middle panel shows the mRNA levels of S6K2 after knockdown (because of the weak activity of S6K2 antibodies). (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in the presence of control siRNA was set at 1.0. (D) Knockdown of Tsc1 and Tsc2 led to enhanced Mdm2 S163 phosphorylation in MEFs. Tsc1 and Tsc2 were knocked down by siRNA in MEFs for 48 h before addition of Dox. The phosphorylation of Mdm2 S163, Mdm2, and Tsc1/2 were analysed by western blot. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in the presence of control siRNA was set at 1.0. (E) Tumour kidney tissues from four TSC patients (A–D) showed enhanced Hdm2 S166 phosphorylation. TSC tumour samples and matched normal tissues (four each) were homogenized and the phosphorylation of Hdm2 S166 and S6K1 T389 was analysed by western blot. (F) Co-expression of S6K1 led to Mdm2 phosphorylation on S163. Empty vector or S6K1 was co-transfected with Mdm2 in 293T cells, which were treated with Dox for 4 h. Mdm2 was immunoprecipitated and phosphorylation at S163 or S183 (upper arrow) was analysed by western blot. (G) In vitro kinase assay showed that S6K1 could directly phosphorylate Mdm2. HA-tagged S6K1 or HA-tagged S6K2 was expressed in 293T cells and was immunoprecipitated from the cells. GST-Mdm2, which was expressed and purified from bacteria, was used as substrate to incubate with immunoprecipitated S6K1 or S6K2. Phosphorylation of Mdm2 S163 was determined by western blot. (H) Requirement for S6K1 T389 for Mdm2 S163 phosphorylation. S6K1 or S6K1 T389A mutant was expressed together with Mdm2 in 293T cells, and then Mdm2 was immunoprecipitated and its S163 phosphorylation was determined by western blot.
To identify the major kinase(s) responsible for the Dox-induced S163 phosphorylation, we screened a kinase inhibitors library (94 in total) from BIOMOL international, and found that a number of inhibitors could markedly hinder this phosphorylation (data not shown). These include the inhibitors for EGFR, PDGFR, PI-3K, MAPK, IKK2, PKC, and mTOR, pointing to the involvement of the RTK-PI-3K-AKT/MAPK-mTOR pathway in Mdm2 S163 phosphorylation. To pinpoint the downstream kinases of this pathway, we pretreated MEFs with mTOR inhibitor rapamycin or the ATP site-specific inhibitor of mTOR, Torin1, and then challenged the cells with Dox (Thoreen and Sabatini, 2009; Thoreen et al, 2009). It was found that Dox-induced Mdm2 S163 phosphorylation was markedly reduced by rapamycin or Torin1 (Figure 1A and B). However, both S163 and 183 lie in a RXRXXS motif, which matches the consensus recognition sequence of S6K1, the major mTOR downstream kinase, but not that of mTOR (Ruvinsky and Meyuhas, 2006). We then knocked down S6K1 and its homologue S6K2 with siRNA and found that S6K knockdown markedly inhibited Dox-induced S163 phosphorylation, without sufficiently altering the protein levels of Mdm2 (Figure 1C). There is a considerable portion of S163 phosphorylated Mdm2 that were not responding to mTOR inhibition or S6K downregulation, which could exist in a different pool or protein complex and their phosphorylation could be carried out by other kinases, suggesting that Mdm2 phosphorylation is regulated in a rather complicated manner. Nevertheless, these results indicate that mTOR-S6K1/2 activity is necessary for optimal Mdm2 S163 phosphorylation, especially in response to DNA damage.
We then tested whether Mdm2 is a direct substrate of S6K1/2. We found that knockdown of Tsc1 and Tsc2 could promote Mdm2 S163 phosphorylation in MEFs (Figure 1D). Similarly, tumour kidney tissues from TSC patients showed a drastic increase in Hdm2 S166 phosphorylation, accompanied with a slight increase in the protein levels of Hdm2, compared with the matched normal tissues (Figure 1E). In addition, co-expression of mTOR, S6K1, or S6K2 with Mdm2 in 293T cells led to Mdm2 S163 phosphorylation as well as S183 phosphorylation (Figure 1F and data not shown), suggesting that S6K activation is sufficient for Mdm2 S163 phosphorylation in vivo. Furthermore, immunoprecipitated S6K1 or 2 could phosphorylate S163 of bacteria-expressed GST-Mdm2 fusion protein in vitro (Figure 1G). These results indicate that S6K1 is both necessary and sufficient for optimal Mdm2 S163 phosphorylation and that Mdm2 is a substrate of S6K1. S6K1 is phosphorylated on T389 by mTOR, which promotes S6K1 activation and generates a interaction surface that favours PDK1 binding (Frodin et al, 2002), but disfavours eIF3 binding (Holz et al, 2005). Here, we found that T389 is essential for Mdm2 S163 phosphorylation (Figure 1H).
Genotoxic stress activates mTOR-S6K1 through the p38α MAPK pathway
Consistent with these findings, Dox or HU could activate S6K1 in MEFs, justified by an increase in S6K1 phosphorylation on T389, T421, and S424, which were all blocked by rapamycin pretreatment (Figure 1A; Supplementary Figure S1). Dox-induced T389 phosphorylation was rather transient, compared with Mdm2 S163 phosphorylation (Figure 1A, right upper panel). Dox also activated mTOR at S2481 phosphorylation, an autophosphorylation site and an indication of mTOR activation (Supplementary Figure S1A). Although a few studies reported that Akt1 could directly phosphorylate Mdm2 at S163, several lines of evidence suggest that Mdm2 phosphorylation was not directly carried out by Akt1 in our experimental settings. First, inactivation of the mTOR-S6K signalling by rapamycin or S6K1/2 knockdown activated Akt1 through feedback regulation on IRS-1, as previously reported (Figure 1A and data not shown) (Harrington et al, 2004; Shah et al, 2004). However, Mdm2 S163 phosphorylation was decreased under these conditions. Second, growth factors and serum have been reported to enhance Mdm2 phosphorylation on S163 (Zhou et al, 2001). Yet, we found that serum-induced Mdm2 S163 phosphorylation could be largely inhibited by rapamycin pretreatment (Supplementary Figure S3A). Under this condition, S163 phosphorylation was accompanied with a very transient upregulation of Mdm2, which quickly returned to the basal level, suggesting that Mdm2 phosphorylation might be able to transiently stabilize Mdm2. Third, co-expression of Akt1 and Mdm2 in 293T cells led to S163 phosphorylation of Mdm2, but this phosphorylation could be blocked by treatment with rapamycin (Supplementary Figure S3B). These results, taken together, suggest that S6K1, rather than Akt, is a major kinase responsible for Mdm2 S163 phosphorylation in our experimental settings.
Among other kinases that are reported to phosphorylate Mdm2 S163 are Erks and p38 MAPK, both of which are activated by genotoxic stress (Supplementary Figure S1). p38 MAPK has been reported to be activated by genotoxic stress, especially by UV, and it phosphorylates Cdc25 and p53 to regulate cell proliferation and survival (Thornton and Rincon, 2009). We found that inhibition of p38 with SB203580 impeded the basal and Dox-induced Mdm2 S163 phosphorylation (Figure 2A, left panel), whereas inhibition of Erks with U0126 even slightly enhanced this phosphorylation (Supplementary Figure S4A and B). As Mdm2 S163 phosphorylation is mainly mediated by S6K1 under genotoxic stress and several studies suggest that p38 MAPK interacts with the mTOR upstream regulator Tsc proteins (Li et al, 2003; Tee et al, 2003), we tested the possibility that the mTOR-S6K pathway might mediate the effect of p38 MAPK on Mdm2 S163 phosphorylation. It was indeed found that inhibition of p38 MAPK decreased the activation of both mTOR and S6K1, accompanied by a decrease in Akt1 S473 phosphorylation (but not S308) (Figure 2A, left panel). To substantiate these findings, we used primary p38α−/− MEFs and found that p38 deficiency similarly inhibited Dox-induced activation of S6K1, mTOR, and Akt1, and phosphorylation of Mdm2 (Figure 2B). However, in Tsc2−/− MEFs, p38 MAPK inhibitors failed to repress S6K1 activation or Mdm2 S163 phosphorylation (Figure 2A, right upper panel). Moreover, the defect in Mdm2 S163 phosphorylation observed in p38α−/− MEFs could be rescued by retroviral expression of S6K1 (Figure 2C), which could be activated by growth factors in the serum. Finally, combinational use of rapamycin and SB203580 did not show an additive effect on the suppression of Mdm2 S163 phosphorylation compared with single treatment (Figure 2D). Taken together, these data support the concept that Dox-activated p38 MAPK regulates Mdm2 S163 phosphorylation through TSC-mTOR-S6K1. Several studies have reported that p38 MAPK, MK2, and Akt1 form a complex, which promotes p38 MAPK-mediated Akt1 activation (Rane et al, 2001; Kim et al, 2008).
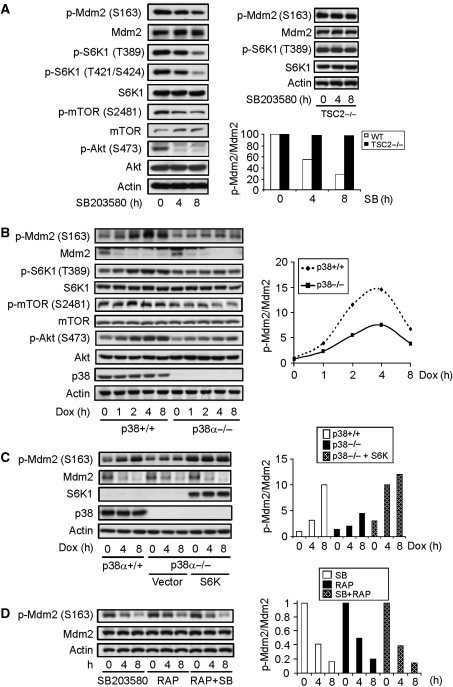
Figure 2.
Genotoxic stress activates p38α-mTOR-S6K pathway to phosphorylate Mdm2 on S163. (A) Inhibition of p38α with an inhibitor decreased Mdm2 S163 phosphorylation, S6K1 T389 phosphorylation, and mTOR S2481 phosphorylation in normal MEFs (left panel) but not in Tsc2−/− MEFs (right upper panel). Normal and Tsc2−/− MEFs were treated with p38 MAPK inhibitor SB203580 for different periods of time and Mdm2 phosphorylation and S6K1 activation were analysed with western blot. (Right bottom panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in the presence of SB was set at 1.0. (B) p38α deficiency also resulted in decreased Mdm2 S163 phosphorylation and S6K1 T389 phosphorylation. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in control WT MEFs was set at 1.0. (C) Ectopic expression of S6K rescued the Mdm2 S163 phosphorylation defect in p38α−/− MEFs. p38α-deficient MEFs infected with control retrovirus or virus expressing S6K1 were treated with Dox for different periods of time. Mdm2 S163 phosphorylation and Mdm2 were analysed with western blot. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in control WT MEFs was set at 1.0. (D) Combinational use of rapamycin and SB203580 showed no additive effect on the suppression of Mdm2 S163 phosphorylation. MEFs were treated with p38 inhibitor, Rapamycin, or both for different periods of time. Mdm2 phosphorylation was analysed with western blot. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in presence of SB was set at 1.0.
p38α MAPK is known to be activated under genotoxic stress, yet how it is activated remains a mystery. A couple of studies suggest that Atr might have a function in p38 MAPK activation under UV-induced genotoxic stress. However, we found that Atm deficiency, Atr knockdown, or both showed little effect on p38 MAPK activation or Mdm2 phosphorylation on S163, in primary MEFs (Supplementary Figure S5). These findings suggest that there exists a p38-Akt-mTOR-S6K-Mdm2 pathway, which is activated by DNA damage in an Atm/Atr-independent manner. Further investigation will be needed to understand how p38 MAPK is activated in response to genotoxic stress.
Genotoxic stress promotes S6K1–Mdm2 interaction independent of Mdm2 S163 phosphorylation
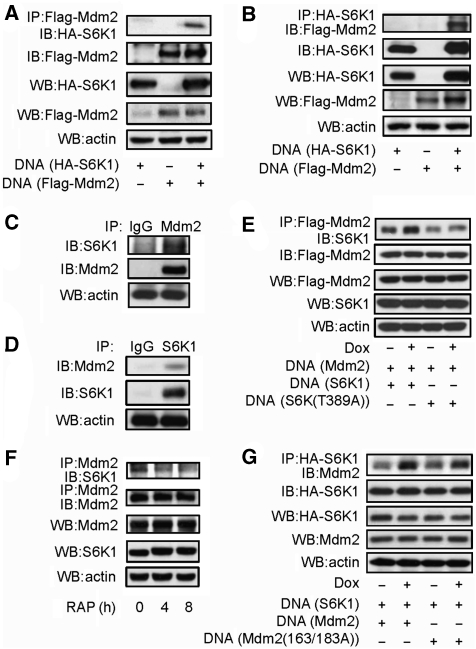
We then sought to determine whether Mdm2 and S6K interact with each other. Co-immunoprecipitation assays showed that in cells ectopically expressing S6K1 and Mdm2, immunoprecipitated Mdm2 could bring down S6K1, and vice versa, immunoprecipitated S6K1 could bring down Mdm2 as well (Figure 3A and B). This was further validated by the co-immunoprecipitation of endogenous Mdm2 and S6K1 in MEFs (Figure 3C and D). These results show that Mdm2 is not only a substrate of S6K1 but also an interacting partner. Moreover, this interaction was enhanced by genotoxic stress (Figure 3E) and T389A mutation reduced this interaction at the basal level and in response to genotoxic stress (Figure 3E). Note that under normal growth conditions, a considerable portion of S6K1 is phosphorylated on T389, likely because of the growth factors and nutrients in the serum. Consistently, rapamycin treatment decreased the complex formation of endogenous S6K1 and Mdm2 (Figure 3F). However, S163/183A mutant Mdm2 still showed enhanced interaction with S6K1 in response to genotoxic stress (Figure 3G). Taken together, these results indicate that the enhanced S6K1–Mdm2 interaction requires S6K1 activation/T389 phosphorylation but not Mdm2 S163/183 phosphorylation.
Figure 3.
Mdm2 interacts with S6K1. (A) Interaction between S6K1 and Mdm2. HA-S6K1, Flag-Mdm2, or both was expressed in 293T cells. Flag-Mdm2 was immunoprecipitated with anti-Flag antibodies and Mdm2-associated S6K1 was detected by western blot. (B) Same as Figure 3A, except that HA-S6K1 was immunoprecipitated with anti-HA antibodies and S6K1-associated Mdm2 was detected by western blot. (C) Interaction of endogenous S6K and Mdm2 in MEFs. Endogenous Mdm2 was immunoprecipitated with antibodies against Mdm2 and Protein A plus G beads. Mdm2-associated S6K1 was detected by western blot. (D) Endogenous S6K1 was immunoprecipitated with antibodies against S6K1 and Protein A plus G beads. S6K1-associated Mdm2 was detected by western blot. (E) Genotoxic stress enhanced Mdm2–S6K1 interaction, which required T389 phosphorylation of S6K1. HA-S6K1 or HA-mutant S6K (T389A) together with Flag-Mdm2 was expressed in 293T cells. Transfected cells were treated with Dox for 2 h before preparation of cell lyse, then Flag-Mdm2 was immunoprecipitated with anti-Flag antibodies and Mdm2-associated S6K1 was detected by western blot. (F) Endogenous S6K1–Mdm2 interaction was disrupted by rapamycin. MEFs were treated with rapamycin for different periods of time, then endogenous Mdm2 was immunoprecipitated with antibodies against Mdm2 and Protein A plus G beads. Mdm2-associated S6K1 was detected by western blot. (G) Genotoxic stress-enhanced Mdm2–S6K1 interaction did not require S163/183 phosphorylation of Mdm2. HA-S6K1 together with Flag-Mdm2 or Flag-mutant Mdm2 (S163A/S183A) was expressed in 293T cells. Transfected cells were treated with Dox for 2 h before preparation of cell lyse, then HA-S6K1 was immunoprecipitated with anti-HA antibodies and S6K1-associated Mdm2 was detected by western blot.
S6K1 activation inhibits Mdm2 nuclear entry
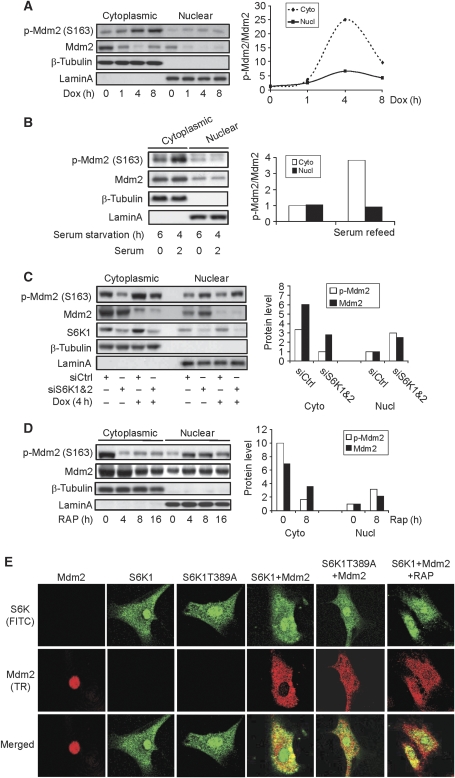
We then tested whether S6K1 regulates Mdm2 nuclear entry. Both S6K1 and Hdm2 are found in the cytoplasm and the nucleus, with more being localized in the cytoplasm (Figure 4C; Supplementary Figure S6). Thus, nuclear translocation of Mdm2 is important in downregulation of p53, as p53 ubiquitination by Mdm2 mainly occurs in the nucleus. However, genotoxic stress or serum addition led to marked increase in Mdm2 S163 phosphorylation in the cytoplasm, but much less in the nuclear fraction (Figure 4A and B). Under this condition, the cytoplasmic and nuclear Mdm2 molecules were similarly downregulated. But no obvious Mdm2 nuclear transport was observed. Moreover, tumour kidney tissues from TSC patients showed a drastic increase in Hdm2 S166 phosphorylation, but it did not markedly affect the localization of Hdm2 in tumour samples (Supplementary Figure S6). These results suggest that S6K1 activation and Mdm2 S163 phosphorylation are not sufficient in triggering Mdm2 translocation.
Figure 4.
S6K activation is required for Mdm2 cytoplasmic retention. (A) Genotoxic stress led to Mdm2 S163 phosphorylation mainly in the cytoplasm but not Mdm2 nuclear translocation. MEFs were treated with Dox for different periods of time, the cytoplasmic and nuclear proteins were fractionated and S163 phosphorylated Mdm2 and the protein levels of Mdm2 were analysed by western blot. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at time 0 in cytoplasmic fraction was set at 1.0. (B) Serum led to Mdm2 S163 phosphorylation but not much Mdm2 nuclear translocation. MEFs were starved for 4 h and refeeded with 10% FCS medium for 2 h. The cytoplasmic and nuclear proteins were fractionated. (Right panel) Quantitation data of Mdm2 S163 phosphorylation normalized to Mdm2 protein levels. The value of p-Mdm2 S163 at MEFs with 6 h serum starvation was set at 1.0. (C) Knockdown of S6K led to nuclear accumulation of Mdm2. S6K1 and S6K2 were knocked down in MEFs for 2 days, which were treated with Dox for 4 h. The cytoplasmic and nuclear proteins were fractionated, 20 μl out of 200 μl cytoplasmic fraction or 20 μl out of 80 μl nuclear fraction was separated by electrophoresis on SDS–PAGE gels (cytoplasmic to nuclear per cell ratio is 1:2.5) and Mdm2 S163 phosphorylation and Mdm2 protein levels were analysed by western blot. (Right panel) Quantitation data of Mdm2 and p-Mdm2 S163 of untreated MEFs. The values of nuclear Mdm2 and p-Mdm2 S163 were set at 1.0. (D) Inhibition of mTOR by rapamycin also led to Mdm2 nuclear transport. MEFs were treated with rapamycin for different periods of time and were fractionated into the cytoplasmic and nuclear portions. A measure of 20 of 200 μl cytoplasmic fraction or 20 of 80 μl nuclear fraction was separated by electrophoresis on SDS–PAGE gels (cytoplasmic to nuclear per cell ratio is 1:2.5). Mdm2 S163 phosphorylation and Mdm2 protein levels were analysed by western blot. (Right panel) Quantitation data of Mdm2 and p-Mdm2 S163 at 0 and 8 h rapamycin treatment. The values of nuclear Mdm2 and p-Mdm2 S163 were set at 1.0. (E) S6K activation has a function in Mdm2 cytoplasmic retention. Mdm2 was expressed by transient transfection in MEFs that stably expressed S6K1 or S6K1 T389A mutant with retrovirus. Cells were fixed, permeated with 0.1% Triton X-100, and blocked with 2% BSA. Primary antibodies (rabbit anti-S6K1 and mouse anti-Mdm2) were added, followed by incubation of Texas red-conjugated goat anti-mouse secondary antibody and FITC-conjugated goat anti-rabbit secondary antibody.
However, we found that S6K1/2 knockdown by siRNA or inhibition of S6K with rapamycin led to a nuclear translocation of Mdm2 (Figure 4C and D), as the levels of cytoplasmic Mdm2 went down, whereas the levels of nuclear Mdm2 went up and the levels of total Mdm2 showed no change. The remaining S163 phosphorylation in the absence of S6K activity is likely to be carried out by other kinases, or because of a slow S163 dephosphorylation process. In addition, we found that ectopically expressed Mdm2 was mainly localized in the nucleus of MEFs (Figure 4E). Co-expression of S6K1 led to Mdm2 cytoplasmic localization while T389A S6K1 mutant showed a much less effect. In Mdm2 and S6K1 co-transfected MEFs, rapamycin treatment led to more Mdm2 nuclear localization (Figure 4E). These results, together with the fractionation data (Figure 4C and D), support a necessary function of S6K1 activation in Mdm2 cytoplasmic retention. This is well correlated with the observation that Mdm2 forms a much tighter complex with activated/T389 phosphorylated S6K1. The observation that DNA damage activated S6K1 but failed to promote further Mdm2 cytoplasmic accumulation could be attributable to the saturated effect on Mdm2 (which is expressed at low levels) by S6K1, which is readily activated by growth factors and nutrients in the medium. Interestingly, the fractionation experiments show that rapamycin or S6K knockdown-induced Mdm2 nuclear translocation was accompanied with an increase in S163 phosphorylated Mdm2 (Figure 4C and D), suggesting that S163/183 phosphorylated Mdm2 might tend to enter nucleus under this condition, consistent with the finding that Akt1-mediated Mdm2 S163 phosphorylation promotes Mdm2 nuclear import (Mayo and Donner, 2001).
S6K inhibits Mdm2-mediated p53 ubiquitination and degradation
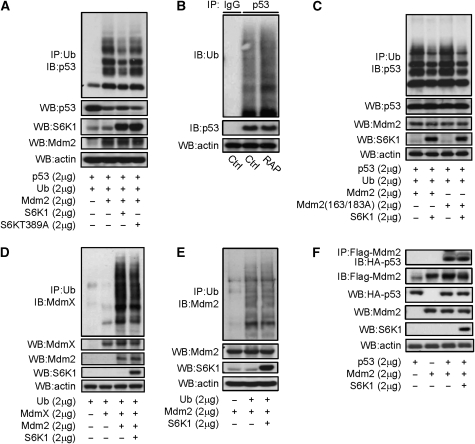
In addition, we found that S6K1 affected Mdm2-mediated p53 ubiquitination. S6K1 expression inhibited Mdm2-mediated p53 ubiquitination, whereas the S6K1 T389A mutant showed a much less inhibitory effect, in H1299 cells, which do not express p53 (Figure 5A). Moreover, treatment with rapamycin led to an increase in the ubiquitination of endogenous p53 in MEFs (Figure 5B). However, S163/183A mutant Mdm2 was able to mediate p53 ubiquitination, which could still be inhibited by S6K1 (Figure 5C), indicating that the inhibitory effect of S6K1 on Mdm2-mediated p53 ubiquitination is dependent on S6K1 activity/T389 phosphorylation, but independent of Mdm2 S163 phosphorylation. Moreover, S6K1 also slightly inhibited Mdm2-mediated MdmX ubiquitination (Figure 5D), yet it did not affect the auto-ubiquitination of Mdm2 (Figure 5E) or the interaction between Mdm2 and p53 (Figure 5F).
Figure 5.
S6K is a negative regulator of Mdm2-mediated p53 ubiquitination. (A) S6K1, but not T389A mutant S6K1, inhibited Mdm2-mediated p53 ubiquitination in H1299 cells. A measure of 2 μg of Flag-p53, HA-ubiquitin, and Mdm2, together with S6K1 or mutant S6KT389A, were expressed in H1299 cells, which were then treated with 10 μM of MG132 for 4 h before being harvested. Ubiquitinated proteins were immunoprecipitated with anti-HA (Ub) antibodies. Ubiquitinated p53 was detected by western blot using anti-p53 antibodies. (B) Inhibition of mTOR-S6K by rapamycin enhanced endogenous p53 ubiquitination in MEFs. MEFs were treated with or without rapamycin for 4 h, then followed by 10 μM of MG132 for 4 h before being harvested. Endogenous p53 was immunoprecipitated with antibodies against p53. Ubiquitinated p53 was detected with anti-ubiquitin antibodies. (C) S163/183A mutant Mdm2 could mediate p53 ubiquitination, which was inhibited by co-expression of S6K1 in H1299 cells. (D) S6K1 expression modestly inhibited Mdm2-mediated MdmX ubiquitination in H1299 cells. A measure of 2 μg each of HA-ubiquitin, MdmX and Mdm2, together with S6K1 were expressed in H1299 cells, which were then treated with 10 μM of MG132 for 4 h before being harvested. Ubiquitinated proteins were immunoprecipitated with anti-HA antibodies. Ubiquitinated MdmX was detected by western blot using anti-MdmX antibodies. (E) S6K1 expression showed no effect on Mdm2 self-ubiquitination in H1299 cells. A measure of 2 μg each of HA-ubiquitin and Mdm2, together with S6K1 were expressed in H1299 cells, which were then treated with 10 μM of MG132 for 4 h before being harvested. Ubiquitinated proteins were immunoprecipitated with anti-HA antibodies. Ubiquitinated Mdm2 was detected by western blot using anti-Mdm2 antibodies. (F) S6K1 did not show much effect on Mdm2–p53 interaction. Flag-Mdm2 and HA-p53 were expressed together with or without S6K1 in 293T cells, and Flag-Mdm2 was then immunoprecipitated and the associated p53 was determined by western blot.
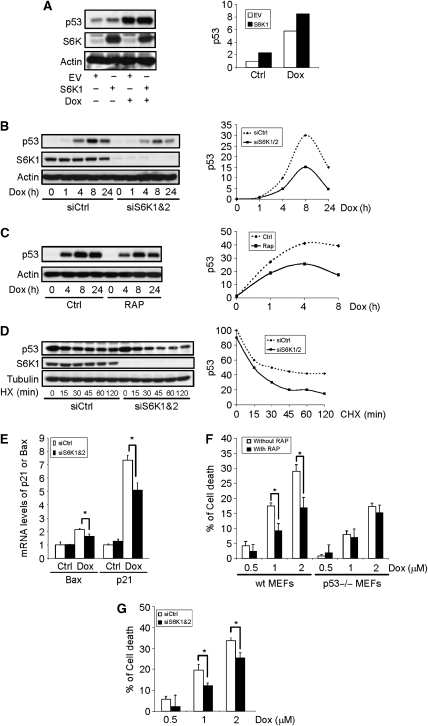
The observation that S6K1 activation is necessary to retain Mdm2 in the cytoplasm and to inhibit Mdm2-mediated p53 ubiquitination suggests that S6K is a negative regulator of Mdm2 but a positive regulator of p53. Indeed, we found that ectopic expression of S6K1 in primary MEFs cells led to a modest increase in the p53 protein levels and potentiated genotoxic stress-induced p53 expression, whereas S6K1 T389A failed to do so (Figures 1H and 6A). We then knocked down S6K1 and S6K2 with siRNA in primary MEFs to test whether these two proteins regulate p53 induction by Dox. It was obvious that genotoxic stress-induced p53 accumulation was impeded under this condition (Figure 6B). Moreover, pretreatment of MEFs with mTOR inhibitor rapamycin or Torin1 produced similar results (Figures 1B and 6C). Under these conditions, Dox treatment led to a drastic reduction in Mdm2, yet more S163 phosphorylated Mdm2 were found in the nucleus (Figure 4C and data not shown), which could promote p53 degradation. Indeed, we found that in S6K1/2 knockdown cells the half-life of p53 was shortened (Figure 6D). These results indicate that S6K participates in cell response to genotoxic stress by regulating p53 stability, consistent with our observation that S6K negatively regulates the function of Mdm2. This conclusion is further supported by our findings that S6K1/2 knockdown or inactivation did not alter p53 mRNA levels or p53 translation rate at the basal level and in response to DNA damage (Supplementary Figure S7A and B). A previous study reported that deficiency of Tsc2 led to upregulation of p53 because of enhanced translation of p53 mRNA (Lee et al, 2007). Our data show that the enhanced translation is not likely to be mediated by S6K1.
Figure 6.
S6K1 regulates p53 induction and cell response to genotoxic stress. (A) Expression of S6K1 led to an increase in p53 protein. S6K1 (or empty vector) was stably expressed in MEFs with retrovirus. p53 and S6K1 protein levels were analysed by western blot. (Right panel) Quantitation data. The value of p53 at MEFs with empty vector expression in absence of Dox was set at 1.0. (B) Knockdown of S6K compromised DNA damage-induced p53 upregulation. S6K1 and S6K2 were knocked down in MEFs for 48 h and followed by Dox treatment. p53 and S6K1 protein levels were analysed by western blot. (Right panel) Quantitation data. The value of p53 in the absence of Dox was set at 1.0. (C) Inhibition of mTOR also compromised DNA damage-induced p53 accumulation. MEFs were pretreated with rapamycin for 1 h before addition of Dox. p53 protein levels were analysed by western blot. (Right panel) Quantitation data. The value of p53 in the absence of Dox was set at 1.0. (D) S6K1/2 knockdown led to increased p53 turnover. S6K1 and S6K2 were knocked down in MEFs, which was followed by 10 μM cycloheximide (CHX) treatment for different periods of time, and p53 and S6K1 protein levels were analysed by western blot. (Right panel) Quantitation data. The value of p53 at 1 h in the presence of siS6K was set at 1.0. (E) S6K knockdown compromised the induction of p53 target genes Bax and p21. S6K1 and S6K2 were knocked down in MEFs, which received Dox treatment for 4 h. Total RNA was extracted and p21 and Bax mRNA levels were determined by real-time PCR. (F) Inhibition of S6K with rapamycin rendered MEFs more resistance to Dox-induced cell death in p53-dependent manner. WT and p53−/− MEFs were pretreated with rapamycin for 1 h, and followed by treatment with different concentrations of Dox for 24 h. Cells survival rates were determined by WST-1 assay. (G) S6K1/2 downregulation rendered MEFs more resistance to Dox-induced cell death. S6K1 and S6K2 were knocked down in MEFs (see Figure 1B for knockdown of S6K2 mRNA), and followed by treatment with different concentrations of Dox for 24 h. Cell viability was determined by WST-1 assay.
S6K1 regulates p53-mediated cell death response to genotoxic stress
Genotoxic stress leads to upregulation of p53 target genes p21 and Bax, which was compromised in the presence of S6K1/2 knockdown (Figure 6E), consistent with the compromised induction of p53. We also found that pretreatment of MEFs with rapamycin or knockdown of S6K1/2 rendered the cells more resistance to Dox-induced cell death (Figure 6F and G). However, in p53−/− MEFs, inhibition of mTOR with rapamycin did not significantly alter the cell death response to Dox (Figure 6F), indicating that mTOR-S6K1 signalling has a p53-dependent pro-apoptotic function in cell response to DNA damage. Thus, inhibition of the mTOR-S6K pathway downregulates p53 expression and render the cells more resistance to genotoxic stress, this may help to defer the aging process, as observed in S6K1−/− mice and mice administrated with rapamycin (Harrison et al, 2009; Selman et al, 2009). Surprisingly, the suppressive effect of rapamycin on apoptosis was not observed in cancer cell lines. Consistent with previously published results, tumour cell lines HeLa and HCT116 showed an increase in Dox-induced cell death rate when pretreated with rapamycin (data not shown) (Easton and Houghton, 2006; Inoki and Guan, 2009). This is likely due to the alterations in the DNA damage response pathway, which frequently occur in the process of cell immortalization and transformation.
The mTOR-S6K pathway connects cell nutrient status to DNA damage response
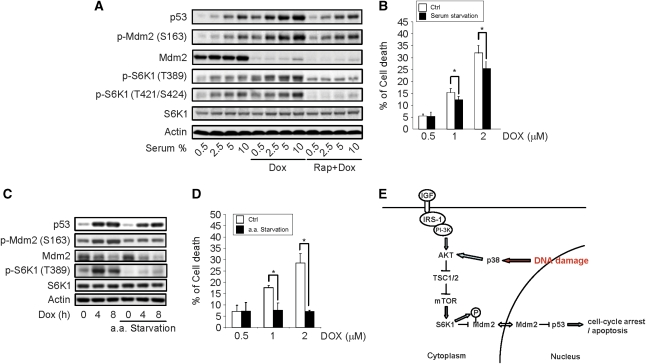
One prediction of these findings is that growth condition and energetic levels, which are sensed by mTOR-S6K, should also have an influence on cell response to DNA damage by regulating Mdm2. Indeed, we found that cells grown in higher concentrations of serum showed more S6K1 activation and stronger p53 induction at the basal level and in response to genotoxic stress. Pretreatment of cells with rapamycin could blunt these effects (Figure 7A). The dose-dependent effects of serum on S6K1 activation and p53 induction suggest that the mTOR-S6K pathway may regulate p53 induction in a quantitative way. Moreover, serum starvation could render the cells more resistance to Dox-induced cell death (Figure 7B). In addition, amino-acid starvation led to a decrease in S6K1 activation, which was accompanied with a reduction in p53 induction and reduced cell death rates upon Dox treatment (Figure 7C and D). Moreover, cell cultures with higher cellular densities showed lower S6K1 activation and reduced p53 induction (without medium change before adding Dox). Similar results have been reported previously (Bar et al, 2004). Taken together, these results suggest that cells use the S6K1–Mdm2 link to adjust its response to DNA damage, thus revealing a novel function for the mTOR-S6K1 pathway.
Figure 7.
The mTOR-S6K pathway connects metabolic status to DNA damage response. (A) Lower amounts of serum compromised p53 induction by genotoxic stress, which was blunted by rapamycin pretreatment. MEFs were growth in medium with different concentrations of serum treated with rapamycin for 1 h, and followed by 4 h of Dox treatment. The value of p53 at 0.5% serum was set at 1.0. (B) Serum starvation rendered MEFs more resistance to Dox-induced cell death. MEFs were treated with different concentration of Dox in either normal or serum-free medium for 24 h. Cell viability was determined by WST-1 assay. (C) Amino-acid starvation compromised p53 induction by genotoxic stress. MEFs were grown in normal medium or amino-acid starvation medium (HBSS) for 1 h and were followed by Dox treatment. (D) Amino-acid starvation rendered MEFs more resistance to Dox-induced cell death. MEFs were treated with different concentration of Dox in either normal medium or amino-acid starvation (HBSS) for 24 h. Cell viability was determined by WST-1 assay. (E) A schematic diagram showing how S6K1 might regulate Mdm2 and p53.
Discussion
Cells respond to DNA damage by activating pathways that involve Chk2/Chk1, p38 MAPK-MK2, and p53 to halt cell proliferation and prevent tumorigenesis. Here, by studying Mdm2 S163 phosphorylation, we identified another pathway, p38-Akt-mTOR-S6K1-Mdm2, that contributes to the tight regulation of p53 in response to DNA damage. S6K1 regulates several aspects of Mdm2 action, including 163/183 phosphorylation, nuclearcytoplasmic shuttling, and Mdm2-mediated ubiquitination of its substrates, likely through interaction between these two proteins. As such, S6K regulates the stability of p53, the best-studied Mdm2 substrate, cell death response under genotoxic stress in normal cells. Thus, the mTOR-S6K pathway has a function in DNA damage response by transmitting pro-apoptotic signals and may also regulate tumorigenesis. Previous studies indicate that p38 MAPK can directly act on Cdc25 and p53, this study reveals that p38 MAPK can also regulate p53 stability through the mTOR-S6K-Mdm2 pathway, further highlighting the importance of p38 MAPK in DNA damage response. Moreover, as a sensor of the nutrient and energy status, mTOR-S6K signalling can be activated by PI-3K-Akt and AMPK. This integrates metabolic signals and DNA damage response. Enriched environment will lead to S6K1 T389 phosphorylation and activation, enhanced S6K1-Mdm2 complex formation, and Mdm2 cytoplasmic retention, allowing maximal p53 induction upon genotoxic stress, which might be needed to counteract the strong mitogenic signals and enhanced protein synthesis mediated by the mTOR-S6K pathway (Figure 7). On the other hand, poor environment will downplay mTOR-S6K signalling, leading to reduced p53 induction upon genotoxic stress. This might be sufficient to cause cell-cycle arrest in the presence of weak mitogenic signals. Therefore, S6K1–Mdm2 interaction may provide the link between cells' status (nutrients, energy, and growth factors) and their response to DNA damage.
This study establishes that S6K1 is not only a kinase for Mdm2 S163 phosphorylation but also a physical interacting partner under genotoxic stress. S6K1 can phosphorylate Mdm2 on S163 in vivo and in vitro and is required for S163 phosphorylation in response to DNA damage. TSC tumour samples also show a massive increase in S6K1 T389 and Hdm2 S166 phosphorylation. Moreover, S6K1 forms a complex with Mdm2, which can be enhanced upon S6K activation in response to DNA damage. These findings, taken together, indicate that Mdm2 is a bona fide substrate of S6K1 in the cytoplasm. Although it has been reported that Mdm2 phosphorylation on S163/183 could be carried out by activated Akt1, MAPK, or its downstream kinases, and Pim kinases (Weber et al, 2005; Abe et al, 2008; Hogan et al, 2008), our study clearly shows that S6K1 has a dominant function in Mdm2 S163 phosphorylation in cell response to genotoxic stress.
Our results indicate that S6K1 regulates several aspects of Mdm2 function that may not involve S163/183 phosphorylation. First, S6K1 have an important function in Mdm2 cytoplasmic retention, likely through S6K1–Mdm2 interaction, which is independent of S163/183 phosphorylation. Inactivation or knockdown of S6K1 leads to Mdm2 nuclear translocation. Co-expression of S6K1 promotes Mdm2 cytoplasmic localization. This function of S6K1 seems to require T389 phosphorylation, as T389A mutation inhibits S6K1–Mdm2 interaction and reduces S6K1's ability to promote Mdm2 cytoplasmic localization. Second, S6K1 inhibits Mdm2-mediated p53 ubiquitination in a T389 phosphorylation-dependent manner, and inhibition of S6K1 activation enhances Mdm2-mediated p53 ubiquitination. Phosphorylation of S163 and S183 has no significant function in Mdm2-mediated p53 ubiquitination either. Theoretically, S6K1-mediated Mdm2 cytoplasmic retention and inhibition of Mdm2-mediated p53 ubiquitination would both contribute to maximal p53 stabilization. However, under normal culture conditions, nutrients and growth factors-activated S6K1 might be sufficient to retain Mmd2 in the cytoplasm. Thus, DNA damage-induced S6K1 activation may not be able to induce further Mdm2 cytoplasmic translocation. Taking all these into consideration, we propose that in primary cells growing in laboratory culture conditions, S6K1 may regulate genotoxic stress-induced p53 upregulation in two ways. One is through inhibition of Mdm2-mediated p53 ubiquitination by interacting with Mdm2, which is enhanced under genotoxic stress. The other is through retaining Mdm2 in the cytoplasm, which is mainly attributable to nutrient/growth factor-mediated mTOR-S6K1 activation.
On the other hand, conditions such as poor environment, the presence of mTOR-S6K1 inhibitors, or even the niches that maintain stem cells in a quiescent state, may downplay mTOR-S6K1 signalling and lead to Mdm2 nuclear translocation, which might be facilitated by S163/183 phosphorylation (carried out by other kinases), as observed in Figure 4 and in the presence of Akt1 activation (Mayo and Donner, 2001). Moreover, under these conditions, S6K1 shows reduced interaction with Mdm2 and thus loses its inhibitory effect on Mdm2-mediated p53 ubiquitination. These would lead to p53 degradation, which might be further enhanced by S163/183 phosphorylation as this phosphorylation has been reported to inhibit Mdm2–p19ARF interaction in the nucleus and promote p53 degradation (Zhou et al, 2001). In addition, we found that there was a correlation between S163/183 phosphorylation and Mdm2 upregulation, yet the effect of S163/183 phosphorylation on Mdm2 stabilization, if any, is very transient and modest. Further studies will be needed to determine whether this is a direct effect of S163/183 phosphorylation.
Our findings suggest that Mdm2 nuclearcytoplasmic shuttling can be controlled by S6K1 activation/T389 phosphorylation and Mdm2 S163 phosphorylation in opposite ways. As both events can be caused by multiple pathways, S6K1–Mdm2 interaction might provide a platform to integrate various signals to determine Mdm2 localization (Feng et al, 2007; Budanov and Karin, 2008). This might be the reason that inconsistent conclusions were drawn regarding the function of S163/183 phosphorylation in various studies, with some showing that Mdm2 S163 phosphorylation promotes Mdm2 nuclear import, whereas the others failed to observe such a phenomenon (Mayo and Donner, 2001; Zhou et al, 2001; Weber et al, 2005; Malmlof et al, 2007; Hogan et al, 2008). This inconsistency could be due to the use of different growth factors or reagents that differentially activate S6Ks and p163/183 phosphorylation in various cell lines.
In summary, this study reveals another genotoxic stress-responsive pathway, p38-Akt-mTOR-S6K1–Mdm2, which helps to regulate p53 stability and p53-mediated cell death. This pathway can also sense the cells' nutrient and energy status and transmits signals to fine-tune cells' response to DNA damage. The interaction between S6K1 and Mdm2 also links two of the prominent pathways that control aging at the cell and organism levels, the mTOR-S6K pathway and the Mdm2–p53 pathway. Further investigation will be needed to determine whether p53 participates in mTOR-S6K1-mediated aging process. These findings might be of help in developing strategies to retain Mdm2 in the cytoplasm to facilitate p53-based cancer therapy.
Materials and methods
Chemicals and antibodies
Dox, rapamycin, U0126, SB203580, and antibody against Mdm2 were purchased from Calbiochem. Antibodies against p70S6K (S6K1), T389 phosphorylated p70S6K, T421/S424 phosphorylated p70S6K, p44/42 MAPK, T202/Y204 phosphorylated p44/42, p38, T180/Y182 phosphorylated p38, mTOR, S2481 phosphorylated mTOR, Akt1, S473 phosphorylated Akt, p53, S15 phosphorylated p53, Tsc1, Tsc2, S163 phosphorylated Mdm2, lamin, β-tubulin, and actin were purchased from Cell Signaling.
Tumour samples, cell cultures, and transfection
TSC patients with angiomyolipoma and normal kidney tissues were obtained from the Brain and Tissue Bank for Development Disorders (University of Maryland). This study has been approved by The Institutional Review Board of The University of Texas Health Science Center at San Antonio, TX. MEFs were prepared as previously described (Li et al, 2004). MEFs and 293T cells were cultured in DMEM supplemented with 10% FCS. H1299 cells were cultured in RPMI supplemented with 10% FCS. To transiently express a given protein, 293T or H1299 cells were transfected with lipofectamine following the manufacturer's instructions (Invitrogen).
To stably express a given protein in MEFs, packaging cells PlatE were transfected with Fugene transfection reagent following the manufacturer's instructions (Roche). Viruses were harvested and were used for the transduction of MEFs. For knockdown experiments, S6K1/S6K2 (siGENOME), Tsc1/Tsc2, or Mdm2 (ON-TARGETplus SMART pool) siRNA were used. To perform amino-acid starvation, MEFs were preincubated in Hank's balanced salt solution (HBSS).
Cell viability analysis
To measure cell death rates, cells were plated in 96-well plates at the density of 1 × 104 per well, after which the cells were treated with different concentrations of Dox (0–2 μM) for 24 h. Cell proliferation reagent WST-1 (Roche) was added to each well and was further incubated for 3 h at 37°C. The cell viability was determined by measuring the absorbance against background control by microplate (ELISA) reader at wavelength 430 nm. The reference wavelength is 650 nm.
Ubiquitination assay
Cells were transfected with Flag-tagged p53 and HA-tagged ubiquitin together with various constructs; and then exposed to 10 μM MG132 for 4 h to inhibit proteasome-mediated degradation of ubiquitinated proteins before preparation of cell lysates. The cells lysates were immunoprecipitated with anti-HA affinity matrix (ROCHE), eluted with 2 × SDS–PAGE sample buffer and followed by western blot analysis of target protein p53.
For endogenous ubiquitination assay, MEFs were treated with MG132 for 4 h, endogenous p53 were immunoprecipitated with antibodies against p53 and Protein A plus G beads. Ubiquitinated p53 was detected with anti-ubiquitin antibodies.
p53 Translation rate determination
To determine p53 translation rate, cells were labelled with 100 μCi/ml 35S-methionine for 30 min. p53 was then immunoprecipitated from cells using antibody against p53 and Protein A plus G beads. The reaction was terminated by addition of 2 × SDS–PAGE sample buffer and boiling for 5 min. Proteins were analysed by SDS–PAGE, and the 35S-methionine-labelled p53 were detected by auto-radiography.
Immunoprecipitation, cells fractionation, and western blot
Cells were washed with PBS and lysed in RIPA buffer containing phosphatase and protease inhibitors. To fractionate cytoplasmic and nuclear protein, cells or tumour samples were lysed in NE-PER Nuclear and Cytoplasmic Extraction Reagent (Thermo SCIENTIFIC) following the manufacturer's instruction. Protein concentration was determined by Bio-Rad assay. Equal amounts of protein (50 μg) were separated by electrophoresis on SDS–PAGE gels, and then transferred onto a nitrocellulose membrane (Millipore), which were probed with primary and secondary antibodies, and visualized using ECL kit (GE Healthcare). For immunoprecipitation, antibodies were added to the cell lysate (1 mg) at 4°C for overnight incubation, followed by incubation with Protein A plus G beads at 4°C for 4 h. The immunoprecipitated proteins were released from the beads by boiling in 2 × sample buffer for 5 min and subsequently analysed by western blot.
In vitro kinase assay for S6K1/2
HA-tagged S6K1 or HA-tagged S6K2 was expressed in 293T cell and immunoprecipitated from cell lysates using anti-HA affinity matrix (ROCHE). Matrix-bound S6K1 or S6K2 was washed with Buffer A (1% Igepal CA-630, 0.5% sodium deoxychloate, 100 mM NaCl, 10 mM Tris, pH 7.2, 1 mM EDTA, 1 mM sodium orthovanadate, 1 mM DTT, 10 μg/ml Leupeptin, 10 μg/ml pepstatin, 40 μg/ml PMSF), Buffer B (1 M NaCl, 0.1% Igepal CA-630, 10 mM Tris, pH 7.2, 1 mM sodium orthovanadate, 1 mM DTT, 10 μg/ml Leupeptin, 10 μg/ml pepstatin, 40 μg/ml PMSF) and Buffer ST (150 mM NaCl, 50 mM Tris/HCl, pH 7.2, 50 mM Tris-base, 1 mM sodium orthovanadate, 1 mM DTT, 10 μg/ml Leupeptin, 10 μg/ml pepstatin, 40 μg/ml PMSF). Purified GST-Mdm2 (1 μg) was mixed with matrix-bound S6K1 or S6K2 in kinase assay buffer (20 mM Hepes, 10 mM MgCl2, 100 μg/ml BSA, 100 μM DTT, 50 μM ATP, 3 ng/μl protein kinase inhibitor) and incubated at 37°C for 15 min. The reaction was terminated by addition of 2 × SDS–PAGE sample buffer and boiling for 5 min. Proteins were analysed by SDS–PAGE, and the active and S163 phosphorylated form of GST-Mdm2 were detected by western blot.
RNA isolation and real-time PCR
Total RNA was extracted from cells with TRIzol® reagent (Invitrogen) following the manufacturer's instruction. cDNAs were synthesized from 1 μg of total RNA using iScript cDNA Synthesis Kit (Bio-Rad). The detection and quantification of target mRNA were performed with LightCycler 480® real-time PCR (ROCHE).
Image acquisition, quantitation of western blot, and statistical analysis
All the presented data have been repeated at least three times, with similar results being obtained. The results from one experiment were presented. For data quantitation, western blot results were scanned with a Molecular Dynamics scanning densitometer. Statistical analysis was performed using Student's t-test. Significant association was defined when P<0.05 as compared with control.
Supplementary Material
Acknowledgments
We thank Drs Alan Porter and Huiyi Kua for helpful discussions; June Lin, Grace Khoo, Delia Chua, Bijin Au, and Teo Huan Qing for technical support; Dr Q Yu for providing expression constructs, cell lines, and reagents; Professor Koji Okamoto for providing MdmX expression constructs; and Drs Nathanael S Gray and David M Sabatini for providing Torin1. This work was supported by the Agency for Science, Technology, and Research of the Republic of Singapore.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe Y, Oda-Sato E, Tobiume K, Kawauchi K, Taya Y, Okamoto K, Oren M, Tanaka N (2008) Hedgehog signaling overrides p53-mediated tumor suppression by activating Mdm2. Proc Natl Acad Sci USA 105: 4838–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar V, Alliouachene S, Sotiropoulos A, Sobering A, Athea Y, Djouadi F, Miraux S, Thiaudiere E, Foretz M, Viollet B, Diolez P, Bastin J, Benit P, Rustin P, Carling D, Sandri M, Ventura-Clapier R, Pende M (2007) S6 kinase deletion suppresses muscle growth adaptations to nutrient availability by activating AMP kinase. Cell Metab 5: 476–487 [DOI] [PubMed] [Google Scholar]
- Averous J, Proud CG (2006) When translation meets transformation: the mTOR story. Oncogene 25: 6423–6435 [DOI] [PubMed] [Google Scholar]
- Bar J, Cohen-Noyman E, Geiger B, Oren M (2004) Attenuation of the p53 response to DNA damage by high cell density. Oncogene 23: 2128–2137 [DOI] [PubMed] [Google Scholar]
- Budanov AV, Karin M (2008) p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134: 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dann SG, Selvaraj A, Thomas G (2007) mTOR Complex1-S6K1 signaling: at the crossroads of obesity, diabetes and cancer. Trends Mol Med 13: 252–259 [DOI] [PubMed] [Google Scholar]
- Easton JB, Houghton PJ (2006) mTOR and cancer therapy. Oncogene 25: 6436–6446 [DOI] [PubMed] [Google Scholar]
- Feng Z, Hu W, de Stanchina E, Teresky AK, Jin S, Lowe S, Levine AJ (2007) The regulation of AMPK beta1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res 67: 3043–3053 [DOI] [PubMed] [Google Scholar]
- Frodin M, Antal TL, Dummler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM (2002) A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J 21: 5396–5407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM (2007) Defining the role of mTOR in cancer. Cancer Cell 12: 9–22 [DOI] [PubMed] [Google Scholar]
- Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP, Lamb RF (2004) The TSC1-2 tumor suppressor controls insulin-PI3K signaling via regulation of IRS proteins. J Cell Biol 166: 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, Pahor M, Javors MA, Fernandez E, Miller RA (2009) Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan C, Hutchison C, Marcar L, Milne D, Saville M, Goodlad J, Kernohan N, Meek D (2008) Elevated levels of oncogenic protein kinase Pim-1 induce the p53 pathway in cultured cells and correlate with increased Mdm2 in mantle cell lymphoma. J Biol Chem 283: 18012–18023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holz MK, Ballif BA, Gygi SP, Blenis J (2005) mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123: 569–580 [DOI] [PubMed] [Google Scholar]
- Inoki K, Guan KL (2009) Tuberous sclerosis complex, implication from a rare genetic disease to common cancer treatment. Hum Mol Genet 18: R94–R100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Byun JY, Yun CH, Park IC, Lee KH, Lee SJ (2008) c-Src-p38 mitogen-activated protein kinase signaling is required for Akt activation in response to ionizing radiation. Mol Cancer Res 6: 1872–1880 [DOI] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL (2007) Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J 26: 4812–4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Wang X, Rasheed N, Hu Y, Boast S, Ishii T, Nakayama K, Nakayama KI, Goff SP (2004) Distinct roles of c-Abl and Atm in oxidative stress response are mediated by protein kinase C delta. Genes Dev 18: 1824–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Inoki K, Vacratsis P, Guan KL (2003) The p38 and MK2 kinase cascade phosphorylates tuberin, the tuberous sclerosis 2 gene product, and enhances its interaction with 14-3-3. J Biol Chem 278: 13663–13671 [DOI] [PubMed] [Google Scholar]
- Ma XM, Blenis J (2009) Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 10: 307–318 [DOI] [PubMed] [Google Scholar]
- Malmlof M, Roudier E, Hogberg J, Stenius U (2007) MEK-ERK-mediated phosphorylation of Mdm2 at Ser-166 in hepatocytes. Mdm2 is activated in response to inhibited Akt signaling. J Biol Chem 282: 2288–2296 [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N (2006) mTOR, translation initiation and cancer. Oncogene 25: 6416–6422 [DOI] [PubMed] [Google Scholar]
- Mayo LD, Donner DB (2001) A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA 98: 11598–11603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meek DW, Knippschild U (2003) Posttranslational modification of MDM2. Mol Cancer Res 1: 1017–1026 [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G (2004) S6K1(−/−)/S6K2(−/−) mice exhibit perinatal lethality and rapamycin-sensitive 5′-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol 24: 3112–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane MJ, Coxon PY, Powell DW, Webster R, Klein JB, Pierce W, Ping P, McLeish KR (2001) p38 Kinase-dependent MAPKAPK-2 activation functions as 3-phosphoinositide-dependent kinase-2 for Akt in human neutrophils. J Biol Chem 276: 3517–3523 [DOI] [PubMed] [Google Scholar]
- Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB (2007) p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11: 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O (2006) Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31: 342–348 [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al Qassab H, Carmignac D, Ramadani F, Woods A, Robinson IC, Schuster E, Batterham RL, Kozma SC, Thomas G, Carling D, Okkenhaug K, Thornton JM, Partridge L et al. (2009) Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326: 140–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M, Blasco MA (2007) Cancer and ageing: convergent and divergent mechanisms. Nat Rev Mol Cell Biol 8: 715–722 [DOI] [PubMed] [Google Scholar]
- Shah OJ, Wang Z, Hunter T (2004) Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 14: 1650–1656 [DOI] [PubMed] [Google Scholar]
- Shinozaki T, Nota A, Taya Y, Okamoto K (2003) Functional role of Mdm2 phosphorylation by ATR in attenuation of p53 nuclear export. Oncogene 22: 8870–8880 [DOI] [PubMed] [Google Scholar]
- Tee AR, Anjum R, Blenis J (2003) Inactivation of the tuberous sclerosis complex-1 and -2 gene products occurs by phosphoinositide 3-kinase/Akt-dependent and -independent phosphorylation of tuberin. J Biol Chem 278: 37288–37296 [DOI] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284: 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Sabatini DM (2009) Rapamycin inhibits mTORC1, but not completely. Autophagy 5: 725–726 [DOI] [PubMed] [Google Scholar]
- Thornton TM, Rincon M (2009) Non-classical p38 map kinase functions: cell cycle checkpoints and survival. Int J Biol Sci 5: 44–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um SH, D'Alessio D, Thomas G (2006) Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab 3: 393–402 [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408: 307–310 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X (2002) Live or let die: the cell's response to p53. Nat Rev Cancer 2: 594–604 [DOI] [PubMed] [Google Scholar]
- Weber HO, Ludwig RL, Morrison D, Kotlyarov A, Gaestel M, Vousden KH (2005) HDM2 phosphorylation by MAPKAP kinase 2. Oncogene 24: 1965–1972 [DOI] [PubMed] [Google Scholar]
- Whibley C, Pharoah PD, Hollstein M (2009) p53 polymorphisms: cancer implications. Nat Rev Cancer 9: 95–107 [DOI] [PubMed] [Google Scholar]
- Woods SC, Seeley RJ, Cota D (2008) Regulation of food intake through hypothalamic signaling networks involving mTOR. Annu Rev Nutr 28: 295–311 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Stephen CW, Lane DP (2001) Cocompartmentalization of p53 and Mdm2 is a major determinant for Mdm2-mediated degradation of p53. Exp Cell Res 270: 66–77 [DOI] [PubMed] [Google Scholar]
- Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Hung MC (2001) HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol 3: 973–982 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.