Calmodulin dissociation regulates Myo5 recruitment and function at endocytic sites
The type I myosin Myo5 promotes actin-dependent membrane remodelling. Calmodulin binds to Myo5 and maintains it in a closed and inactive conformation. Calmodulin dissociation from Myo5 leads to local activation of actin polymerization.
Keywords: Arp2/3 complex, calmodulin, endocytosis, myosin-I, WIP-Vrp1
Abstract
Myosins-I are conserved proteins that bear an N-terminal motor head followed by a Tail Homology 1 (TH1) lipid-binding domain. Some myosins-I have an additional C-terminal extension (Cext) that promotes Arp2/3 complex-dependent actin polymerization. The head and the tail are separated by a neck that binds calmodulin or calmodulin-related light chains. Myosins-I are known to participate in actin-dependent membrane remodelling. However, the molecular mechanisms controlling their recruitment and their biochemical activities in vivo are far from being understood. In this study, we provided evidence suggesting the existence of an inhibitory interaction between the TH1 domain of the yeast myosin-I Myo5 and its Cext. The TH1 domain prevented binding of the Myo5 Cext to the yeast WIP homologue Vrp1, Myo5 Cext-induced actin polymerization and recruitment of the Myo5 Cext to endocytic sites. Our data also indicated that calmodulin dissociation from Myo5 weakened the interaction between the neck and TH1 domains and the Cext. Concomitantly, calmodulin dissociation triggered Myo5 binding to Vrp1, extended the myosin-I lifespan at endocytic sites and activated Myo5-induced actin polymerization.
Introduction
Myosins-I are widely expressed actin-dependent molecular motors, which share a conserved structural organization. Besides the N-terminal actin-activated ATPase that translocates actin filaments, all myosins-I bear a Tail Homology 1 (TH1) domain that binds acidic phospholipids in vitro (Pollard et al, 1991). The motor head and the TH1 domain are separated by an α helical neck that associates with calmodulin or calmodulin-related light chains (Pollard et al, 1991). In addition, the so-called long-tailed myosins-I bear a C-terminal extension (Cext), which can trigger Arp2/3 complex-dependent actin polymerization. The Cext includes a Tail Homology 2 (TH2) domain, which binds filamentous actin (Pollard et al, 1991) and a Src Homology 3 (SH3) domain, which associates with proline-rich motifs (Kuriyan and Cowburn, 1997). The fungal myosins-I also bear an acidic peptide, which directly participates in the activation of the Arp2/3 complex (Evangelista et al, 2000; Lechler et al, 2000, 2001; Lee et al, 2000; Idrissi et al, 2002; Sun et al, 2006) (Figure 1B). The protozoal and the long-tailed mammalian myosins-I lack this acidic peptide but might indirectly activate the Arp2/3 complex through association with CARMIL (Jung et al, 2001).
Figure 1.
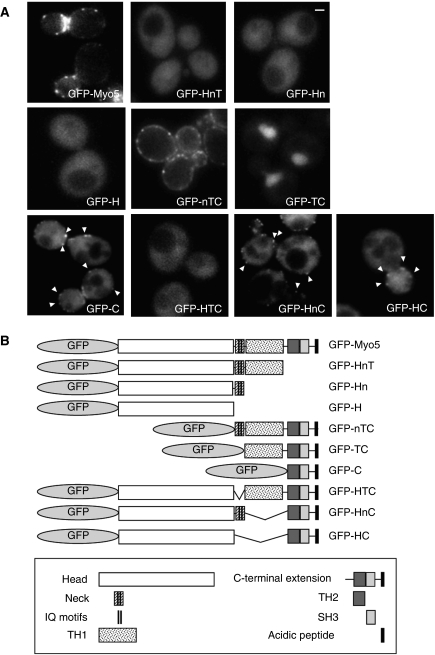
Subcellular localization of GFP-Myo5 constructs. (A) Fluorescence micrographs of live myo5Δ cells (SCMIG275), expressing the indicated GFP fusion proteins represented in (B) expressed from centromeric plasmids under the control of the MYO5 promoter. Cells were grown to mid-log phase at 25°C and observed by conventional fluorescence microscopy. Arrowheads indicate cortical patches. Bar=1 μm.
Myosins-I participate in a number of processes requiring actin-dependent remodelling or movement of cellular membranes. These include clathrin-dependent endocytic budding in yeast and mammals (Geli and Riezman, 1996; Sun et al, 2006; Krendel et al, 2007), vacuole contraction, cell motility, phagocytosis and pinocytosis in protozoa (Jung and Hammer, 1990; Wessels et al, 1991; Doberstein et al, 1993; Titus et al, 1993; Novak et al, 1995; Novak and Titus, 1997), and membrane traffic along the endocytic and secretory pathways in protozoa and higher eukaryotes (Fath et al, 1994; Durrbach et al, 1996, 2000; Temesvari et al, 1996; Raposo et al, 1999; Huber et al, 2000; Neuhaus and Soldati, 2000; Cordonnier et al, 2001; Bose et al, 2002). Thus, myosin-I function requires its precise targeting and/or activation at particular membrane subdomains. Despite intense research on these proteins, the mechanisms that spatially and temporally define their recruitment and/or regulate their biochemical activities in vivo are poorly understood.
The yeast long-tailed myosin-I Myo5, and its homologue Myo3, are well-characterized members of the myosin-I family. Myo5 and Myo3 participate in the formation of endocytic vesicles at the plasma membrane (PM) (Geli and Riezman, 1996). Myo5 is recruited to endocytic sites labelled with clathrin, stays anchored at the PM for 10 to 15 s and disappears around the time the endocytic coat moves into the cytosol (Jonsdottir and Li, 2004; Sun et al, 2006). During the transient association of the myosins-I with the endocytic sites, their mechanochemical activity and their actin polymerization promoting activity contribute to vesicle budding (Sun et al, 2006; Galletta et al, 2008; Idrissi et al, 2008).
Myo5 recruitment at cortical patches is at least partially dependent on its SH3 domain. Mutation of this domain or deletion of the SH3 interacting partners Vrp1/WIP (WASP-interacting protein) or Las17/WASP (Wiskott–Aldrich syndrome protein) causes partial mislocalization of the myosin to the cytosol (Anderson et al, 1998; Sun et al, 2006). Intriguingly, Vrp1 and Las17 arrive significantly earlier than the myosin at the endocytic sites, indicating that their association with Myo5 might be regulated (Jonsdottir and Li, 2004; Sun et al, 2006). On the other hand, domains other than the SH3 are involved in the cortical recruitment of Myo5 as the SH3 domain alone does not localize to endocytic patches (Anderson et al, 1998).
In this study, we provided evidence indicating the existence of an inhibitory interaction between the TH1 domain and the Cext, which prevented Myo5 binding to Vrp1. Interestingly, we found that dissociation of Cmd1 from the Myo5 neck weakened the interaction between a fragment containing the neck and TH1 domain and the Cext. Concomitantly, Cmd1 dissociation promoted binding of Myo5 to Vrp1. Consistent with previous observations indicating that Vrp1 participates in Myo5 recruitment to cortical endocytic sites (Anderson et al, 1998; Sun et al, 2006) and that Vrp1 is required to trigger Myo5-induced actin polymerization (Geli et al, 2000; Lechler et al, 2001; Sun et al, 2006), we found that Cmd1 dissociation from the myosin elongated its lifespan at cortical endocytic sites and triggered Myo5-induced actin polymerization in vivo and in vitro.
Results
The Myo5 TH1 domain interacts with the Cext and negatively regulates association with Vrp1 and recruitment to endocytic sites
To analyse the function of different domains in the recruitment of Myo5 to cortical patches, we investigated the localization of GFP-tagged full-length or truncated Myo5 by fluorescence microscopy (Figure 1). As previously shown, full-length Myo5 (GFP-Myo5) concentrated at cortical patches that partially colocalized with endocytic markers (Figure 1A; Supplementary Figure S1A) (Anderson et al, 1998; Jonsdottir and Li, 2004; Sun et al, 2006). In addition, a diffuse PM and a weak cytosolic staining were observed. Consistent with the observation that the SH3 domain is required for Myo5 localization (Anderson et al, 1998), truncation of the Cext (GFP-HnT) impaired cortical patch recruitment (Figure 1A). On the other hand, we found that the complete Cext (GFP-C) was recruited to endocytic sites (Figure 1A; Supplementary Figure S1A), even though a strong cytosolic signal was also observed for this construct. The overexpression of GFP-C compared with GFP-Myo5 could account for the partial mislocalization of the Cext (Supplementary Figure S1B). In addition, the absence of the TH1 lipid-binding domain, which targets other myosins-I to specific cellular membranes, could partially impair proper recruitment of Myo5 to the PM. However, we observed that addition of amino acids 774 to 996 of the TH1 domain to the Cext (GFP-TC) did not improve Myo5 localization but rather mistargeted the myosin to the cytosol and the nucleus (Figure 1A). Interestingly, cortical patch localization was recovered with addition of the neck and 15 amino acids of the N-terminus of the TH1 domain (GFP-nTC) (Figure 1A; Supplementary Figure S1A). These results suggested that the Myo5 neck might be required to release an inhibitory interaction between the TH1 domain and the Cext. Consistent with this hypothesis, a GFP fusion protein lacking only the neck (GFP-HTC) lost the cortical patch localization, which was partially recovered by deletion of the TH1 (GFP-HC) (Figure 1A; Supplementary Figure S1A). Conversely, a GFP fusion protein missing only the TH1 domain (GFP-HnC) was still recruited to endocytic sites (Figure 1A; Supplementary Figure S1A), although the absence of the TH1 domain again increased the cytosolic signal.
Binding of the Myo5 Cext to Vrp1 participates in the myosin recruitment to endocytic patches (Anderson et al, 1998; Sun et al, 2006). Therefore, we next investigated whether the presence of the TH1 domain interfered with Myo5 binding to Vrp1 in the absence of the neck. Extracts of yeast cells expressing different Protein A (PA)-tagged Myo5 constructs and haemagglutinin (HA)-tagged Vrp1 (Vrp1-HA) were subjected to IgG pull downs. The presence of Vrp1-HA in the precipitates was detected by immunoblotting. As shown in Figures 1A and 2A, a correlation between the cortical patch localization and Vrp1-HA binding was observed for the different constructs. Thus, a construct bearing the Myo5 Cext (PA-C) pulled down Vrp1-HA but addition of a TH1 domain (PA-TC) blocked the interaction. Further, the presence of the Myo5 neck and 15 amino acids of the N-terminus of the TH1 domain (PA-nTC) recovered Vrp1-HA binding to levels similar to the wild-type (wt) myosin. Also consistent with our model, deletion of the TH1 domain (PA-HnC) significantly increased binding of Myo5 to Vrp1-HA, whereas deletion of the neck (PA-HTC) completely abolished the interaction. Deletion of the neck did not seem to grossly affect the structure of the tail as the PA-HTC construct still bound to Rvs167 similar to the wt protein (Geli et al, 2000; Tong et al, 2002) (Figure 2B).
Figure 2.
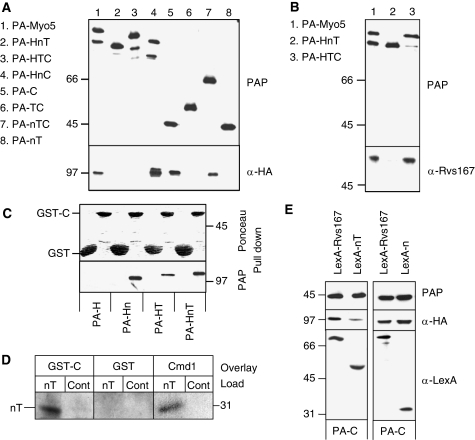
The Myo5 TH1 domain blocks the interaction of the Myo5 Cext with Vrp1. (A, B, E) Immunoblots of IgG-Sepharose pull downs from myo5Δ vrp1Δ cells (SCMIG304), expressing the indicated PA-Myo5 constructs and Vrp1-HA, either overexpressing (E) or not (A, B) the indicated LexA fusion proteins (E). PAP, a peroxidase-conjugated anti-HA antibody and anti-Rvs167 or anti-Cmd1 antibodies, combined with the adequate peroxidase-conjugated secondary antibody, were used for detection of the PA-Myo5 constructs, Vrp1-HA, Rvs167 and Cmd1, respectively. All PA-Myo5 constructs and Vrp1-HA were expressed from centromeric plasmids under the control of their own promoters. (C) Immunoblot of glutathione-Sepharose pull downs of GST-C or GST, incubated with extracts of a myo5Δ strain (SCMIG275), expressing the indicated PA-Myo5 constructs from centromeric plasmids under the MYO5 promoter, decorated with Ponceau red to detect GST-C and GST or PAP to detect the PA-Myo5 constructs. (D) Overlay assay of an Myo5 nT fragment, partially renatured after SDS–PAGE and western blot, incubated with 20 mM GST or GST fused to the Myo5 Cext (GST-C) or 60 mM Cmd1. GST and Cmd1 were detected using anti-GST and anti-Cmd1 antibodies and the adequate peroxidase-conjugated secondary antibodies. Control lanes (Cont) were loaded with residual proteins that might elute from the IgG-Sepharose under the conditions used for the nT purification, derived from a strain not expressing the PA-nT fragment.
Previous results suggested an intramolecular interaction between the TH1 domain and the Cext of the protozoal long-tailed myosins-I (Jontes et al, 1998; Lee et al, 1999; Ishikawa et al, 2004; Hwang et al, 2007). We failed to express significant amounts of the TH1 domain alone, but pull-down experiments using the Cext fused to GST (GST-C) and yeast extracts expressing PA-tagged Myo5 truncations bearing the H, Hn, HT and HnT fragments demonstrated a specific interaction between the Cext and the TH1 domain, which seemed to be stabilized by the presence of the neck (Figure 2C). The interaction between the nT fragment and the Cext was direct, as the nT fragment could be overlaid with GST-C purified from Escherichia coli (Figure 2D).
To analyse whether the nT fragment interfered with the Cext-dependent Myo5/Vrp1 interaction, PA-C was pulled down from yeast expressing Vrp1-HA and overexpressing LexA fused to the Myo5 neck and TH1 domains (LexA-nT) or LexA fused to Rvs167 as a control. As shown in Figure 2E, LexA-nT was pulled down with the Cext and significantly interfered with the Myo5/Vrp1 interaction when compared with LexA-Rvs167. The Vrp1-HA signal was reduced 6.57±1.02-fold (n=3) in the pull downs from cells expressing LexA-nT with respect to those expressing LexA-Rvs167. The inhibitory domain was contained within the TH1 domain as a LexA-n construct still interacted with PA-C but failed to interfere with Vrp1-HA binding (Figure 2E).
Cmd1 dissociation from the Myo5 neck weakens the neck-TH1/Cext interaction and promotes Myo5 binding to Vrp1
Our results suggested that the neck and the TH1 domains interact with the Myo5 Cext, that the TH1 domain interferes with Myo5 binding to Vrp1 and that the adjacent Myo5 neck modulates the release of the neck-TH1/Cext interaction. To investigate whether calmodulin association/dissociation from the Myo5 neck might influence Myo5 binding to Vrp1, PA-Myo5 was precipitated with IgG-Sepharose from an extract of yeast expressing Vrp1-HA in the absence or presence of 5 mM CaCl2. The Myo5 neck contains two IQ motifs, which bind calmodulin in the absence of Ca2+. Addition of 5 mM CaCl2 triggers dissociation of calmodulin (Geli et al, 1998). As shown in Figure 3A, addition of calcium significantly reduced Cmd1 association to Myo5 and concomitantly increased the Myo5/Vrp1 interaction. Vrp1 specifically bound PA-Myo5 as deletion of the Cext (PA-HnT) completely eliminated the Vrp1-HA signal (Figure 3A). To demonstrate that Cmd1 dissociation and not the CaCl2 treatment per se increased Vrp1 binding, we used myosin and calmodulin mutations that specifically disrupt the Cmd1/Myo5 interaction in the absence of calcium: myo5-iqΔ and cmd1-226 (Geli et al, 1998). The IQ motifs required for Cmd1 binding (IQRAWRRFLQR and IQRTIRERKEG) are deleted in the Myo5-iqΔ mutant. The mutant loses its capacity to interact with Cmd1 but, in contrast to the HTC construct, it retains a portion of the neck. The Cmd1-226 mutant bears an F92A substitution, which specifically disrupts the Cmd1/Myo5 interaction (Ohya and Botstein, 1994; Geli et al, 1998). Consistent with our hypothesis, the cmd1-226 mutation significantly decreased association of PA-Myo5 with Cmd1 and concomitantly increased binding to Vrp1, as compared with the wt CMD1 or with the cmd1-231 mutation, which does not affect the Myo5/Cmd1 interaction (Figure 3B) (Geli et al, 1998). The Myo5-iqΔ mutant did not bind Cmd1 and it coprecipitated significant amounts of Vrp1 in the absence or presence of calcium (Figure 3A). However, Myo5-iqΔ bound slightly less Vrp1-HA than the intact calmodulin-stripped Myo5. The Vrp1-HA signal appeared 3±0.7-fold increased in the Myo5-iqΔ pull downs compared with the Cmd1-bound Myo5 (n=3), whereas calmodulin stripping from the wt Myo5 resulted in a 9±3.4-fold increase of the Vrp1-HA signal (n=3). The result suggested that an intact neck might be required to completely release the nT/C interaction. Deletion of individual IQ motifs did not seem to have a major influence in Myo5 binding to Vrp1, suggesting that full activation required dissociation of Cmd1 from both IQ motifs (Supplementary Figure S2A).
Figure 3.
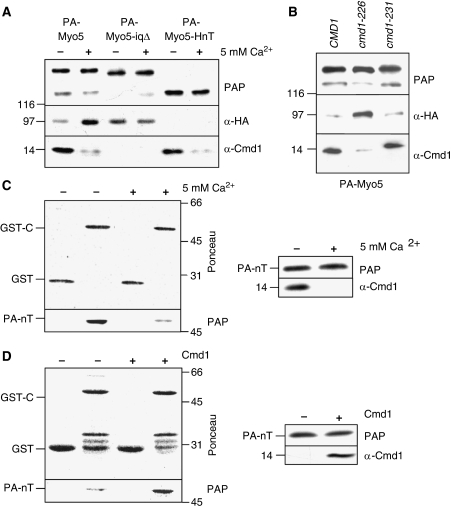
Cmd1 dissociation promotes Myo5 binding to Vrp1 and weakens the nT/C interaction. (A, B) Immunoblots of IgG-Sepharose pull downs from myo5Δ vrp1Δ cells (SCMIG304) (A) or wt (CMD1, SCMIG947), cmd1-226 (SCMIG182) and cmd1-231 (SCMIG187) strains (B), expressing the indicated PA-Myo5 constructs and Vrp1-HA in the absence (A(−), B) or in the presence 5 mM CaCl2 (A (+)) to trigger Cmd1 dissociation. PAP, a peroxidase-conjugated anti-HA antibody and an anti-Cmd1 antibody combined with the adequate peroxidase-conjugated secondary antibody were used for detection of the PA-Myo5 constructs, Vrp1-HA and Cmd1, respectively. All PA-Myo5 constructs and Vrp1-HA were expressed from centromeric plasmids under the control of their own promoters. (C) Immunoblot of glutathione-Sepharose pull downs of GST-C or GST, incubated with PA-nT, purified in the absence (−) or presence (+) of 5 mM CaCl2 to trigger Cmd1 dissociation. Ponceau, PAP and an anti-Cmd1 antibody, combined with the appropriate secondary antibody, were used for detection of GST-C and GST, PA-nT and Cmd1, respectively. (D) Immunoblot of GST-C or GST incubated with Cmd1-free PA-nT, in the absence (−) or presence (+) of 50 nM of recombinant purified Cmd1, processed as in (C).
To investigate whether association of Cmd1 to the myosin neck could influence the affinity of the nT fragment and the Cext, purified PA-nT was incubated with GST fused to the Cext (GST-C), in the presence or absence of 5 mM CaCl2 (Figure 3C). In the presence of 5 mM CaCl2 Cmd1 dissociated from the nT fragment (Figure 3C) (Geli et al, 1998). Concomitantly, the presence of CaCl2 triggered dissociation of PA-nT from the Cext (Figure 3C). Dissociation of Cmd1 also disrupted the interaction between the PA-Hn and PA-HnT fragments and the Cext but did not have an influence on the PA-HT/C or the PA-HnT-iqΔ/C interactions (Supplementary Figure S3A).
To demonstrate that association of Cmd1 to the myosin neck could reversibly influence the affinity between the Myo5 fragments, Cmd1-free PA-nT was incubated with GST-C-coated beads, in the presence or absence of recombinant purified Cmd1. As shown in Figure 3D, Cmd1 binding favoured the interaction of PA-nT with the Cext.
Cmd1 dissociation from Myo5 extends the myosin lifespan at cortical patches
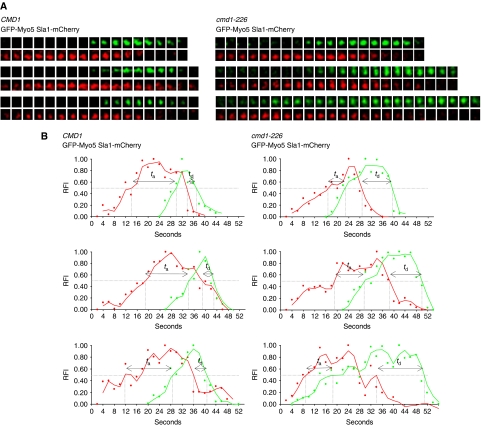
As Vrp1 participates in the recruitment of Myo5 to cortical endocytic patches in vivo, we next investigated whether Cmd1 also controlled Myo5 association to endocytic sites using live-cell imaging. Under the experimental conditions used, no significant endocytic defects are observed in the myo5-iqΔ and cmd1-226 mutants (Geli et al, 1998) and consistently, no significant difference in the dynamics of the clathrin-coat component Sla1-mCherry at cortical patches could be observed for the different mutants, as compared with the isogenic wts (Table 1, SCMIG1079 pGFP-MYO5 versus SCMIG1079 pGFP-myo5-iqΔ and SCMIG1077 pGFP-MYO5 versus SCMIG1078 pGFP-MYO5, mCherry channel). In contrast, the lifespan of GFP-Myo5 significantly increased in mutants that disrupted the Myo5/Cmd1 interaction (GFP-myo5-iqΔ and cmd1-226, Table 1, GFP channel and Figure 4A and B). The average lifespan of GFP-Myo5 in different strain backgrounds expressing the wt Cmd1 was 12 to 13.1 s, whereas it was extended up to 19.5 s in strains where the Myo5/Cmd1 interaction was disrupted (Table 1, GFP channel and Figure 4A and B). A fraction of the GFP-Myo5 cortical patches still exhibited wt dynamics under conditions that disrupted the Myo5/Cmd1 interaction. However, about 50% of the patches had lifespans longer than 16 s in the cmd1-226 mutant (versus only <8% in the isogenic wts) (Figure 4C and D). Consistent with the observation that an intact neck was required to fully promote binding of Myo5 to Vrp1, the Myo5-iqΔ mutant was slightly less efficient than the cmd1-226 mutant in extending the Myo5 lifespan at cortical sites (Table 1, the lifespan of Myo5-iqΔ was 14.7 s, versus >15.9 s for the wt Myo5 in the cmd1-226 mutants). The extension of the Myo5 lifespan at cortical sites was specific for the conditions that disrupted the Myo5/Cmd1 interaction as deletion of the individual IQ motifs or the cmd1-231 mutation did not have any significant effect on the Myo5 dynamics (Table 1).
Table 1. Lifespans of GFP-Myo5, Sla1-mCherry and Abp1-RFP patches in mutants that disrupt the Myo5/Cmd1 interaction.
| Channel | RFP/mCherry | GFP | ||||||
|---|---|---|---|---|---|---|---|---|
| Strain | Avg | s.d. | n | P | Avg | s.d. | n | P |
| GFP-MYO5 (Y06549 pGFP-MYO5) | — | — | — | 12.1 | 3.5 | 80 | ||
| GFP-myo5-iq1Δ (Y06549 pGFP-myo5-iq1Δ) | 13.7 | 3.4 | 80 | 0.003 | ||||
| GFP-myo5-iq2Δ (Y06549 pGFP-myo5-iq2Δ) | 12.8 | 3.3 | 80 | 0.1 | ||||
| GFP-myo5-iqΔ (Y06549 pGFP-myo5-iqΔ) | — | — | — | — | 14.7 | 5.2 | 80 | 1 × 10−4 |
| CMD1 GFP-MYO5 (SCMIG1057 pGFP-MYO5) | — | — | — | 12.2 | 3.7 | 80 | ||
| cmd1-226 GFP-MYO5 (SCMIG1058 pGFP-MYO5) | — | — | — | — | 15.9 | 4.9 | 80 | 1 × 10−4 |
| cmd1-231 GFP-MYO5 (SCMIG1059 pGFP-MYO5) | — | — | — | — | 13.2 | 4.6 | 80 | 0.2 |
| GFP-MYO5 SLA1-mCherry (SCMIG1079 pGFP-MYO5) | 23.3 | 4.9 | 57 | ND | ND | ND | ||
| GFP-myo5-iqΔ SLA1-mCherry (SCMIG1079 pGFP-myo5-iqΔ) | 21.7 | 3.9 | 57 | 0.3147 | ND | ND | ND | ND |
| CMD1 GFP-MYO5 SLA1-mCherry (SCMIG1077 pGFP-MYO5) | 24.6 | 4.4 | 51 | 13.1 | 3.3 | 51 | ||
| cmd1-226 GFP-MYO5 SLA1-mCherry (SCMIG1078 pGFP-MYO5) | 24.12 | 4.4 | 51 | 0.5310 | 18.4 | 6.5 | 51 | 2 × 10−6 |
| GFP-MYO5 ABP1-RFP (SCMIG880 pGFP-MYO5) | 14.1 | 3.2 | 52 | ND | ND | ND | ||
| GFP-myo5-iqΔ ABP1-RFP (SCMIG880 pGFP-myo5-iqΔ) | 21.6 | 7.3 | 54 | 7 × 10−10 | ND | ND | ND | ND |
| CMD1 GFP-MYO5 ABP1-RFP (SCMIG1063 pGFP-MYO5) | 13.4 | 4.5 | 56 | 12.3 | 4.5 | 59 | ||
| cmd1-226 GFP-MYO5 ABP1-RFP (SCMIG1064 pGFP-MYO5) | 18.5 | 5.7 | 58 | 8 × 10−7 | 19.5 | 5.7 | 52 | 8 × 10−11 |
| The average (avg) and standard deviation (s.d.) are indicated in seconds. ‘n' represents the number of cortical patches analysed for the indicated strains under the adequate conditions to visualize RFP and mCherry or GFP. ‘P' represents P-value of the Student's t-test, compared with the isogenic wt. P-values under 0.001 indicate statistically significant differences. ND denotes not determined. | ||||||||
Figure 4.
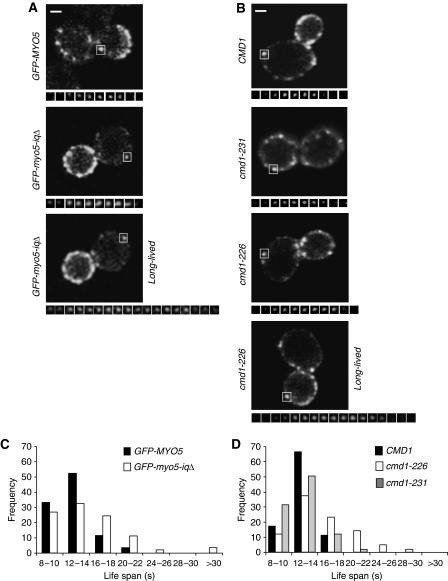
Cmd1 dissociation from Myo5 expands its lifespan at cortical patches. (A, B) Fluorescence micrographs of consecutive frames from time-lapse movies of cortical patches (lower panels) of GFP-Myo5 or GFP-Myo5-iqΔ, expressed in a myo5Δ strain (Y06549) (A) or GFP-Myo5 expressed in a cmd1Δ myo5Δ strain, expressing the wt CMD1 (SCMIG1057) or the cmd1-231 (SCMIG1059) or cmd1-226 (SCMIG1058) mutants (B). The upper panels show a single frame of the imaged cells. The white square frames the cortical patch under analysis. Frames were recorded every 2 s. Constructs were expressed on centromeric plasmids under the MYO5 promoter. (C, D) Frequency distribution for the lifespan of the 80 GFP-Myo5 cortical patches analysed for the strains described in (A, B), respectively. Bar=1 μm.
Next, we investigated whether Cmd1 dissociation promoted early arrival of Myo5 to the endocytic sites and/or slowed down its dissociation from the PM. For this purpose, we examined the dynamics of long-lived (lifespan longer than 16 s) GFP-Myo5 patches relative to Sla1, in the cmd1-226 mutant. The lifespan of the Sla1-mCherry was unaltered in the subpopulation of long-lived GFP-Myo5 patches (Table 2, lifespan Sla1-mCherry, SCMIG1077 pGFP-MYO5 versus SCMIG1078 pGFP-MYO5). However, the myosin arrived on average 5.5 s earlier in the cmd1-226 mutant, as compared with the CMD1 strain (Table 2, ta GFP-Myo5 and Figure 5). This result indicated that Cmd1 dissociation increased the probability of Myo5 meeting available binding partners at cortical patches. Interestingly also, GFP-Myo5 remained at the PM for a longer time (5.7 s on average) after departure of the endocytic coat in the cmd1-226 mutant (Table 2, td GFP-Myo5 and Figure 5). This observation suggested that Cmd1 reassociation to Myo5 might contribute to bring back the cortical myosin to the cytosol after vesicle fission occurs.
Table 2. GFP-Myo5 arrives earlier to and departs later from the endocytic sites in the cmd1-226 mutant.
| Strain Parameter | CMD1 GFP-MYO5 SLA1-mCherry (SCMIG1077 pGFP-MYO5) | cmd1-226 GFP-MYO5 SLA1-mCherry (SCMIG1078 pGFP-MYO5) | P | ||||
|---|---|---|---|---|---|---|---|
| Avg | s.d. | n | Avg | s.d. | n | ||
| Ls Sla1-mCherry | 26.0 | 3.3 | 20 | 24.9a | 4.6a | 20 | 0.3889 |
| Ls GFP-Myo5 | 13.1 | 2.2 | 20 | 23.8a | 4.6a | 20 | 1 × 10−5 |
| ta GFP-Myo5 | 17.7 | 4.0 | 20 | 12.3a | 4.6a | 20 | 2.5 × 10−4 |
| td GFP-Myo5 | 4.8 | 2.9 | 20 | 10.5a | 3.7a | 20 | 1 × 10−5 |
| Lifespans (Ls) of 20 correlative Sla1-mCherry and GFP-Myo5 patches and arrival (ta) and departure times (td) of GFP-Myo5 relative to Sla1-mCherry, analysed in the indicated strains (see Materials and methods and Figure 5). The average (avg) and standard deviation (s.d.) are indicated in seconds. ‘n' represents the number of pairs of cortical patches analysed for the indicated yeast strains. ‘P' represents the P-value of the Student's t-test. P-values under 0.001 indicate statistically significant differences. | |||||||
| aSubpopulation associated with GFP-Myo5 cortical patches with lifespans longer than 16 s. | |||||||
Figure 5.
GFP-Myo5 arrives earlier to and departs later from the endocytic sites in the cmd1-226 mutant. (A) Fluorescence micrographs of consecutive frames from three representative double-colour time-lapse movies of cortical patches in cmd1Δ myo5Δ strains with mCherry-tagged SLA1, expressing GFP-Myo5 from a centromeric plasmid under the MYO5 promoter and either the wt CMD1 (SCMIG1077) or the cmd1-226 mutant (SCMIG1078). Frames were recorded every 2 s. GFP-Myo5 patches with lifespans longer than 16 s were analysed for the cmd1-226 strain. (B) Graphs demonstrating the relative fluorescence intensity (RFI) plotted against time for the Sla1-mCherry (red) and GFP-Myo5 (green) double-colour time-lapse movies shown in (A). ta and td indicate the arrival and departure times of GFP-Myo5 relative to Sla1-mCherry, respectively.
Cmd1 dissociation from Myo5 triggers actin polymerization in vitro and the assembly of actin at endocytic sites in vivo
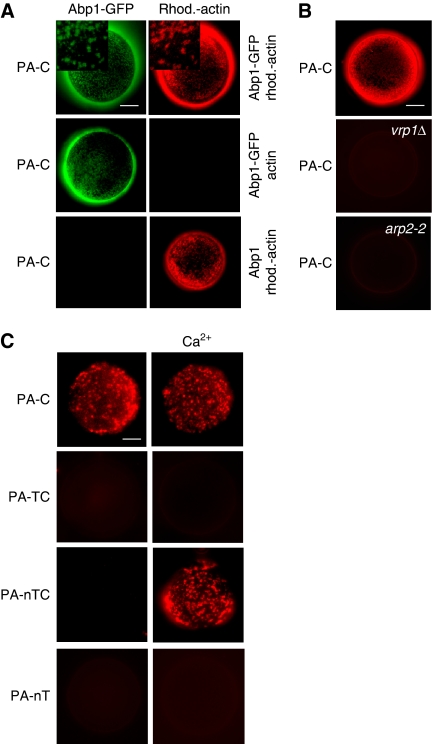
Vrp1 is required to drive Myo5-induced actin polymerization (Geli et al, 2000; Lechler et al, 2001; Sun et al, 2006). Thus, we next investigated whether Cmd1 might also modulate the capacity of the Myo5 Cext to trigger the assembly of endocytic actin structures. We have previously demonstrated that the Myo5 Cext fused to GST and immobilized on the surface of glutathione-Sepharose beads induces the formation of complex actin structures in the presence of yeast extracts (Geli et al, 2000; Idrissi et al, 2002). The actin structures can be labelled with proteins involved in the endocytic uptake and its assembly requires the presence of Vrp1 and the Arp2/3 complex (Geli et al, 2000; Idrissi et al, 2002). PA-tagged Myo5 fragments containing either the Cext alone (PA-C) or together with either the TH1 domain (PA-TC) or the neck and the TH1 domains (PA-nTC) were purified from yeast and immobilized on the surface of IgG-Sepharose beads. Similar to GST-C, PA-C induced assembly of actin structures, which could be visualized under the fluorescence microscope using rhodamine-labelled actin (Figure 6A). The structures could be labelled with the endocytic protein Abp1 (Figure 6A), and their formation was dependent on the presence of functional Vrp1 and Arp2/3 complex (Figure 6B). Consistent with the observation that the TH1 domain prevented binding of the Cext to Vrp1 (Figure 2A), the presence of the TH1 domain (PA-TC) completely abolished the capacity of the Myo5 Cext to induce the formation of actin patches (Figure 6C). Consistent with the hypothesis that Cmd1 dissociation might regulate this autoinhibitory interaction, we found that the construct bearing the neck (PA-nTC) could induce formation of the in vitro generated actin structures, only when it was purified under conditions that dissociated Cmd1 from the neck (5 mM CaCl2; Figure 6C). The neck was required to promote calcium-induced actin polymerization in the presence of the TH1 domain as the CaCl2 treatment did not alter the actin polymerization promoting capacity of PA-TC (Figure 6C). Finally, actin polymerization induced by Cmd1 dissociation was triggered by the Cext as PA-nT failed to trigger the formation of actin patches under any of the experimental conditions assayed (Figure 6C).
Figure 6.
Cmd1 dissociation from Myo5 triggers actin polymerization in vitro. (A–C) Fluorescence micrographs of IgG-Sepharose beads coated with equimolar amounts of the indicated PA-Myo5 constructs incubated for 5 min at 26°C in the presence of yeast extracts from either abp1Δ strains, expressing Abp1-GFP (Y071BP19 pABP1-GFP) or untagged Abp1 (Y071BP19 pABP1) (A), wt (SCMIG19), vrp1Δ (SCMIG40) or arp2-2 (HR4156) strains (B) or a wt strain (BY4741) (C), and 1 μM rhodamine-labelled actin (A, rhod.-actin, B, C) or unlabelled actin (A, actin). Insets show higher magnifications of the Abp1-GFP and rhodamine-actin patches. Bar=10 μm.
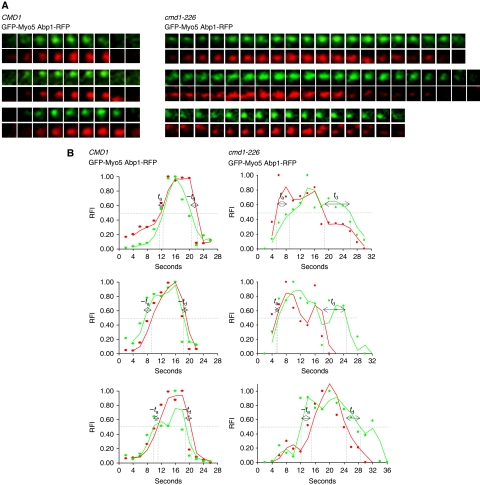
Live-cell imaging of Abp1-RFP in strains bearing myosin-I mutations revealed that these molecular motors, together with Las17, have an essential function in promoting the assembly of complex actin structures at endocytic sites (Sun et al, 2006; Galletta et al, 2008). To investigate whether Cmd1 dissociation from Myo5 might regulate the assembly of the Abp1-containing actin structures in vivo, the dynamics of Abp1-RFP was analysed under conditions that disrupted the Myo5/Cmd1 interaction. In contrast to Sla1-mCherry, the Abp1-RFP lifespan at the endocytic patches was significantly expanded in cells expressing the Myo5-iqΔ or the Cmd1-226 mutants, as compared with the isogenic wts (Table 1, SCMIG880 pGFP-MYO5 versus SCMIG880 pGFP-myo5-iqΔ and SCMIG1063 pGFP-MYO5 versus SCMIG1064 pGFP-MYO5, RFP channel). When considering the subpopulation of GFP-Myo5 patches with lifespans longer than 16 s in the cmd1-226 mutant, no significant differences were observed in the arrival time of GFP-Myo5 relative to Abp1-RFP, as compared with the isogenic wt (Table 3, ta GFP-Myo5 and Figure 7). Abp1-RFP recruitment closely followed Myo5 recruitment in both strains (Table 3, ta GFP-Myo5 and Figure 7). As Myo5 arrived earlier at the endocytic sites in the cmd1-226 mutant (Table 2, ta GFP-Myo5 and Figure 5), the results indicated that Cmd1 dissociation from Myo5 favoured early arrival of Myo5 followed by early onset of actin polymerization. On the other hand, consistent with the observation that Abp1 travels into the cytosol together with the clathrin coat, Myo5 dissociation from the PM was delayed on average 5 s after departure of Abp1-RFP in the cmd1-226 strain (Table 3, td GFP-Myo5 and Figure 7; and Table 2, td GFP-Myo5 and Figure 5).
Table 3. Early arrival of GFP-Myo5 to the endocytic sites in the cmd1-226 is accompanied by early recruitment of Abp1-RFP.
| Strain | CMD1 GFP-MYO5 Abp1-RFP (SCMIG1063 pGFP-MYO5) | cmd1-226 GFP-MYO5 Abp1-RFP (SCMIG1064 pGFP-MYO5) | P | ||||
|---|---|---|---|---|---|---|---|
| Parameter | Avg | s.d. | n | Avg | s.d. | n | |
| Ls Abp1-RFP | 13.5 | 2.5 | 20 | 20.7a | 5.2a | 20 | 4 × 10−4 |
| Ls GFP-Myo5 | 11.7 | 1.9 | 20 | 24.3a | 5.2a | 20 | 2 × 10−6 |
| ta GFP-Myo5 | −0.05 | 2.8 | 20 | −0.08a | 3.4a | 20 | 0.6436 |
| td GFP-Myo5 | −1.9 | 2.4 | 20 | 3.5a | 4.25a | 20 | 4 × 10−3 |
| Lifespans (Ls) of 20 correlative Abp1-RFP and GFP-Myo5 patches and arrival (ta) and departure times (td) of GFP-Myo5 relative to Abp1-RFP, analysed in the indicated strains (see Materials and methods and Figure 7). The average (avg) and standard deviation (s.d.) are indicated in seconds. ‘n' represents the number of pairs of cortical patches analysed for the indicated yeast strains. ‘P' represents the P-value of the Student's t-test. P-values under 0.001 indicate statistically significant differences. | |||||||
| aSubpopulation associated with GFP-Myo5 cortical patches with lifespans longer than 16 s. | |||||||
Figure 7.
Cmd1 dissociation from Myo5 triggers actin assembly at endocytic sites. (A) Fluorescence micrographs of consecutive frames from double-colour time-lapse movies of three representative cortical patches in cmd1Δ myo5Δ strains with RFP-tagged ABP1, expressing GFP-Myo5 from a centromeric plasmid under the MYO5 promoter and either the wt CMD1 (SCMIG1063) or the cmd1-226 mutant (SCMIG1064). Frames were recorded every 2 s. GFP-Myo5 patches with lifespans longer than 16 s were analysed for the cmd1-226 strain. (B) Graphs demonstrating the relative fluorescence intensity (RFI) plotted against time for the Abp1-RFP (red) and GFP-Myo5 (green) double-colour time-lapse movies shown in panel (A). ta and td indicate the arrival and departure times of GFP-Myo5 relative to ABP1-RFP, respectively.
Differential binding of cytosolic and PM-associated Myo5 to Cmd1 and Vrp1
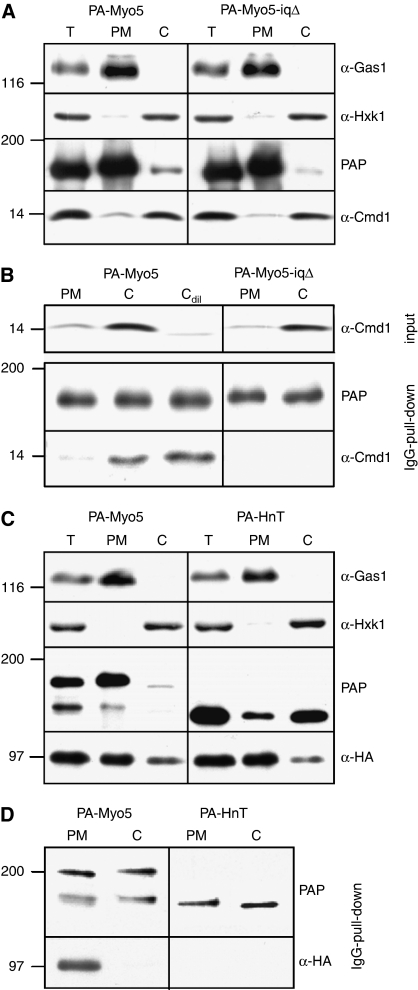
Our data indicated that disruption of the Myo5/Cmd1 interaction promotes Myo5 binding to Vrp1, expands the myosin lifespan at cortical patches and activates myosin-I-induced actin polymerization in vitro and in vivo. Thus, the results strongly suggested that a mechanism involving Cmd1 dissociation from Myo5 regulates its recruitment and activity at endocytic sites. If this hypothesis was correct, the PM-associated and cytosolic Myo5 should reciprocally bind Cmd1 and Vrp1.
To test our hypothesis, we prepared cytosolic (C) and PM fractions from yeast cells expressing PA-Myo5 and analysed the amount of Cmd1 coprecipitating with the myosin-I in IgG-Sepharose pull downs. The GPI-anchored protein Gas1 and hexokinase (Hxk1) were used as PM and cytosolic markers, respectively (Figure 8A and C). As shown in Figure 8B, the PM-associated Myo5 (PM) clearly coprecipitated much less Cmd1 than the cytosolic Myo5 (C). The associated Cmd1 specifically bound to the Myo5 neck as the signal disappeared when PA-Myo5-iqΔ was used instead of the wt (Figure 8B). The lack of association of Myo5 with Cmd1 at the PM was not merely due to the low concentration of Cmd1 in this fraction (Figure 8A), as a 1 to 10 dilution of the cytosol did not alter binding of the cytosolic Myo5 to Cmd1 (Cdil; Figure 8B).
Figure 8.
Differential binding of cytosolic and PM-associated Myo5 to Cmd1 and Vrp1. (A, C) Immunoblots of 10 μg of total (T), PM or cytosolic (C) fractions prepared from myo5Δ cells (SCMIG275) (A) or myo5Δ vrp1Δ cells (SCMIG304) expressing Vrp1-HA (C), expressing the indicated PA-Myo5 constructs. PAP, a peroxidase-conjugated anti-HA antibody and antibodies against Cmd1 or the PM and cytosolic markers Gas1 and Hxk1, combined with the appropriate peroxidase-conjugated secondary antibody, were used for detection of the PA-Myo5 constructs, Vrp1-HA, Cmd1, Hxk1 or Gas1, respectively. (B, D) Immunoblots of IgG pull downs from yeast PM and cytosolic (C) fractions from myo5Δ cells (SCMIG275) (B) or myo5Δ vrp1Δ cells (SCMIG304) expressing Vrp1-HA (D), expressing the indicated PA-Myo5 constructs. Membranes were processed as described in (A, C).
We next precipitated PA-Myo5 with IgG-Sepharose from cytosolic and PM fractions of yeast cells expressing Vrp1-HA (Figure 8C). As shown in Figure 8D, the PM-associated Myo5 (PM) coprecipitated much more Vrp1-HA than the cytosolic myosin (C). Vrp1 specifically bound to the PM-associated Myo5 as deletion of the Cext (PA-HnT) completely eliminated Vrp1 from the precipitates (Figure 8D). The experiments indicated that, compared with the cytosolic Myo5, the PM-associated Myo5 is poorly bound to Cmd1 but strongly interacts with Vrp1. Consistent also with previous immunolocalization results (Brockerhoff and Davis, 1992), which failed to demonstrate the presence of Cmd1 at actin patches, we did not observe colocalization of Cmd1-mCherry and GFP-Myo5 at endocytic sites in live cells (Supplementary Figure S4). In contrast, Myo5 and Vrp1 overlap for 10 to 15 s at endocytic patches in analogous experiments (Sun et al, 2006).
Discussion
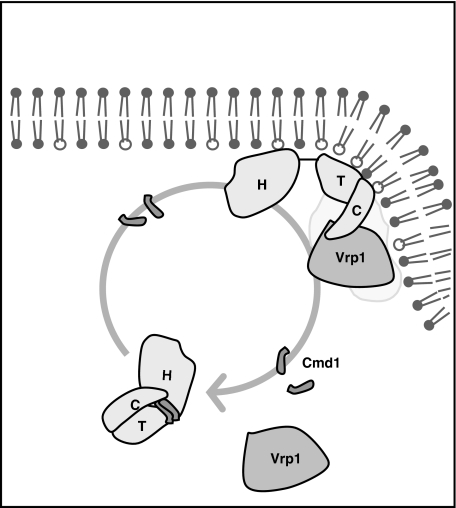
On the basis of our observations and the available literature, we propose the model depicted in Figure 9. According to this model, the cytosolic myosin bound to Cmd1 maintains a closed conformation where the TH1 domain interacts with the Cext preventing association with Vrp1 and Myo5-induced actin polymerization. At the PM, a still unidentified mechanism releases the tight calmodulin/myosin-I interaction. As a consequence, the intramolecular inhibitory interaction is disrupted, allowing Myo5 binding to Vrp1 and local activation of Myo5-induced actin polymerization. Finally, Cmd1 reassociation to Myo5 contributes to dissociate it from the PM after vesicle fission.
Figure 9.
A model for the regulation of Myo5 recruitment and activation at the cortical endocytic patch. Cytosolic Myo5 interacts with Cmd1, which stabilizes a conformation where the Cext (C) folds back and binds to the TH1 (T) domain. Under this circumstance, the SH3 domain is not accessible for binding to Vrp1. An intracellular signal at the PM causes Cmd1 dissociation from the Myo5 neck and releases the inhibitory interaction. SH3-mediated binding of Myo5 to Vrp1 concentrates Myo5 at the cortical endocytic patches and triggers Myo5-induced actin polymerization. Cmd1 reassociation to the Myo5 neck contributes to dissociate it from the PM.
Robust evidence for a direct intramolecular interaction between the myosin TH1 domain and the Cext was already present in the literature for the long-tailed protozoal myosins-I. Similar to what we found for Myo5, pull-down experiments with purified components demonstrated such interaction for the Acanthamoeba castellanii myosin-IA (Lee et al, 1999). In addition, structural data on the A. castellanii myosin-IB and C showed a surprising globular structure of the tail, where the SH3 domain folds back on the TH1 domain (Jontes et al, 1998; Ishikawa et al, 2004). More recently, Hwang et al (2007) have provided NMR data supporting such an intramolecular interaction. We have now revealed the physiological significance of this interaction and described a molecular mechanism that might contribute to its modulation.
We showed that the TH1 domain prevented interaction of the Myo5 Cext with Vrp1 in immunoprecipitation experiments. Consistent with the cellular function described for the Myo5/Vrp1 interaction (Anderson et al, 1998; Geli et al, 2000; Lechler et al, 2001; Sun et al, 2006), we also found that the TH1 domain impairs recruitment of the Myo5 Cext to cortical patches in vivo and abrogated its activity as an actin polymerization promoting factor in vitro. The TH1 domain most likely exerted its inhibitory activity intramolecularly, as we failed to demonstrate oligomerization of the myosin under the same experimental conditions that showed the Myo5/Vrp1 and Myo5/Cmd1 interactions (Supplementary Figure S5).
An autoinhibitory activity contained within the TH1 domain was not described before. On the contrary, the TH1 domain has been shown to bind acidic phospholipids (Adams and Pollard, 1989; Hayden et al, 1990; Doberstein and Pollard, 1992) and to contribute to membrane targeting of different myosins-I (Miyata et al, 1989; Lee et al, 2000; Yamashita et al, 2000; Oberholzer et al, 2004). The TH1 domain of Myo5 also binds to acidic phospholipids (data not shown) and its deletion causes partial mislocalization of the myosin to the cytosol (Figure 1; GFP-HnC). Thus, in addition to the autoinhibitory activity, the TH1 domain probably exerts a positive function by recruiting Myo5 to the inner leaflet of the PM.
On the other hand, our work also revealed that Cmd1 dissociation promoted Myo5 binding to Vrp1. Again in agreement with previously published data on the function of the Myo5/Vrp1 interaction (Anderson et al, 1998; Geli et al, 2000; Lechler et al, 2001; Jonsdottir and Li, 2004; Sun et al, 2006), we found that dissociation of Cmd1 from Myo5 significantly extended the myosin lifespan at cortical patches, enhanced actin polymerization at endocytic sites and triggered Myo5-induced actin polymerization in vitro. Concomitantly, Cmd1 dissociation weakened the interaction between a fragment containing the neck and TH1 domains and the Cext. Considering all the data, it is reasonable to propose that Cmd1 dissociation promotes Myo5 binding to Vrp1 by releasing the autoinhibitory interaction between the TH1 domain and the Cext.
We made the novel observation that the Cmd1-bound but not the Cmd1-stripped Myo5 neck strongly interacts with the Cext and most likely contributes to stabilize the inhibitory interaction between the adjacent TH1 domain and the Cext (Figure 2C; Supplementary Figure S3A). On the basis of the Myo5 IQ motifs sequences, it appears likely that Cmd1 is bound in an extended conformation (Terrak et al, 2005), with one lobe free to interact with the Cext. Thus, it is tempting to propose a simple model where Cmd1 works as a clamp that holds the TH1 domain folded onto the Cext. However, our results indicated that the activation mechanism might have a second level of complexity. On the one hand, we failed to detect a direct interaction between free Cmd1 and the Cext (Supplementary Figure S3B and C). This raises the interesting possibility that Cmd1 might need to interact first to the Myo5 neck in order to cross-link the nT and C fragments. On the other hand, our results suggested that the presence of an intact neck is important to release the autoinhibitory interaction when the TH1 is covalently linked to the Cext. This was inferred from the observation that deletion of the neck in the presence of the TH1 domain completely abolished Cext-dependent recruitment of Myo5 to cortical patches, Myo5 interaction with Vrp1 and Myo5-induced actin polymerization. Deletion of the neck might cause missfolding of the tail, thereby preventing Vrp1 association. However, this is probably not the case because the HTC construct binds to Rvs167 similar to the wt Myo5 (Figure 2B) and partial Trypsin digestion experiments indicated an analogous conformation for the Cmd1-bound wt Myo5 and for the HTC mutant (Supplementary Figure S6, see below). Further evidence supporting the active function of the neck in releasing the autoinhibition derives from the Myo5-iqΔ mutant. We found that this Myo5 mutant, which lacks a portion of the neck, was less efficient than the cmd1-226 mutant in extending the Myo5 lifespan at cortical sites and in promoting Myo5 binding to Vrp1.
Interestingly, a major conformational change induced in the TH1 domain of the short-tailed Brush Border myosin-I can be observed upon calmodulin dissociation from the myosin neck (Whittaker and Milligan, 1997). We hypothesize that dissociation of Cmd1 could transmit a conformational change from the neck to the TH1 domain, which could influence its affinity for the Cext. Consistent with this view, we have found that Cmd1 dissociation was accompanied by the exposure of a Trypsin digestion site within the TH1 domain and that exposure of this additional site required the presence of an intact neck (Supplementary Figure S6). Thus, treatment of the wt Myo5 with 5 mM CaCl2 exposed a Trypsin digestion site that generated an N-terminal 116 kDa fragment (Supplementary Figure S6, Myo5 Cmd1, CaCl2). This fragment was also present after digestion of the wt Myo5 purified from the cmd1-226 mutant, independently of the presence of CaCl2 (Supplementary Figure S6). The Trypsin digestion site seemed to be less exposed in the Myo5-iqΔ mutant and it could not be detected in the HTC construct, which lacks the complete neck (Supplementary Figure S6). Alternatively or in addition to a putative conformational change transmitted from the neck to the TH1 domain, the Cmd1-stripped neck could bind other molecules that sterically hinder the TH1/Cext interaction (Swanljung-Collins and Collins, 1992; Cyr et al, 2002; Tang et al, 2002; Hirono et al, 2004; Phillips et al, 2006) or it might recruit enzymes that modify the Myo5 tail.
Intramolecular interactions regulating protein localization and function of a number of factors that promote actin polymerization have been described before. An interaction between the N- and C-terminus of the mammalian formin formin-related gene in leukocytes α (FRLα) modulates its PM localization and actin assembly activity (Seth et al, 2006). Similarly, induction of Arp2/3 complex-dependent actin polymerization by the C-terminal portion of neuronal WASP is autoinhibited by the N-terminal fragment (Rohatgi et al, 2000). Local activation of actin polymerization promoting factors can be achieved by direct interaction with signal transducers such as Cdc42 and PIns(4,5)P2, which release the autoinhibitory interactions (Rohatgi et al, 2000; Seth et al, 2006). Downregulating futile cycles of actin polymerization might be essential to prevent energy waste and the assembly of actin structures that could interfere with a number of cellular functions.
A key question that follows from our work is the nature of the signal triggering Cmd1 release and activation of Myo5 at endocytic sites. Even though increasing evidence points to a function of calcium and calmodulin regulating endocytic uptake in mammals (Wu et al, 2009), we do not favour the idea that calcium triggers Cmd1 dissociation from Myo5 at endocytic sites because a Cmd1 mutant that does not bind calcium (cmd1-3) (Geiser et al, 1991) does not exhibit an uptake defect in yeast (Kubler et al, 1994). Nevertheless, further experiments would be required to rule out this possibility. On the contrary, a number of results indicate that local production of PIns(4,5)P2 at endocytic sites could contribute to trigger Cmd1 dissociation from Myo5. First, it was recently shown that myosin-I mutants exhibit synthetic lethal interactions with mutations in the Mss4 PIns4P-5 kinase responsible for the production of PIns(4,5)P2 at endocytic sites (Audhya et al, 2004). Further, the lifespan of Myo5 is significantly expanded in a yeast synaptojanin mutant defective in PIns(4,5)P2 consumption (Sun et al, 2007). Finally, Hayden et al (1990) demonstrated that association of the Brush Border myosin-I with liposomes containing PIns(4,5)P2 displaces calmodulin from the myosin neck. In addition, other molecular mechanisms including protein kinase C-dependent phosphorylation of the myosin-I neck and/or actin and nucleotide binding to the motor head (Lieto-Trivedi and Coluccio, 2008) might contribute to lower the tight myosin-I/calmodulin interaction. Further studies will now be required to investigate these possibilities.
Materials and methods
Strains and genetic techniques
Yeast strains used are listed in Table 4. Details on strain construction are available upon request. Strains without plasmids were grown in complete yeast peptone dextrose and strains with plasmids were selected on synthetic dextrose complete (SDC) lacking the appropriate nutrient (Dulic et al, 1991) at 25°C. Scoring of genetic markers was performed as described (Sherman et al, 1974). Transformation of yeast was accomplished by the lithium acetate method (Ito et al, 1983). All tagged versions of yeast genes used in this study complemented the growth defects of the corresponding knock-out strains.
Table 4. Yeast strains.
| Strain | Genotype | Reference |
|---|---|---|
| SCMIG182 | MATa ade2 ade3::cmd1-226::TRP1 his3 leu2 lys2 trp1 ura3 cmd1Δ::HIS3 bar1Δ::LYS2 | Geli et al (1998) |
| SCMIG947 | MATa ade2 ade3::CMD1::TRP1 his3 leu2 lys2 trp1 ura3 cmd1Δ::HIS3 bar1Δ::LYS2 | Geli et al (1998) |
| SCMIG187 | MATa ade2 ade3:: cmd1-231::TRP1 his3 leu2 lys2 trp1 ura3 cmd1Δ::HIS3bar1::LYS2 | Geli et al (1998) |
| SCMIG19 | MATa his3 leu2 trp1 ura3 bar1 | Idrissi et al (2002) |
| SCMIG275 | MATa his3 leu2 lys2 trp1 ura3 bar1 myo5Δ::TRP1 | Idrissi et al (2002) |
| SCMIG40 | MATa vrp1Δ::URA3 ura3 leu2 his3 lys2 trp1 ade2 bar1 | Geli et al (2000) |
| RH4165 | MATa arp2-2::URA3 ura3 leu2 his3 trp1 ade2 bar1 | Idrissi et al (2002) |
| BY4741 | MATa leu2 his3 ura3 met15 | Euroscarf |
| Y06549 | MATa myo5Δ::KMX leu2 his3 ura3 met15 | Euroscarf |
| Y07199 | MATa abp1Δ::KMX leu2 his3 ura3 met15 | Euroscarf |
| SCMIG304 | MATa his3 leu2 lys2 trp1 ura3 bar1 vrp1Δ::URA3 myo5Δ::TRP1 + pURA3MYO5 | Geli et al (2000) |
| SCMIG880 | MATα ura3 leu2 his3 trp1 myo5Δ::TRP1 ABP1-RFP::KMX bar1? | This study |
| SCMIG1047 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX pURA3CMD1 | This study |
| SCMIG1057 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX pTRP1CMD1 | This study |
| SCMIG1058 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX pTRP1 cmd1-226 | This study |
| SCMIG1059 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX pTRP1 cmd1-231 | This study |
| SCMIG1061 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX ABP1-RFP::KMX pURA3CMD1 | This study |
| SCMIG1063 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX ABP1-RFP::KMX pTRP1CMD1 | This study |
| SCMIG1064 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX ABP1-RFP::KMX pTRP1 cmd1-226 | This study |
| SCMIG1076 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX SLA1-mCherry::HIS3MX pURA3CMD1 | This study |
| SCMIG1077 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX SLA1-mCherry::HIS3MX pTRP1CMD1 | This study |
| SCMIG1078 | MATa his3 leu2 trp1 ura3 met15 cmd1Δ::KMX myo5Δ::KMX SLA1-mCherry::HIS3MX pTRP1 cmd1-226 | This study |
| SCMIG1079 | MATa myo5Δ::KMX leu2 his3 ura3 met15 SLA1-mCherry::HIS3MX | This study |
| SCMIG1080 | MATa his3 leu2 ura3 met15 myo5Δ::KMX CMD1-mCherry::HIS3MX | This study |
DNA techniques and plasmid construction
Plasmids used are listed in Table 5. Details on plasmid construction are available upon request. DNA manipulations were performed as described (Sambrook et al, 1989). Enzymes for molecular biology were obtained from New England Biolabs. Plasmids were purified with the Nucleospin plasmid purification kit (Macherey-Nagel). DNA was purified from agarose gels using the gel extraction kit from Qiagen. Transformation of E. coli was performed by electroporation (Dower et al, 1988). PCRs were performed with a Vent polymerase (New England Biolabs) and a TRIO-thermoblock (Biometra GmbH). Oligonucleotides were synthesized by Interactiva.
Table 5. Plasmids.
| Plasmid | Marker | Ori | Insert | Reference |
|---|---|---|---|---|
| pMYO5 | URA3 | CEN4 | MYO5 | Geli and Riezman (1996) |
| pHTC | URA3 | CEN4 | myo5 (aa 1–1219)–(aa 705–773)Δ | This study |
| pHnC | URA3 | CEN4 | myo5 (aa 1–1219)–(aa 774–905)Δ | This study |
| pmycMYO5 | LEU2 | CEN4 | myc-MYO5 | This study |
| pMYO5-HA3 | URA3 | CEN4 | MYO5 + 3HA | Idrissi et al (2002) |
| pGEX-5X-3 | — | — | GST | Pharmacia |
| pGST-C | — | — | GST-myo5 (aa 982–1219) | Geli et al (2000) |
| pGST-CMD1 | — | — | GST-CMD1 | This study |
| pGFP-MYO5.LEU | LEU2 | CEN4 | GFP-MYO5 | This study |
| pGFP-MYO5 | URA3 | CEN4 | GFP-MYO5 | This study |
| pGFP-myo5-iqΔ | URA3 | CEN4 | GFP-myo5 (aa 1–1219)–(aa 725–735)Δ (aa 743–753)Δ | This study |
| pGFP-myo5-iq1Δ | URA3 | CEN4 | GFP-myo5 (aa 1–1219)–(aa 725–735)Δ | This study |
| pGFP-myo5-iq2Δ | URA3 | CEN4 | GFP-myo5 (aa 1–1219)–(aa 743–753)Δ | This study |
| pGFP-H | URA3 | CEN4 | GFP-myo5 (aa 1–704) | This study |
| pGFP-Hn | URA3 | CEN4 | GFP-myo5 (aa 1–773) | This study |
| pGFP-HnT | URA3 | CEN4 | GFP-myo5 (aa 1–996) | This study |
| pGFP-HTC | URA3 | CEN4 | GFP-myo5 (aa 1–1219)–(aa 705–773)Δ | This study |
| pGFP-HnC | URA3 | CEN4 | GFP-myo5 (aa 1–1219)–(aa 774–905)Δ | This study |
| pGFP-nT | URA3 | CEN4 | GFP-myo5 (aa 705–996) | This study |
| pGFP-nTC | URA3 | CEN4 | GFP-myo5 (aa 705–1219) | This study |
| pGFP-TC | URA3 | CEN4 | GFP-myo5 (aa 774–1219) | This study |
| pGFP-C | URA3 | CEN4 | GFP-myo5 (aa 882–1219) | This study |
| pGFP-HC | URA3 | CEN4 | GFP-myo5 (aa 705–905)Δ | This study |
| pABP1-GFP | URA3 | CEN4 | ABP1-GFP | This study |
| pABP1 | URA3 | CEN4 | ABP1-GFP | This study |
| pVRP1-GFP | LEU2 | CEN4 | VRP1-GFP | This study |
| pPA-MYO5 | URA3 | CEN4 | Protein A-MYO5 | This study |
| pPA-myo5-iqΔ | URA3 | CEN4 | Protein A-myo5 (aa 1–1219)–(aa 725–735)Δ (aa 743–753)Δ | This study |
| pPA-myo5-iq1Δ | URA3 | CEN4 | Protein A-myo5 (aa 1–1219)–(aa 725–735)Δ | This study |
| pPA-myo5-iq2Δ | URA3 | CEN4 | Protein A-myo5 (aa 1–1219)–(aa 743–753)Δ | This study |
| pPA-H | URA3 | CEN4 | Protein A-myo5 (aa 1–704) | This study |
| pPA-Hn | URA3 | CEN4 | Protein A-myo5 (aa 1–773) | This study |
| pPA-HnT | URA3 | CEN4 | Protein A-myo5 (aa 1–996) | This study |
| pPA-HTC | URA3 | CEN4 | Protein A-myo5 (aa 1–1219)–(aa 705–773)Δ | This study |
| pPA-HnC | URA3 | CEN4 | Protein A-myo5 (aa 1–1219)–(aa 774–905)Δ | This study |
| pPA-nT | URA3 | CEN4 | Protein A-myo5 (aa 705–996) | This study |
| pPA-nTC | URA3 | CEN4 | Protein A-myo5 (aa 705–1219) | This study |
| pPA-TC | URA3 | CEN4 | Protein A-myo5 (aa 774–1219) | This study |
| pPA-C | URA3 | CEN4 | Protein A-myo5 (aa 882–1219) | This study |
| pPA-HT | URA3 | CEN4 | Protein A-myo5 (aa 1–996)–(aa 705–773)Δ | This study |
| pVRP1-HA | LEU2 | CEN4 | VRP1 + 3HA | This study |
| pLexA-RVS167 | HIS3 | 2 μ | LexA + RVS167 | This study |
| pLexA-nT | HIS3 | 2 μ | LexA + myo5 (aa 705–996) | This study |
| pLexA-n | HIS3 | 2 μ | LexA + myo5 (aa 705–773) | This study |
SDS–PAGE, immunoblots and antibodies
SDS–PAGE was performed as described (Laemmli, 1970) using a minigel system (Bio-Rad Laboratories). High and low range SDS–PAGE molecular weight standards (Bio-Rad Laboratories) were used. Coomassie brilliant blue (Harlow and Lane, 1988) was used for detection of total protein. Protein concentration was determined with the Bio-Rad Protein Assay (Bio-Rad Laboratories). Immunoblots were performed as described (Geli and Riezman, 1998) except that transfer of Gas1 was done overnight in the absence of methanol. Antibodies used are listed in Table 6. Nitrocellulose membranes were stained with Ponceau Red for detection of total protein (Harlow and Lane, 1988). An enhanced chemoluminescence (ECL) detection kit (Amersham Biosciences) was used for detection of peroxidase-conjugated antibodies.
Table 6. Antibodies.
| Antigen | Source | Modification | Reference | Dilution |
|---|---|---|---|---|
| Peroxidase | Rabbit antibody | Peroxidase conjugated, peroxidase complex (PAP) | DAKO A/S | 1:1000 |
| Gas1 | Rabbit serum | Muniz et al (2001) | 1:50 000 | |
| Hxk1 | Rabbit serum | van Tuinen and Riezman (1987) | 1.2000 | |
| Cmd1 | Rabbit serum | Geli et al (1998) | 1:1000 | |
| HA | Rat monoclonal antibody (3F10) | Roche | 1:1000 | |
| HA | Rat monoclonal antibody (3F10) | Peroxidase conjugated | Roche | 1:500 |
| MYC | Mouse monoclonal antibody (9E10) | Roche | 1:2000 | |
| GST | Goat serum | Amersham Biosciences | 1:2000 | |
| Rvs167 | Rabbit | H. Riezman | 1/10 000 | |
| LexA | Mouse monoclonal antibody | Santa Cruz | 1:2000 | |
| GFP | Rabbit IgG | Invitrogen | 1:1700 | |
| Mouse IgG | Goat antibody | Peroxidase conjugated | Sigma | 1:4000 |
| Rabbit IgG | Goat antibody | Peroxidase conjugated | Sigma | 1:5000 |
| Rat IgG | Goat antibody | Peroxidase conjugated | Sigma | 1:5000 |
| Goat IgG | Rat IgG | Peroxidase conjugated | Sigma | 1:2000 |
Protein purification from yeast and E. coli
Purification of recombinant GST fusion proteins from BL21 E. coli (Novagen) was performed as described (Geli et al, 2000). Cmd1 was released from the GST-Cmd1-coated glutathione-Sepharose beads by digestion with thrombin-agarose (Thrombin clean cleavage kit, Sigma). Glutathione-Sepharose beads were from Amersham Biosciences. PA-nT was purified from SCMIG275 pPA-nT using IgG-Sepharose (Amersham-Bioscience). After glass bead-lysis in 100 μl of IP buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA) in the presence of protease inhibitors (PIs) (0.5 mM PMSF, 1 μg/ml aprotinin, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml antipain), NaCl and Triton X-100 were adjusted to 0.5 M and 1% final concentrations, respectively. Unbroken cells were eliminated by centrifugation at 2500 g for 5 min. After centrifugation at 10 000 g for 10 min at 4°C, the supernatant was incubated with IgG-Sepharose for 2 h at 4°C. Beads were collected and stepwise eluted with 20 μl of 0.5 M acetic acid pH 3.4 (adjusted with ammonium acetate), 0.5 M NaCl, 0.1% Tween-20. The eluate was mixed with 10 μl of 1 M Tris pH 9 for pH neutralization. For the purification of the Cmd1-free PA-nT, 5 mM CaCl2 was added to the extract before addition of the Sepharose.
Overlay, pull downs and immunoprecipitations
For the glutathione-Sepharose pull downs, 1 μg of bead-bound GST-C or GST were incubated with 30 ng of PA-nT bound to Cmd1 or Cmd1-free PA-nT in 1 ml TBS-TB (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20, 1.5% BSA) for 3 h at 4°C. Where indicated, 5 mM CaCl2 was added to promote Cmd1 dissociation and 1 μg of Cmd1 was added in the presence of 1 mM EGTA to promote reassociation. Beads were collected, washed with TBS-TB and TBS-T (10 mM Tris pH 8.0, 150 mM NaCl, 0.1% Tween-20) and eluted in SDS-sample buffer. For the IgG pull downs from yeast extracts, cells were glass bead-lysed in LB (25 mM Tris, pH 8.5, 5 mM EDTA) in the presence of PIs. Unbroken cells were eliminated at 700 g for 10 min at 4°C. The supernatant was diluted with the same volume of BB (10 mM Tris pH 7.5, 0.2 mM EDTA, 0.2 mM DTT) containing PIs. After centrifugation at 700 g for 10 min at 4°C, the extract was adjusted to 10 mg of protein/ml and Triton X-100 and NaCl were added to 1% and 150 mM final concentrations, respectively. A measure of 1 ml of extract was incubated with 30 μl of 50% IgG-Sepharose for 2 h at 4°C. Beads were washed with LB:BB (1:1), 1% Triton X-100, 150 mM NaCl, and subsequently with LB:BB (1:1) and boiled in 25 μl SDS-sample buffer. Four 1:2 dilutions were loaded for each construct and immunoblotted for PA and Vrp1-HA. The corresponding signals were compared within the linear range. Quantifications were performed with imageJ from at least three independent experiments. The Vrp1-HA signal was normalized to the PAP signal for each sample.
For IgG-Sepharose pull downs from PM and cytosolic fractions, 0.5 ml of the different fractions were diluted with 0.5 ml of LB:BB (1:1) in the presence of PIs. Triton X-100 and NaCl were added to 1% and 150 mM final concentration, respectively. Samples were incubated with 40 μl of 50% IgG-Sepharose for 2 h at 4°C. Beads were washed with LB:BB (1:1), 1% Triton X-100, 150 mM NaCl and eluted with 25 μl SDS-sample buffer.
For the overlay assay, 0.15 μg of the nT fragment was separated by SDS–PAGE and transferred to a nitrocellulose filter in 30 mM Tris, 240 mM Glycine. Proteins were then partially renaturated in 10 mM Tris pH 8, 150 mM NaCl, 0.05% Tween-20 and the membrane was blocked over night at 4°C in blocking buffer (10 mM Tris pH 8, 150 mM NaCl, 0.05% Tween-20, 3% BSA). The membrane was then overlaid with 20 mM purified GST, GST-C or 60 mM Cmd1 for 3 h at 4°C, then with the goat anti-GST antibody or the rabbit anti-Cmd1 for 1.5 h at 4°C and finally with a peroxidase-conjugated antibody against goat IgG for 1 h in blocking buffer. After washing with blocking buffer and subsequently with 10 mM Tris pH 8, 150 mM NaCl, 0.05% Tween-20, peroxidase was detected with an ECL detection kit. For excision of the nT fragment from the PA-nT-coated beads, the PA-nT-coated IgG-Sepharose beads were incubated in 30 μl of 50 mM Tris pH 8, 0.5 mM EDTA and 10 Units (1 μl) of AcTEV (Invitrogen) over night at 4°C. The supernatant was discarded and the protein, which was still bound to the Sepharose beads, was eluted by incubating in 30 μl of 50 mM Tris pH 8, 1 M NaCl, 0.5 mM EDTA for 30 min with shaking at 4°C.
For the pull-down experiments (Figure 2C), 1 μg of bead-bound GST-C or GST were incubated with protein extracts of the strains expressing PA-H, PA-Hn, PA-HT and PA-HnT prepared in IP buffer 1% Triton X-100 in the presence of PIs. After incubation for 1 h at 4°C, beads were recovered, washed three times with IP buffer 1% Triton X-100 and twice with IP buffer. Beads were resuspended in 30 μl of SDS-sample buffer and extracted proteins were separated by SDS–PAGE and transferred to a nitrocellulose filter. GST-C and GST were detected by staining with Ponceau red and the PA-Myo5 constructs were detected using PAP. In all, 1/125 of the total protein extracts were loaded as input.
Purification of yeast PM and cytosol
The isolation of PM and cytosolic fractions were performed based on (Serrano, 1988). For further detail see supplementary data.
In vitro actin polymerization assay
The rhodamine-actin polymerization assay on Sepharose beads was performed as described (Idrissi et al, 2002) except that IgG-Sepharose beads coated with PA-tagged Myo5 constructs purified from yeast were used instead of the GST construct purified from E. coli.
Fluorescence microscopy
Fluorescence micrographs in Figure 1 were obtained with an Axiophot fluorescence microscope (Zeiss) equipped with a GFP filter (excitation 470/40, LP520) and an Olympus DP70 camera. Cells were grown to mid-log phase, harvested, diluted in a small volume of SDC medium and visualized on poly-lysine-coated slides. Rhodamine-actin in the in vitro polymerization assay was observed using a rhodamine filter (excitation BP545/30, emission LP590). For the time-lapse fluorescence microscopy, yeast expressing GFP-Myo5 were grown to mid-log phase in SDC medium. Time-lapse videos were collected from cells immobilized in 0.8% low-melt agarose prepared in complete synthetic medium. A Perkin Elmer UltraView ERS Spinning-Disk microscope, equipped with Argon (488, 514 nm), Argon/Krypton (568 nm) and HeNe (405, 440, 640 nm) Lasers, a 405/488/568/640 Dichroic and a Hamamatsu C9100-50 EMCCD camera was used to collect images. A × 63 1.4NA Oil DIC Plan-Apochromat objective was used for all movies. Time-lapse microscopy images were taken every 2 s and were acquired/processed using Volocity from Improvision (Perkin Elmer). Volocity files were converted to multi-tiff for image processing and to csv for relative intensity fluorescence analysis using Microsoft Excel. Average, standard deviation and P-values for the Student's t-test were calculated with Microsoft Excel. All images were collected with identical sensitivity and exposure times of 600 ms, 500 ms and 1 s for GFP-Myo5, Abp1-RFP and Sla1-mCherry, respectively. Only bright, well-defined patches were considered for further analysis.
For analysis of arrival and departure times of GFP-Myo5 relative to Sla1-mCherry or Abp1-RFP, relative fluorescence intensity (RFI) curves plotted against time (s) were calculated for each pair and the time point when Sla1-mCherry or Abp1-RFP reached RFI=0.5 was subtracted from the time point when GFP-Myo5 reached RFI=0.5 either before (arrival) or after (departure) the time point when Sla1-mCherry or Abp1-RFP reached RFI=1 (see Figures 5 and 7).
Supplementary Material
Acknowledgments
We thank M Pons, J Pérez and M Cid for technical assistance. This work was supported by BUF2005-04089, BFU2008-03500 and CSD2009-00016 (MIG) and NIH R01 GM055796 (SKL) grants. HG was a recipient of a predoctoral fellowship from the Generalitat de Catalunya. F-ZI is a Ramon y Cajal postdoctoral fellow. JG and VR are supported by JAE postdoctoral and technician contracts from the CSIC, respectively. JRC was supported by a NIH training grant T32-HL07188 and an American Heart Association postdoctoral fellowship 6-6253Y.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams RJ, Pollard TD (1989) Binding of myosin I to membrane lipids. Nature 340: 565–568 [DOI] [PubMed] [Google Scholar]
- Anderson BL, Boldogh I, Evangelista M, Boone C, Greene LA, Pon LA (1998) The Src homology domain 3 (SH3) of a yeast type I myosin, Myo5p, binds to verprolin and is required for targeting to sites of actin polarization. J Cell Biol 141: 1357–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Loewith R, Parsons AB, Gao L, Tabuchi M, Zhou H, Boone C, Hall MN, Emr SD (2004) Genome-wide lethality screen identifies new PI4,5P2 effectors that regulate the actin cytoskeleton. EMBO J 23: 3747–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose A, Guilherme A, Robida SI, Nicoloro SM, Zhou QL, Jiang ZY, Pomerleau DP, Czech MP (2002) Glucose transporter recycling in response to insulin is facilitated by myosin Myo1c. Nature 420: 821–824 [DOI] [PubMed] [Google Scholar]
- Brockerhoff SE, Davis TN (1992) Calmodulin concentrates at regions of cell growth in Saccharomyces cerevisiae. J Cell Biol 118: 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordonnier MN, Dauzonne D, Louvard D, Coudrier E (2001) Actin filaments and myosin I alpha cooperate with microtubules for the movement of lysosomes. Mol Biol Cell 12: 4013–4029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyr JL, Dumont RA, Gillespie PG (2002) Myosin-1c interacts with hair-cell receptors through its calmodulin-binding IQ domains. J Neurosci 22: 2487–2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doberstein SK, Baines IC, Wiegand G, Korn ED, Pollard TD (1993) Inhibition of contractile vacuole function in vivo by antibodies against myosin-I. Nature 365: 841–843 [DOI] [PubMed] [Google Scholar]
- Doberstein SK, Pollard TD (1992) Localization and specificity of the phospholipid and actin binding sites on the tail of Acanthamoeba myosin IC. J Cell Biol 117: 1241–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower WJ, Miller JF, Ragsdale CW (1988) High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16: 6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulic V, Egerton M, Elguindi I, Raths S, Singer B, Riezman H (1991) Yeast endocytosis assays. Methods Enzymol 194: 697–710 [DOI] [PubMed] [Google Scholar]
- Durrbach A, Collins K, Matsudaira P, Louvard D, Coudrier E (1996) Brush border myosin-I truncated in the motor domain impairs the distribution and the function of endocytic compartments in an hepatoma cell line. Proc Natl Acad Sci USA 93: 7053–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrbach A, Raposo G, Tenza D, Louvard D, Coudrier E (2000) Truncated brush border myosin I affects membrane traffic in polarized epithelial cells. Traffic 1: 411–424 [DOI] [PubMed] [Google Scholar]
- Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Thomas DY, Boone C (2000) A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol 148: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fath KR, Trimbur GM, Burgess DR (1994) Molecular motors are differentially distributed on Golgi membranes from polarized epithelial cells. J Cell Biol 126: 661–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galletta BJ, Chuang DY, Cooper JA (2008) Distinct roles for Arp2/3 regulators in actin assembly and endocytosis. PLoS Biol 6: e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, van Tuinen D, Brockerhoff SE, Neff MM, Davis TN (1991) Can calmodulin function without binding calcium? Cell 65: 949–959 [DOI] [PubMed] [Google Scholar]
- Geli MI, Lombardi R, Schmelzl B, Riezman H (2000) An intact SH3 domain is required for myosin I-induced actin polymerization. EMBO J 19: 4281–4291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geli MI, Riezman H (1996) Role of type I myosins in receptor-mediated endocytosis in yeast. Science 272: 533–535 [DOI] [PubMed] [Google Scholar]
- Geli MI, Riezman H (1998) Endocytic internalization in yeast and animal cells: similar and different. J Cell Sci 111 (Part 8): 1031–1037 [DOI] [PubMed] [Google Scholar]
- Geli MI, Wesp A, Riezman H (1998) Distinct functions of calmodulin are required for the uptake step of receptor-mediated endocytosis in yeast: the type I myosin Myo5p is one of the calmodulin targets. EMBO J 17: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies, a Laboratory Manual. NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Hayden SM, Wolenski JS, Mooseker MS (1990) Binding of brush border myosin I to phospholipid vesicles. J Cell Biol 111: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirono M, Denis CS, Richardson GP, Gillespie PG (2004) Hair cells require phosphatidylinositol 4,5-bisphosphate for mechanical transduction and adaptation. Neuron 44: 309–320 [DOI] [PubMed] [Google Scholar]
- Huber LA, Fialka I, Paiha K, Hunziker W, Sacks DB, Bahler M, Way M, Gagescu R, Gruenberg J (2000) Both calmodulin and the unconventional myosin Myr4 regulate membrane trafficking along the recycling pathway of MDCK cells. Traffic 1: 494–503 [DOI] [PubMed] [Google Scholar]
- Hwang KJ, Mahmoodian F, Ferretti JA, Korn ED, Gruschus JM (2007) Intramolecular interaction in the tail of Acanthamoeba myosin IC between the SH3 domain and a putative pleckstrin homology domain. Proc Natl Acad Sci USA 104: 784–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi FZ, Grotsch H, Fernandez-Golbano IM, Presciatto-Baschong C, Riezman H, Geli MI (2008) Distinct acto/myosin-I structures associate with endocytic profiles at the plasma membrane. J Cell Biol 180: 1219–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idrissi FZ, Wolf BL, Geli MI (2002) Cofilin, but not profilin, is required for myosin-I-induced actin polymerization and the endocytic uptake in yeast. Mol Biol Cell 13: 4074–4087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Cheng N, Liu X, Korn ED, Steven AC (2004) Subdomain organization of the Acanthamoeba myosin IC tail from cryo-electron microscopy. Proc Natl Acad Sci USA 101: 12189–12194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153: 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsdottir GA, Li R (2004) Dynamics of yeast Myosin I: evidence for a possible role in scission of endocytic vesicles. Curr Biol 14: 1604–1609 [DOI] [PubMed] [Google Scholar]
- Jontes JD, Ostap EM, Pollard TD, Milligan RA (1998) Three-dimensional structure of Acanthamoeba castellanii myosin-IB (MIB) determined by cryoelectron microscopy of decorated actin filaments. J Cell Biol 141: 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Hammer JA III (1990) Generation and characterization of Dictyostelium cells deficient in a myosin I heavy chain isoform. J Cell Biol 110: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA III (2001) The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol 153: 1479–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Osterweil EK, Mooseker MS (2007) Myosin 1E interacts with synaptojanin-1 and dynamin and is involved in endocytosis. FEBS Lett 581: 644–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler E, Schimmoller F, Riezman H (1994) Calcium-independent calmodulin requirement for endocytosis in yeast. EMBO J 13: 5539–5546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyan J, Cowburn D (1997) Modular peptide recognition domains in eukaryotic signaling. Annu Rev Biophys Biomol Struct 26: 259–288 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lechler T, Jonsdottir GA, Klee SK, Pellman D, Li R (2001) A two-tiered mechanism by which Cdc42 controls the localization and activation of an Arp2/3-activating motor complex in yeast. J Cell Biol 155: 261–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechler T, Shevchenko A, Li R (2000) Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol 148: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Bezanilla M, Pollard TD (2000) Fission yeast myosin-I, Myo1p, stimulates actin assembly by Arp2/3 complex and shares functions with WASp. J Cell Biol 151: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WL, Ostap EM, Zot HG, Pollard TD (1999) Organization and ligand binding properties of the tail of Acanthamoeba myosin-IA. Identification of an actin-binding site in the basic (tail homology-1) domain. J Biol Chem 274: 35159–35171 [DOI] [PubMed] [Google Scholar]
- Lieto-Trivedi A, Coluccio LM (2008) Calcium, nucleotide, and actin affect the interaction of mammalian Myo1c with its light chain calmodulin. Biochemistry 47: 10218–10226 [DOI] [PubMed] [Google Scholar]
- Miyata H, Bowers B, Korn ED (1989) Plasma membrane association of Acanthamoeba myosin I. J Cell Biol 109: 1519–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniz M, Morsomme P, Riezman H (2001) Protein sorting upon exit from the endoplasmic reticulum. Cell 104: 313–320 [DOI] [PubMed] [Google Scholar]
- Neuhaus EM, Soldati T (2000) A myosin I is involved in membrane recycling from early endosomes. J Cell Biol 150: 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Peterson MD, Reedy MC, Titus MA (1995) Dictyostelium myosin I double mutants exhibit conditional defects in pinocytosis. J Cell Biol 131: 1205–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak KD, Titus MA (1997) Myosin I overexpression impairs cell migration. J Cell Biol 136: 633–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Iouk TL, Thomas DY, Whiteway M (2004) Functional characterization of myosin I tail regions in Candida albicans. Eukaryotic cell 3: 1272–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y, Botstein D (1994) Structure-based systematic isolation of conditional-lethal mutations in the single yeast calmodulin gene. Genetics 138: 1041–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips KR, Tong S, Goodyear R, Richardson GP, Cyr JL (2006) Stereociliary myosin-1c receptors are sensitive to calcium chelation and absent from cadherin 23 mutant mice. J Neurosci 26: 10777–10788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Doberstein SK, Zot HG (1991) Myosin-I. Annu Rev Physiol 53: 653–681 [DOI] [PubMed] [Google Scholar]
- Raposo G, Cordonnier MN, Tenza D, Menichi B, Durrbach A, Louvard D, Coudrier E (1999) Association of myosin I alpha with endosomes and lysosomes in mammalian cells. Mol Biol Cell 10: 1477–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ho HY, Kirschner MW (2000) Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol 150: 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press [Google Scholar]
- Serrano R (1988) H+-ATPase from plasma membranes of Saccharomyces cerevisiae and Avena sativa roots: purification and reconstitution. Methods Enzymol 157: 533–544 [DOI] [PubMed] [Google Scholar]
- Seth A, Otomo C, Rosen MK (2006) Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLalpha and mDia1. J Cell Biol 174: 701–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman S, Fink G, Lawrence C (1974) Methods in Yeast Genetics. Cols Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sun Y, Carroll S, Kaksonen M, Toshima JY, Drubin DG (2007) PtdIns(4,5)P2 turnover is required for multiple stages during clathrin- and actin-dependent endocytic internalization. J Cell Biol 177: 355–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Martin AC, Drubin DG (2006) Endocytic internalization in budding yeast requires coordinated actin nucleation and myosin motor activity. Dev Cell 11: 33–46 [DOI] [PubMed] [Google Scholar]
- Swanljung-Collins H, Collins JH (1992) Phosphorylation of brush border myosin I by protein kinase C is regulated by Ca(2+)-stimulated binding of myosin I to phosphatidylserine concerted with calmodulin dissociation. J Biol Chem 267: 3445–3454 [PubMed] [Google Scholar]
- Tang N, Lin T, Ostap EM (2002) Dynamics of myo1c (myosin-ibeta) lipid binding and dissociation. J Biol Chem 277: 42763–42768 [DOI] [PubMed] [Google Scholar]
- Temesvari LA, Bush JM, Peterson MD, Novak KD, Titus MA, Cardelli JA (1996) Examination of the endosomal and lysosomal pathways in Dictyostelium discoideum myosin I mutants. J Cell Sci 109(Part 3): 663–673 [DOI] [PubMed] [Google Scholar]
- Terrak M, Rebowski G, Lu RC, Grabarek Z, Dominguez R (2005) Structure of the light chain-binding domain of myosin V. Proc Natl Acad Sci USA 102: 12718–12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titus MA, Wessels D, Spudich JA, Soll D (1993) The unconventional myosin encoded by the myoA gene plays a role in Dictyostelium motility. Mol Biol Cell 4: 233–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong AH, Drees B, Nardelli G, Bader GD, Brannetti B, Castagnoli L, Evangelista M, Ferracuti S, Nelson B, Paoluzi S, Quondam M, Zucconi A, Hogue CW, Fields S, Boone C, Cesareni G (2002) A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295: 321–324 [DOI] [PubMed] [Google Scholar]
- van Tuinen E, Riezman H (1987) Immunolocalization of glyceraldehyde-3-phosphate dehydrogenase, hexokinase, and carboxypeptidase Y in yeast cells at the ultrastructural level. J Histochem Cytochem 35: 327–333 [DOI] [PubMed] [Google Scholar]
- Wessels D, Murray J, Jung G, Hammer JA III, Soll DR (1991) Myosin IB null mutants of Dictyostelium exhibit abnormalities in motility. Cell Motil Cytoskeleton 20: 301–315 [DOI] [PubMed] [Google Scholar]
- Whittaker M, Milligan RA (1997) Conformational changes due to calcium-induced calmodulin dissociation in brush border myosin I-decorated F-actin revealed by cryoelectron microscopy and image analysis. J Mol Biol 269: 548–557 [DOI] [PubMed] [Google Scholar]
- Wu XS, McNeil BD, Xu J, Fan J, Xue L, Melicoff E, Adachi R, Bai L, Wu LG (2009) Ca(2+) and calmodulin initiate all forms of endocytosis during depolarization at a nerve terminal. Nature Neurosci 12: 1003–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita RA, Osherov N, May GS (2000) Localization of wild type and mutant class I myosin proteins in Aspergillus nidulans using GFP-fusion proteins. Cell Motil Cytoskeleton 45: 163–172 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.