Integrating anaerobic/aerobic sensing and the general stress response via the ArcZ small RNA
The alternative bacterial sigma factor RpoS is upregulated in response to various cellular stresses and instigates large-scale changes in gene expression required for adaptation. In this study, Gottesman and colleagues identify the ArcZ small RNA, which is regulated by the ArcA–ArcB two-component system, as a positive regulator of rpoS translation.
Keywords: ArcA, ArcZ, Hfq, RpoS
Abstract
The alternative sigma factor RpoS responds to multiple stresses and activates a large number of genes that allow bacteria to adapt to changing environments. The accumulation of RpoS is regulated at multiple levels, including the regulation of its translation by small regulatory RNAs (sRNAs). A library of plasmids expressing each of 26 Escherichia coli sRNAs that bind Hfq was created to globally and rapidly analyse regulation of an rpoS–lacZ translational fusion. The approach can be easily applied to any gene of interest. When overexpressed, four sRNAs, including OxyS, previously shown to repress rpoS, were observed to repress the expression of the rpoS–lacZ fusion. Along with DsrA and RprA, two previously defined activators of rpoS translation, a third new sRNA activator, ArcZ, was identified. The expression of arcZ is repressed by the aerobic/anaerobic-sensing ArcA–ArcB two-component system under anaerobic conditions and adds translational regulation to the ArcA–ArcB regulon. ArcZ directly represses, and is repressed by, arcB transcription, providing a negative feedback loop that may affect functioning of the ArcA–ArcB regulon.
Introduction
Small regulatory RNAs (sRNAs) have important roles in all kingdoms of life. Close to 100 sRNAs have been identified thus far in Escherichia coli (Sharma and Vogel, 2009). In bacteria, many sRNAs act by base-pairing to specific target mRNAs and changing their translation and/or stability (Gottesman, 2004; Waters and Storz, 2009). Most of these pairing sRNAs need the RNA chaperone Hfq for their action (Brennan and Link, 2007).
RpoS (σS), the master regulator of the general stress response in E. coli, is a sigma subunit of RNA polymerase that is expressed at high levels in various stress conditions, as well as in stationary phase (Hengge-Aronis, 2002). Almost 500 genes are, directly or indirectly, under the control of RpoS, highlighting its importance in controlling the stress response in the cell (Weber et al, 2005). RpoS is unique in helping the cell respond to a wide array of stresses, and levels of RpoS are regulated in response to these stresses (Hengge-Aronis, 2002). Much of the regulation of rpoS expression occurs at the post-transcriptional level through translational regulation of the rpoS mRNA by sRNAs and control of RpoS proteolysis by adaptor/anti-adaptor mechanisms (Repoila et al, 2003; Bougdour et al, 2006, 2008).
The translation of rpoS is severely diminished in an hfq mutant (Brown and Elliott, 1996; Muffler et al, 1996). Mutations that restored translation to an hfq mutant defined an inhibitory stem-loop in the rpoS mRNA leader blocking ribosome binding (Brown and Elliott, 1997). Two Hfq-binding sRNAs, DsrA and RprA, were subsequently observed to positively regulate translation of rpoS by base-pairing to the upstream part of this translation inhibitory stem loop, freeing the ribosome-binding site (RBS; Sledjeski et al, 1996; Majdalani et al, 1998, 2001; Soper and Woodson, 2008; Updegrove et al, 2008). In addition, OxyS, an sRNA expressed under oxidative stress, was shown to repress rpoS expression by a mechanism not fully understood, but requiring binding of the sRNA to Hfq (Altuvia et al, 1997; Zhang et al, 1998).
We have been developing experimental approaches for rapidly screening targets of interest for regulation by sRNAs (Mandin and Gottesman, 2009). In this study, we take advantage of the close-to-saturation identification of Hfq-binding sRNAs in E. coli to ask globally regarding the effects of these sRNAs on rpoS, both as an mRNA already known to be regulated by three sRNAs, and as a target gene of interest, given its central role in developmental shifts and stress responses in E. coli and related bacteria. Our results identify several sRNAs that negatively regulate rpoS and a third positive regulator of rpoS translation, ArcZ. The ArcZ sRNA has been previously identified in at least two genomic searches for sRNAs in E. coli (originally named RyhA and SraH; Argaman et al, 2001; Wassarman et al, 2001), and was recently renamed ArcZ because it is encoded convergently with and overlapping arcB (see below); it was demonstrated to have broad effects on gene expression in Salmonella enterica (Papenfort et al, 2009).
Results
Construction of an overexpression library dedicated to the Hfq-binding sRNAs of E. coli
sRNAs that bind Hfq are very likely to pair with and regulate mRNA stability and translation (for review, see Gottesman et al (2006)). As immunoprecipitation with Hfq, coupled with microarrays or deep sequencing, allows sensitive detection of this class of sRNAs (Zhang et al, 2003; Sittka et al, 2008), we may be close to identifying all of the Hfq-binding sRNAs in E. coli. A total of 30 Hfq-binding sRNAs have been identified thus far in E. coli (Zhang et al, 2003).
To focus directly on the Hfq-binding sRNAs and how they regulate selected target mRNAs, a set of 26 Hfq-binding sRNAs were cloned into a pBR-plac plasmid (Guillier and Gottesman, 2006) so that they will be expressed from their native 5′-end under the control of the Plac promoter (see Table I and Materials and methods section). Those not included in the set had not had their 5′-ends mapped at the time this study began. After induction, each sRNA was overexpressed more than 10-fold as compared with the chromosomal copy (Supplementary Figure S1). The detected sizes corresponded to the predicted or previously observed sizes of the respective sRNAs.
Table 1. Hfq-binding sRNAs in the plasmid librarya.
| Name | Flanking genes | Orientations | Size (nt) | Reference |
|---|---|---|---|---|
| SgrSb (RyaA) | sgrR/setA | < >> > | ∼220 | Vanderpool and Gottesman (2004) |
| ChiXb (MicM/RybC/SroB) | ybaK/ybaP | < >> < | 88 | Mandin and Gottesman (2009) |
| RybBc | ybjK/ybjL | > << < | 80 | Thompson et al (2007); Coornaert et al (2010) |
| FnrS (RydD) | ydaN/dbpA | > >> > | 122 | Durand and Storz (2010) |
| MicCc (ISO63) | ompN/ydbK | < >> < | 109 | Chen et al (2004); Coornaert et al (2010) |
| RydC | cybB/ydcA | > << > | 61 | Antal et al (2005) |
| MgrRb | yneM/ydeH | > << < | 98 | Moon and Gottesman (2009) |
| RprA | ydiK/ydiL | > >> > | 105 | Majdalani et al (2001) |
| RyeB | pphA/yebY | < << < | 104, 74 | Vogel et al (2003) |
| CyaRb (RyeE) | yegQ/orgK | > > < | 86 | De Lay and Gottesman (2009) |
| MicF | ompC/yojN | < >> > | 93 | Mizuno et al (1984) |
| GlmY (tke1, SroF) | yfhK/purL | < << < | 150, 180 | Urban and Vogel (2008) |
| MicAc (SraD) | luxS(ygaG)/gshA | < >> < | ∼70 | Udekwu et al (2005); Coornaert et al (2010) |
| GcvB | gcvA/ygdI | < >> < | 205 | Urbanowski et al (2000) |
| OmrAb (rygA/sraE) | aas/galR | < << > | 88 | Guillier and Gottesman (2006) |
| OmrBb (rygB) | aas/galR | < << > | 82 | Guillier and Gottesman (2006) |
| ArcZ (RyhA/SraH) | elbB/arcB | < >> < | ∼55, 88, 120 | Papenfort et al (2009) |
| RyhB (SraI) | yhhX/yhhY | < << > | 90 | Massé and Gottesman (2002) |
| GadY (IS183) | gadW/gadX | < >> < | 105, 90, 59 | Opdyke et al (2004) |
| GlmZ (RyiA/SraJ) | aslA/hemY | < >> < | 210 | Urban and Vogel (2008) |
| OxyS | argH/oxyR | > << > | 109 | Altuvia et al (1997) |
| DicF | rzpQ/dicB | > >> > | 53 | Bouché and Bouché (1989) |
| DsrA | dsrB/yedP | > << > | 85 | Sledjeski et al (1996) |
| Spot42 (spf) | polA/yihA | > >> < | 109 | Møller et al (2002) |
| RseX | yedR/yedS | < >> > | 91 | Douchin et al (2006) |
| IS118 | yfdI/tfaS | > << > | 194 | Zhang et al (2003); K Moon, personal communication |
| asRNAs listed here were cloned into the pBRplac plasmid (Guillier and Gottesman, 2006) from their natural transcriptional start site. | ||||
| bPlasmids that were previously available in the laboratory (see reference). | ||||
| cKind gift of M Guillier (Coornaert et al, 2010). All other plasmids were cloned in this study (see Materials and methods section). Orientation of the sRNA relative to its neighbouring genes on the chromosome is indicated in bold. | ||||
Use of the sRNA library reveals multiple sRNAs controlling the expression of the rpoS–lacZ fusion
To study translational regulation of rpoS, we constructed a strain, PM1409, containing an rpoS–lacZ translational fusion under the control of the arabinose-inducible PBAD promoter, by recombineering in PM1205, a strain designed to simplify construction of translational fusions, as described previously (Mandin and Gottesman, 2009). The 5′-end of the rpoS–lacZ fusion transcript corresponds to the rpoS major transcriptional start site (Lange et al, 1995), and retains the long 5′-UTR necessary for post-transcriptional regulation by Hfq (Brown and Elliott, 1996; Soper and Woodson, 2008; Updegrove et al, 2008). The coding sequence of rpoS was fused at its tenth amino acid to lacZ; thus, the translational fusion lacks the RpoS region required for RpoS degradation by ClpX/P (Studemann et al, 2003). In summary, effects of the overexpression of the sRNAs on the rpoS–lacZ fusion should only reflect changes in translation or mRNA stability of rpoS, as other sequences for regulation have been deleted; however, sRNA effects that require sequences beyond the tenth codon will not be detected.
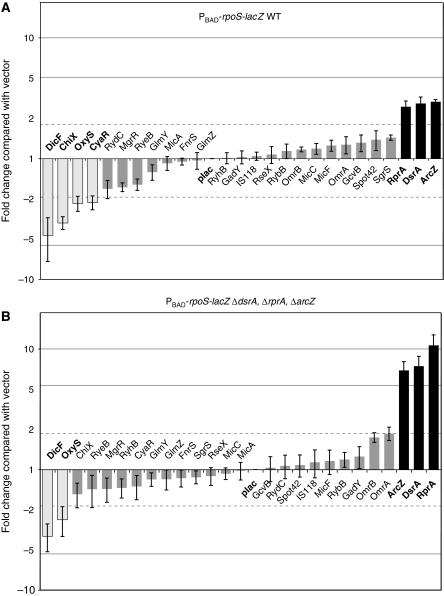
Each of the sRNA-expressing plasmids and a vector control were used to transform the strain carrying the PBAD–rpoS–lacZ fusion and assayed as described in the Materials and methods section (Figure 1). The majority of the plasmids (18/26) had less than a two-fold effect on rpoS–lac expression; thus, not every Hfq-binding sRNA affects rpoS expression, even when overexpressed. As expected, DsrA and RprA were observed to activate the rpoS–lacZ fusion by 2.6-fold each, and OxyS repressed by 2.4-fold, thus confirming the validity of our technique. However, in addition to the known rpoS regulators listed above, one additional sRNA, ArcZ, upregulated and three sRNAs, CyaR, ChiX, and DicF, downregulated the expression of the rpoS–lacZ fusion (Figure 1A). To test whether these latter sRNAs acted indirectly, through the positively acting sRNAs, a strain was constructed in which the genes encoding DsrA, RprA and ArcZ were deleted from the chromosome, and the full set of plasmids were again introduced into the strain and assayed (Figure 1B). The basal level in this case was significantly lower (≈30 specific units), so a higher concentration of arabinose was used, resulting in a basal level of 78 specific units. In this strain, the positive effects of ArcZ, DsrA, and RprA were even stronger (>5 × stimulation), both confirming the original results and demonstrating that none of these three sRNAs acts indirectly through the other two. The mode of action of ArcZ is investigated further below.
Figure 1.
Use of a dedicated library of sRNAs to study regulation of an rpoS–lacZ translational fusion. (A) Screening of the sRNA library on the PBAD–rpoS–lacZ fusion (PM1409). The effect of the overexpression of each sRNA on the rpoS–lacZ fusion was plotted as a function of the fold change compared with the basal activity of PM1409 containing a pBR-plac control vector. Fold changes greater than two were considered significant. Grey bars represent sRNAs for which effects were not considered significant; black and light grey bars indicate sRNAs having an activating or a repressing effect, respectively. (B) sRNA overexpression effect on the PBAD–rpoS–lacZ fusion independent of the known positively acting sRNAs. As above, but with PM1417, a dsrA rprA arcZ triple deletion mutant derivative of PM1409.
Of the four negatively regulating sRNAs, only two (DicF and OxyS, both of which show toxicity when overproduced, data not shown) still had a significant effect in the triple deletion strain, suggesting that CyaR and ChiX may act indirectly, via effects on expression and/or stability of the chromosomally encoded positively acting sRNAs. We observed no evidence for direct pairing of any of these four negatively acting sRNAs with the rpoS leader; a predicted pairing of ChiX with rpoS was not supported by the behaviour of mutations in the potential pairing site (data not shown). In additional experiments, CyaR and ChiX were able to repress the rpoS–lacZ fusion when only one of the activating sRNAs was deleted from the chromosome (Supplementary Figure S2). The results suggest that CyaR and ChiX, when overexpressed, can downregulate rpoS expression indirectly by counteracting the activation by DsrA, RprA or ArcZ. Possibly CyaR and ChiX, which are abundant in the cell under these conditions, bind Hfq and keep it from binding the positively regulating sRNAs or rpoS mRNA.
In a control experiment, the same plasmid library was introduced into a strain carrying a PBAD–lacZ fusion; the effects of the plasmids on the fusion were generally modest (Supplementary Figure S3A); normalizing the results in Figure 1 to those in Supplementary Figure S3A did not change the identification of plasmids with changes of >2-fold (Supplementary Figure S3B).
ArcZ base-pairs directly to and activates translation by opening the stem loop structure in the rpoS mRNA 5′-UTR
If ArcZ directly regulates rpoS translation, we would expect to see base-pairing between ArcZ and the rpoS mRNA. Using the mfold program, we were able to predict base-pairing between the two RNAs (Figure 2B). The sequence of the rpoS mRNA predicted to base-pair with ArcZ (from nt 446 to 472 in the rpoS mRNA 5′-UTR) is the same region in which base-pairing with DsrA and RprA occurs (Figure 2B; Majdalani et al, 1998, 2002). This region is in the upper stem of the rpoS mRNA 5′-UTR stem-loop structure, thus suggesting that ArcZ acts by a similar mechanism as DsrA and RprA to open this stem.
Figure 2.
ArcZ direct pairing with the rpoS leader. (A) The rpoS mRNA hairpin, adapted from Soper and Woodson (2008), is shown. The region of the rpoS leader involved in base-pairing with the sRNAs is shaded in grey in this panel and (B). Nucleotides are numbered from the +1 of the rpoS mRNA. The AUG translation start codon is boxed, as are the nucleotides mutated in the experiments in Figure 2C. RBS, ribosome binding site. (B) Predicted base-pairings of each of three sRNAs with the shaded portion of the rpoS leader are shown. Mutated nucleotides are boxed and the changes made in them are shown; positions are numbered according to the transcriptional start site. The sequence in the striped box in ArcZ is not present in the short processed form of the RNA (see text). (C) Effect of the wild-type sRNAs or the C-to-G mutations in each of the sRNAs were tested on the wild-type PBAD–rpoS–lacZ fusion (PM1409), rpoS(G463C)–lacZ fusion (PM1433), rpoS(C561G)–lacZ fusion (PM1438), and rpoS(G463C-C561G)–lacZ fusion (PM1439). Note the difference in the y-axis in the left- and right-hand panels.
ArcZ base-pairing to rpoS mRNA was tested by introducing mutations that are expected to disrupt or decrease pairing, as well as compensating mutations; these are shown in Figure 2A and B. We focussed on G463 of the rpoS leader, a position that both pairs with the bottom part of the stem at position C561, and is predicted to pair with a C in each of the sRNAs (Figure 2B). The effect of expressing wild-type or mutant forms of DsrA, RprA, or ArcZ was tested on the wild-type rpoS fusion, a fusion with a C561G or a G463C mutation (each mutation abolishing the inhibitory G:C base-pair), and a fusion with G463C and C561G, restoring base-pairing within the stem (Figure 2A).
The rpoS-G463C mutation, disrupting the inhibitory stem, increases the basal activity of the fusion, and neither wild-type nor mutant forms of the sRNAs stimulated expression any further (Figure 2C). Opening the stem-loop by introducing the C561G mutation on the lower half of the hairpin also gave a fusion that was not stimulated by wild-type or mutant sRNAs, although the basal level of expression was much decreased (see different y-axis for panels of Figure 2C). The C561G mutation is close to the rpoS RBS, and apparently changes overall translation of the message. However, these results clearly demonstrate that disruption of this single base-pair in the rpoS hairpin abolishes translational repression and the need for sRNA activation of translation. Restoring base-pairing by combining these two mutations lowers the basal level significantly (Figure 2C, G463C-C561G results).
For each of the sRNAs, introduction of a C-to-G point mutation that disrupts base-pairing to G463 was sufficient to inhibit their positive effect on the wild-type rpoS–lacZ activity (Figure 2C, left-most panel). However, as predicted, these mutant sRNAs are able to activate the G463C:C561G fusion (Figure 2C, right-most panel). Notably, however, although wild-type RprA and ArcZ were unable to activate this mutant fusion, wild-type DsrA could, indicating that it may be less dependent on pairing at this position. Overall, these data confirm that ArcZ pairs with rpoS in this region, and demonstrates that opening up the hairpin abrogates the role of all three stimulatory sRNAs.
ArcZ expression from the chromosome contributes to rpoS translation
If the activation by ArcZ is physiologically relevant, we would expect a deletion to impinge on RpoS synthesis. The contribution of each of the three sRNAs was tested by measuring expression of the PBAD–rpoS–lacZ fusion in LB (Figure 3A) or minimal medium (Figure 3B) at 37°C. Deletion of arcZ had a modest, although consistent, effect on the expression of the rpoS–lacZ fusion in LB. Deletion of rprA had no effect on the fusion, expected as RprA is not expressed under these growth conditions (data not shown). Deletion of dsrA had the largest effect on the rpoS–lacZ fusion. Combining the arcZ deletion with either a dsrA or an rprA mutant had additive effects, further confirming that each sRNA contributes independently to post-transcriptional regulation of rpoS expression. However, we note that effects are not strictly additive, possibly suggesting that each sRNA may affect expression or stability of the others.
Figure 3.
Role of DsrA, RprA, and ArcZ in expression of rpoS. (A) Isogenic PBAD–rpoS–lacZ strains deleted either for dsrA (PM1411), rprA (PM1412), arcZ (PM1413), or carrying combinations of double or triple mutations in each of the three sRNAs (see Supplementary Table S1) were grown at 37°C in LB containing 0.002% arabinose to stationary phase. Samples were taken and β-galactosidase activity was measured as described previously (Majdalani et al, 1998). An hfq mutant (PM1419) was also tested. The wild-type strain (PM1409) is shown in black; all mutants are dark grey. (B) A subset of the strains in (A), carrying either single mutations or the triple mutation, were grown in minimal medium containing 0.2% arabinose and assayed for rpoS expression.
In minimal medium (Figure 3B), ArcZ provided the largest contribution to rpoS expression; deleting arcZ reduced the expression by 40%, whereas a dsrA mutation reduced it by 30%. As discussed below, regulatory signals for arcZ suggest that it is best expressed under aerobic growth conditions; however, we find a contribution of ArcZ to the expression of RpoS even under microaerobic growth conditions (see below, Figure 5). Thus, ArcZ has a role in RpoS translation under a variety of growth conditions.
Figure 5.
Effect of ArcA on expression of ArcZ and RpoS. (A) Expression of the ParcZ–lacZ transcriptional fusion as a function of oxygen. Wild-type cells containing the ParcZ(-100)–lacZ fusion (PM1450) and isogenic derivatives deleted for arcA (PM1453) or arcB (PM1456) were grown in minimal medium overnight in presence (dark bars) or absence (light bars) of oxygen before samples were removed and assayed for β-galactosidase activity. (B) Northern blot showing expression of ArcZ as a function of oxygen. Strain NM525 and its arcA∷kan derivative PM1493 were grown overnight at 37°C in minimal medium before samples were removed and the RNA extracted. Northern blots were subsequently performed and probed with the ArcZ-NB4 probe (see Supplementary Table S2). (C) Expression of the PBAD–rpoS–lacZ fusion as a function of arcZ and arcA. Left panel: Experiments are as in (A), but using cells carrying the rpoS–lacZ translational fusion (PM1409) and its arcA (PM1480), arcZ (PM1413), and arcA, arcZ (PM1481) mutant derivatives grown in minimal medium containing 0.2% arabinose. As in Figures 1, 2 and 3, the promoter of this fusion is PBAD and the fusion protein is not subject to RpoS-specific degradation. Right panel: Assay of a control PBAD–lacZ fusion (lacZ under control of the lacZ RBS) (PM1051) under anaerobic and aerobic conditions as for the left panel. (D) Strain SG30013, carrying an rpoS–lacZ translational fusion containing the promoter region of rpoS and enough of the rpoS gene to be subject to RssB-dependent degradation, and its arcA∷kan (PM1620), arcZ∷tet (PM1621), and arcA∷kan, arcZ∷tet double mutant (PM1622) derivatives were grown in LB at 37°C until the indicated OD. Cells were then lysed and β-galactosidase activity was measured as described in the Materials and methods section.
The basal activity of the PBAD–rpoS–lacZ fusion in a triple dsrA, rprA, and arcZ mutant background was higher than that of an hfq deletion mutant, in which all known Hfq-dependent sRNA activity is thought to be impaired (Figure 3A and B). There may be a role for Hfq in translation of RpoS even in the absence of sRNAs, possibly by affecting rpoS mRNA, or another Hfq-dependent sRNA, not present in the library, is able to activate rpoS translation. Consistent with this, an RNA region between the pstA and pstB genes was observed to activate rpoS expression, possibly through a mechanism similar to DsrA and RprA (Ruiz and Silhavy, 2003; Schurdell et al, 2007).
ArcZ is a conserved sRNA observed in three forms
The arcZ gene is encoded in an intergenic region between elbB and arcB (Figure 4A). The sRNA gene is encoded next to and convergent with arcB, encoding a histidine kinase of a two-component system involved in regulation of aerobic-to-anaerobic growth transition (see Figure 4A). On the basis of that linkage, the sRNA was recently renamed ArcZ for ArcB-associated RNA in a study of the homologous sRNA in S. enterica (Papenfort et al, 2009). The stop codon of the arcB open reading frame is located inside the arcZ gene; mapping of the 3′-ends of the arcB mRNA (arrows in Figure 4A) are consistent with an overlap between the two transcripts of >25 nt.
Figure 4.
ArcZ sRNA. (A) Alignment of arcZ in various enterobacterial strains. Flanking genes are shown; the start codon for elbB and stop codon for arcB are boxed. Sequence identities are shaded in grey. The transcription start site (+1) and the deduced arcZ promoter elements are labelled. A putative ArcA binding site overlapping the −35 element is indicated and residues close to the ArcA box consensus, as defined by McGuire et al (1999), are shown with stars. The boxed sequence in the sRNA is the sequence predicted to be involved in pairing with the rpoS leader. The * and ** show minor and major sRNA cleavage sites, respectively; cleavage is between A and T for the major site of processing. The 5′ and 3′ probes used for the northern blot in Figure 4B detect the underlined sequences. Arrows indicate the position of 3′ RACE clones for arcB mRNA, and the number of isolates for each position. The 3′-end was determined in a mutant not expressing ArcZ (PM1520); no clones could be obtained from the wild-type cells. (B) Northern blot experiments showing the pattern of expression of ArcZ sRNA. An overnight culture of the WT strain MG1655 was diluted 500-fold in LB and grown at 37°C; samples were collected at the indicated OD600. RNA was extracted and northern blots were performed with either a probe directed against the 5′-end (ArcZNB1, left panel) or the 3′-end (ArcZ NB4, right panel) of the sRNA. (C) β-galactosidase activity of the ParcZ (-100)–lacZ fusion as a function of growth. Overnight cultures of PM1450, containing a ParcZ(-100)–lacZ fusion (indicated by black diamonds) and PM1451, containing the same promoter fusion with a TG-to-CC mutation at position −14/15 of the promoter (empty squares) were diluted 500-fold in LB medium at 37°C. The t0 sample was taken after 2 h of growth; then samples were taken at regular intervals and β-galactosidase activity determined (solid lines, y-axis on the left) and OD600nm measured (dashed lines, y-axis on the right).
arcZ is well conserved in its 3′-half, from nt 64 to 120, but is more divergent in its 5′-end (Figure 4A). The sequence of ArcZ predicted to be involved in base-pairing with rpoS is located in the highly conserved 3′-end of the sRNA (boxed sequence in Figure 4A). A recent study observed that this region of the sRNA was involved in base-pairing with at least three other targets (Papenfort et al, 2009).
Another study has shown that ArcZ is processed to a shorter form (Argaman et al, 2001; Papenfort et al, 2009). We confirmed this by northern blot with probes directed against either the 5′- or the 3′-half of the ArcZ RNA (see Figure 4A). The full-length form was 120 nt (labelled FL); two shorter forms were detected, a low-abundance 88 nt form (labelled *), and a 55 nt form, detected only with a probe directed against the 3′-end (labelled **; Figure 4B). All forms of ArcZ peaked in stationary phase. The 5′-end of this product as determined by 5′ RACE is shown in Figure 4A (**) and is consistent with previous findings (Argaman et al, 2001).
Given its accumulation, this processed product may be the active form of ArcZ. One prediction of this would be that only nucleotides present in the processed form would participate in pairing for regulation. The predicted pairing of ArcZ with rpoS extends a few nucleotides 5′ to the processing site, and RprA and DsrA pair with rpoS in this region as well, allowing us to test this model (Supplementary Figure S4B). Mutation of rpoSU468 to C, changing a G:U base-pair in the rpoS stem to a G:C base-pair, greatly decreases the basal level of expression of the fusion, is insensitive to wild-type DsrA or RprA, but is stimulated by DsrA or RprA carrying a compensating mutation (Supplementary Figure S4C, right panel; (Majdalani et al, 1998, 2002)). Although a mutation in DsrA reduces expression of the wild-type fusion, a mutation in arcZ in the comparable position has no effect on regulation of the wild-type rpoS∷lacZ fusion (Supplementary Figure S4C, left side). The ArcZ mutant is also unable to activate the mutant form of rpoS∷lacZ (Supplementary Figure S4C, right side). Therefore, positions outside the processed form do not affect regulation. These results are consistent with our recent in vivo and in vitro observations showing that the processed form of ArcZ is sufficient for regulation and that the processed form but not the full-length ArcZ can anneal to rpoS in the presence of Hfq (Soper et al, 2010).
A multicopy library screen identifies ArcA/ArcB as negatively regulating arcZ expression
Alignment of the promoter region of arcZ shows a well-conserved extended −10 (TGCTATCTT) and −35 (TGTAAC) elements, both of which are close to consensus, separated by 17 nt. To search for signals governing ArcZ expression, we constructed a lacZ transcriptional fusion with the arcZ promoter, starting from −100 nt upstream of the transcriptional start site and extending to +1 nt inside arcZ (PM1450). Promoter activity could be detected as soon as mid-log phase and peaked in stationary phase during growth in LB at 37°C, in line with the northern blot data shown above (Figure 4B and C). Mutation at position −14/−15 of the promoter should eliminate the extended −10 TG motif (Figure 4A). Such a mutation reduced the activity of the transcriptional fusion modestly (Figure 4C); therefore, the extended TG is not required for arcZ expression, consistent with the presence of a good −35 region. Given the increase in expression in stationary phase, a possible role for RpoS in regulating arcZ was considered. However, introduction of an rpoS∷tet mutation had no effect on the expression of either the fusion or ArcZ sRNA (data not shown).
A multicopy genomic library was introduced into cells containing the fusion and screened on MacConkey lactose ampicillin plates to search for genes that, when overexpressed, would activate or repress the ParcZ promoter. Eight validated plasmids were identified out of 10 000 screened colonies, four of which activated and four of which repressed. The genes present in each plasmid were determined by sequencing (Table II). Only single examples of each region were observed, strongly suggesting that we have not saturated the search for regions impinging on arcZ regulation.
Table 2. Clones identified by screening the ParcZ–lacZ fusion.
| Plasmid name | Insert boundaries | Genes contained in the insert | Phenotype in the WT fusiona | Phenotype in the arcA∷kan fusiona | Phenotype in the arcB∷kan fusiona |
|---|---|---|---|---|---|
| pHDB3 | NA | NA | + | +++ | ++ |
| 4.0.1 | 2509299–2510946 | ‘ypeC (>), mntH (<) | − | +++ | ++ |
| 4.0.2 | 3349955–3351692 | ‘arcB (<), yhcC' (<) | − | +++ | ++ |
| 4.3.2 | 772534–775105 | ‘cydB (>), ybgT (>), ybgE (>), ybgC (>), tolQ (>) | − | +++ | ++ |
| 4.10.1 | 4451944–4455510 | ‘yjfF (>), fbp (<), mpl (>), yjgA' (<) | − | +++ | ++ |
| 4.3.1 | 4475389–4477428 | ‘argI (<), rraB (>), yjgM' (<) | +++ | +++ | ++++ |
| 4.2.2 | 888192–891290 | ‘ybjL (<), ybjM (>), grxA (<), ybjC (>), nfsA (>), rimK'(>) | +++ | +++ | ++ |
| 3.10.1 | 4354828–4358893 | ‘cadAb (<), cadBb (<) | +++ | +++ | +++ |
| 4.2.1 | 2403330–2407115 | ‘nuoAb (<), lrhA(<), ybfQ (>) | +++ | +++ | +++ |
| a− and + signs indicate Lac phenotype of the ParcZ-lacZ fusion as seen on MacConkey plates in the WT strain (PM1450), or in the isogenic arcA∷kan (PM1453) and arcB∷kan (PM1456) derivatives. | |||||
| bGenes previously shown to be directly regulated by ArcA. | |||||
Interestingly, one of the negatively regulating plasmids, p4.0.2, contained a portion of the arcB gene, encoding the histidine kinase of the ArcA/ArcB two-component system. The arcB fragment in plasmid p4.0.2 contains less than half of the ORF (372 out of 778 amino acids), and is unlikely to be functional, but may be capable of interacting with chromosomally encoded wild-type ArcB. Moreover, two of the positively regulating plasmids, p3.10.1 and p4.2.1, contain genes that have been shown to be directly regulated by ArcA binding to their promoters (cadBA, encoding subunits of the lysine decarboxylase and nuoA, encoding the subunit 1 of the ubiquinone oxydoreductase; Bongaerts et al, 1995; Reams et al, 1997).
This combination of genes on the plasmids suggested the possibility that the ArcB/ArcA two-component system might negatively regulate ParcZ. If so, plasmids that increased the activity of the ArcB sensor kinase or directly activated ArcA would be expected to downregulate the ParcZ fusion, whereas plasmids that led to less ArcA activity or titrated ArcA from ParcZ would be expected to upregulate the fusion. Indeed, deletion of arcA increased the expression of the fusion in the presence of a vector (Table II, first line), abolished the negative effect of overexpression of arcB in plasmid p4.0.2, and abolished repression by other negatively regulating plasmids (Table II). Deletion of arcB also suppressed the repressing effect of these plasmids. Therefore, the four repressing plasmids seem to activate ArcB signalling to ArcA, resulting in increased repression of arcZ.
Of the four positively regulating plasmids, the two containing known ArcA-binding sites (p3.10.1 and p4.2.1) are unchanged in an arcA or arcB mutant (Table II), consistent with titration of ArcA, making the strain phenotypically ArcA−. Of the other two, plasmid 4.3.1 activates more when ArcB is absent, suggesting that it acts downstream of ArcB; plasmid 4.2.2 shows less activity when ArcB is absent, consistent with it acting to make ArcB a more active phosphatase (rather than kinase). These plasmids have not been further studied.
Taken together, these data strongly suggested that arcZ expression was repressed, directly or indirectly, by the ArcA/ArcB two-component system, and that the plasmids that increase arcZ–lacZ expression are titrating ArcA from the arcZ promoter. The failure to find any plasmids that are not dependent on ArcA for their action supports ArcA as a possible direct regulator.
The ArcA/ArcB two-component system represses ArcZ expression under anaerobic growth
The ArcA/ArcB two-component system is involved in the regulation of various genes implicated in respiratory or fermentative metabolism (for review, see Gunsalus and Park (1994)). The ArcB sensor kinase autophosphorylates under anaerobic conditions, in turn phosphorylating the ArcA response regulator, that is then able to repress or activate various genes. Oxidized quinones act as a signal to inhibit ArcB phosphorylation (Georgellis et al, 2001; Malpica et al, 2004). Consistent with expectations for an ArcA-repressed sRNA, arcZ expression was severely diminished when cells were grown in minimal medium under anaerobic growth conditions, whether measured by activity of the ParcZ fusion or by northern blot (Figure 5A and B). Deleting arcA from the chromosome had a modest activating effect on arcZ expression during aerobic growth but completely abolished anaerobic repression (Figure 5A and B). A previous study did not report any effect of a mutant in arcB on ArcZ levels during oxygenated growth (Papenfort et al, 2009). Consistent with their results, we also observed that deleting arcB from the chromosome had practically no effect on expression from the ParcZ–lacZ fusion under aerobic conditions (Figure 5A). However, this mutation partially abolished repression under anaerobic growth. These mild phenotypes are predicted in mutants of histidine kinase of two-component systems, as the response regulator may by activated by small molecules or other histidine kinases. The results clearly demonstrate that ArcA represses expression of arcZ during anaerobic growth, the expected condition for ArcA to be active.
These results do not distinguish direct repression by ArcA from an indirect effect. A possible ArcA-binding site overlaps the −35 region of the arcZ promoter (McGuire et al, 1999; Figure 4A), but mutational changes in this region did not abrogate ArcA repression (Supplementary Figure S5). However, the following observations support a direct effect of ArcA: (1) ArcA-dependent repression is seen with a fusion containing only 43 nt upstream of the +1, making it unlikely there is an upstream positive regulator, repressed by ArcA (data not shown); (2) Mutations in the spacer have little effect on regulation, inconsistent with a regulator interacting with a site there (Supplementary Figure S5); (3) one mutation in the −35 region, consistent with increasing similarity to an ArcA consensus, increased repression (Supplementary Figure S5).
It has been reported that ArcA/ArcB represses rpoS expression both at the transcriptional level, by binding to the rpoS promoter, and at the post-translational level, by phosphorylating RssB, increasing RpoS degradation by ClpX/P (Mika and Hengge, 2005). As we show here, ArcZ activates rpoS translation, and ArcZ is repressed by ArcA. Thus, our data suggest that ArcA should also repress rpoS translation through ArcZ.
We first tested whether the deletion mutants of arcZ and/or arcA had an effect on the PBAD–rpoS–lacZ fusion, which should reflect only translational regulation of rpoS, in cells grown in minimal medium with or without oxygen (Figure 5C). As expected, deletion of arcA had no effect on the activity of the PBAD–rpoS–lacZ fusion under aerobic growth conditions, whereas deletion of arcZ reduced expression of the fusion. A double arcA arcZ mutant had the expression level of an arcZ mutant, as expected. Under anaerobic conditions, the overall level of expression of the fusion was lower, partially due to a two-fold decrease in the activity of the PBAD promoter under anaerobic conditions (Figure 5C, right panel). However, the effects of ArcA and ArcZ were as expected. Deletion of arcA increased rpoS–lacZ activity, deletion of arcZ decreased activity, and the double arcA arcZ mutation was similar to arcZ alone. Therefore, there is significant ArcA-dependent repression of RpoS, at the level of translation, and this repression is due to ArcZ.
To assess the contribution of ArcZ to RpoS accumulation in the natural context (transcriptional and proteolytic degradation pathways intact), two experiments were done. In one, the expression of an rpoS–lacZ translational fusion under the control of its native promoter, and subject to regulated degradation, was measured in the presence and absence of arcA and arcZ at early log, mid-log, and late stages of growth (Figure 5D); in the second test, the level of RpoS protein was determined by western blot (Supplementary Figure S6). The results generally agree.
Mika and Hengge (2005) had observed an increase in RpoS in an arcA mutant early in exponential phase during aerobic growth, when RpoS levels are low, and our results confirm that observation (Figure 5D and Supplementary Figure S6). The effects of deleting arcA are modest, suggesting that, in spite of the multiple levels of regulation, RpoS levels are robustly controlled by other mechanisms even in the absence of ArcA. An arcA mutant led to a modest (2–3-fold) increase in RpoS in early exponential growth under aerobic conditions; this increase was abolished in an arcA arcZ double mutant (Figure 5D). The differences between the effect of an arcA mutant in Figure 5C and in Figure 5D or Supplementary Figure S6 presumably reflect effects of ArcA at the level of rpoS transcription and degradation of the RpoS protein (Mika and Hengge, 2005).
In aerobic stationary phase growth, an arcA mutation or an arcZ mutation led to modestly lower levels of RpoS (less than a two-fold effect), with a stronger effect for the arcZ mutation (Figure 5D and Supplementary Figure S6). An effect of ArcA in contributing to RpoS accumulation in stationary phase has been reported previously (Mika and Hengge, 2005).
In anaerobic growth conditions, RpoS was not detectable in exponential phase, even in the absence of ArcA (Supplementary Figure S6), and the fusion was too low to measure. In stationary phase, arcA mutants led to a modest increase in RpoS accumulation (almost two-fold) and this increase was abolished in the arcA arcZ double mutant, consistent with the effects seen with the fusion (Supplementary Figure S6).
These results confirm that the ArcA/ArcB two-component system negatively regulates RpoS, not only through transcription repression and proteolysis (Mika and Hengge, 2005), but also at the level of rpoS translation through repression of arcZ.
An autoregulatory loop: effects of ArcZ on ArcB and vice versa
As we observed that ArcA/ArcB represses arcZ at the level of transcription (Figure 5) and that arcZ is encoded next to and overlapping the arcB gene (Figure 4A), we wondered whether ArcZ itself could regulate arcB expression. However, we were not able to detect the arcB message by northern blot analysis in any of the conditions tested, even in anaerobic conditions (data not shown).
To be able to detect and control the arcB message, the arcB promoter was replaced by a PBAD promoter linked to a chloramphenicol cassette (Figure 6A; Morita et al, 2004). This promoter replacement was first introduced into the ParcZ–lacZ fusion strain. Induction of the expression of arcB from the PBAD promoter with 0.2% arabinose repressed the arcZ fusion in an arcA-dependent manner on MacConkey lactose plates, suggesting that increased ArcB activates ArcA repression (data not shown). Furthermore, arcB mRNA could be detected by northern blot analysis after 15 min of induction, but not without induction (Figure 6B, compare lane 4 to lane 2). To look at effects of ArcZ on arcB expression without affecting the sequence complementarity between the two genes, we constructed an arcZ promoter mutant by replacing the ParcZ −10 element (TATCTT) with a run of six Cs. This promoter mutation, previously described to shut down gene expression (Opdyke et al, 2004), completely abolished ArcZ expression (Figure 6B, lanes 6–10 and lanes 16–20; Figure 6C, lanes 13–18). Expression (Figure 6B) and stability (Figure 6C) of arcB mRNA and ArcZ RNA were compared as a function of expression of the other.
Figure 6.
An ArcZ and ArcA/ArcB regulatory loop. (A) Diagram of overlap of the arcB and arcZ transcripts, and expression of arcB under the control of a PBAD promoter. On the right side, mutations in the arcZ promoter inactivate its expression without disrupting the overlap region. (B) Four isogenic strains were examined for the expression of arcB mRNA and expression of ArcZ. Lanes 1–5: PM1560, in which the arcB promoter has been replaced by an arabinose-inducible PBAD promoter; lanes 6–10: PM1561, a derivative of PM1560 in which the −10 element of the arcZ promoter was inactivated; lanes 11–15, PM1562, an arcA∷kan derivative of PM1560; lanes 16–20, PM1563, an arcA∷kan derivative of PM1561. All strains were grown in LB to OD600=1 and samples were collected (Time 0); cultures were split and 0.2% arabinose was added to one of two parallel cultures. Samples were collected after 15 and 60 min of growth, RNA extracted, and northern blot analyses performed using probes against arcB (upper panels) and ArcZ (second row). Northern blot signals were quantified relative to the SsrA control, and variation in intensity during the course of the experiment was compared by setting the Time 0 point to 1. (C) Strains PM1560 (PBAD–arcB arcZ+) and PM1561 (PBAD–arcB arcZ−) were grown as in Figure 6B. After 15 min of induction (Time: 0; lanes 2, 8, and 14), samples were removed and rifampicin was added at a final concentration of 250 μM. Samples were collected at the described intervals and RNA prepared for northern blots. SsrA levels were used to normalize as in (B).
When arcB is induced in the absence of arcZ expression (Figure 6B, lanes 9–10 and 19–20), arcB mRNA levels rise 30–35-fold within 15 min and remain high at 60 min. However, in cells in which arcZ is expressed (arcA+ arcZ+, Figure 6B, lanes 4 and 5), the initial level of arcB expression is lower, and it decreases significantly by 60 min.
This decrease, in the presence of continued arabinose treatment, reflects at least in part accelerated degradation of arcB mRNA when ArcZ is produced. In Figure 6C, addition of rifampicin in the presence (lanes 8–12) or absence (lanes 14–18) of ArcZ show that arcB mRNA, which has a half-life of about 5 min in the absence of ArcZ, is significantly more unstable (half-life of about 2 min) when ArcZ is present.
Although ArcZ negatively affects arcB accumulation, induction of arcB mRNA negatively affects ArcZ in at least two ways, forming a branched feedback loop. Induction of ArcB leads to the activation of ArcA (measurable by increased repression of the ParcZ–lac and of a Psdh–lacZ fusion; as expected, this is fully ArcA dependent), and therefore tighter repression of arcZ. However, there is also evidence of an ArcA-independent cis effect of the overlapping transcription of arcB and arcZ. Thus, in Figure 6B, lanes 4 and 5 (arcA+), as well as lanes 14 and 15 (arcA−), ArcZ levels are decreased as a function of arcB induction. As noted above arcB levels go down as well. Therefore, this suggests there is mutual negative regulation of arcB and arcZ RNAs, and this is ArcA independent.
Full-length ArcZ is not at all stable, and disappears completely once arcB is induced (Figure 6C, lanes 1–3 and 7–9). However, the processed 55 nt form is quite stable, in the presence or absence of arcB mRNA. Therefore, if the degradation of the arcB transcript and ArcZ are coupled, it is the full-length ArcZ that is sensitive to this degradation, decreasing the appearance of the processed form. Alternatively, transcription of arcB may directly interfere with arcZ promoter activity.
In the experiments above, arcB expression is artificially high and any effects on the natural promoter are lost. We used qPCR to examine arcB under the control of its own promoter (Supplementary Figure S7). In these experiments, arcB mRNA levels are low and are unaffected by either arcZ or arcA mutations under aerobic conditions. However, under anaerobic conditions, arcB levels rise significantly in an arcZ mutant, consistent with the data from Figure 6. However, they also rise in an arcA mutant; this increase is apparently due to ArcZ, as the level of arcB mRNA in an arcZ arcA mutation is similar to that in an arcZ mutation.
An interpretation of these results is that ArcZ has both a negative and positive effect on arcB mRNA levels. As only the negative effect of ArcZ is seen in Figure 6, we suggest that the positive effect is on the arcB promoter, not present in the PBAD–arcB constructs. Under aerobic conditions, the positive and negative effects may counteract each other, explaining the lack of an effect on arcB. Clearly, this positive effect is likely to be indirect, and while the transcriptional regulator has not been identified, one candidate would be RpoS itself, as we show in this study it is positively regulated by ArcZ. If this extra regulatory loop is confirmed, it would suggest that not only do ArcA and ArcB negatively regulate RpoS, but RpoS participates in the negative regulation.
Discussion
In this study, we have used a specific sRNA library to examine translational regulation of the stationary sigma factor rpoS. The known regulators of rpoS were observed, and a number of new sRNA regulators were revealed, including ArcZ, a third positive regulator of rpoS. The expression of arcZ is repressed under anaerobic growth by the ArcA/ArcB two-component system. ArcB, the sensor kinase, is transcribed convergently and overlapping arcZ, and the two transcripts negatively affect each other. Thus, ArcZ both provides a feedback loop for the ArcA/ArcB system and links the RpoS-dependent regulon to ArcA/ArcB through translational regulation (Figure 7). ArcA/ArcB have previously been described as negative regulators of RpoS transcription and stability (Mika and Hengge, 2005). ArcZ adds a third level of ArcA-dependent negative regulation of rpoS, emphasizing the importance of the control of rpoS expression during anaerobic growth.
Figure 7.
Regulatory circuits for ArcA, ArcB, and ArcZ. See text.
Construction of a sRNA library allows simple and rapid screening for sRNAs regulating genes of interest
Construction of a plasmid library allowing the overexpression of 26 sRNAs that bind Hfq is a useful tool for screening targets of interest for regulation. A few very unstable or poorly expressed Hfq-binding sRNAs may still remain undefined in E. coli, and other sRNAs that do not bind Hfq may also function by base-pairing, as seen in other species (for review, see Jousselin et al (2009)). However, we consider this library both a starting point for construction of a broader sRNA library and one that represents a major portion of the most abundant family of sRNAs.
The use of the sRNA library has advantages over the approach we previously took, using a genomic library to find sRNAs regulating dpiB (Mandin and Gottesman, 2009), and is conceptually similar to the experiments of Urban and Vogel (2007), co-expressing sRNAs with plasmids expressing gfp fusion reporters. A dedicated sRNA library allows analysis of the effect of sRNAs that might be poorly expressed from their native promoters and the analysis of the effect of toxic sRNAs. For example, we were not able to isolate transformants for plasmids containing OxyS or DicF when inducer was present in the plate, strongly suggesting that high-level expression of these sRNAs is detrimental to the cell, as previously described (Bouché and Bouché, 1989; Altuvia et al, 1997). Therefore, use of a genomic library might not have allowed identification of such toxic sRNAs as regulators.
A different approach to identifying the Hfq-dependent regulator of a gene of interest was recently described by Papenfort et al (2008), using single mutations in each known sRNA to find the regulator for ompX. That approach, although certainly useful, will be most effective for sRNAs that are well expressed under the growth conditions tested. For instance, the role of RprA in regulating rpoS is not evident from the phenotype of an rprA mutation, unless the regulatory cascade for rprA is activated (Majdalani et al, 2001).
Three sRNA activators of rpoS translation
One of the major findings of this study is the identification of a new sRNA, ArcZ, which enhances rpoS translation, in addition to DsrA and RprA. Noticeably, all three sRNAs bind to the same region of the rpoS mRNA and activate translation by opening the stem-loop structure in the rpoS 5′-UTR, allowing the translation machinery access to the RBS. However, they differ in their overall sequences, in the sequences involved in pairing, and in the position of the pairing sequence within the sRNA. In addition, the in vitro properties of these three sRNAs differ significantly (Soper et al, 2010). These observations may suggest independent evolution of these three regulatory RNAs.
Why does the cell use three different sRNAs to perform the same function for rpoS? In addition to providing the cells with the ability to respond to different signals, two advantages can be suggested. First, it is likely that the three sRNAs have different strengths in their ability to open the rpoS structure, as we suggest in a recent study (Soper et al, 2010), and thus may induce rpoS to different levels. Second, it is clear that each sRNA has, in addition to rpoS, other genes that it will co-regulate, and thus will activate a different global response. For example, in addition to rpoS, DsrA, transcription of which is enhanced at low temperature, is known to repress the expression of at least one gene, hns, encoding a histone-like protein, itself involved in the transcriptional regulation of many genes (Lease et al, 1998); several other targets have been predicted for DsrA. RprA also has a set of unique (not affected by DsrA) negative targets (Majdalani et al, in preparation). In Salmonella, overexpression of ArcZ alters the levels of many transcripts, with at least three direct ArcZ targets (Papenfort et al, 2009). Thus, the use of specifically regulated sRNAs, each activating rpoS translation, allows the differentiation of the general RpoS stress response into a more tailored response to unique stress signals.
The ArcA regulon, RpoS, and the role of ArcZ
The ArcA/ArcB system is one of the central regulators in E. coli and many other bacteria, controlling a large number of functions in response to oxygen availability (Malpica et al, 2006). Direct positive and negative regulation of many genes has been shown, generally with the biggest effect of ArcA seen under anaerobic conditions when ArcB is actively phosphorylating ArcA. Our results with ArcZ are consistent with this general picture; ArcZ is well expressed under aerobic growth conditions and less well under anaerobic growth. Mutations in arcA relieve the anaerobic repression. Although we have not demonstrated direct binding of ArcA to the arcZ promoter, we have no evidence for another regulator of this sRNA.
The existence of an ArcA-repressed activator of RpoS is consistent with previous observations suggesting that ArcA and ArcB negatively regulate RpoS (Mika and Hengge, 2005). In those studies, evidence of both ArcA repression of rpoS transcription and ArcB stimulation of RpoS degradation was found. ArcA repression of ArcZ provides a third level of negative regulation, and our studies with arcA and arcZ mutations demonstrate that this regulation has a significant effect on translation of RpoS.
ArcZ clearly has targets other than RpoS. Papenfort et al (2009) demonstrated direct pairing of ArcZ with the RNAs for tpx, encoding a lipid hydroperoxide peroxidase, and sdaC, encoding a putative serine transporter in Salmonella; both pairings are conserved in E. coli. In addition, they observed wide-spread changes in gene expression on overexpression of ArcZ. Some of these effects are probably due to increased RpoS, as they note, but others may reflect other direct targets of ArcZ. Thus far, the direct targets of ArcZ do not provide a clear understanding of the physiological significance of this regulation. tpx has been reported to be negatively regulated by ArcA (Kim et al, 1999); as regulation by ArcZ is also negative, this regulatory circuit would suggest interlocking mechanisms for keeping tpx expression relatively low under most conditions; it would be transcriptionally repressed under anaerobic growth conditions by ArcA, and translationally repressed under aerobic conditions by ArcZ. It seems likely that a subset of genes that have been assigned to the ArcA/ArcB regulon are in fact indirectly regulated by ArcA, through ArcZ, but the complexity of the effects of ArcA, RpoS, and the apparent broad effects of ArcZ leave this as an interesting future direction.
An additional complexity of this network is the feedback loop we have demonstrated, in which ArcZ represses and is directly repressed by arcB expression (Figure 7). Given the overlap between the arcZ and arcB transcripts, and that ArcZ destabilizes the arcB mRNA, the likely mechanism for this is annealing of the complementary RNAs followed by RNase degradation. ArcA has been shown to negatively regulate arcB mRNA levels, particularly at low oxygen, (Shalel-Levanon et al, 2005); based on our observations (Figure 6 and Supplementary Figure S7), we would suggest that this is through ArcZ.
Although ArcZ destabilizes the arcB mRNA, we did not find any evidence that the abundantly processed ArcZ itself was destabilized by arcB mRNA, even though levels of the sRNA drastically decreased upon arcB induction. We favour a model in which the arcB antisense RNA destabilizes the full-length arcZ transcript, reducing the population of RNAs that can be processed to the 55 nt form. Once processed, ArcZ would become insensitive to degradation. These results indicate that ArcZ is repressed by the two-component system in at least two different ways: by ArcA-mediated repression and directly by arcB transcription. The nucleases involved in this mutual destruction have not been identified, although RNase E is known to have a role in degradation of arcB mRNA (Aiso and Ohki, 2003).
Functionally, the purpose for this regulatory feedback loop may be to provide a homeostatic regulation of the ArcA/ArcB regulon: when ArcZ is highly produced (aerobic growth), it represses ArcA activation by downregulating the levels of ArcB. The consequence of this is maintenance of ArcZ expression (Figure 7). Conversely, when ArcA is activated (anaerobic conditions), it represses arcZ expression, therefore allowing higher ArcB expression and thus its own activation. This double regulation resembles a bi-stable system in which once one of the actors (ArcZ or ArcA/ArcB) is expressed or activated sufficiently, it favours its own expression and represses the other. In this model, the signal allowing the switch from one ‘mode' to another would be the state of ArcA phosphorylation, which is itself driven by ArcB sensing of aerobic or anaerobic conditions. How these two systems communicate to regulate gene expression will be of great interest to investigate in the future.
Materials and methods
Strains and plasmids
E. coli strains used in this study are derivatives of strain MG1655 and are listed in Supplementary Table S1; their construction is described in Supplementary data. arcA∷kan, arcB∷kan, and arcZ∷tet mutations were moved into strains by P1 transduction (Silhavy et al, 1984).
For the sRNA library, plasmids that were not previously available (Table I) were constructed by PCR amplifying the sRNA genes from strain MG1655 using primers described in Supplementary Table S2. sRNAs genes were amplified from their described transcriptional start site to >50 nt downstream of their predicted or identified transcriptional terminator. The PCR products were then digested with AatII and EcoRI and ligated into the digested pBR-plac vector (Guillier and Gottesman, 2006). The ligation products were transformed into strain NM525 and plasmids were selected on ampicillin-containing plates.
Site-directed mutants in pDsrA, pRprA, and pArcZ were constructed using the Quickchange II site-directed mutagenesis kit (Stratagene) following the manufacturer's instructions with primers described in Supplementary Table S1.
β-galactosidase activity measurements
Determination of the β-galactosidase activity of the ParcZ–lacZ transcriptional fusion was determined using the standard assay described by Miller (1992). In all other cases, β-galactosidase measurements were performed as described previously (Majdalani et al, 1998). Calculated specific activities correspond to kinetic measurements of Vmax/OD600 as read on a SpectraMax 250 microtiter plate reader (Molecular Devices).
Library screen
The library screen was carried out in microtiter dish format. The TSS transformation (Chung and Miller, 1988) was used, followed by spotting cells for selection of transformants on LB ampicillin plates. These spots were used directly to inoculate media for growth and β-galactosidase assay. The details of the procedure are described in Supplementary data.
RNA extraction and northern blot experiments
Overnight cultures of the strains to be analysed were grown in LB-ampicillin, diluted 500-fold in fresh medium containing ampicillin (100 μg/ml) and IPTG (100 μM) when indicated and incubated at 37°C with agitation. At the indicated OD600, 800 μl samples were removed from each culture and RNA was extracted from the samples using the hot phenol method (Massé et al, 2003). Northern blots were performed with 3–10 μg total RNA as described previously (Mandin and Gottesman, 2009). Oligonucleotides used as probes are described in Supplementary Table S2.
5′ and 3′ rapid amplification of cDNA ends
Rapid amplification of 5′ complementary DNA ends was carried out as described previously (Mandin and Gottesman, 2009). The PBAD-RNA adaptor was used, and the arcZ RNA was reverse transcribed and amplified using oligonucleotide ArcZ-5′RACE 1 and PBAD-DNA (Supplementary Table S2). Rapid amplification of 3′ complementary DNA ends was carried out as described previously (Argaman et al, 2001), using RNA prepared from PM1490 or its arcZ mutant derivative PM1520; clones were only obtained from the arcZ mutant. The 3′-end of arcB was reverse-transcribed using oligonucleotides 3′ E1 DNA adaptor and arcB 3′ RACE-1.
In each case, the subsequent PCR products were separated on gels and directly cloned in the pCR4-TOPO vector (Invitrogen). Plasmids were prepared from randomly chosen colonies and inserts were sequenced using oligonucleotides M13-for and M13-rev.
Supplementary Material
Acknowledgments
We thank Maude Guillier and Kyung Moon for providing plasmids. We thank members of our laboratory and Gisela Storz for comments on the paper. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aiso T, Ohki R (2003) Instability of sensory histidine kinase mRNAs in Escherichia coli. Genes Cells 8: 179–187 [DOI] [PubMed] [Google Scholar]
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G (1997) A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90: 43–53 [DOI] [PubMed] [Google Scholar]
- Antal M, Bordeau V, Douchin V, Felden B (2005) A small bacterial RNA regulates a putative ABC transporter. J Biol Chem 280: 7901–7908 [DOI] [PubMed] [Google Scholar]
- Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S (2001) Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11: 941–950 [DOI] [PubMed] [Google Scholar]
- Bongaerts J, Zoske S, Weidner U, Unden G (1995) Transcriptional regulation of the proton translocating NADH dehydrogenase genes (nuoA–N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol 16: 521–534 [DOI] [PubMed] [Google Scholar]
- Bouché F, Bouché JP (1989) Genetic evidence that DicF, a second division inhibitor encoded by the Escherichia coli dicB operon, is probably RNA. Mol Microbiol 3: 991–994 [DOI] [PubMed] [Google Scholar]
- Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S (2008) Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol 68: 298–313 [DOI] [PubMed] [Google Scholar]
- Bougdour A, Wickner S, Gottesman S (2006) Modulating RssB activity: IraP, a novel regulator of σS stability in Escherichia coli. Genes Dev 20: 884–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RG, Link TM (2007) Hfq structure, function and ligand binding. Curr Opin Microbiol 10: 125–133 [DOI] [PubMed] [Google Scholar]
- Brown L, Elliott T (1996) Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J Bacteriol 178: 3763–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown L, Elliott T (1997) Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol 179: 656–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang A, Blyn LB, Storz G (2004) MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol 186: 6689–6697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CT, Miller RH (1988) A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res 16: 3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M (2010) MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 76: 467–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Gottesman S (2009) The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191: 461–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douchin V, Bohn C, Bouloc P (2006) Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J Biol Chem 281: 12253–12259 [DOI] [PubMed] [Google Scholar]
- Durand S, Storz G (2010) Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 75: 1215–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D, Kwon O, Lin EC (2001) Quinones as the redox signal for the arc two-component system of bacteria. Science 292: 2314–2316 [DOI] [PubMed] [Google Scholar]
- Gottesman S (2004) The small RNA regulators of Escherichia coli: roles and mechanisms. Annu Rev Microbiol 58: 303–328 [DOI] [PubMed] [Google Scholar]
- Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ (2006) Small RNA regulators and the bacterial response to stress. Cold Spring Harb Symp Quant Biol 71: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillier M, Gottesman S (2006) Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59: 231–247 [DOI] [PubMed] [Google Scholar]
- Gunsalus RP, Park SJ (1994) Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res Microbiol 145: 437–450 [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R (2002) Signal transduction and regulatory mechanisms involved in control of the σS (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev 66: 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin A, Metzinger L, Felden B (2009) On the facultative requirement of the bacterial RNA chaperone, Hfq. Trends Microbiol 17: 399–405 [DOI] [PubMed] [Google Scholar]
- Kim SJ, Han YH, Kim IH, Kim HK (1999) Involvement of ArcA and Fnr in expression of Escherichia coli thiol peroxidase gene. IUBMB Life 48: 215–218 [DOI] [PubMed] [Google Scholar]
- Lange R, Fischer D, Hengge-Aronis R (1995) Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA polymerase in Escherichia coli. J Bacteriol 177: 4676–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease RA, Cusick M, Belfort M (1998) Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA 95: 12456–12461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K, Gottesman S (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 39: 1382–1394 [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA 95: 12462–12467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majdalani N, Hernandez D, Gottesman S (2002) Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol 46: 813–826 [DOI] [PubMed] [Google Scholar]
- Malpica R, Franco B, Rodriguez C, Kwon O, Georgellis D (2004) Identification of a quinone-sensitive redox switch in the ArcB sensor kinase. Proc Natl Acad Sci USA 101: 13318–13323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpica R, Sandoval GR, Rodriguez C, Franco B, Georgellis D (2006) Signaling by the arc two-component system provides a link between the redox state of the quinone pool and gene expression. Antioxid Redox Signal 8: 781–795 [DOI] [PubMed] [Google Scholar]
- Mandin P, Gottesman S (2009) A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA–DpiB two-component system. Mol Microbiol 72: 551–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Escorcia FE, Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massé E, Gottesman S (2002) A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci USA 99: 4620–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AM, De Wulf P, Church GM, Lin EC (1999) A weight matrix for binding recognition by the redox-response regulator ArcA-P of Escherichia coli. Mol Microbiol 32: 219–221 [DOI] [PubMed] [Google Scholar]
- Mika F, Hengge R (2005) A two-component phosphotransfer network involving ArcB, ArcA, and RssB coordinates synthesis and proteolysis of σS (RpoS) in E. coli. Genes Dev 19: 2770–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J (1992) A Short Course in Bacterial Genetics: A laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Mizuno T, Chou MY, Inouye M (1984) A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci USA 81: 1966–1970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller T, Franch T, Udesen C, Gerdes K, Valentin-Hansen P (2002) Spot 42 RNA mediates discoordinate expression of the E. coli galactose operon. Genes Dev 16: 1696–1706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K, Gottesman S (2009) A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74: 1314–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita T, Kawamoto H, Mizota T, Inada T, Aiba H (2004) Enolase in the RNA degradosome plays a crucial role in the rapid decay of glucose transporter mRNA in the response to phosphosugar stress in Escherichia coli. Mol Microbiol 54: 1063–1075 [DOI] [PubMed] [Google Scholar]
- Muffler A, Fischer D, Hengge-Aronis R (1996) The RNA-binding protein HF-I, known as a host factor for phage Qbeta RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev 10: 1143–1151 [DOI] [PubMed] [Google Scholar]
- Opdyke JA, Kang JG, Storz G (2004) GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186: 6698–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J (2008) Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68: 890–906 [DOI] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158 [DOI] [PubMed] [Google Scholar]
- Reams SG, Lee N, Mat-Jan F, Clark DP (1997) Effect of chelating agents and respiratory inhibitors on regulation of the cadA gene in Escherichia coli. Arch Microbiol 167: 209–216 [DOI] [PubMed] [Google Scholar]
- Repoila F, Majdalani N, Gottesman S (2003) Small non-coding RNAs, co-ordinators of adaptation processes in Escherichia coli: the RpoS paradigm. Mol Microbiol 48: 855–861 [DOI] [PubMed] [Google Scholar]
- Ruiz N, Silhavy TJ (2003) Constitutive activation of the Escherichia coli Pho regulon upregulates rpoS translation in an Hfq-dependent fashion. J Bacteriol 185: 5984–5992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurdell MS, Woodbury GM, McCleary WR (2007) Genetic evidence suggests that the intergenic region between pstA and pstB plays a role in the regulation of rpoS translation during phosphate limitation. J Bacteriol 189: 1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalel-Levanon S, San KY, Bennett GN (2005) Effect of ArcA and FNR on the expression of genes related to the oxygen regulation and the glycolysis pathway in Escherichia coli under microaerobic growth conditions. Biotechnol Bioeng 92: 147–159 [DOI] [PubMed] [Google Scholar]
- Sharma CM, Vogel J (2009) Experimental approaches for the discovery and characterization of regulatory small RNA. Curr Opin Microbiol 12: 536–546 [DOI] [PubMed] [Google Scholar]
- Silhavy T, Berman M, Enquist L (1984) Experiments with Gene Fusions. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J (2008) Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4: e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski DD, Gupta A, Gottesman S (1996) The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J 15: 3993–4000 [PMC free article] [PubMed] [Google Scholar]
- Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA (2010) Positive regulation by small RNAs and the role of Hfq. Proc Natl Acad Sci USA 107: 9602–9607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper TJ, Woodson SA (2008) The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14: 1907–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studemann A, Noirclerc-Savoye M, Klauck E, Becker G, Schneider D, Hengge R (2003) Sequential recognition of two distinct sites in σS by the proteolytic targeting factor RssB and ClpX. EMBO J 22: 4111–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KM, Rhodius VA, Gottesman S (2007) σE regulates and is regulated by a small RNA in Escherichia coli. J Bacteriol 189: 4243–4256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udekwu KI, Darfeuille F, Vogel J, Reimegard J, Holmqvist E, Wagner EG (2005) Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev 19: 2355–2366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegrove T, Wilf N, Sun X, Wartell RM (2008) Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry 47: 11184–11195 [DOI] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2007) Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res 35: 1018–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban JH, Vogel J (2008) Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol 6: e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanowski ML, Stauffer LT, Stauffer GV (2000) The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol 37: 856–868 [DOI] [PubMed] [Google Scholar]
- Vanderpool CK, Gottesman S (2004) Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076–1089 [DOI] [PubMed] [Google Scholar]
- Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG (2003) RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res 31: 6435–6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S (2001) Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15: 1637–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G (2009) Regulatory RNAs in bacteria. Cell 136: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R (2005) Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187: 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G (1998) The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J 17: 6061–6068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S (2003) Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50: 1111–1124 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.