Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication
A new functional interaction of BLM helicase with Rif1, the homolog of a telomere length regulator in yeast, sheds light on one of BLM's less-understood functions, the facilitation of stalled replication fork restart.
Keywords: BLM, Bloom syndrome, replication, Rif1, RMI
Abstract
BLM, the helicase defective in Bloom syndrome, is part of a multiprotein complex that protects genome stability. Here, we show that Rif1 is a novel component of the BLM complex and works with BLM to promote recovery of stalled replication forks. First, Rif1 physically interacts with the BLM complex through a conserved C-terminal domain, and the stability of Rif1 depends on the presence of the BLM complex. Second, Rif1 and BLM are recruited with similar kinetics to stalled replication forks, and the Rif1 recruitment is delayed in BLM-deficient cells. Third, genetic analyses in vertebrate DT40 cells suggest that BLM and Rif1 work in a common pathway to resist replication stress and promote recovery of stalled forks. Importantly, vertebrate Rif1 contains a DNA-binding domain that resembles the αCTD domain of bacterial RNA polymerase α; and this domain preferentially binds fork and Holliday junction (HJ) DNA in vitro and is required for Rif1 to resist replication stress in vivo. Our data suggest that Rif1 provides a new DNA-binding interface for the BLM complex to restart stalled replication forks.
Introduction
Bloom syndrome (BS) is an autosomal recessive disease characterized by growth retardation, immunological defects, reduced fertility, sensitivity to sunlight, and predisposition to a wide variety of cancers (German, 1993; Bachrati and Hickson, 2008). Cells from BS patients display genomic instability, characterized by elevated frequencies of sister chromatid exchanges (SCEs), chromosomal breaks, interchanges between homologous chromosomes, and sensitivity to DNA-damaging agents. BLM, the gene product defective in BS, belongs to the RecQ helicase family, which also includes RECQL1, WRN, RECQL4, and RECQL5 in mammals. Mutations in WRN and RECQL4 cause Werner's syndrome and Rothmund–Thomson syndrome, respectively; both of which resemble BS in genomic instability and cancer predisposition (Bachrati and Hickson, 2008). Mutations in RecQ helicases in yeast (Watt et al, 1996; Myung et al, 2001) and Drosophila (McVey et al, 2007; Wu et al, 2008) also lead to genomic instability and cellular sensitivity to replication stress. These findings underscore the importance of RecQ helicases in protecting genome integrity in all eukaryotes.
BLM possesses a 3′ to 5′ DNA unwinding activity and is capable of resolving a variety of DNA structures, including replication forks, Holliday junctions (HJs), D-loops, and G4 DNA (Sun et al, 1998; Karow et al, 2000; Bachrati et al, 2006; Ralf et al, 2006). In addition, BLM and its orthologs contain a DNA strand-exchange activity, which is required for suppression of hyper-recombination in yeast (Chen and Brill, 2010). Increasing evidence has shown that BLM regulates several steps of homologous recombination (HR)-dependent repair of double-strand DNA breaks (DSBs). For example, BLM can upregulate this process by stimulating resection of DNA ends at the DSBs and/or by promoting the primer extension step after formation of D-loops (Bugreev et al, 2007; Gravel et al, 2008). Alternatively, BLM can downregulate the process by disrupting the RAD51-coated presynaptic filament and D-loops (Bugreev et al, 2007). Moreover, BLM associates with topoisomerase 3α (Topo 3α), RMI1, and RMI2, to form a conserved complex, named BTR, which works coordinately to resolve double HJ (dHJ) in a way that suppresses crossover recombination (Wu and Hickson, 2003; Raynard et al, 2006; Wu et al, 2006; Xu et al, 2008). Defects in any BTR components result in increased SCE frequency, the hallmark feature of BLM-deficient cells.
In addition to its functions in HR-dependent DNA repair, BLM also facilitates restart of stalled replication forks, possibly by promoting reversal of stalled forks into HJs, which may be subsequently repaired through a template switching mechanism (Ralf et al, 2006). Cells deficient in BLM have impaired fork velocity, reduced efficiency of recovering stalled replication forks, and display hypersensitivity to several drugs that induce replication stress (Davies et al, 2007; Rao et al, 2007).
Rif1 is a highly conserved protein present from yeast to mammals. It was originally discovered in budding yeast as a protein that associates with the telomeric DNA-binding protein Rap1p and negatively regulates telomere length (Hardy et al, 1992). Rif1 in mammals, however, does not regulate length of normal telomeres (Silverman et al, 2004; Xu and Blackburn, 2004; Buonomo et al, 2009). Rather, it localizes to DNA damage sites, and its depletion results in cellular sensitivity to ionizing radiation, reduced HR-dependent repair of DSBs, and defective intra-S-phase checkpoint (Silverman et al, 2004; Xu and Blackburn, 2004; Buonomo et al, 2009; Wang et al, 2009). Most recently, a study of Rif1-knockout mice suggested that it has a function in the repair of stalled replication forks by facilitating HR-dependent DNA repair (Buonomo et al, 2009). Moreover, Rif1 mutations have been detected in several human cancer cell lines (Sjoblom et al, 2006; Howarth et al, 2008). Unfortunately, no recognizable domains or biochemical activities have been described for Rif1, so that its mechanism of action remains unclear.
We have previously purified three BLM-containing complexes from HeLa nuclear extracts and identified most of the components (Meetei et al, 2003). Several components, BLM, Topo 3α, RMI1, and RMI2, are common to all BLM complexes (Singh et al, 2008; Xu et al, 2008). Other components are present only in specific complexes. These include the Fanconi anemia core complex proteins (FANCA, FANCB, FANCC, FANCF, FANCG, FANCL, FANCM, FAAP100, and FAAP24), replication protein A (RPA), MLH1, and an uncharacterized 250 kDa polypeptide referred to as BLAP250 (BLM-associated 250 kDa protein) (Meetei et al, 2003). Here, we identify BLAP250 as Rif1, and show that it works with BLM to promote recovery of stalled replication forks and to resist replication stress in vertebrate DT40 cells. Importantly, vertebrate (but not yeast) Rif1 contains a DNA-binding domain that resembles the αCTD domain of bacterial RNA polymerase α and preferentially binds fork or HJ DNA. We demonstrate that this DNA-binding activity is required for Rif1 to prevent accumulation of stalled replication forks and to resist replication stress. Thus, Rif1 confers a new DNA interface for the BLM complex to maintain normal replication.
Results
Rif1 is a novel component of BLM complex
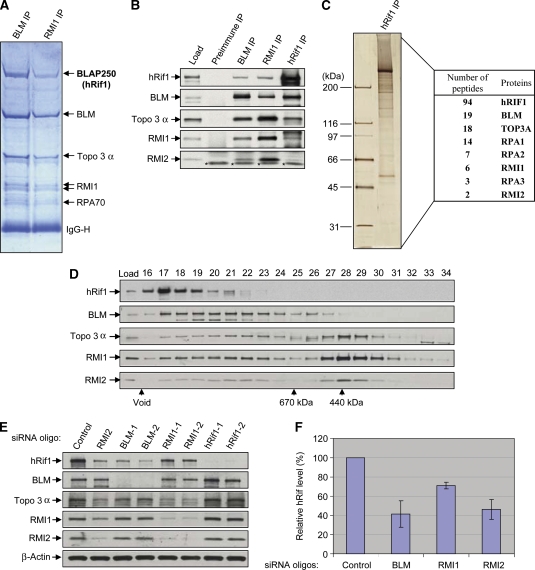
We previously immunopurified BLM complexes from HeLa nuclear extracts using BLM and RMI1 (BLAP75) antibodies and detected a 250 kDa polypeptide at a level similar to other BLM complex components, as revealed by SDS–PAGE and Coomassie staining (Figure 1A) (Meetei et al, 2003; Yin et al, 2005). This polypeptide, referred to as BLAP250, was identified by mass spectrometry as the human ortholog of yeast Rif1 (hRif1; data not shown). An antibody against hRif1 recognized this polypeptide in immunoblotting assays, confirming that BLAP250 is hRif1 (Figure 1B).
Figure 1.
hRif1 is a new component of BLM complex and its stability depends on the other components of the complex. (A) A Coomassie blue-stained SDS gel showing that hRif1 (previously named BLAP250) is present in complexes immunoprecipitated by both BLM and RMI1 antibodies. The major polypeptides on the gel (marked with arrows) were indentified by mass spectrometry. This figure is reproduced from Figure 1 of a previous publication (Yin et al, 2005) for readers' convenience. (B) Immunoblotting shows that hRif1 coimmunoprecipitates with other components of the BLM complex. Preimmune serum was used as a negative control. The nuclear extract input is shown as load. A cross-reactive polypeptide is indicated with asterisks. (C) A silver-stained SDS gel showing the polypeptides immunopurified by an hRif1 antibody from peak fractions of Rif1 after fractionation of HeLa nuclear extract by Superose 6 column (see D). The proteins identified by mass spectrometry and the number of peptides discovered for each protein are listed in the table on the right. (D) Immunoblotting shows that the Superose 6 fractionation profile of hRif1 overlaps with those of the BLM complex components. The fractions corresponding to molecular weight standards are indicated at the bottom. The profiles of the BLM complex components (the bottom four panels) are reproduced from Supplementary Figure S1 of a previous publication (Xu et al, 2008) for comparison. (E, F) Immunoblotting (E) and its quantification (F) show that the hRif1 protein levels are reduced in HeLa cells depleted of BLM, RMI1, or RMI2 by siRNA. Immunoblotting of β-actin was used as a loading control. (F) The relative hRif1 levels were shown, with that of control siRNA-treated cells being set as 100%. Quantification was done using data from at least four independent experiments for each protein. The data represent the mean values, and the error bars represent standard errors.
The fact that the hRif1 was isolated by antibodies against two different BLM complex components suggests that it could be a subunit of the same complex. To independently confirm this, we performed an unbiased immunopurification of hRif1-associated proteins using an hRif1 antibody. Silver staining revealed that the most abundant polypeptide in the immunoprecipitate was a 250-kDa protein (Figure 1C), which was identified by mass spectrometry as hRif1 (data not shown). The other major polypeptides were similarly identified as degradation products of hRif1 (data not shown). The presence of these degradation products precluded us from visualizing potential BLM complex components by silver staining. We therefore analysed the entire hRif1 immunoprecipitate by mass spectrometry and identified peptides derived from many BLM complex components, including BLM, Topo 3α, RMI1, RMI2, and RPA (Figure 1C, right table). Immunoblotting also confirmed the presence of these proteins in the hRif1 immunoprecipitate (Figure 1B), indicating that hRif1 is an integral component of the BLM complex.
We found the mRNA levels of hRif1 and BLM are significantly correlated (P<0.001) in a microarray screening of 60 human cancer cell lines (Supplementary Figure S1A). Moreover, the correlation coefficient between the two is the highest than that between all other BLM complex components (Supplementary Figure S1B and data not shown). This correlation provides indirect evidence for hRif1 being a component of the BLM complex, so that their expression might be coregulated in some cells.
A fraction of hRif1 associates with BLM
Fractionation of HeLa nuclear extract by Superpose 6 gel-filtration chromatography revealed that hRif1 fractionated in a sharp peak near the exclusion volume of the column, which corresponds to a complex of about 2 MDa (Figure 1D). This molecular weight is much larger than the calculated mass of hRif1 (about 250 kDa), suggesting that hRif1 is part of a large complex. As the most abundant polypeptides in the immunoprecipitate are derived from hRif1 (Figure 1C), the complex most likely consists of more than one copy of hRif1. Indeed, our co-IP analyses of hRif1-deletion mutants and studies of hRif1 recombinant proteins showed that it can self-associate through its C-terminal domain both in vitro and in vivo (Supplementary Figure S2).
Unlike hRif1, which has a single peak by Superose fractionation, the other components of the BLM complex fractionated in several peaks, reflecting the existence of different BLM complexes (Figure 1D) (Meetei et al, 2003). The hRif1 peak only partially overlaps with BLM complex components, implying that only a subset of hRif1 complex contains BLM, and vice versa. To further examine this issue, we quantitatively depleted the BLM complex from HeLa nuclear extract with a BLM antibody, and found about 50% of hRif1 was codepleted (Supplementary Figure S3). This suggests that about half of hRif1 in cells is associated with the BLM complex, whereas the other half is not, hinting that hRif1 may function both with and without BLM.
The stability of hRif1 is dependent on the BLM complex
The stability of one protein within a multisubunit complex often depends on other components, as proper folding of one protein may require interactions with its partners. This interdependence for stability can be used to verify whether two proteins are components of the same complex in vivo. Analysis of hRif1 revealed that its protein level was reduced in HeLa or HEK293 cells depleted of BLM, RMI1, and RMI2 by siRNA (Figure 1E and F; Supplementary Figure S4), which is consistent with the previous findings in which the stability of various BLM complex components is reported to depend on each other (Yin et al, 2005; Xu et al, 2008). These data support the notion that hRif1 is part of the BLM complex.
hRif1 and the BLM complex are recruited to the same replication origin in response to HU-induced replication stress
Rif1, 53BP1, and a fraction of BLM have been found to colocalize in nuclear foci induced by aphidicolin (an inhibitor of DNA polymerase α) (Buonomo et al, 2009), hinting that they may be recruited to stalled replication forks as a complex. Our mass spectrometry analyses failed to detect the presence of 53BP1 in the purified BLM complex (data not shown), arguing that 53BP1 is not part of the BLM complex and may be recruited separately.
To precisely define whether hRif1 and BLM bind to the same locus on DNA, we used a CHIP assay (Thangavel et al, 2010) and found that the two proteins plus RMI1 were all recruited to a replication origin at the human lamin B2 locus in cells under hydroxyurea (HU)-induced replication stress, but not under normal growth conditions (Supplementary Figure S5; HU reduces the cellular nucleotide pool by inhibiting ribonucleotide reductase). These data are consistent with the findings that hRif1 is a component of the BLM complex (Figure 1), and imply that hRif1 may be recruited to the stalled forks at the replication origin as part of this complex.
Several BLM complex components have been found to colocalize in nuclear foci in cells treated with drugs that induce interstrand cross-links (ICLs), which are absolute blocks of DNA replication (Yin et al, 2005; Xu et al, 2008). Using a new Chromatin-IP assay, eCHIP (Shen et al, 2009), we detected hRif1 recruitment to a site-specific psoralen ICL on an episomal plasmid transfected into cells (Supplementary Figure S6). Moreover, the hRif1 recruitment was stimulated by about three-fold when the plasmid was allowed to replicate, supporting the notion that hRif1 acts at stalled replication forks.
hRif1 and BLM are recruited to DNA ICLs with the same kinetics and a majority of them colocalize
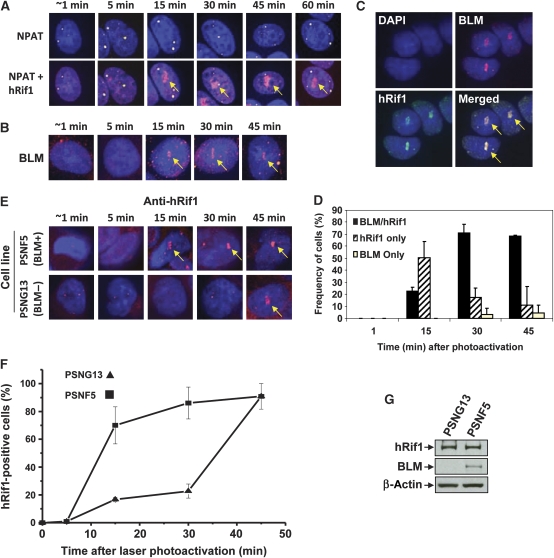
We next examined whether recruitment of BLM and hRif1 occurs at the same time and depends on each other. We used laser-activated psoralen conjugates to induce ICLs within a localized area in the nuclei of human SW480 cells (Thazhathveetil et al, 2007; Muniandy et al, 2009), and found that hRif1 and BLM are recruited to the target area with the same kinetics: the recruitment occurred within 15 min of photoactivation, and persisted without apparent diminution for at least 45 min (Figure 2A and B). Moreover, the two proteins colocalized in about 70% of cells at 30 or 45 min after photoactivation (Figure 2C and D). These results support the biochemical data that significant fractions of BLM and hRif1 are present in the same complex (Figure 1), and imply that they may be recruited to ICLs together as a complex.
Figure 2.
hRif1 and BLM are recruited to stalled replication forks with similar kinetics, and the Rif1 recruitment is delayed in BLM-deficient cells. (A) Immunofluorescence showing that hRif1 is strongly recruited to ICLs induced by laser-activated psoralen in S-phase cells. The arrows indicate the recruitment of hRif1 to the laser-targeted area. Costaining with the cell cycle marker (NPAT) distinguishes the S-phase cells (which have three to four yellow dots) from G1 cells (which have two yellow dots). As a control, hRif1 was not recruited to the targeted area in cells treated with laser alone (data not shown). (B) Immunofluorescence showing that BLM is recruited to ICLs generated by laser-activated psoralen with kinetics similar to hRif1 as shown in (A). (C) Representative immunofluorescence images showing colocalization of hRif1 and BLM at ICLs induced by laser-activated psoralen. (D) A graph showing percentage of SW480 cells that have recruitment of hRif1, BLM, or both proteins to DNA interstrand cross-links induced by laser-activated psoralen. (E, F) Representative images and statistical analyses showing that recruitment of hRif1 is delayed in the BLM-deficient cell line (PSNG13) compared with the same line complemented by reexpression of BLM (PSNG5). (G) Immunoblotting shows that the hRif1 protein level is comparable in a Bloom syndrome patient derived cell line (PSNG13) and the same line complemented by BLM (PSNF5).
We also observed that in some cells, only one protein was recruited. For example, 30 min following photoactivation, about 20% of the cells had recruitment of hRif1 but no BLM, whereas about 5% of the cells had recruitment of BLM but no hRif1 (Figure 2D). The results are consistent with the biochemistry data that a portion of hRif1 and BLM can form distinct complexes (Figure 1D), which could be independently recruited to ICLs.
Recruitment of Rif1 to ICLs is delayed in BLM-deficient cells
A prior study showed that Rif1 is not required for BLM recruitment to replication stress-induced foci (Buonomo et al, 2009). This prompted us to examine the opposite scenario--whether BLM is required for hRif1 recruitment, using a pair of isogenic cell lines derived from a BS patient (Gaymes et al, 2002). In the BLM-proficient cells (PSNF5, which was complemented by exogenous BLM), the hRif1 recruitment to the ICL occurred within 15 min after photoactivation (Figure 2E and F), similar to that observed in SW480 cells (Figure 2A). In contrast, in the BLM-deficient cells (pSNG13), the hRif1 recruitment was delayed until 45 min (Figure 2E and F). A control experiment showed that the hRif1 protein level was comparable in both cell lines (Figure 2G). The data suggest that hRif1 is dependent on BLM for its normal recruitment to stalled forks.
The result that Rif1 level in the BLM-deficient cell line was comparable to that of the BLM-complemented line differs from the siRNA data, which showed reduction of Rif1 level in BLM-depleted HeLa or HEK293 cells (Figure 1E and F; Supplementary Figure S4). Analyses of BLM−/− chicken DT40 cell line also showed a normal level of Rif1 (Supplementary Figure S7), which is in agreement with the findings from human BLM-deficient cells but different from those of siRNA studies. One explanation for this difference is that the siRNA method allows observation of the acute effect of BLM depletion—the reduced Rif1 stability. In contrast, the BLM-deficient cells have been cultured for many generations to yield enough quantity for western analyses. During this long period, cells may have increased production of Rif1 to compensate for its reduced stability. This compensatory mechanism may be important to overcome the growth disadvantage of cells that lack Rif1, as Rif1−/− mice are embryonic lethal (Buonomo et al, 2009), and chicken Rif1−/−/BLM−/− DT40 cells have reduced proliferation rate (see below).
Rif1 and BLM work in the same pathway in vertebrates to tolerate replication stress caused by FUdR and aphidicolin
Although previous work has shown that Rif1 contributes to cellular resistance to replication stress (Buonomo et al, 2009), it remains unclear whether Rif1 works in the same or a different pathway as does BLM. To examine this issue, we performed epistasis analyses using chicken DT40 cells. These cells have high gene-targeting efficiency and have been widely used for genetic studies of DNA damage response factors, including the BLM complex (Wang et al, 2000; Seki et al, 2006; Xu et al, 2008). We inactivated the Rif1 gene in both wild-type and BLM−/− DT40 cells (Supplementary Figure S8 and data not shown), and found that Rif1−/− cells had a growth rate indistinguishable from that of wild-type cells (Supplementary Figure S9). This is in contrast to BLM−/− cells that have a slower rate (Wang et al, 2000). BLM−/−/Rif1−/− cells had a slower growth rate than BLM−/− cells, suggesting that Rif1 has a function in cell proliferation in the absence of BLM.
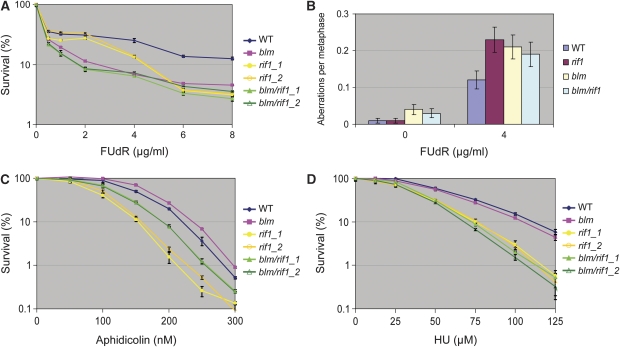
Both Rif1−/− and BLM−/− cells are sensitive to high concentration of 5-fluorodeoxyuridine (FUdR), which stalls replication by inhibiting thymidylate synthase to block incorporation of thymidine nucleotides into DNA (Figure 3A). Moreover, both cells displayed increased levels of chromosomal aberration in the presence of FUdR (Figure 3B). Notably, BLM−/−/Rif1−/− double-mutant cells resembled the single-mutant cells in sensitivity to high dose of FUdR and chromosomal aberration levels (Figure 3A and B), suggesting that Rif1 and BLM work in a common pathway to resist replication stress caused by high dose of FUdR.
Figure 3.
Rif1 and BLM function in the same pathway in DT40 cells to resist replication stress induced by FUdR and aphidicolin. (A, C, D) Sensitivity curves of different DT40 cells to DNA replication inhibitors, FUdR (A), aphidicolin (C), and HU (D). Mean and s.d. from three independent experiments are shown. Two different clones are tested for the rif1 mutants. (B) Histograms showing the chromosomal aberrations of different DT40 cells untreated or treated with FUdR (4 μg/ml) for 12 h. In all, 200 cells were scored for each preparation. Error bars were shown as the standard error of the mean.
Rif1−/− DT40 cells also exhibited sensitivity to replication inhibitors, aphidicolin, and HU (Figure 3C and D), consistent with findings from a recent study using Rif1−/− mouse cells (Buonomo et al, 2009). Interestingly, BLM−/− cells lacked obvious sensitivity to aphidicolin, and BLM−/−/Rif1−/− cells had lower sensitivity (or increased resistance) than Rif1−/− cells (Figure 3C). The fact that Rif1−/− cells depend on BLM to exhibit aphidicolin sensitivity provides additional evidence for these two genes working in the same pathway.
Conversely, BLM−/− cells lacked sensitivity to HU (Figure 3D), and BLM−/−/Rif1−/− cells showed similar HU sensitivity as Rif1−/− cells, indicating that while Rif1 is required for cellular resistance to HU-induced replication stress, BLM is dispensable. Taken together, these data suggest that Rif1 and BLM work in the same pathway to resist replication stress induced by FUdR and aphidicolin, but not HU.
Rif1 and BLM work in the same pathway to prevent accumulation of stalled replication forks
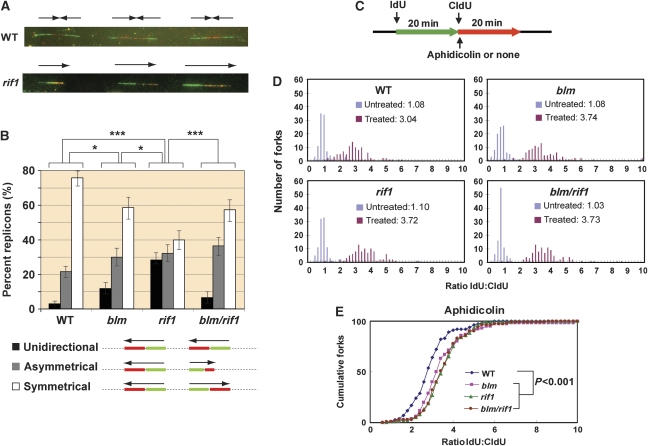
The findings that Rif1−/− cells are sensitive to replication stress implicate a defect in dealing with stalled forks, which may lead to their accumulation. We investigated this possibility by single DNA molecule analysis, which has been previously used to show accumulation of stalled forks in BLM-deficient cells (Rao et al, 2007). In this assay, cellular DNA was sequentially pulse labelled, first with iododeoxyuridine (IdU), and then with chlorodeoxyuridine (CldU), for equal times. The newly synthesized DNA was subsequently visualized after combing by immunofluorescence microscopy with specific antibodies against IdU and CldU (Figure 4A). A normal replication bubble is reflected by a symmetrical fluorescent signal because its two forks, which are stained in green (IdU) and red (CldU), move in opposite directions with the same velocity (Figure 4B, bottom panels) (Conti et al, 2007). However, if one of the two forks is stalled prior to or during the pulse labelling, a unidirectional or asymmetrical fluorescent signal will be observed, respectively.
Figure 4.
Rif1 and BLM work in the same pathway in chicken DT40 cells to prevent accumulation of stalled replication forks. (A) Representative images from single DNA molecule assays of wild-type (WT) and Rif1−/− DT40 cells. (Top) WT—the replication forks move bidirectionally from the origins (located between the green IdU signals) and progress symmetrically until they merge in the red (CldU) signals. (Bottom) Example of unidirectional signal (frequently observed in Rif1−/− cells). The single-replication forks move unidirectionally from left to right. (B) Histogram reporting the percentages of unidirectional (black), asymmetrical (grey), and symmetrical (white) forks in the four cell lines analysed. Histograms represent the sum of replicons measured and analysed in three independent experiments; the error bars indicate 95% confidence intervals for each set of data. Conditions that show significant differences of P-value <0.05 or P-value <0.001 are shown as one or three asterisks, respectively. (Bottom) Schematic representation of the different types of signals analysed. The primary data analysis of asymmetrical/symmetrical forks is shown in Supplementary Figure S10. (C) Schematic representation of the DNA fibre analyses to measure replication recovery (top). Where required, aphidicolin (100 ng/ml) was added after IdU labelling. (D) Histograms showing replication stalling in DT40 cells of various genotypes untreated or treated with aphidicolin. The mean ratio is shown in the centre. The higher ratio reflects more stalled forks or less recovery. (E) The data of the aphidicolin-treated DT40 cells were replotted as a cumulative percentage of forks at each ratio. The P-value for the ratio distribution of each mutant compared with wild type is shown.
The percentage of total stalled forks (unidirectional plus asymmetrical fluorescent signals among total signals) in BLM−/− DT40 cells (about 40%) was higher than that of wild-type cells (24%) (Figure 4B; Supplementary Figure S10), consistent with the previous data of human BLM-deficient cells (Rao et al, 2007). The percentage of total stalled forks in Rif1−/− cells (60%) was not only higher than that of wild-type cells, but also BLM−/− cells (Figure 4B). In particular, the level of stalled forks represented by the unidirectional signal in Rif1−/− cells (about 30%) was about 10 times higher than that of wild-type cells (about 3%) and three times higher than that of BLM−/− cells (about 10%), suggesting that Rif1 could have a function more important than BLM in preventing accumulation of stalled forks. Notably, the percentage of total stalled forks or those represented by unidirectional signals in BLM−/−/Rif1−/− cells (about 40 and 7%, respectively) was lower than those of Rif1−/− cells (60 and 30%, respectively), but similar to that of BLM−/− cells. The data suggest that the BLM complex without Rif1 may be more detrimental to cells than complete absence of the BLM–Rif1 complex. The fact that Rif1−/− cells depend on BLM to accumulate increased levels of stalled forks supports the notion that the two genes work in the same pathway.
Rif1 and BLM act in the same pathway to promote recovery of stalled forks
The increased accumulation of stalled forks may be due to defective recovery of these forks, as has been observed in human BLM-deficient cells treated with either aphidicolin or HU (Davies et al, 2007; Shimura et al, 2008). This prompted us to investigate whether Rif1−/− DT40 cells have the same defect using a similar single DNA molecule assay (Edmunds et al, 2008) (Figure 4C). In this method, cells were sequentially pulse labelled with IdU and CldU as described above. One important difference is that the CldU labelling was performed in the presence of aphidicolin, which is expected to stall replication forks and inhibit the CldU incorporation (Shimura et al, 2008). Recovery of these stalled forks will allow CldU labelling to continue, and the efficiency of this recovery is negatively correlated with the ratio between the IdU and CldU tracks (the higher ratio represents more defective recovery of stalled forks).
Without aphidicolin treatment, the mean ratio of IdU:CldU was close to 1.0 in both wild-type and mutant DT40 cells (Figure 4D). Following aphidicolin treatment, the ratio was increased to about 3.0 in wild-type cells, which reflects the normal efficiency of the cellular machinery to respond to forks stalled by aphidicolin. In BLM−/− or Rif1−/− cells, this ratio of IdU:CldU was increased to about 3.7, which is significantly higher than that of the wild-type cells (P-value<0.001) and suggests a reduced efficiency of stalled fork recovery of both cells. Notably, the ratio in BLM−/−/Rif1−/− cells was similar to that in BLM−/− or Rif1−/− cells. This similarity was further revealed by a plot of the cumulative percentage forks versus each IdU:CldU ratio, which clearly shows that the IdU:CldU ratio of all mutant cells is similar to each other but is higher than that of the wild-type cells (Figure 4E). The data suggest that Rif1 and BLM work in the same pathway to promote recovery of stalled forks.
Vertebrate Rif1 is predicted to contain a HEAT-repeat domain and a DNA-binding domain
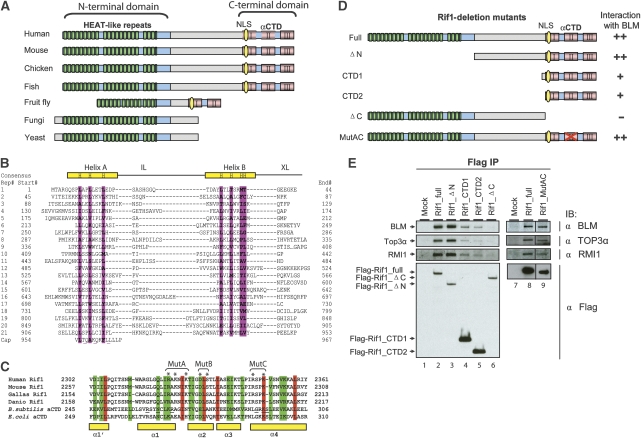
Bioinformatic analyses revealed that Rif1 contains two conserved domains at each terminus and a non-conserved middle region (Figure 5A). The N-terminal domain is conserved in all eukaryotes and is predicted to consist of 14–21 tandem HEAT-like (Huntingtin, Elongation factor 3, A subunit of protein phosphatase 2A, and Tor1) repeats, whose lengths vary among different species (Figure 5B; Supplementary Figure S11A). HEAT repeats have been found in proteins of diverse functions, including DNA damage response molecules ATM, ATR, and DNA-PKcs (Perry and Kleckner, 2003), where they mediate critical intermolecular protein–protein interactions (You et al, 2005).
Figure 5.
Vertebrate Rif1 consists of a HEAT-repeat domain and a C-terminal domain; the latter contains a DNA-binding domain and interacts with the BLM complex. (A) Schematic representation of Rif1 in different eukaryotic species. Conserved and non-conserved regions are labelled by blue and grey, respectively. The green blocks show the arrangement of predicted HEAT repeats. The three conserved C-terminal subdomains are marked by red blocks. The C-II subdomain resembles αCTD domain of the bacterial RNA polymerase α. The potential nuclear localization signal (NLS) is marked by yellow oval. The Rif1 orthologs are identified by using the BLASTP algorithm to search the NR database maintained at NCBI. They are from mouse (Mus musculus NP_780447), chicken (Gallus gallus XP_422162), zebrafish (Danio rerio XP_695378), fruit fly (Drosophila melanogaster NP_725497), fungi (Penicillium marneffei XP_002150486), and yeast (Schizosaccharomyces pombe NP_593910). (B) Structure-based sequence alignment of the human Rif1 HEAT-repeat domain. Repeat numbers and corresponding starting amino acids are shown on the left. Structural positions with strong preferences for hydrophobic amino acids are shaded purple and represented on the ‘Consensus' line with an ‘H'. Two gaps between repeats 9–10 and repeats 18–19 are shown with ‘…'. IL, intraunit loop; XL, external or interunit loop. (C) Sequence alignment and secondary structure elements of Rif1 and RNA polymerase αCTD domains. The sequences shown include Rif1 from four species in (A) and RNA polymerase α subunit of Bacillus subtilis (P20429) and Escherichia coli (ACI76960). The residues mutated in MutA, B, and C are marked with asterisks. (D) Schematic representation of different Rif1-deletion mutants (left), and their ability to coimmunoprecipitate with BLM from HEK293 extract (right). (E) IP-western testing whether Rif1-deletion mutants in (D) and a point mutant (AC) coimmunoprecipitate with different BLM complex components. The Flag-tagged full-length (FL) or different mutants of Rif1 were transfected into HEK293 cells, and coimmunoprecipitation was performed using the Flag antibody. The data on the right (lanes 7–9) are spliced and assembled together from the same immunoblotting images (Supplementary Figure S14). The black lines indicate where the splicing and assembly occur.
Unlike HEAT repeats that are conserved in all eukaryotes, the C-terminal domain is conserved only in vertebrates, partially conserved in fruit flies, and completely absent in yeast (Figure 5A). The domain can be divided into three subdomains: C-I, C-II, and C-III. Interestingly, the C-II subdomain shows strong similarity to the carboxyl-terminal domain of bacterial RNA polymerase α subunit (αCTD domain) (Figure 5C; Supplementary Figure S11B). The αCTD domain has two important activities: to bind promoter DNA and to interact with transcriptional activators (Ross et al, 2001; Benoff et al, 2002; Newberry et al, 2005). The similarity between C-II and the αCTD domain hinted that Rif1 may possess DNA-binding activity through its C-terminal domain.
The conserved C-terminal domain of hRif1 is required for its association with the BLM complex
We generated a series of Flag-tagged deletion mutants of hRif1 to map the domain that interacts with the BLM complex (Figure 5D). Transfection and IP-western using HEK293 cells showed that a mutant deleted of HEAT repeats (ΔN) retained normal association with BLM, Topo 3α, and RMI1 (Figure 5E, lanes 2 and 3), whereas a mutant deleted of the C-terminal domain had no detectable association (Figure 5E, lane 6), suggesting that the BLM-interaction domain lies within the C-terminal domain. Two mutants with only the C-terminal domain (CTD1 and CTD2) retained association with BLM complex components, albeit at reduced levels (Figure 5E, compare lanes 4, 5 with 2 and 3). The results suggest that the C-terminal domain is the minimal domain required for association with BLM, whereas other regions of Rif1, such as the middle non-conserved region, can enhance this association.
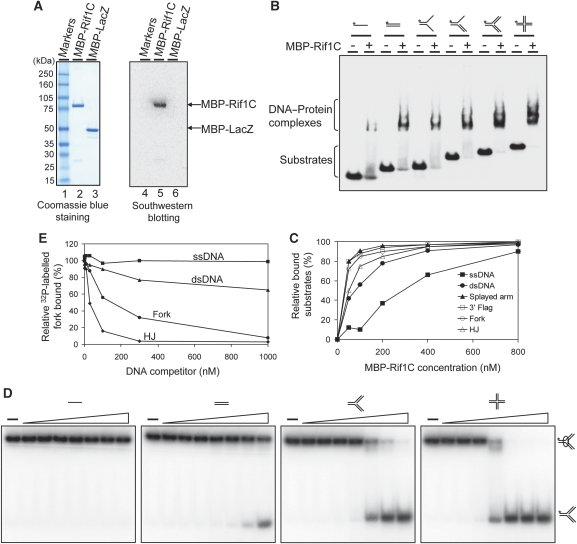
The conserved C-terminal domain of hRif1 possesses DNA-binding activity
The similarity between the C-II subdomain of hRif1 and the αCTD domain of bacterial RNA polymerase prompted us to investigate whether the C-terminal domain of hRif1 possesses DNA-binding activity. We expressed and purified a recombinant protein containing this domain (which includes C-I, C-II, and C-III) fused to the maltose-binding protein (MBP-Rif1C) (Figure 6A, left panel), and found that it binds a variety of DNA substrates, including ssDNA, dsDNA, splayed arm, 3′ flag, fork, and HJs, in gel-shift assays (Figure 6B). A titration experiment with increasing concentrations of the protein revealed that MBP-Rif1C had stronger binding activity for substrates containing dsDNA than ssDNA (Figure 6C; Supplementary Figure S12). Competition analyses using fork DNA as the probe further showed that HJ and fork are the strongest competitors, followed by dsDNA, with ssDNA as the weakest (Figure 6D and E). These data suggest that the C-terminal domain of Rif1 preferentially binds HJs and forks, and has intermediate affinity for dsDNA and lowest affinity for ssDNA.
Figure 6.
The conserved C-terminal region of Rif1 preferentially binds fork and HJ DNA over ssDNA. (A) Coomassie blue staining (left) and southwestern blotting (right) show that the purified MBP-Rif1C protein (MBP-fused to Rif1-C-terminal domain) has DNA-binding activity. A 32P-labelled HJ DNA was used as a probe. MBP-lacZ was included as a negative control. (B) Gel-shift assay shows that MBP-Rif1C has DNA-binding activity to various synthetic DNA substrates illustrated at the top. The 32P-labelled probe is denoted with an asterisk. Reactions contained 1 nM of the indicated 32P-labelled substrates and 400 nM purified MBP-Rif1C. The protein–DNA complexes were analysed by 3–15% polyacrylamide gels. (C) A titration experiment shows that MBP-Rif1C has strong DNA-binding affinity for substrates containing dsDNA compared with ssDNA. The primary data are presented in Supplementary Figure S12. (D, E) Competition gel-shift assay and its quantification to show which DNA substrates are preferentially recognized by MBP-Rif1C. In all, 400 nM MBP-Rif1C was incubated with 1 nM of 32P-labelled fork DNA in the presence of the indicated amounts of non-labelled DNA competitors as described in Materials and methods.
It was noted that in the competition assay, a 200-fold excess of non-labelled fork DNA did not fully eliminate the binding of MBP-Rif1C to the labelled probe. This could be due to slow dissociation rate of the preformed MBP-Rif1C-labelled DNA complex, as the competitor DNA was added 15 min after formation of the complex.
We investigated whether the observed DNA-binding activity is derived from MBP-Rif1C itself but not a contaminant by South–Western assay using the HJ DNA as the probe. The DNA probe specifically recognized the MBP-Rif1C protein but not any other contaminants, as evidenced by the fact that the single band on the audioradiograph matched exactly with the position of MBP-Rif1C protein on the Coomassie-stained gel (Figure 6A, compare lane 2 with lane 5). As a control, the probe did not react with any of the 12 marker proteins (Figure 6A, lanes 1 and 4) or MBP-LacZ (Figure 6A, lanes 3 and 6). The latter control indicates that the observed DNA-binding activity is derived from Rif1C but not MBP.
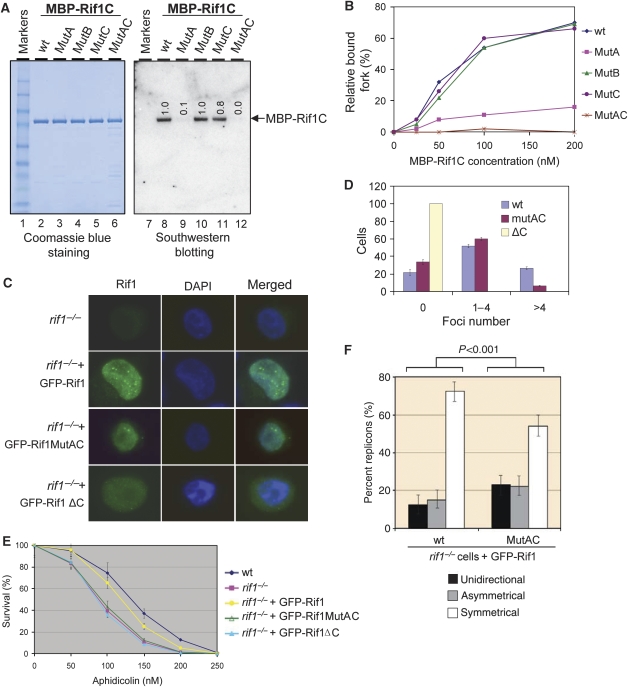
Mutations within the αCTD-like domain of hRif1 abrogates its DNA-binding activity
To study whether the αCTD-like domain (C-II) is responsible for the observed DNA-binding activity, we generated three point mutations within this domain by substituting three clusters of conserved amino-acid residues with alanine (see mutants A, B, and C in Figure 5C; Figure 7A, lanes 3–5). Previous studies of the αCTD domain of Bacillus subtilis RNA polymerase revealed that the residues near mutants A and C can directly contact DNA (Figure 5C, underlined residues) (Newberry et al, 2005), hinting that these two mutants may be defective in DNA binding. Consistent with this prediction, South–Western analyses showed that the DNA-binding activity hRif1 was normal in mutant B, reduced in mutant A (>90%) and C (about 20%), and completely deficient in the AC double mutant (carrying simultaneous mutations of residues in both mutants A and C) (Figure 7A, compare lanes 9–12 with 8). One caveat of this assay is that the proteins need to be denatured and renatured prior to the analysis. Therefore, one explanation of the above data is that renaturation rather than DNA-binding activity of the protein was affected in the mutants. To circumvent this problem, we performed gel-shift analysis using non-denatured proteins and found that the DNA-binding activity was strongly reduced in mutant A and was completely lost in the AC double mutant (Figure 7B; Supplementary Figure S13). These data demonstrate that hRif1 has DNA-binding activity derived from its αCTD domain.
Figure 7.
The DNA-binding activity of Rif1 is required for its efficient recruitment to HU-induced nuclear foci and for cellular resistance to replication stress. (A) Coomassie blue staining (left) and southwestern blotting (right) to show that the DNA-binding activity of purified MBP-Rif1C is derived from its αCTD motif. The mutated residues in mutants A, B, and C were shown in Figure 5C. The numbers in the autoradiograph indicate the relative-binding affinity of each mutant compared with the wild-type protein. (B) A graph showing the DNA-binding activity of MBP-Rif1C wild-type (WT) and its various point mutants (MutA, B, C, and the AC double mutant) by gel-shift assays. The raw data are shown in Supplementary Figure S13. Reaction mixtures included the indicated proteins (at 0, 25, 50, 100, and 200 nM) and 1 nM 32P-labelled fork DNA. The protein–DNA complexes were analysed by 4–12% TBE gels. (C, D) Representative immunofluorescence images and statistical analyses show that recruitment of Rif1 to HU-induced nuclear foci is reduced in MutAC and eliminated in the C-terminal deletion mutant of GFP-tagged Rif1 (ΔC). The cells were treated with 2 mM HU 16 h before fixing and staining. More than 200 cells were scored for each preparation. (E) Sensitivity curves of the Rif1−/− DT40 cells complemented by wild-type, MutAC, or ΔC mutant of GFP-Rif1. Mean and s.d. from three independent experiments are shown. (F) Single-molecule analysis showed that the Rif1−/− DT40 cells complemented by GFP-Rif1-MutAC have an increased level of stalled replication forks compared with cells complemented by wild-type GFP-Rif1. This is reflected by the increase of unidirectional or asymmetrical forks and a simultaneous decrease of symmetrical forks. Histograms represent the sum of replicons measured and analysed in two independent experiments; the error bars indicate 95% confidence intervals for each set of data. A detailed description of the assay is described in legend for Figure 4B.
The DNA-binding activity of the αCTD domain is required for Rif1 to prevent accumulation of stalled forks and to resist replication stress
The data from this and other studies have shown that Rif1 is recruited to stalled replication forks and is required for cellular survival to replication stress (Silverman et al, 2004; Buonomo et al, 2009) (Figures 2, 3 and 4). We investigated whether the DNA-binding activity of the Rif1-αCTD domain is required for these functions by testing the αCTD mutants (AC and ΔC) in Rif1−/− DT40 cells. For wild-type hRif1-complemented cells, Rif1 formed nuclear foci in response to HU (Figure 7C and D), and their cellular sensitivity to aphidicolin was largely corrected (Figure 7E). For mutant AC-complemented Rif1−/− cells, the percentage of cells with more than four Rif1 foci was reduced by four-fold (Figure 7C and D), and their aphidicolin sensitivity was comparable to that of Rif1−/− null cells (Figure 7E). Moreover, single-molecule analyses of untreated cells revealed that these cells displayed a higher level of stalled replication forks (represented by the sum of unidirectional and asymmetrical forks) compared with cells complemented with wild-type hRif1 (Figure 7F). A control experiment showed that mutant AC has normal association with the BLM complex (Figure 5E, lanes 7–9; Supplementary Figure S14). The data suggest that the DNA-binding activity of the αCTD domain is important for efficient recruitment of Rif1 to stalled replication forks, where it can promote recovery of these forks, leading to cellular resistance to replication stress.
Optimal Rif1 recruitment to stalled forks requires both αCTD DNA-binding activity and BLM association
The findings that the mutant AC can form residual amounts of nuclear foci indicate that Rif1 can be recruited to stalled forks through a mechanism other than its αCTD-associated DNA binding. We hypothesize that this alternative mechanism might be its BLM association based on the data that hRif1 recruitment to ICLs is delayed in BLM-deficient cells (Figure 2E and F). We therefore examined Rif1−/− cells complemented by the mutant ΔC, which lacks both DNA-binding activity and BLM association (Figure 5E). The aphidicolin sensitivity of these cells was comparable to that of Rif1−/− null cells or mutant AC-complemented cells (Figure 7E), supporting the notion that the αCTD-associated DNA-binding activity is required for cellular survival to replication stress. Notably, the mutant ΔC-complemented cells had no detectable Rif1 foci (Figure 7C and D), unlike the mutant AC-complemented cells that had reduced but detectable amounts of Rif1 foci. Together with the data of Figure 2, these findings suggest that both BLM association and αCTD-associated DNA binding are required for optimal recruitment of Rif1 to stalled forks; and disruption of either activity can reduce the efficiency or rate of the recruitment, whereas inactivation of both can eliminate the recruitment.
One unique defect of mutant ΔC is that it is localized in both the nucleus and cytoplasm, whereas wild-type and mutant AC are present exclusively in the nucleus (Figure 7C; similar data were found in human cells (unpublished)). A nuclear localization sequence has been detected within the C-terminal domain of Rif1 (Silverman et al, 2004). Deletion of this sequence in mutant ΔC may cause its inefficient nuclear localization, contributing to its failure to relocalize to stalled forks and to complement aphidicolin sensitivity.
Yeast Rif1, which lacks the C-terminal domain, acts in a pathway separate from Sgs1 (BLM) to resist DNA replication stress
The data above demonstrate that the C-terminal domain of vertebrate Rif1 is important for BLM complex association, DNA binding, and cellular resistance to replication stress. However, this domain is missing in yeast Rif1, which suggests that in yeast, Rif1 and its BLM homolog (Sgs1) may work independently to promote resistance to replication stress. We investigated this possibility by performing epistasis analyses in yeast. Yeast sgs1Δ, rif1Δ, and sgs1Δ rif1Δ double mutants were constructed, diluted, and spotted onto solid media containing HU or MMS (Supplementary Figure S15). As previously shown (Mullen et al, 2001), the growth of sgs1Δ mutants was slightly inhibited in the presence of HU or MMS. The rif1Δ single mutant also displayed a slight sensitivity to these drugs. However, the sensitivity of sgs1Δ rif1Δ double mutants was greater than that of either single mutant. Thus, although yeast Rif1 is required for resistance to DNA replication stress, it acts in a pathway that is separate from Sgs1.
Discussion
Vertebrate Rif1 works with BLM to promote recovery of stalled replication forks
BLM and its associated complex have critical functions in maintaining genome stability through at least three mechanisms: suppression of crossover recombination, facilitating HR-dependent DNA repair, and promoting recovery of stalled replication forks. Rif1 has been found to participate in DNA damage response, contribute to resistance to replication stress, and colocalize with BLM in nuclear foci induced by the replication stress (Silverman et al, 2004; Xu and Blackburn, 2004; Buonomo et al, 2009; Wang et al, 2009). However, Rif1 has not been found in any multiprotein complexes, and there is no evidence for physical or functional interactions between Rif1 and BLM. Moreover, no known domains or biochemical activity have been associated with Rif1, so that its mechanism of action is unknown.
Here, we present evidence that Rif1 is a new component of the BLM complex, and it cooperates with BLM to prevent accumulation of stalled replication forks. First, Rif1 physically interacts with and is stabilized by the BLM complex. Second, Rif1 and BLM are recruited with similar kinetics to replication forks stalled by ICLs; and the Rif1 recruitment is delayed in BLM-deficient cells. Third, our genetic analyses in DT40 cells suggest that BLM and Rif1 work in a common pathway to resist replication stress induced by FUdR or aphidicolin, and to promote recovery of replication forks stalled by aphidicolin treatment. Importantly, we have identified two known domains in Rif1, HEAT repeats and αCTD domain, and shown that the latter can preferentially bind fork and HJ DNA in vitro and is required for Rif1 to resist replication stress in vivo. The interactions between Rif1 and these DNA structures may augment the ability of the BLM complex to process DNA intermediates generated during recovery of stalled replication forks.
Although Rif1 and BLM work in the same pathway to resist FUdR and aphidicolin-induced replication stress, the two proteins exhibited a complicated relationship in response to these and other drugs (Table I). For example, BLM−/−, Rif1−/−, and the BLM−/−/Rif1−/− double-mutant DT40 cells exhibit similar sensitivity to high dose of FUdR. In contrast, BLM mutation suppresses aphidicolin sensitivity of the Rif1−/− DT40 cells. Moreover, Rif1−/−, but not BLM−/−, DT40 cells showed sensitivity to HU, whereas the double mutants showed the same sensitivity as the BLM−/− cells. The underlying mechanism for this difference is unclear, but it may be related to the different replication stress induced by these drugs: FUdR inhibits incorporation of thymidine nucleotides into DNA; aphidicolin directly inhibits DNA polymerase I; and HU treatment depletes dNTP pools in cells by inhibiting ribonucleotide reductase. Future experiments with other drugs may help explain the differences.
Table 1. Genetic interactions between BLM (SGS1) and Rif1.
| Assay | WT | Rif1−/− | Blm−/− (sgs1Δ) | Rif1−/−/Blm−/− (rif1Δ/sgs1Δ) | Genetic interactions |
|---|---|---|---|---|---|
| DT40 | |||||
| FUdR sensitivity (high concentration) | − | ++ | ++ | ++ | Epistatic |
| Aphidicolin sensitivity | − | ++ | − | + | Epistatic; blm suppresses |
| HU sensitivity | − | + | − | + | No interaction |
| Chromosome aberration after FUdR treatment | + | ++ | ++ | ++ | Epistatic |
| Spontaneous stalled replication fork | − | ++ | + | + | Epistatic; blm suppresses |
| Fork stalling after aphidicolin treatment | − | + | + | + | Epistatic |
| SCE | − | − | ++ | ++ | No interaction |
| Yeast | |||||
| MMS sensitivity | − | + | + | ++ | Non-epistatic |
| HU sensitivity | − | + | + | ++ | Non-epistatic |
| Summary of the phenotypes of wild type, rif1−/− single mutant, blm−/−(sgs1Δ) single mutant, and the double mutant in the indicated assays. Their epistatic relationships are summarized on the right. Symbols: −, cellular response absent; +, cellular response present; ++, cellular response enhanced. | |||||
Rif1 is dispensable for the suppression of SCE and HR-dependent DSB repair in DT40 cells
Although other components of the BLM complex are essential for the suppression of SCE (Chaganti et al, 1974; Wang et al, 2000; Wu and Hickson, 2003; Yin et al, 2005; Otsuki et al, 2007; Xu et al, 2008), Rif1 is dispensable for this process. This is evidenced by the observation that the SCE frequency is not significantly increased when Rif1 is inactivated either in wild-type DT40 cells or in those lacking BLM or its associated proteins (RMI2, FANCC, and FANCM) (Supplementary Figure S16A). A recent finding that mouse Rif1−/− cells have normal SCE frequency is consistent with this notion (Buonomo et al, 2009).
Rif1 has been suggested to promote HR-dependent DSB repair based on siRNA studies (Buonomo et al, 2009; Wang et al, 2009). In our hands, the efficiency of HR-dependent DSB repair is reduced by only 10–15% in Rif1-depleted U2OS cells and remains largely normal in Rif1-depleted HEK293 cells or Rif1−/− DT40 cells (Supplementary Figure S16B–D). Surprisingly, the HR-dependent gene-targeting efficiency in Rif1−/− DT40 cells is not decreased, but rather increased, compared with that of the wild-type cells, suggesting that Rif1 negatively regulates this process (Supplementary Figure S16E). This result is reminiscent to the observation that Rif1 mutation in yeast rescues telomere recombination defects in Sgs1 or Top3 mutants (Tsai et al, 2006), implying that Rif1 inhibits Sgs1/Top3-dependent HR at telomeres. Thus, the function of Rif1 in HR-dependent processes could be indirect, and its effects may be positive, negative, or insignificant, depending on the cell type and the process.
Structural basis for distinct functions of Rif1 in human and yeast
Rif1 was originally identified in budding yeast for its function in regulating telomere length through interaction with RAP1 (Hardy et al, 1992). However, mammalian Rif1 neither binds normal telomeres nor contributes to telomere homeostasis (Silverman et al, 2004; Xu and Blackburn, 2004). We found that the telomere lengths in Rif1−/− DT40 cells are indistinguishable from those of wild-type cells when they are cultured over 100 doubling times (data not shown), suggesting that Rif1 is dispensable for normal telomere maintenance in vertebrates.
What is the underlying cause for the functional difference between vertebrate and yeast Rif1? We found a key difference in Rif1 sequences from the above species: Rif1 from vertebrates, but not yeast, contains a C-terminal domain that mediates two critical interactions: to associate with the BLM complex and to preferentially bind DNA substrates that mimic HJ and forks. We propose that through these interactions, this domain can target vertebrate Rif1 to stalled replication forks and promote their restart. Yeast Rif1 lacks this domain so that its recruitment to stalled forks should be independent of Sgs1. In agreement with our hypothesis, Rif1 recruitment to ICLs is delayed in BLM-deficient cells, and Rif1 redistribution to HU-induced nuclear foci is completely inactivated when the entire C-terminal domain is deleted. Moreover, loss of yeast RIF1 gene in the sgs1Δ mutant background revealed that these two genes work in separate pathways to resist replication stress caused by HU and MMS (Supplementary Figure S15). The lack of a C-terminal domain that binds BLM/Sgs1 (and DNA) may explain why yeast Rif1, unlike vertebrate Rif1, functions independently from Sgs1 to resist replication stress.
It should be noted that in yeast, Rif1 has a function in the alternative lengthening of telomeres (ALT) pathway that depends on Sgs1–Top3 and involves HR (Tsai et al, 2006). In mammals, this pathway similarly depends on BLM-Topo 3α; and Rif1 has also been detected at telomeric DNA in ALT cells (Stavropoulos et al, 2002; Silverman et al, 2004; Temime-Smaali et al, 2008). It is therefore possible that Rif1 and BLM may work together in telomere recombination in both species. If this is the case, we would predict that the telomeric function of Rif1 is mediated through its HEAT repeats, which are conserved in all species, and that the physical interactions between Rif1 and BLM/Sgs1 may be dispensable for this telomeric function.
Rif1 provides a new DNA-binding surface for BLM complex to maintain normal replication
Our bioinformatic analyses predict that vertebrate Rif1 has a C-terminal domain that includes a DNA-binding motif similar to that of the αCTD domain of bacterial polymerases. We showed that the C-terminal domain has three important functions: (1) it is required for nuclear localization of Rif1; (2) it is essential for assembly of Rif1 into the BLM complex; and (3) it has a DNA-binding activity with highest affinity for fork and HJ DNA, intermediate affinity for dsDNA, and low affinity for ssDNA. This binding specificity differs from that of RPA, another component of the BLM complex with high affinity for ssDNA. Complementation analyses showed that the DNA-binding activity of Rif1-αCTD is required for Rif1 to relocalize efficiently to stalled forks, to prevent accumulation of stalled forks, and to promote cellular resistance to replication stress. Our data suggest that the Rif1-αCTD domain could provide a new DNA-binding surface for the BLM complex that appears to be specifically important for recovery of stalled replication forks.
How does Rif1 promote the recovery of stalled forks within the BLM complex? One possibility is that Rif1 may bind stalled forks through its αCTD domain, and then recruits BLM to promote its fork reversal activity. We do not favour this model because we are unable to detect significant effects of Rif1 on DNA binding, replication fork reversal, or DNA-stimulated ATPase activities of BLM (data not shown). Another possibility is that BLM binds and remodels the stalled forks first (this may generate HJ or ‘chickenfoot' structures), and then recruits Rif1 through protein–protein interactions. Rif1 could then bind the remodelled forks through its αCTD domain, and recruit additional repair proteins through its HEAT repeats, which are known to serve as a flexible platform for other factors to assemble (Andrade et al, 2001). The binding between the Rif1-αCTD domain and the remodelled forks may also stabilize the retention of Rif1 at the stalled forks, so that disruption of this binding could result in Rif1 dissociation from the forks, as observed in the Rif1-AC mutant.
Several lines of evidence lead us to favour the second model. First, Rif1 recruitment to stalled forks is delayed in BLM-deficient cells. Second, the Rif1-AC mutant that lacks DNA-binding activity can still be recruited to stalled forks, whereas the Rif1-ΔC mutant that lacks both DNA-binding activity and BLM association is completely deficient. These data suggest that BLM is needed for both the rate and efficiency of Rif1 recruitment to the stalled forks. Third, the aphidicolin sensitivity and increased accumulation of stalled forks in Rif1−/− cells are suppressed by inactivation of BLM. This implies that without Rif1, BLM complex may generate remodelled forks that cannot be properly processed, so that they become targets to nuclease attacks, leading to broken forks and other detrimental effects on cell survival. Mutation of BLM disrupts generation of these forks and thus suppresses the defects in rif1 mutants, as cells could then use alternative pathways mediated by other remodelling enzymes (such as a different RecQ helicase) to recover the stalled forks. Rif1 may still be able to bind the remodelled forks through its αCTD domain, but this binding could be slower and inefficient as Rif1 has no direct association with these other enzymes, so that its binding should be an intermolecular reaction, rather than the intramolecular reaction when BLM was present.
Interestingly, the data that several Rif1 mutant phenotypes are suppressed by BLM mutation mimic the earlier findings in which defects associated with the loss of two other BLM/Sgs1 complex components (Top3 or Rmi1) are also suppressed by inactivation of BLM/Sgs1 (Mullen et al, 2005). As such, BLM appears to be a master regulator that is needed by Rif1 as well as by the TOP3/RMI complex to execute diverse functions in order to maintain normal replication and genome stability.
Materials and methods
Cell culture and transfection
Hela and HEK293 cells were cultured at 37°C, 5% CO2, in DMEM supplemented with 10% fetal calf serum. DT40 cells were cultured at 39.5°C, 5% CO2 in RPMI 1640 medium supplemented with 10% fetal calf serum, 1% chicken serum, 10 mM Hepes, and 1% penicillin–streptomycin mixture. Transfection was carried out by electroporation using the Amaxa Nucleofector2 in Solution T. For selection, growth medium containing G418 (2 mg/ml) or puromycin (0.5 μg/ml) was used.
siRNA knockdown
hRif1 siRNA oligos (NNGCAGCUUAUGACUACUAAA and NNGCUUGGUGAAGUCAGUUAC) were obtained from Dharmacon. BLM, RMI1, RMI2, and control siRNA oligos were described previously (Yin et al, 2005; Xu et al, 2008). Transfections of siRNAs were carried out with Oligofectamine following the manufacturer's protocols (Invitrogen).
Antibodies
A rabbit hRif1 polyclonal antibody was made against a fusion protein containing a region of hRif1 (residues 1367–1461) fused to the maltose-binding protein (New England Biolabs). Polyclonal antibodies against BLM, Topo 3α, RMI1, or RMI2 were described previously (Xu et al, 2008).
Fractionation, gel-filtration analysis, and immunoprecipitation
These experiments were performed as described previously (Guo et al, 2009).
Immunofluorescence localization of Rif1 at laser-activated psoralen
This experiment followed a previous protocol (Muniandy et al, 2009).
Generation of the DT40 knockout strains
To generate Rif1 knockout constructs, the 5′ and 3′ arms of Rif1 genomic DNA was amplified from genomic DNA using primer pairs: CCTCGCGGCCGCTCTCCTTCCTGTGTTTG/TCATGGATCCTTTCACACTGCAGCTTCAC and CTGTGGATCCTGTATGCTAAATGTAGGAG/ACCAGGTACCAAGTCTCTCCGAAAAG, respectively. These arms were inserted into NotI/BamHI and BamHI/KpnI sites of pBluescript vector, successively. The resistant gene cassettes were subsequently inserted into BamHI site. The primers ggaacttcagaaattgttttctac/ATCTATTAGACTCTTCCATGCAATG were used for RT–PCR.
The DT40 FANCC−, FANCM−/−, BLM−/−, and RMI2−/− cells have been described previously (Hirano et al, 2005; Mosedale et al, 2005; Otsuki et al, 2007; Xu et al, 2008). BLM−/−/Rif1−/− and FANCM−/−/Rif1−/− cells were generated by inactivating Rif1 gene in BLM−/− and FANCM−/− cells, respectively. RMI2−/−/Rif1−/− and FANCC−/Rif1−/− cells were generated by inactivating RMI2 gene and FANCC gene in Rif1−/− cells, respectively. For complementation experiments, the resistant gene cassettes were first removed from Rif1−/− cells by lox sites recombination, and then these cells were transfected with pEGFPc1-Rif1 (Xu and Blackburn, 2004) vector and selected.
To generate the DR-GFP-contained DT40 or Rif1−/− cells, the vector pOVA-DRGFP (OVALBUMIN locus knock-in construct contained a DR-GFP reporter, a gift from Dr S Takeda) was linearized and transfected to wild-type or Rif1−/− DT40 cells. The targeted cells carrying the DR-GFP knocked-in at the OVALBUMIN locus were confirmed by PCR. The DRGFP-containing FANCC− cells were generated by inactivating FANCC gene in wild-type DT40 cells carrying the DRGFP reporter knocked-in at the OVALBUMIN locus.
SCE assay, growth curve analysis, and sensitivity assay
These assays were performed as described previously (Xu et al, 2008). One exception is the sensitivity assay to aphidicolin and HU, which used cell density of 300–1000 per well and 72 h incubation.
Recombinant protein purification
The MBP-Rif1C wild-type and mutant fusion proteins were expressed in Escherichia coli (Rosetta2 cells, Novagen) using pMALc2E vector (New England Biolabs). Cells were grown at 32°C until OD600=0.8, and were induced with 0.1 mM IPTG at 20°C for 4.0 h. The cell pellet from 1-l culture was lysed in 30 ml B-PER buffer (PIECE) supplemented with 10% glycerol, 300 units DNase I, protease inhibitor cocktail and 1 mM PMSF. The mixture was incubated at room temperature for 15 min and centrifuged at 27 000g for 30 min. The supernatant was added with imidazole to 30 mM and incubated with 1 ml Ni Sepharose 6 Fast Flow beads (GE Healthcare) at 4°C for 2 h. The beads were washed three times with 50 ml PBS buffer (10 mM phosphate, pH 7.4, 138 mM NaCl, and 2.7 mM KCl) supplemented with 30 mM imidazole, 350 mM NaCl, 10% glycerol, and 0.1% Triton X-100. The beads were then poured into a Bio-Rad 2-ml disposable column, and the bound proteins were eluted with 150 mM imidazole in PBSG buffer (PBS buffer supplemented with 10% glycerol and 0.1% Triton X-100). The peak fractions were incubated with 1 ml Amylose Resin (New England Biolabs) at 4°C 1 h. The resin was washed three times with 50 ml PBSG and poured into a Bio-Rad 2-ml disposable column. The bound protein MBP-Rif1C was eluted with 10 mM maltose in PBSG buffer.
MBP-LacZ was expressed and purified with the same method as MBP-Rif1C except that the step of Ni Sepharose 6 Fast Flow beads was omitted.
DNA-binding assay and competition assay
The DNA substrates were made as described previously (Xue et al, 2008). The indicated amount of proteins and 1 nM 32P-labelled DNA subtracts were incubated at 25°C in 10 μl reaction buffer (25 mM Tris–HCl at pH 7.5, 4 mM MgCl2, 50 mM KCl, 1 mM DTT, 100 μg/ml BSA, and 5% glycerol) for 15 min. For competition assay, the indicated amount of cold subtracts were added for another 30 min. The reaction mixture was loaded and resolved on a 6% DNA Retardation Gel (Invitrogen) or 3–15% TBE gel.
Southwestern blotting
Purified MBP-Rif1C or MBP-LacZ proteins (1 μg) were incubated at 37°C in Laemmli sample buffer (Bio-Rad) for 10 min and loaded on 8–16% Tris–Glycine Gel (Invitrogen). The proteins were transferred to nitrocellulose membrane and renatured in blocking buffer (25 mM Tris–HCl at pH 7.5, 4 mM MgCl2, 150 mM KCl, 1 mM DTT, 1% BSA, 10% glycerol, and 0.1% Triton X-100) for 6 h at 4°C. The membrane was incubated with 1 nM 32P-labelled HJ in hybrid buffer (25 mM Tris–HCl at pH 7.5, 4 mM MgCl2, 50 mM KCl, 1 mM DTT, 0.1% BSA, and 10% glycerol) overnight at 4°C. After washing three times with PBS buffer, the membrane was analysed by PhosphorImaging.
Other Materials and methods are described in Supplementary data.
Supplementary Material
Acknowledgments
We thank Drs E Blackburn and L Xu for hRif1 cDNA, I Hickson for Topo 3α antibody, S Takeda for Ovalbumin-DRGFP plasmid, M Takata for FANCC-targeting vector, KJ Patel for FANCM−/− DT40 cells, S Buonomo for Rif1−/− mouse cells, and S Fuggman for reagents and advice. This work was supported in part by the Intramural Research Program of the National Institute on Aging (Z01 AG000657-08), National Institute of Health, and NIH grant GM071268 (to SJB). It is also supported by an AIRC grant (to AV).
Footnotes
The authors declare that they have no conflict of interest.
References
- Andrade MA, Perez-Iratxeta C, Ponting CP (2001) Protein repeats: structures, functions, and evolution. J Struct Biol 134: 117–131 [DOI] [PubMed] [Google Scholar]
- Bachrati CZ, Borts RH, Hickson ID (2006) Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res 34: 2269–2279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachrati CZ, Hickson ID (2008) RecQ helicases: guardian angels of the DNA replication fork. Chromosoma 117: 219–233 [DOI] [PubMed] [Google Scholar]
- Benoff B, Yang H, Lawson CL, Parkinson G, Liu J, Blatter E, Ebright YW, Berman HM, Ebright RH (2002) Structural basis of transcription activation: the CAP-alpha CTD-DNA complex. Science 297: 1562–1566 [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Yu X, Egelman EH, Mazin AV (2007) Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev 21: 3085–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Wu Y, Ferguson D, de Lange T (2009) Mammalian Rif1 contributes to replication stress survival and homology-directed repair. J Cell Biol 187: 385–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaganti RS, Schonberg S, German J (1974) A manyfold increase in sister chromatid exchanges in Bloom's syndrome lymphocytes. Proc Natl Acad Sci USA 71: 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CF, Brill SJ (2010) An essential DNA strand-exchange activity is conserved in the divergent N-termini of BLM orthologs. EMBO J 29: 1713–1725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti C, Seiler JA, Pommier Y (2007) The mammalian DNA replication elongation checkpoint: implication of Chk1 and relationship with origin firing as determined by single DNA molecule and single cell analyses. Cell Cycle 6: 2760–2767 [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Hickson ID (2007) Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat Struct Mol Biol 14: 677–679 [DOI] [PubMed] [Google Scholar]
- Edmunds CE, Simpson LJ, Sale JE (2008) PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol Cell 30: 519–529 [DOI] [PubMed] [Google Scholar]
- Gaymes TJ, North PS, Brady N, Hickson ID, Mufti GJ, Rassool FV (2002) Increased error-prone non homologous DNA end-joining--a proposed mechanism of chromosomal instability in Bloom's syndrome. Oncogene 21: 2525–2533 [DOI] [PubMed] [Google Scholar]
- German J (1993) Bloom syndrome: a mendelian prototype of somatic mutational disease. Medicine (Baltimore) 72: 393–406 [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP (2008) DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R, Xu D, Wang W (2009) Identification and analysis of new proteins involved in the DNA damage response network of Fanconi anemia and Bloom syndrome. Methods 48: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Sussel L, Shore D (1992) A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev 6: 801–814 [DOI] [PubMed] [Google Scholar]
- Hirano S, Yamamoto K, Ishiai M, Yamazoe M, Seki M, Matsushita N, Ohzeki M, Yamashita YM, Arakawa H, Buerstedde JM, Enomoto T, Takeda S, Thompson LH, Takata M (2005) Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J 24: 418–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth KD, Blood KA, Ng BL, Beavis JC, Chua Y, Cooke SL, Raby S, Ichimura K, Collins VP, Carter NP, Edwards PA (2008) Array painting reveals a high frequency of balanced translocations in breast cancer cell lines that break in cancer-relevant genes. Oncogene 27: 3345–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Constantinou A, Li JL, West SC, Hickson ID (2000) The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc Natl Acad Sci USA 97: 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVey M, Andersen SL, Broze Y, Sekelsky J (2007) Multiple functions of Drosophila BLM helicase in maintenance of genome stability. Genetics 176: 1979–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Sechi S, Wallisch M, Yang D, Young MK, Joenje H, Hoatlin ME, Wang W (2003) A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol Cell Biol 23: 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol 12: 763–771 [DOI] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ (2005) Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol 25: 4476–4487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muniandy PA, Thapa D, Thazhathveetil AK, Liu ST, Seidman MM (2009) Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. J Biol Chem 284: 27908–27917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Chen C, Kolodner RD (2001) SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat Genet 27: 113–116 [DOI] [PubMed] [Google Scholar]
- Newberry KJ, Nakano S, Zuber P, Brennan RG (2005) Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the alpha C-terminal domain of RNA polymerase. Proc Natl Acad Sci USA 102: 15839–15844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuki M, Seki M, Inoue E, Yoshimura A, Kato G, Yamanouchi S, Kawabe Y, Tada S, Shinohara A, Komura J, Ono T, Takeda S, Ishii Y, Enomoto T (2007) Functional interactions between BLM and XRCC3 in the cell. J Cell Biol 179: 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J, Kleckner N (2003) The ATRs, ATMs, and TORs are giant HEAT repeat proteins. Cell 112: 151–155 [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L (2006) The Bloom's syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281: 22839–22846 [DOI] [PubMed] [Google Scholar]
- Rao VA, Conti C, Guirouilh-Barbat J, Nakamura A, Miao ZH, Davies SL, Sacca B, Hickson ID, Bensimon A, Pommier Y (2007) Endogenous gamma-H2AX-ATM-Chk2 checkpoint activation in Bloom's syndrome helicase deficient cells is related to DNA replication arrested forks. Mol Cancer Res 5: 713–724 [DOI] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P (2006) A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIalpha, and BLAP75. J Biol Chem 281: 13861–13864 [DOI] [PubMed] [Google Scholar]
- Ross W, Ernst A, Gourse RL (2001) Fine structure of E. coli RNA polymerase-promoter interactions: alpha subunit binding to the UP element minor groove. Genes Dev 15: 491–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Nakagawa T, Seki T, Kato G, Tada S, Takahashi Y, Yoshimura A, Kobayashi T, Aoki A, Otsuki M, Habermann FA, Tanabe H, Ishii Y, Enomoto T (2006) Bloom helicase and DNA topoisomerase IIIalpha are involved in the dissolution of sister chromatids. Mol Cell Biol 26: 6299–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Do H, Li Y, Chung WH, Tomasz M, de Winter JP, Xia B, Elledge SJ, Wang W, Li L (2009) Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell 35: 716–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura T, Torres MJ, Martin MM, Rao VA, Pommier Y, Katsura M, Miyagawa K, Aladjem MI (2008) Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. J Mol Biol 375: 1152–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman J, Takai H, Buonomo SB, Eisenhaber F, de Lange T (2004) Human Rif1, ortholog of a yeast telomeric protein, is regulated by ATM and 53BP1 and functions in the S-phase checkpoint. Genes Dev 18: 2108–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR (2008) BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev 22: 2856–2868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274 [DOI] [PubMed] [Google Scholar]
- Stavropoulos DJ, Bradshaw PS, Li X, Pasic I, Truong K, Ikura M, Ungrin M, Meyn MS (2002) The Bloom syndrome helicase BLM interacts with TRF2 in ALT cells and promotes telomeric DNA synthesis. Hum Mol Genet 11: 3135–3144 [DOI] [PubMed] [Google Scholar]
- Sun H, Karow JK, Hickson ID, Maizels N (1998) The Bloom's syndrome helicase unwinds G4 DNA. J Biol Chem 273: 27587–27592 [DOI] [PubMed] [Google Scholar]
- Temime-Smaali N, Guittat L, Wenner T, Bayart E, Douarre C, Gomez D, Giraud-Panis MJ, Londono-Vallejo A, Gilson E, Amor-Gueret M, Riou JF (2008) Topoisomerase IIIalpha is required for normal proliferation and telomere stability in alternative lengthening of telomeres. EMBO J 27: 1513–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangavel S, Mendoza-Maldonado R, Tissino E, Sidorova JM, Yin J, Wang W, Monnat RJ Jr, Falaschi A, Vindigni A (2010) The human RECQ1 and RECQ4 helicases play distinct roles in DNA replication initiation. Mol Cell Biol 30: 1382–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thazhathveetil AK, Liu ST, Indig FE, Seidman MM (2007) Psoralen conjugates for visualization of genomic interstrand cross-links localized by laser photoactivation. Bioconjug Chem 18: 431–437 [DOI] [PubMed] [Google Scholar]
- Tsai HJ, Huang WH, Li TK, Tsai YL, Wu KJ, Tseng SF, Teng SC (2006) Involvement of topoisomerase III in telomere-telomere recombination. J Biol Chem 281: 13717–13723 [DOI] [PubMed] [Google Scholar]
- Wang H, Zhao A, Chen L, Zhong X, Liao J, Gao M, Cai M, Lee DH, Li J, Chowdhury D, Yang YG, Pfeifer GP, Yen Y, Xu X (2009) Human RIF1 encodes an anti-apoptotic factor required for DNA repair. Carcinogenesis 30: 1314–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T (2000) Possible association of BLM in decreasing DNA double strand breaks during DNA replication. EMBO J 19: 3428–3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ (1996) SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Capp C, Feng L, Hsieh TS (2008) Drosophila homologue of the Rothmund-Thomson syndrome gene: essential function in DNA replication during development. Dev Biol 323: 130–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID (2006) BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci USA 103: 4068–4073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID (2003) The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874 [DOI] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W (2008) RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 22: 2843–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Blackburn EH (2004) Human Rif1 protein binds aberrant telomeres and aligns along anaphase midzone microtubules. J Cell Biol 167: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Li Y, Guo R, Ling C, Wang W (2008) FANCM of the Fanconi anemia core complex is required for both monoubiquitination and DNA repair. Hum Mol Genet 17: 1641–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W (2005) BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity. EMBO J 24: 1465–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Chahwan C, Bailis J, Hunter T, Russell P (2005) ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol Cell Biol 25: 5363–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.