Dynamin GTPase regulation is altered by PH domain mutations found in centronuclear myopathy patients
Dynamin GTPase is a key player in membrane fission events. Based on disease-relevant dynamin mutants that have been linked to centronuclear myopathy (CNM) Mark Lemmon's lab put forward a model that the dynamin PH domain allosterically couples lipid binding.
Keywords: dynamin, endocytosis, GTPase, PH domain, phosphoinositide
Abstract
The large GTPase dynamin has an important membrane scission function in receptor-mediated endocytosis and other cellular processes. Self-assembly on phosphoinositide-containing membranes stimulates dynamin GTPase activity, which is crucial for its function. Although the pleckstrin-homology (PH) domain is known to mediate phosphoinositide binding by dynamin, it remains unclear how this promotes activation. Here, we describe studies of dynamin PH domain mutations found in centronuclear myopathy (CNM) that increase dynamin's GTPase activity without altering phosphoinositide binding. CNM mutations in the PH domain C-terminal α-helix appear to cause conformational changes in dynamin that alter control of the GTP hydrolysis cycle. These mutations either ‘sensitize' dynamin to lipid stimulation or elevate basal GTPase rates by promoting self-assembly and thus rendering dynamin no longer lipid responsive. We also describe a low-resolution structure of dimeric dynamin from small-angle X-ray scattering that reveals conformational changes induced by CNM mutations, and defines requirements for domain rearrangement upon dynamin self-assembly at membrane surfaces. Our data suggest that changes in the PH domain may couple lipid binding to dynamin GTPase activation at sites of vesicle invagination.
Introduction
Dynamins and related large GTPases drive vesicle scission and fusion events that regulate numerous cellular processes (Praefcke and McMahon, 2004). The functions of dynamins themselves have been most well documented in receptor-mediated endocytosis, largely because of its importance in regulating cell signalling events that control cell proliferation and differentiation (Vieira et al, 1996; Mosesson et al, 2008). Owing to the complexity of endocytosis, which uses an array of factors including clathrin, BAR domain containing membrane modulators, and the actin-polymerization machinery (Kaksonen et al, 2006; Benmerah and Lamaze, 2007; Ferguson et al, 2009), a mechanistic understanding of how dynamin drives and/or coordinates vesicle scission is only now slowly emerging.
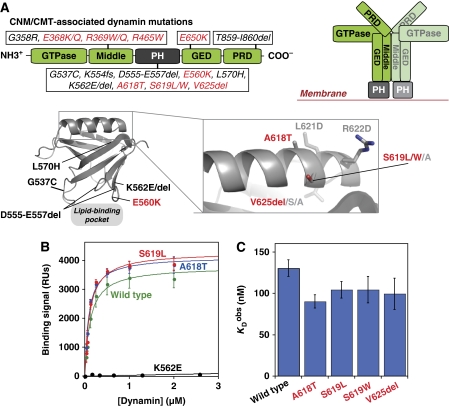
Three dynamin isoforms are expressed differentially in human cells, but all share the same basic function and domain architecture (Figure 1A). An N-terminal GTPase domain and a more C-terminal GTPase effector domain (GED) together control GTP hydrolysis in dynamin assemblies (Muhlberg et al, 1997). A ‘middle' domain (along with the GED domain) drives dynamin oligomerization (Muhlberg et al, 1997; Smirnova et al, 1999; Ramachandran et al, 2007). The pleckstrin-homology (PH) domain binds phosphatidylinositol-(4,5)-bisphosphate (PtdIns(4,5)P2) in the plasma membrane (Tuma et al, 1993; Salim et al, 1996; Zheng et al, 1996). Finally, a C-terminal proline/arginine-rich domain (PRD) recruits endocytic accessory factors and directs dynamin to endocytic sites (Gout et al, 1993; Shpetner et al, 1996). Dynamin has been observed as a helical oligomer at the neck of budding vesicles in cells (van der Bliek et al, 1993; Takei et al, 1995) and forms similar helical coils on synthetic lipid tubules in vitro, the pitch of which varies with guanine-nucleotide-binding state (Sweitzer and Hinshaw, 1998; Stowell et al, 1999; Zhang and Hinshaw, 2001). These observations led to the long-held view that dynamin assembles into helical ‘collars' that constrict during its GTPase cycle and thereby ‘pinch' budding endocytic vesicles off the membrane. Indeed, the relatively high basal GTPase activity of dynamin (kobs∼1 min−1) is increased over 200-fold when it assembles on lipid substrates, suggesting the activation of a scission ‘machine' in this context (Tuma et al, 1993). In addition to a function for dynamin assemblies in membrane scission, unassembled dynamin tetramers appear to participate in early stages of vesicle formation and maturation. In this guise, dynamin's function appears more similar to that of traditional signalling GTPases whose activity is regulated by the nucleotide-bound state (Sever et al, 1999; Loerke et al, 2009). These studies and others have pointed towards the GED as a critical regulatory region for dynamin GTPase activity.
Figure 1.
Disease-related mutations in the dynamin PH domain and effects on phosphoinositide binding. (A) Dynamin domain architecture is shown (left) as a linear cartoon representation with the location of centronuclear myopathy (CNM) and Charcot–Marie–Tooth (CMT) disease-related mutations notated for each domain. On the right, a cartoon of the repeating dimer unit seen in EM reconstructions of dynamin tubules (Zhang and Hinshaw, 2001) illustrates the proposed spatial arrangement of constituent domains. In the lower panel, the structure of the dynamin PH domain (PDB 1DYN; Ferguson et al, 1994) is shown, together with a magnified view of the C-terminal α-helix. Residues mutated in CNM are labelled in red. Additional residues mutated in this study are labelled in light grey. (B) SPR-binding curves for association of wild-type and selected dynamin variants with membranes containing 3% PtdIns(4,5)P2 in a DOPC background. Response units (RUs) were corrected for binding to a 100% DOPC control surface. (C) Comparison of KDobs values for CNM dynamin variants, showing that CNM mutations do not impair lipid binding. Values presented are averages±s.d. for fits to at least three independent binding isotherms using at least two independent protein preparations.
In this paper, we focus on the function of the PH domain in dynamin action and regulation. Recent reports (Bethoney et al, 2009; Ramachandran et al, 2009) contradict the initial view that the dynamin PH domain is a simple membrane-targeting device with no direct function in dynamin's mechanism of action (Salim et al, 1996; Klein et al, 1998). Indeed, studies of several mutations argue that the PH domain is required for dynamin to function even after it is localized to coated pit necks (Bethoney et al, 2009; Ramachandran et al, 2009) and suggest that this domain has an active ‘effector' function in dynamin-mediated membrane scission. The PH domain could promote vesicle scission directly by mediating the clustering of membrane phosphoinositides (Bethoney et al, 2009) and/or by penetrating and thus distorting membranes (Ramachandran et al, 2009). We set out to test an alternative—but not exclusive—hypothesis that the PH domain coordinates the regulation of dynamin's GTPase activity with its membrane association at coated pit necks. Indeed, several pieces of evidence suggest a conformational coupling between the PH and GTPase domains of dynamin (Muhlberg et al, 1997; Solomaha and Palfrey, 2005; Ramachandran and Schmid, 2008). There are also several precedents, especially among Rho family and Arf guanine-nucleotide exchange factors (GEFs) (Rossman and Sondek, 2005; DiNitto et al, 2007), for PH domain engagement by membrane phosphoinositides allosterically controlling the activity of adjacent GTPase-regulating domains (Lemmon, 2004). With this putative conformational coupling in mind, we first considered human disease-related dynamin mutations linked either to centronuclear myopathy (CNM) or Charcot–Marie–Tooth (CMT) neuropathy (Durieux et al, 2010). Dynamin mutations in CNM and CMT patients are predominantly located in the PH domain (Figure 1A). CNM mutations cluster in the C-terminal α-helix of the dynamin PH domain and are particularly interesting. They are distant from the lipid-binding site, suggesting that their effects might not arise from interference with PtdIns(4,5)P2 binding. Moreover, the CNM mutations lie in a region shown in other PH domains to be important for binding to small G-proteins and/or modulating the GTPase activity of their binding partners (Jin et al, 2005; DiNitto et al, 2007; Bunney et al, 2009). We present biochemical and biophysical analyses of dynamin CNM variants that suggest a coupling of phospholipid binding to the dynamin PH domain with stimulation of its GTPase activity. Mutations in the C-terminal α-helix of the PH domain do not affect PtdIns(4,5)P2 binding, but instead elevate the basal GTPase activity of dynamin. Certain CNM mutations promote substantial activation of GTP hydrolysis and dynamin oligomerization even in the absence of lipid. In others, this effect is partial and the CNM variants respond more readily to lipid substrate. On the basis of these findings, we suggest a mechanism in which membrane binding optimizes inter- and/or intra-molecular interactions between dynamin's constituent domains to promote stimulation of its GTPase activity.
Results
CNM mutations in the α-helix of the dynamin PH domain do not reduce lipid affinity
To assess the functional consequences of CNM mutations in the C-terminal α-helix of the dynamin-1 PH domain (A618T, S619L, S619W, and V625del; Figure 1A), we first asked whether they impair phosphoinositide binding in a quantitative surface plasmon resonance (SPR) assay (Yu and Lemmon, 2001). Increasing concentrations of purified dynamin variants were flowed over a biosensor surface bearing 3% (mol /mol) PtdIns(4,5)P2 in a DOPC background, and affinity constants (KDobs) were derived from fits to equilibrium-binding isotherms (Figure 1B). Wild-type dynamin-1 bound to the lipid surface with a KDobs value of 130±10 nM (Table I), in agreement with previous reports (Bethoney et al, 2009). Lipid binding was essentially abolished by the K562E mutation found in CMT patients (Zuchner et al, 2005), which directly alters the PH domain lipid-binding pocket (Figure 1A and B). In contrast, all CNM mutants investigated here bound PtdIns(4,5)P2 with affinities equivalent to wild-type protein (Figure 1B and C; Table I). Thus, defects in dynamin function caused by these CNM mutations do not arise from impaired lipid binding (or gross misfolding), arguing that some other biochemical activity of dynamin must be perturbed.
Table 1. Lipid-binding affinity, basal GTP hydrolysis kinetic parameters, and PH domain folding stability for dynamin variantsa.
| Dynamin 1 variantb | KDobs (PtdIns(4,5)P2) (nM)c | Basal kobs (min−1)d | KMapp (μM) | ΔGuH20 (kcal mol−1)e |
|---|---|---|---|---|
| Wild type | 130±10 | 1.1±0.1 | 123±33 | 6.1±0.2 |
| A618T | 90±8 | 1.9±0.2 | 73±6 | 5.0±0.2 |
| S619L | 104±10 | 78±4 | 42±15 | 3.2±0.6 |
| S619W | 104±16 | 83±13 | 51±11 | 3.3±0.5 |
| V625del | 99±19 | 117±24 | 52±22 | 1.9±0.1 |
| K562E | >10 000 | 1.0±0.1 | 128±10 | 8.7±0.7 |
| S619A | 130±37 | 1.2±0.3 | 94±15 | 7.1±0.3 |
| L621D | 67±31 | 3.3±0.9 | 71±9 | 4.2±0.2 |
| R622D | 86±9 | 1.9±0.2 | 85±7 | 5.1±0.3 |
| A618T/L621D/R622D | 104±10 | 67±6 | 63±22 | 2.1±0.4 |
| aAll values for full-length dynamin constructs represent averages from a minimum of three independent experiments using at least two distinct protein preparations. | ||||
| bMutations above the horizontal line have all been found in CNM or CMT patients; those below the line are artificial mutations generated for this study. | ||||
| cKDobs values are for binding of full-length dynamin-1 variants to lipid vesicles composed of 3% PtdIns(4,5)P2 in a DOPC background, as determined from surface plasmon resonance experiments such as those shown in Figure 1B. | ||||
| dKMapp and kobs values for GTP hydrolysis are calculated by fitting data to saturation kinetics curves, as exemplified by Figure 2A and C. | ||||
| eΔGu values were determined for isolated PH domain variants at 37°C and calculated from fits to equilibrium unfolding urea chemical melts as shown for three examples in Figure 5B. Errors plotted reflect s.d. for a minimum of three independent repeats. | ||||
Dynamin PH domain mutations alter basal GTPase activity
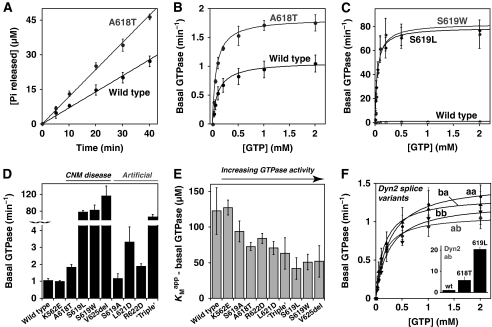
The CNM mutations studied here lie in a C-terminal motif known in several other PH domains (including those from Trio, p63RhoGEF, Exo84, Grp1, and PLC-γ) to modulate regulatory interactions with associated G-proteins through diverse direct and indirect mechanisms (Jin et al, 2005; DiNitto et al, 2007; Lutz et al, 2007; Rojas et al, 2007; Bunney et al, 2009). By analogy, the equivalent region of the dynamin PH domain might have a function in regulating dynamin's GTPase activity. To investigate this possibility, we examined the basal GTPase activity of each dynamin CNM variant in the absence of lipid. To measure GTP hydrolysis rates, we followed time-dependent release of free inorganic phosphate using a colorimetric assay (Figure 2A; see Materials and methods). Wild-type dynamin-1 exhibited a relatively low basal GTPase activity (kobs=1.1 min−1; Figure 2A and B; Table I) in agreement with the previous reports for unassembled dynamins (Warnock et al, 1997; Song et al, 2004a), as did the lipid-binding defective K562E mutant (kobs=1.0 min−1). In contrast, all CNM mutations in the PH domain C-terminal α-helix elevate basal GTPase activities (Figure 2A–D; Table I) by factors ranging from ∼2-fold (for A618T) to over 100-fold (V625del). To ensure that these elevated basal rates reflect only intrinsic dynamin activity, and not co-purified contaminants, an additional GTPase domain mutation (T65A) was introduced into S619L-mutated dynamin-1 (Song et al, 2004a). This mutation diminished GTP hydrolysis to the same low level (kobs=0.09 min−1) seen for T65A-mutated dynamin-1 with a wild-type PH domain (kobs=0.07 min−1, data not shown). Interestingly, basal GTPase activities of the S619L, S619W, and V625del CNM mutants were all comparable with the rapid lipid-stimulated hydrolysis activity seen for wild-type dynamin-1 (Tuma et al, 1993). This suggests either that the PH domain normally maintains dynamin's GTPase in a non-activated (autoinhibited) state when not bound to membranes, and/or that the CNM mutations alter inter- or intra-molecular interactions in a similar manner to membrane binding.
Figure 2.
Mutations in the C-terminal α-helix of dynamin's PH domain alter basal GTPase activity. (A) Basal rates of GTP hydrolysis by dynamin variants (0.5 μM) were determined as described in Materials and methods. Initial rates from these experiments were plotted as a function of GTP concentration (1 mM in this panel) and fit to a simple saturation kinetics model. (B) The A618T CNM mutation causes a small but significant increase in basal GTPase activity relative to wild-type dynamin. (C) Dramatically elevated basal GTPase rates for the S619L and S619W variants are shown, with wild-type dynamin re-plotted for comparison. Kinetic parameters for all variants are listed in Table I. (D) Comparison of kobs values for basal GTP hydrolysis by additional dynamin variants with mutations in the C-terminal PH domain α-helix. ‘Triple' signifies the A618T/L621D/R622D mutation. (E) KMapp values are reduced below the wild-type value for dynamin variants with elevated basal GTPase rates. (F) All four dynamin 2 (Dyn2) splice isoforms (aa, ab, ba, and bb) have similar basal GTPase rates to that of the Dyn1 ba isoform focused on in this study. The inset shows aberrant regulation of this basal GTPase activity when the A618T or S619L mutations are introduced into Dyn2 ab.
We also introduced several artificial mutations into the C-terminal α-helix of dynamin's PH domain, to test the hypothesis that structural alterations in this region lead to dysregulated basal GTPase activity. Aspartate substitutions at either L621 or R622—on the solvent-exposed surface of this helix (Figure 1A)—caused small elevations in basal GTPase rates, to 3.3 and 1.9 min−1, respectively (Figure 2D; Table I), comparable in magnitude to the effect of the CNM A618T mutation. Moreover, a triple mutant that contains three individually mild GTPase mutations in the PH domain C-terminal helix (A618T/L621D/R622D) showed substantially elevated (∼60-fold) basal GTPase activity (kobs=67 min−1) similar to that seen with the most activated CNM variants. In contrast, a more conservative substitution at serine 619 (with alanine instead of leucine or tryptophan as seen in CNM) had little effect on basal GTP hydrolysis rates (kobs=1.2 min−1; Figure 2D; Table I). These findings further implicate the C-terminal α-helix of the PH domain in modulating dynamin GTPase activity. Interestingly, the dynamin variants with increased basal kobs values also tend to have reduced apparent enzymatic dissociation constants (KMapp), as shown in Figure 2E and Table I, so are saturated with GTP substrate more readily. This could reflect higher GTP-binding affinities for the more rapidly hydrolysing CNM-mutated dynamin-1 variants, or alternatively their enhanced assembly (which promotes GTP binding). In either case, our data show that, rather than affecting the phospholipid-binding properties of dynamin, CNM mutations in the C-terminal region of the PH domain enhance dynamin's GTPase activity.
As patient-derived CNM and CMT mutations occur in the ubiquitously expressed dynamin-2 (rather than dynamin-1 studied here), it was important to confirm (or otherwise) similar effects in dynamin-2, which is 79% identical to dynamin-1. As shown in Figure 2F (Table II), all four dynamin-2 splice variants, denoted aa, ab, ba, and bb (Sontag et al, 1994; Cao et al, 1998), exhibit similar basal GTP hydrolysis rates (average kobs=1.3±0.2) and KMapp values for GTP (average KMapp=215±59 μM). These are equivalent to the parameters measured for dynamin-1 (analogous to the ba splice variant). Moreover, the A618T and S619L mutations had the same effects on basal GTPase activity in dynamin-2 as seen for dynamin-1, increasing kobs to 5.8 and 20 min−1, respectively (Figure 2F, insert).
Table 2. Steady-state GTP hydrolysis parameters for dynamin 2 variantsa.
| Dynamin 2 variantb | Basal kobs (min−1) | KMapp (μM) |
|---|---|---|
| Wild-type isoform aa | 1.5±0.3 | 267±25 |
| Wild-type isoform ab | 1.1±0.1 | 155±41 |
| Wild-type isoform ba | 1.3±0.3 | 158±34 |
| Wild-type isoform bb | 1.2±0.2 | 280±24 |
| A618T isoform ab | 5.8±1 | 68±5 |
| S619L isoform ab | 20±1 | 171±63 |
| aAll values represent averages from a minimum of three independent experiments using at least two distinct protein preparations. KMapp and kobs values for GTP hydrolysis are calculated from fits of data to a saturation kinetics model as shown in Figure 2A and F. | ||
| bResidue changes of the dynamin 2 splice variants are fully described in Cao et al (1998). | ||
CNM mutations in dynamin's PH domain cause aberrant lipid stimulation of GTPase activity
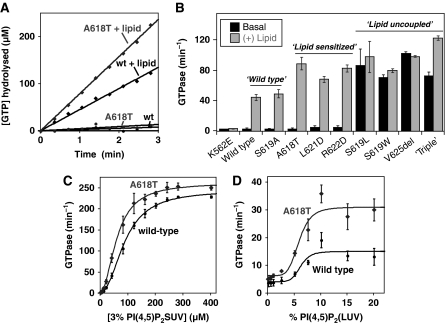
Upon binding and assembling into higher-order oligomers on lipid membranes, the GTPase activity of dynamin-1 increases by over two orders of magnitude (Tuma et al, 1993). Several CNM mutations cause similar elevations of GTPase activity even in the absence of membranes. We hypothesized that these CNM mutations might mimic the normal mechanism of dynamin activation upon lipid binding (and dynamin assembly), possibly by altering the orientation of the PH domain and/or its inter- or intra-molecular interactions with other domains within dynamin. To test this hypothesis, we analysed the response of all PH domain-mutated CNM variants to lipid binding, reasoning that they should respond to lipid more readily if an activating conformational change is favoured by the CNM mutation (in variants with slightly elevated basal activity) or may not respond at all if the mutation severely disrupts regulatory interactions (in variants with already high basal activity). As described above, each of the CNM variants retains wild-type PtdIns(4,5)P2-binding affinity. We measured GTPase activities for the CNM dynamin variants and other mutants described above in the presence of small unilamellar lipid vesicles (SUVs) composed of 3% (mol/mol) PtdIns(4,5)P2 (at a constant total lipid concentration of 40 μM). Three distinct classes of behaviour were observed, which were directly related to basal GTPase activities: ‘wild type', ‘lipid uncoupled', and ‘lipid sensitized' (Figure 3). The wild-type class, exemplified by the S619A mutant (which exhibits wild-type basal GTPase activity and is not found in CNM), responds robustly to lipid substrate and hydrolyses GTP at an average rate of 47±2 min−1 in the presence of PtdIns(4,5)P2-containing vesicles (Figure 3B). In stark contrast, GTP hydrolysis by the lipid-uncoupled variants (including the S619L, S619W, and V625del CNM variants) does not increase past the already rapid basal rate (Figure 3B), even when excess lipid is present (data not shown). Lipid-sensitized variants are exemplified by the A618T CNM variant (and the artificial L621D and R622D mutants). They exhibit elevated lipid-stimulated GTPase activities that are 1.5–2-fold higher than seen for wild type (Figure 3A and B), paralleling the ∼2-fold increase in their basal activities (Table I). For example, A618T dynamin hydrolysed GTP at 90 min−1 in the presence of PtdIns(4,5)P2-containing vesicles—twice the rate seen for wild-type dynamin under the same conditions (46±4 min−1). This conservation of an approximately two-fold increase in both basal and lipid-mediated GTP hydrolysis rates for lipid-sensitized variants of dynamin suggests that the two activities may be regulated by a common mechanism involving alterations in its structure or oligomerization state.
Figure 3.
Aberrant response of dynamin PH domain variants to stimulation with lipid vesicles. (A) GTP hydrolysis by dynamin variants (0.5 μM) was monitored over time in the presence of 0.1 μm extruded lipid vesicles composed of 3% PtdIns(4,5)P2 in a DOPC background (40 μM total lipid concentration). The example shown here shows two-fold activation of the A618T variant relative to wild-type dynamin. (B) Summary of lipid-mediated GTPase activation of all dynamin variants in this study, determined as in (A) with wild-type and K562E dynamin as positive and negative controls, respectively. Mutated variants segregate into three classes that are directly related to their basal GTPase activities. Variants with wild-type basal GTPase rates also exhibit ‘wild-type' response to lipid. Variants with mildly elevated basal GTPase activity respond more robustly to lipid (‘lipid sensitized'). Variants with greatly elevated basal GTPase rates show little or no response to lipid past their already-stimulated GTPase activity (‘lipid uncoupled'). (C) Activation of wild type and the ‘lipid-sensitized' A618T variant dynamins (0.5 μM) with increasing concentrations of small unilamellar vesicles (SUVs) composed of 3% PtdIns(4,5)P2/97% DOPC. (D) Activation of A618T and wild-type dynamin by large unilamellar vesicles (LUVs) with increasing percentages of PtdIns(4,5)P2, at constant total lipid concentration of 50 μM. Errors are from averages of at least three repeat experiments with at least two different protein preparations.
To probe the enhanced response of the lipid-sensitized class further, we analysed GTP hydrolysis rates of wild-type and A618T dynamin in the presence of increasing PtdIns(4,5)P2 levels in experiments using SUVs (average diameter 0.1 μm). In titrations with SUVs containing 3% PtdIns(4,5)P2, both wild-type and A618T dynamin exhibited similar degrees of cooperative activation, with a cooperativity coefficient (n) of ∼2. However, the A618T CNM variant showed higher rates of stimulated GTPase activity at all lipid concentrations (Figure 3C). Specifically, A618T dynamin-1 was ∼two-fold more active than wild type up to the lipid concentration required for half-maximal activation (K0.5), and the values then converged such that A618T is ∼7% more active than wild type in the presence of saturating lipid. Thus, A618T dynamin responds more readily to lower lipid concentrations than the wild-type enzyme, and has a higher maximal turnover rate.
As it is not clear whether dynamins deform low-curvature membranes in vivo or require pre-curved membranes for their activation, we also analysed the effect of large unilamellar vesicles (LUVs) on GTPase activity. Specifically, we measured GTPase activation of wild-type and A618T dynamin by LUVs (size range ∼0.1–10 μm) with increasing percentages of PtdIns(4,5)P2 at constant total lipid concentration (50 μM; Figure 3D). The degree of maximal GTPase stimulation was about 10-fold lower for LUVs than for SUVs, as expected from previous reports on the curvature dependence of dynamin activity (Yoshida et al, 2004; Ramachandran and Schmid, 2008; Roux et al, 2010). However, the rate of stimulated GTP hydrolysis by A618T dynamin-1 was again elevated above that seen for wild type at all lipid substrate concentrations. Interestingly, a full ∼two-fold increase in LUV-stimulated GTPase activity was maintained with the A618T variant even at saturating lipid concentrations. Taken together, both our SUV and LUV-based assays argue that the lipid-sensitized class of CNM-derived PH domain mutants ‘sense' and respond to PtdIns(4,5)P2 more readily than wild-type dynamins. The fact that similar elevations in GTPase activity can be afforded either by mutating the PH domain or occupying its PtdIns(4,5)P2-binding site suggests that the dynamin PH domain is ‘poised' to couple lipid binding with enhanced GTPase activity at sites of membrane vesiculation.
Severe GTPase regulation mutants have an increased propensity for self-association
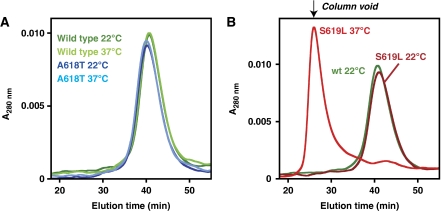
Dynamin activation during endocytosis is thought to involve its self-assembly into GTPase-active collars or tubules, which can be recapitulated in vitro under certain conditions even in the absence of lipid (Hinshaw and Schmid, 1995; Warnock et al, 1996). We asked, therefore, whether CNM mutations from the different lipid-sensitivity classes influence dynamin oligomerization. Similar to wild-type dynamin, all variants eluted as tetramers in the final 4°C gel filtration step during purification (data not shown; see Materials and methods). This observation, together with the studies of lipid binding and GTPase activity described above, argues that none of the variants is grossly misfolded. However, we reasoned that the more active dynamin variants might oligomerize in the 37°C GTPase assay conditions if their mutations destabilize PH domain interactions that normally restrict self-association. To investigate this possibility, we used gel filtration chromatography to separate dynamin tetramers from higher-order oligomers after pre-incubation at different temperatures. As shown in Figure 4A, both wild-type and A618T dynamin, which have modest basal GTPase activities (Table I), elute as tetramers after pre-incubation at 22 or 37°C. In contrast, the S619L variant (Figure 4B) and the S619W and V625del CNM variants (data not shown) all shift from a tetrameric state at 22°C to a higher-order assembly at 37°C that runs in the void volume of the gel filtration column. This migration position suggests assemblies larger than ∼700 kDa (a dimer of tetramers or greater), but smaller than expected for large-scale tubule formation or non-specific protein aggregation. Indeed, such larger aggregates would be removed from the sample by high-speed centrifugation and filtration before column loading, and no such losses were observed. Moreover, the elevated GTPase activity (Figure 2C) of this oligomeric S619L species argues that the overall integrity (and solubility) of the protein is not altered. Nevertheless, the enhanced propensity of the S619L, S619W, and V625del CNM-derived variants to self-associate may account for their elevated basal GTPase activities reported in Figure 2 and Table I. This finding suggests that certain PH domain mutations can increase the oligomerization potential of dynamin—which is important for its activation. These mutations might reverse intra-molecular interactions that normally have an autoinhibitory function in dynamin by preventing its oligomerization—but are disrupted upon membrane binding during activation of wild-type dynamin.
Figure 4.
The ‘lipid-uncoupled' S619L variant, but not the ‘lipid-sensitized' A618T variant, forms higher-ordered oligomers at elevated temperatures. Wild-type and selected mutants (1 μM) were pre-incubated at 22 or 37°C for 10 min, centrifuged at high speed, and then analysed by size exclusion chromatography (SEC). (A) SEC profile of the differently treated wild-type and A618T variants. (B) Analysis of the S619L mutant shows oligomerization after incubation at 37°C (a 22°C wild-type profile is re-plotted for reference).
PH domain C-terminal α-helix as a GTPase modulating motif
The CNM mutations that elevate dynamin's basal GTPase activity are all found towards the end of the PH domain's characteristic C-terminal α-helix. An equivalent motif in other PH domains has been implicated in either direct or indirect regulation of associated GTPases (Jin et al, 2005; Lutz et al, 2007; Rojas et al, 2007; Bunney et al, 2009). For example, the C-terminal part of the Grp1 PH domain contributes to autoinhibitory interactions that restrain guanine-nucleotide exchange activity of its adjacent Sec7 domain (DiNitto et al, 2007). Alterations in the conformation of the dynamin GED domain, which is adjacent to the PH domain, have an important function in regulating GTPase activity (Sever et al, 1999; Chappie et al, 2010). We speculated, therefore, that the conformation of the GED domain might be ‘restrained' or autoinhibited in dynamin tetramers by interacting with a region of the dynamin PH domain that encompasses the C-terminal α-helix motif. GED/PH interactions could occur either within or between dynamin molecules in tetramers.
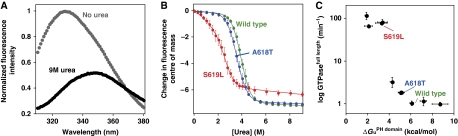
The CNM mutations could impair autoinhibitory PH domain-mediated interactions by disrupting an important binding site or tertiary structural epitope in which the C-terminal α-helix participates. Such mutations might be expected to destabilize tertiary interactions of the C-terminal α-helix with the PH domain core (Figure 1A). To test this hypothesis, we determined the thermodynamic folding energy of purified PH domain variants in isolation from the rest of dynamin using urea chemical melts (Figure 5A and B). The wild-type dynamin PH domain is particularly stable, with an equilibrium unfolding energy (ΔGu) at 37°C of 6.1 kcal/mol. For comparison, ΔGu for the well-studied PH domain from PLC-δ1 cannot be measured at 37°C, and is just 3.2 kcal/mol at 25°C (Supplementary Figure S1). As shown in Figure 5C (Table I), equilibrium folding measurements of isolated dynamin PH domain variants at 37°C illustrated a striking correlation between their thermodynamic stability (ΔGu) and the basal GTPase rates of the corresponding full-length dynamins. For instance, the A618T mutant (ΔGu=5.0 kcal/mol) is destabilized by ∼1.1 kcal/mol compared with wild type, and shows mildly elevated basal GTPase activity in intact dynamin. The S619L mutation is significantly more destabilizing (ΔGu=3.2 kcal/mol) and causes dynamin to be lipid uncoupled (Figure 5C). This stability correlation is consistent with the possibility that the PH domain has a function in maintaining dynamin in an unassembled, non-activated GTPase state. The most destabilizing mutations are all associated with the severely GTPase-dysregulated CNM variants. For instance, the destabilizing V625del mutation (ΔGu=1.9 kcal/mol) removes a valine side chain from the extreme terminus of the α-helix that projects into the hydrophobic core of the PH domain (Figure 1A). Removing this interaction would be expected to destabilize docking of the C-terminal α-helix into the PH domain core, reducing overall helix stability and thereby disrupting interaction sites that encompass the C-terminal α-helix motif. Consistent with this view, substituting V625 with a polar serine residue elevates basal GTPase rates to the same degree seen for the V625del mutation, whereas substitution with an apolar alanine does not (Supplementary Figure S2).
Figure 5.
Basal dynamin GTPase activity correlates with PH domain folding stability. (A) Upon unfolding in high concentrations of urea at 37°C, the tryptophan fluorescence emission spectra of purified dynamin PH domain (excitation at 280 nm) is red shifted and reduced in intensity. (B) Changes in the fluorescence centre of mass from spectra such as those in (A) were compared at increasing urea concentrations, and a six parameter fit was used (see Materials and methods) to extract values for free energy of unfolding (ΔGu). (C) Comparison of ΔGu values determined for isolated PH domain variants with basal GTPase rates determined for full-length variants harbouring the same PH domain mutations. All 10 PH domain variants listed in Table I are presented (some points overlap), with wild-type and the A618T and S619L variants from (B) highlighted for reference.
Given the complexity of protein folding, it is not possible to ascertain from these measurements alone whether the stability perturbations extend beyond the C-terminal α-helix and globally destabilize the PH domain (thereby indirectly destabilizing alternative interaction motifs). However, it is important to point out that all of the mutated PH domains function as stably folded domains. Wild-type PtdIns(4,5)P2-binding affinity is retained in even the most destabilized variants (Figure 1; Table I), and all mutated dynamin PH domains are still more stable in urea melts than the well-studied, high-affinity PtdIns(4,5)P2-binding PH domain from phospholipase-Cδ1 (see Supplementary Figure S1 for a comparison of PLC-δ-PH and the S619L-mutated PH domain). For example the ΔGu value for the S619L-mutated PH domain (one of the most destabilized) indicates that only a small fraction (∼1 in 200 molecules) will be in a non-native state at any given time at 37°C, and this ignores the fact that PH domain stability is almost certainly enhanced by quaternary interactions in the context of full-length dynamin. Thus, the mutations presented here at a minimum do alter the local stability of the C-terminal helix in which they reside, but in the context of a very stable PH domain. Interestingly, NMR studies have revealed that another PH-like domain can maintain the core β-sandwich structure that characterizes the PH fold even with major conformational flexibility of the C-terminal α-helix (Husnjak et al, 2008). We suggest, therefore, that the C-terminal α-helix of dynamin's PH domain may be important in forming contacts that have an important function in GTPase regulation. The effect of CNM mutations in dynamin may be analogous to tumourigenic mutations in the tumour suppressor protein p53, which cause localized structural changes that globally destabilize the DNA-binding domain while maintaining the overall tertiary fold without altering local stability of distal secondary motifs (Wong et al, 1999).
PH domain mutations result in global dynamin shape changes
If the CNM mutations studied here alter PH domain-mediated interactions as suggested above, an accompanying change in dynamin's conformation is to be expected. To test this hypothesis, we used small-angle X-ray scattering (SAXS). This approach necessitated the production of a highly soluble, self-assembly restricted dynamin variant, as SAXS studies of heterogeneous oligomers cannot be interpreted. We introduced R399A and I690K mutations, which have been reported individually to restrict dynamin self-assembly to the formation of dimers or tetramers, respectively (Song et al, 2004b; Ramachandran et al, 2007). We also deleted the unstructured C-terminal PRD domain that is reported to enhance self-assembly (Warnock et al, 1997). The resulting construct yielded protein that is extremely soluble (to ⩾400 μM) compared with wild-type dynamin, and behaves as a monodisperse dimer in sedimentation velocity ultracentrifugation experiments (Supplementary Figure S3).
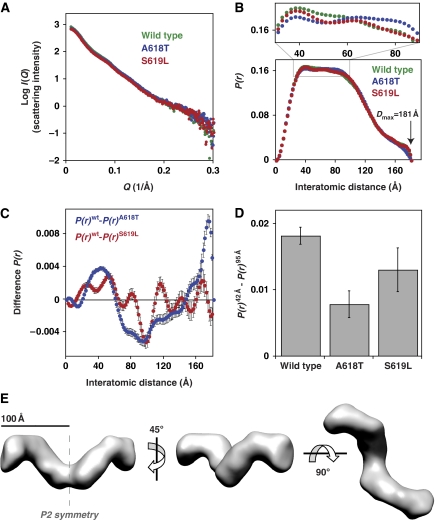
In the context of this dimer variant, we analysed the conformational effects of one CNM mutation from the lipid-sensitized class (A618T) and another from the lipid-uncoupled class (S619L). Importantly, the effects of these mutations on basal GTPase activity described above are retained in the dimer context (Supplementary Figure S4). Parameters derived from the SAXS scattering curves (Figure 6A) revealed a strong similarity in overall solution behaviour of wild-type dimeric dynamin and these two CNM variants (Supplementary Table SI). Their radii of gyration (Rg) values are all the same, within error (mean Rg=60±0.2 Å), as are values for their maximum molecular dimension (average Dmax=181±1 Å) determined from interatomic distance distribution functions (or P(r) curves) obtained by inverse Fourier transform of the scattered intensity. P(r) curves describe the length distribution of all interatomic vectors in a molecule and are sensitive, therefore, to structural alterations. As shown in Figure 6B, measurable differences can be seen between the shapes of the P(r) curves. Specifically, there are significant changes in the population of scattering elements in the 42–95 Å regions, most readily appreciated from a magnification of this region (upper panel in Figure 6B) and in difference P(r) curves individually comparing wild type to the two mutants (Figure 6C). The differences were consistent through numerous independent repeats (⩾14) of the SAXS experiment, using different protein preparations, X-ray exposure times, and synchrotron radiation sources. The differences are also significant (t-test, P<0.0001) when comparing P(r) values at 42 and 95 Å for individual variants (Figure 6D). Furthermore, differences are apparent in the low Q region of the primary scattering data (Supplementary Figure S5), revealing that these conformational changes are sufficiently large to be directly observed even at very low resolution. These data argue that—even in the context of a dynamin dimer—CNM-derived mutations in the C-terminal α-helix of the PH domain alter the overall conformation of dynamin (and increase basal GTPase rates), consistent with our hypothesis that these mutations disrupt autoinhibitory inter-domain interactions. We were surprised to find that the P(r) curves for the A618T (lipid-sensitized) variant differ more from wild-type P(r) curves than those obtained with the S619L (fully lipid-uncoupled) variant. The S619L mutation may disrupt PH domain interactions to an extent that allows the protein to explore multiple conformations (including those similar to wild-type and A618T dynamin), whereas the more subtle A618T lipid-sensitizing mutation might lock the PH domain in a discrete, aberrant position.
Figure 6.
Small-angle X-ray scattering (SAXS) reveals conformational changes associated with PH domain mutations. (A) Example scattering data for wild-type and the A618T and S619L PH domain variants (30 μM) in the context of a dimeric dynamin mutant (ΔPRD/I690K/R399A), plotting Log I(Q) against Q (4π sin(θ)/λ, where 2θ=total scattering angle). (B) Inverse Fourier transformation of scattering curves yields the interatomic distance P(r) curve. Although the overall shape of the P(r) curves and maximum molecular dimensions (Dmax) are similar, magnification reveals subtle differences reflecting conformational changes in the dynamin variants. (C) The difference P(r) curve for the A618T (P(r)wt−P(r)A618T) and S619L (P(r)wt−P(r)S619L) variants highlight significant differences in the population of scattering elements of ∼42 and 95 Å. The difference observed at 175 Å for the A618T variant is highly variable between data sets and thus ignored for comparative purposes. Error bars are propagated from calculated errors in the P(r) curves generated by the program GNOM, reflecting deviation of the P(r) curve fit to the experimental scattering data. (D) Comparison of P(r)42−P(r)95 Å for ⩾14 individual scattering curves reveals that changes in P(r) between wild type and mutants are statistically significant (t-test P<0.0001). (E) Three views of a three-dimensional model (envelope) of the wild-type dynamin dimer calculated by ab initio shape reconstruction from the experimental scattering data and P(r) curve (see Materials and methods). P2 symmetry was imposed on the model (two-fold axis shown in the left-most envelope) for an optimal fit to the experimental data (χ2=2.25), although a similar two-fold symmetric model results even when none is imposed (χ2=3.03; Supplementary Figure S6).
Changes in dynamin conformation upon self-assembly
SAXS data can be translated into a limited range of most probable molecular envelopes (shapes) through ab initio calculations (Svergun, 1999; Svergun et al, 2001). Molecular envelopes reconstructed for the different variants of the dynamin dimer all give similar shapes (Figure 6E; Supplementary Figure S6), indicating that changes in conformation caused by the CNM mutations are smaller than the resolution of such calculations. However, the envelopes for wild-type and mutated proteins provide useful information on the solution conformation of intact dynamin—for which no crystal structure is known. The generated envelopes show clear two-fold symmetry (Supplementary Figure S6), and analysis of χ2 values comparing the calculated envelopes against the experimental data shows that applying P2 symmetry results in optimal envelope reconstruction. Our data led to striking consistency in the overall shape of molecular envelopes generated from (i) scattering curves collected for different dynamin mutants, (ii) alternate data sets for individual variants, and (iii) the use of different ab initio shape determination programs that each impose unique restraints (Supplementary Figure S6). We conclude, therefore, that the envelopes presented in Supplementary Figures 6E and S6 accurately represent the general shape of a dynamin dimer in solution.
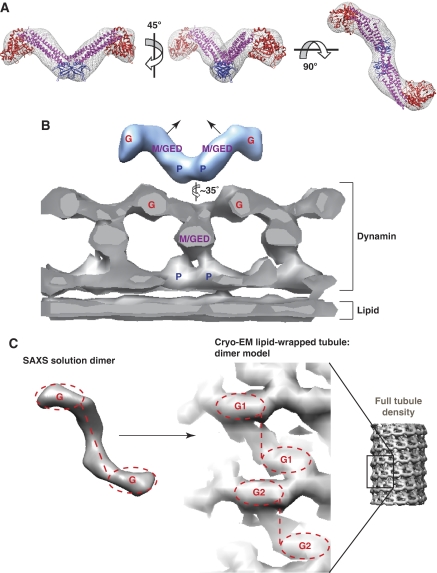
From several orientations, the dynamin dimer envelope resembles a ‘T' shape (Figures 6E and 7A), and shows remarkable similarity to a dimer extracted from the model of dynamin tetramers (Supplementary Figure S7) that was recently proposed by Daumke and co-workers (Gao et al, 2010). A ‘T' shape with different dimensions was also described for a dimeric dynamin unit in cryo-EM reconstructions of dynamin-wrapped lipid tubules (Zhang and Hinshaw, 2001). As shown in Figure 7, a significant conformational change is required to fit the ‘T' observed in our solution SAXS studies into that seen in the EM-based tubule/helices. The ‘T' must be ‘narrowed' to accommodate it in the long dynamin tubule (Figure 7B). Assuming the same assignments of individual domains to the features that were used in interpreting the EM studies (Zhang and Hinshaw, 2001; Mears et al, 2007), this draws the two middle/GED regions of the dimer ‘T' closer together (Figure 7B), altering their relationship with the adjacent PH domain (which would lie at the dimer interface). In the process, the two GTPase domains would be moved away from the PH domain (Figure 7B), but closer to one another in the dimer (Figure 7C). There is a clear precedent for such domain movement mediated by the middle/GED stalk upon membrane assembly. Indeed, EM density seen for the bacterial dynamin-like protein (BDLP) assembled on lipid tubules can only be fit using the crystal structure of a BDLP monomer if substantial conformational changes analogous to those suggested in Figure 7 are applied (Low et al, 2009).
Figure 7.
Hypothetical model of domain organization in SAXS-derived envelopes of a dynamin dimer. (A) Two dynamin PH domains (blue−PDB 1DYN) were fit at the two-fold symmetric interface, dynamin GTPase domains (red—PDB 2AKA) were fit at the far termini of the envelopes, and middle/GED stalks from the dynamin-related MxA protein (magenta—PDB 3LJB) were placed in the connecting volume. This organization fits the SAXS-derived model well, and (B) suggests a model in which conformational changes are required in order to form the tubular assemblies seen in EM reconstructions of dynamin-wrapped lipid tubules in the non-constricted state (Mears et al, 2007). The EM model is, shown as a grey-sliced cross-section depicting the repeating ‘T' dimer structure along one side of the lipid tubule. Locations of the GTPase domain (G), PH domain (P), and the middle/GED (M/GED) stalk, as proposed by Hinshaw and co-workers (Mears et al, 2007) are marked in the EM density. A slight twisting of the SAXS dimer ‘T' envelope is required to adopt the same configuration as the ‘T' inferred from EM studies, necessitating closer apposition of the GTPase domains, with the two stalk (M/GED) regions angling up towards each other. (C) In the SAXS envelope (left) as interpreted here, the GTPase domains angle away from the centre of the envelope (occupied by M/GED and PH regions) in the same manner seen for GTPase domains in the EM dimer model (right). The EM model is shown here as a surface cutout with M/GED density visibly connecting the two GTPase domains in the dimer. Two dimer models (G1 and G2) are depicted to highlight the repetitive dimer unit and to allow comparison with the SAXS-derived dimer envelope (left).
Interestingly, our SAXS-based structural model supports the relative positioning of GTPase domains within the repeating dimeric unit (Figure 7) proposed by Hinshaw and co-workers (Mears et al, 2007). This model argues that there are no direct PH–GTPase interactions in dynamin assemblies, countering the suggestion that the PH domain (or mutations within it) can activate the GTPase domain through direct interactions. The SAXS data are instead consistent with a model in which CNM-derived mutations in the PH domain exert their effects indirectly. They may alter autoinhibitory PH domain/GED interactions (within or between dynamin molecules) in a way that frees the GED domain to promote allosteric activation of the GTPase domain within dynamin oligomers (Sever et al, 1999; Low et al, 2009; Chappie et al, 2010). Alternatively, they could change the relationship between adjacent PH domains in a dynamin assembly to promote optimally activating inter-molecular interactions between the GED and GTPase domains. We suggest that phosphoinositide engagement of the PH domain has a similar influence.
Discussion
Most disease-related dynamin mutations cluster in the middle and PH domains of the molecule (Durieux et al, 2010). Intriguingly, whereas all CMT mutations in the PH domain occur close to the phosphoinositide-binding pocket, every CNM mutation in the PH domain except one (E560K) is found in the C-terminal α-helix. Both CMT and CNM mutations in the dynamin PH domain have generally been assumed to affect phosphoinositide binding. This is indeed true in CMT, for example, in the case of the K562E mutation. However, we show here that PH domain mutations seen in CNM do not affect dynamin's ability to bind PtdIns(4,5)P2. Instead, CNM mutations in the C-terminal α-helix of the PH domain enhance both basal and lipid-dependent GTPase activities of intact dynamin, and cause it to become dysregulated. CNM is clinically distinct from CMT, and the list of CNM mutations in the C-terminal α-helix of dynamin's PH domain is growing as additional patients are studied (Jungbluth et al, 2010; Melberg et al, 2010; Susman et al, 2010); including four newly identified CNM-related mutations within the C-terminal helix (A618D and L621P) or adjacent to it (R522H and P627H). The clustering of CNM-related GTPase-activating mutations in this region suggests that the C-terminal α-helix motif in dynamin's PH domain has an important regulatory function.
Mutations in the PH domain C-terminal α-helix motif fall into two classes. Those in the first class (e.g. A618T) elevate basal GTPase activity mildly and appear to ‘sensitize' dynamin to lipid stimulation. These altered properties are accompanied by a change in dynamin structure that is detectable in SAXS studies of dynamin dimers. PH domain mutations in the second class (e.g. S619L/W and V625del) appear to disrupt the structure even further, and shift dynamin into constitutively GTPase-active homo-oligomers. These findings argue that mutations in the C-terminal α-helix of the PH domain promote interdomain interactions (which may be intra- or inter-molecular) that enhance both self-assembly of dynamin and stimulation of its GTPase activity. As these mutations cause the same activating effects that are seen when dynamin binds to phosphoinositide-containing membranes through its PH domain, we suggest that they may mimic engagement of the phospholipid-binding site.
Our initial focus on mutations in the C-terminal α-helix of dynamin's PH domain stemmed from the fact that the analogous region of other PH domains is frequently involved in modulating (directly or indirectly) the activity of GTPases that associate with their host proteins. Among these, the PH domain of the Grp1 Arf-GTPase GEF is among the closest sequence relatives of the dynamin PH domain. A helical motif that immediately follows the Grp1 PH domain functions as an autoinhibitory element for Grp1 GEF activity mediated by its helical-bundle Sec7 domain (DiNitto et al, 2007). In dynamin, the largely α-helical GED region that follows the PH domain is important for activating the GTPase domain (Sever et al, 1999; Chappie et al, 2010). Specifically, an α-helix from the GED C-terminus interacts with two helices in the GTPase domain to form the ‘bundle signalling element' (Chappie et al, 2010) that has been reported to modulate dynamin function (Chappie et al, 2009). Conformational flexibility in this region may have a function in coupling dynamin's GTPase and PH domains as indicated by earlier studies (Muhlberg et al, 1997; Solomaha and Palfrey, 2005; Ramachandran and Schmid, 2008) and by the analysis of CNM mutations described here. Moreover, the recent structure of a transition-state-dependent dimer of dynamin's GTPase domain (Chappie et al, 2010) indicates that domain rearrangements within dynamin oligomers are intimately linked to regulation of its membrane association and GTP hydrolysis activity. Interactions between the PH domain and its adjacent GED domain could impose restraints on these rearrangements, which might be relieved upon phosphoinositide binding or the introduction of CNM mutations. CNM mutations or phosphoinositide binding may also enhance the ability of the GED (Song et al, 2004b) and the PH domain to promote dynamin oligomerization. This could explain why some CNM variants self-assemble in the absence of lipid, with consequently enhanced GTPase activity (Warnock et al, 1996). On the basis of the recent reports indicating that GTPase activation in dynamin family members results from interactions between adjacent rings within helical assemblies (Chappie et al, 2010, Gao et al, 2010), the minimal active oligomers promoted by CNM mutations would be expected to complete at least a full turn in order to yield activated GTPase domain dimers.
How might PH domain mutations that cause GTPase dysregulation alter cellular events driven by dynamin and thus lead to disease? There are several possibilities reflecting the involvement of dynamins in different cellular processes (Durieux et al, 2010). Defects in control of endocytosis or other trafficking events might be relevant for CNM (Durieux et al, 2010), although the pathological mechanisms remain unclear. Certainly, mutations in numerous other proteins involved in endocytosis and cellular trafficking are also linked to CNM, CMT, and related disorders (Dowling et al, 2008; Nicot and Laporte, 2008). The GTP hydrolysis cycle of dynamin appears to control several distinct stages of endocytosis (Mettlen et al, 2009). If dynamin behaves as a scission machine as commonly suggested, and directly couples GTP hydrolysis to the mechanical severing of budding vesicles (Stowell et al, 1999), then disrupting the coordination of GTPase activity with lipid binding as seen in CNM mutants should dramatically alter its function. Mutations that elevate GTPase rates in the absence of lipid would uncouple mechanical scission from lipid-mediated assembly, resulting in a loss of spatial coordination that would impair efficiency and control of vesicle scission. Indeed, the V625del mutation prevents dynamin-2 from supporting clathrin-mediated endocytosis (Bitoun et al, 2009), as do the V625del and S619L mutations in dynamin-1 (Supplementary Figure S8). The important defect for these and other CNM mutations may not be GTPase activity levels per se, or phosphoinositide binding, but the regulatory interplay between these two functions. Small alterations in activity can have significant functional impact, as illustrated by increased endocytic rates reported for other dynamin mutants that have only slightly altered basal GTPase activities (Song et al, 2004a; Loerke et al, 2009). Moreover, there is experimental precedent for control of endocytic rates by alterations in the C-terminal α-helix of dynamin's PH domain. S-nitrosylation of a cysteine immediately preceding this α-helix causes a two-fold increase in both basal GTP hydrolysis rates and cellular uptake of bacteria (Wang et al, 2006). Furthermore, the rate of nitrosylation in the PH domain is influenced by the guanine-nucleotide-binding state of the GTPase domain, suggesting a conformational link between these two domains. Given the complexity of clathrin-coated pit formation and scission, it is clear that detailed cell-based analyses will be crucial for fully understanding how coupling of dynamin's PH and GTPase domains influences endocytosis, also shedding further light on the aetiology of CNM disease. It is also possible that CNM arises from defects in other cellular functions of dynamin. For example, it is tempting to suggest that the hallmark central nuclei observed in muscle fibres in CNM are linked to impairment of the putative centrosomal activity of dynamin-2 (Thompson et al, 2004; Durieux et al, 2010). In this context, inappropriate regulation of dynamin's GTPase activity could lead to aberrant nuclear positioning.
Materials and methods
Protein production
Full-length dynamin constructs were amplified from either pUHD10-3 Dyn1 (human dynamin-1 ba; Damke et al, 1994) or pCR3.1 (rat dynamin-2 isoforms aa, ab, ba, and bb—gift of M McNiven; Cao et al, 1998) with 5′ NdeI and 3′ BamHI restriction sites and subcloned into the bacterial expression vector pET15b (Novagen). Resulting dynamin constructs have the N-terminal sequence NH3+-MGSS(H)6SSGLVPRGSH preceding the dynamin start methionine. Dynamin-1 PH domain constructs (residues 510–633, PubMed accession code AAH50279) were expressed and purified from a pET11a plasmid as described (Ferguson et al, 1994). All point mutations and the C-terminal PRD deletion were generated by PCR mutagenesis and sequences of alterations were confirmed. Refer to Supplementary data for detailed purification protocols.
PtdIns(4,5)P2-binding experiments
Lipid-binding affinities were determined by SPR assays essentially as described (Yu and Lemmon, 2001), using sensor chips that were coated with 3% (mol/mol) L-α-phosphatidylinositol-(4,5)-bisphosphate (PtdIns(4,5)P2—Cell Signals)/97% (mol/mol) 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC-Avanti Polar Lipids). Binding was assessed using a Biacore 3000 instrument at 25°C with running buffer containing 25 mM HEPES pH 7.5 and 250 mM NaCl.
GTPase assays
Dynamin basal (non-lipid-stimulated) GTPase activities were measured at 37°C essentially as described (Leonard et al, 2005). Briefly, 400 μl reactions were initiated by combining 40 μl of GTP (GE-Amersham Biosciences) at desired 10 × concentration and 360 μl of dynamin variant (0.5 μM final in 400 μl) in Buffer D (25 mM HEPES pH 7.5, 125 mM NaCl, 25 mM KCl, 10 mM MgCl2). Both dynamin and GTP stocks were pre-incubated for 10 min at 37°C before being combined at t=0 min. At given time points, 40 μl of reaction mixture was quenched by addition to 10 μl of 500 mM EDTA in a flat-bottomed, 96-well polystyrene plate. The 0-min control was made by first combining the EDTA and dynamin stocks to allow for Mg++ chelation, followed by addition of the 10 × GTP stock. To measure free inorganic Pi released by GTP hydrolysis, 150 μl of an acidic malachite green/ammonium molybdate mixture was added to the quenched reactions, and absorbance at 650 nm was measured with a Tecan Safire-2 microplate reader. Individual plates contained a phosphate titration series to allow calculation of free Pi in the experimental wells. Initial hydrolysis rates over a range of GTP concentrations were fit to saturation kinetics curves and used to derive the steady-state kinetic parameters kobs and KMapp.
Lipid-stimulated dynamin GTPase activities were measured at 37°C using a pyruvate kinase/lactate dehydrogenase-coupled enzyme system that indirectly reports GTP hydrolysis (GDP production) by directly following oxidation of NADH, essentially as described (Norby, 1988; Ingerman and Nunnari, 2005). A 50-μl stock of 1 μM dynamin diluted in Buffer D was mixed with 50 μl 2 × NADH-coupled mix (final concentrations are 20 units ml–1 pyruvate kinase, 27.5 units ml–1 lactate dehydrogenase, 5 mM phosphoenol pyruvate, 1 mM NADH, and 2.5 mM GTP—all chemicals except GTP from Roche) plus lipid at varying concentrations after 10 min pre-incubations at 37°C. LUVs of varying PtdIns(4,5)P2 and DOPC percentages, and SUVs of 3% PtdIns(4,5)P2/97% DOPC, were prepared as previously described (Narayan and Lemmon, 2006). A total of 90 μl of the combined reaction was pipetted to a pre-warmed Nunc 96-well plate and NADH absorbance at 355 nm was monitored over time to follow the rate of NADH oxidation (and thus indirectly, GTP hydrolysis). Experiments comparing GTPase rates with varying lipid concentrations were fit to the equation:
 |
where n=cooperativity coefficient for binding of dynamin to lipid vesicles, the multiplier for kobslipid accounts for the fraction of vesicle-bound enzyme, and the multiplier for kobsbasal is the remainder of this fraction to account for dynamin populations that convert from basal GTPase activity to lipid-stimulated activity with increasing concentrations of lipid vesicles. Elevated hydrolysis rates calculated from this technique correlate well with those derived from the malachite green method presented above, as expected given its wide use for the determination of enzymatic activity of rapidly hydrolysing NTPases. However, as it is a poor assay for slow NTP hydrolysis rates (⩽2 min−1), presumably because of the high rate of non-enzymatic oxidation of NADH over time, this approach was not used for critical quantitative comparison of basal GTPase rates.
Urea melts
Dynamin-1 PH domain variants were diluted to 0.5 μM in both 0 and 9 M urea solutions containing 50 mM Tris–Cl pH 8.0, 125 mM NaCl, 25 mM KCl, 10 mM MgCl2, and 2 mM DTT, and were then mixed to varying concentrations of urea. Following incubation at either 25°C (Supplementary Figure S1) or 37°C (Figure 5; Table I) for at least 12 h, tryptophan fluorescence emission spectra (excitation λ=280 nm) were measured from 310 to 380 nm using a PTI Spectrofluorometer (dynamin PH domain has four tryptophan residues). Changes in fluorescence centre-of-mass (COM) relative to the 0 M urea value were plotted against increasing concentrations of urea, and fit to obtain the equilibrium unfolding energy ΔGu as described (Santoro and Bolen, 1988). Stability of the PLC-δ1 PH domain was determined identically, but only at 25°C (Supplementary Figure S1), because a stable native baseline could not be obtained at 37°C.
Gel filtration analysis of dynamin variants
Following incubation of full-length dynamin variants (1 μM) at either 22 or 37°C for 10 min in Buffer D, the 500-μl samples were spun at 14 000 rpm (18 000 g) for 20 min at 4°C. Supernatant was then run on a Superose 6 10/300 GL gel filtration column (GE Healthcare) equilibrated with Buffer B (25 mM HEPES pH 7.5, 200 mM NaCl, 2 mM DTT).
Small-angle X-ray scattering
SAXS experiments with 30 μM samples of Dyn1 ΔPRD/I690K/R399A variants (wild-type, A618T, or S619L PH domains) were performed in Buffer C (25 mM HEPES pH 7.5, 250 mM NaCl, 2% glycerol, 10 mM MgCl2, and 2 mM DTT) at 25°C. Data were collected at NSLS beamline X21, and the SSRL/SLAC beamline BL4-2. Data reduction and analysis was performed as previously described (Dawson et al, 2007; Alvarado et al, 2009). All programs were obtained from the EMBL SAXS resource (http://www.embl-hamburg.de/ExternalInfo/Research/Sax/software.html). Scattering intensities from three (NSLS) or five (SSRL) successive runs at increasing exposure times were circularly averaged with beamline software. For NSLS data, the program Primus (Konarev et al, 2003) was used to subtract buffer from sample scattering intensities, and subsequently average intensities for all scattering data that did not exhibit radiation-induced damage according to Guinier plot and Kratky plot (IQ2 versus Q) analysis of individual exposures (this data reduction is performed automatically with SSRL software). The program AutoGNOM (Petoukhov et al, 2007) was used to generate P(r) curves and to determine Dmax and Rg from scattering intensity curve (I(Q) versus Q) in an automatic, unbiased manner, although rounds of manual fitting in GNOM (Svergun, 1992) were used to verify these values. GNOM fits for all experimental data resulted in total estimate calculations of either ‘good' or ‘excellent' (TE≈0.7–0.99). The areas under individual P(r) curves were normalized for differences resulting from changes in protein concentration, exposure times, and calculated I(0) values. Molecular envelopes were generated by ‘averaging' at least 10 individual Dammin (Svergun, 1999) or Gasbor (Svergun et al, 2001) calculations with the Damaver suite of programs (Volkov and Svergun, 2003), and the resulting output files graphically generated using Situs pdb2vol (http://situs.biomachina.org/) and Chimera (http://www.cgl.ucsf.edu/chimera).
Supplementary Material
Acknowledgments
We thank Diego Alvarado, Katarina Moravcevic, Sung Hee Choi, Jeannine Mendrola, Kushol Gupta, Greg Van Duyne, Jim Shorter, Jason Mears, Jenny Hinshaw, Sandra Schmid, and Mark McNiven for advice, helpful discussions, equipment, and material support. Additional thanks to Hiro Tsuruta (SSRL) and Lin Yang (NSLS) for SAXS instrument time and support. Financial support for the NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. The SSRL is a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. This work was funded in part by NIH grant R01-GM078345 (to MAL), and JAK was supported by a Postdoctoral Fellowship from The Jane Coffin Childs Memorial Fund for Medical Research (Fund Project No. 61-1316).
Footnotes
The authors declare that they have no conflict of interest.
References
- Alvarado D, Klein DE, Lemmon MA (2009) ErbB2 resembles an autoinhibited invertebrate epidermal growth factor receptor. Nature 461: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benmerah A, Lamaze C (2007) Clathrin-coated pits: vive la difference? Traffic 8: 970–982 [DOI] [PubMed] [Google Scholar]
- Bethoney KA, King MC, Hinshaw JE, Ostap EM, Lemmon MA (2009) A possible effector role for the pleckstrin homology (PH) domain of dynamin. Proc Natl Acad Sci USA 106: 13359–13364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitoun M, Durieux AC, Prudhon B, Bevilacqua JA, Herledan A, Sakanyan V, Urtizberea A, Cartier L, Romero NB, Guicheney P (2009) Dynamin 2 mutations associated with human diseases impair clathrin-mediated receptor endocytosis. Hum Mutat 30: 1419–1427 [DOI] [PubMed] [Google Scholar]
- Bunney TD, Opaleye O, Roe SM, Vatter P, Baxendale RW, Walliser C, Everett KL, Josephs MB, Christow C, Rodrigues-Lima F, Gierschik P, Pearl LH, Katan M (2009) Structural insights into formation of an active signaling complex between Rac and phospholipase C gamma 2. Mol Cell 34: 223–233 [DOI] [PubMed] [Google Scholar]
- Cao H, Garcia F, McNiven MA (1998) Differential distribution of dynamin isoforms in mammalian cells. Mol Biol Cell 9: 2595–2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F (2010) G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature 465: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Liu YW, Leonard M, Pucadyil TJ, Schmid SL (2009) An intramolecular signaling element that modulates dynamin function in vitro and in vivo. Mol Biol Cell 20: 3561–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H, Baba T, Warnock DE, Schmid SL (1994) Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J Cell Biol 127: 915–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JP, Bu Z, Lemmon MA (2007) Ligand-induced structural transitions in ErbB receptor extracellular domains. Structure 15: 942–954 [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Delprato A, Gabe Lee MT, Cronin TC, Huang S, Guilherme A, Czech MP, Lambright DG (2007) Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol Cell 28: 569–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling JJ, Gibbs EM, Feldman EL (2008) Membrane traffic and muscle: lessons from human disease. Traffic 9: 1035–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux AC, Prudhon B, Guicheney P, Bitoun M (2010) Dynamin 2 and human diseases. J Mol Med 88: 339–350 [DOI] [PubMed] [Google Scholar]
- Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB (1994) Crystal structure at 2.2 A resolution of the pleckstrin homology domain from human dynamin. Cell 79: 199–209 [DOI] [PubMed] [Google Scholar]
- Ferguson S, Raimondi A, Paradise S, Shen H, Mesaki K, Ferguson A, Destaing O, Ko G, Takasaki J, Cremona O, E OT, De Camilli P (2009) Coordinated actions of actin and BAR proteins upstream of dynamin at endocytic clathrin-coated pits. Dev Cell 17: 811–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Paeschke S, Behlke J, Haller O, Kochs G, Daumke O (2010) Structural basis of oligomerization in the stalk region of dynamin-like MxA. Nature 465: 502–506 [DOI] [PubMed] [Google Scholar]
- Gout I, Dhand R, Hiles ID, Fry MJ, Panayotou G, Das P, Truong O, Totty NF, Hsuan J, Booker GW, Campbell ID, Waterfield MD (1993) The GTPase dynamin binds to and is activated by a subset of SH3 domains. Cell 75: 25–36 [PubMed] [Google Scholar]
- Hinshaw JE, Schmid SL (1995) Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature 374: 190–192 [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I (2008) Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Nunnari J (2005) A continuous, regenerative coupled GTPase assay for dynamin-related proteins. Methods Enzymol 404: 611–619 [DOI] [PubMed] [Google Scholar]
- Jin R, Junutula JR, Matern HT, Ervin KE, Scheller RH, Brunger AT (2005) Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. EMBO J 24: 2064–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth H, Cullup T, Lillis S, Zhou H, Abbs S, Sewry C, Muntoni F (2010) Centronuclear myopathy with cataracts due to a novel dynamin 2 (DNM2) mutation. Neuromuscul Disord 20: 49–52 [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Toret CP, Drubin DG (2006) Harnessing actin dynamics for clathrin-mediated endocytosis. Nat Rev Mol Cell Biol 7: 404–414 [DOI] [PubMed] [Google Scholar]
- Klein DE, Lee A, Frank DW, Marks MS, Lemmon MA (1998) The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem 273: 27725–27733 [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI (2003) PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J Appl Cryst 36: 1277–1282 [Google Scholar]
- Lemmon MA (2004) Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32: 707–711 [DOI] [PubMed] [Google Scholar]
- Leonard M, Song BD, Ramachandran R, Schmid SL (2005) Robust colorimetric assays for dynamin's basal and stimulated GTPase activities. Methods Enzymol 404: 490–503 [DOI] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL (2009) Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol 7: e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Sachse C, Amos LA, Lowe J (2009) Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell 139: 1342–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz S, Shankaranarayanan A, Coco C, Ridilla M, Nance MR, Vettel C, Baltus D, Evelyn CR, Neubig RR, Wieland T, Tesmer JJ (2007) Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs. Science 318: 1923–1927 [DOI] [PubMed] [Google Scholar]
- Mears JA, Ray P, Hinshaw JE (2007) A corkscrew model for dynamin constriction. Structure 15: 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melberg A, Kretz C, Kalimo H, Wallgren-Pettersson C, Toussaint A, Bohm J, Stalberg E, Laporte J (2010) Adult course in dynamin 2 dominant centronuclear myopathy with neonatal onset. Neuromuscul Disord 20: 53–56 [DOI] [PubMed] [Google Scholar]
- Mettlen M, Pucadyil T, Ramachandran R, Schmid SL (2009) Dissecting dynamin's role in clathrin-mediated endocytosis. Biochem Soc Trans 37: 1022–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, Yarden Y (2008) Derailed endocytosis: an emerging feature of cancer. Nat Rev Cancer 8: 835–850 [DOI] [PubMed] [Google Scholar]
- Muhlberg AB, Warnock DE, Schmid SL (1997) Domain structure and intramolecular regulation of dynamin GTPase. EMBO J 16: 6676–6683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan K, Lemmon MA (2006) Determining selectivity of phosphoinositide-binding domains. Methods 39: 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicot AS, Laporte J (2008) Endosomal phosphoinositides and human diseases. Traffic 9: 1240–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norby JG (1988) Coupled assay of Na+,K+-ATPase activity. Methods Enzymol 156: 116–119 [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Konarev PV, Kikhney AG, Svergun DI (2007) ATSAS 2.1–towards automated and web-supported small-angle scattering data analysis. J Appl Cryst 40: s223–s228 [Google Scholar]
- Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Nat Rev Mol Cell Biol 5: 133–147 [DOI] [PubMed] [Google Scholar]
- Ramachandran R, Pucadyil TJ, Liu YW, Acharya S, Leonard M, Lukiyanchuk V, Schmid SL (2009) Membrane insertion of the pleckstrin homology domain variable loop 1 is critical for dynamin-catalyzed vesicle scission. Mol Biol Cell 20: 4630–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Schmid SL (2008) Real-time detection reveals that effectors couple dynamin's GTP-dependent conformational changes to the membrane. EMBO J 27: 27–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran R, Surka M, Chappie JS, Fowler DM, Foss TR, Song BD, Schmid SL (2007) The dynamin middle domain is critical for tetramerization and higher-order self-assembly. EMBO J 26: 559–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J (2007) Galphaq directly activates p63RhoGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem 282: 29201–29210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Sondek J (2005) Larger than Dbl: new structural insights into RhoA activation. Trends Biochem Sci 30: 163–165 [DOI] [PubMed] [Google Scholar]
- Roux A, Koster G, Lenz M, Sorre B, Manneville JB, Nassoy P, Bassereau P (2010) Membrane curvature controls dynamin polymerization. Proc Natl Acad Sci USA 107: 4141–4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim K, Bottomley MJ, Querfurth E, Zvelebil MJ, Gout I, Scaife R, Margolis RL, Gigg R, Smith CI, Driscoll PC, Waterfield MD, Panayotou G (1996) Distinct specificity in the recognition of phosphoinositides by the pleckstrin homology domains of dynamin and Bruton's tyrosine kinase. EMBO J 15: 6241–6250 [PMC free article] [PubMed] [Google Scholar]
- Santoro MM, Bolen DW (1988) Unfolding free energy changes determined by the linear extrapolation method. 1. Unfolding of phenylmethanesulfonyl alpha-chymotrypsin using different denaturants. Biochemistry 27: 8063–8068 [DOI] [PubMed] [Google Scholar]
- Sever S, Muhlberg AB, Schmid SL (1999) Impairment of dynamin's GAP domain stimulates receptor-mediated endocytosis. Nature 398: 481–486 [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Herskovits JS, Vallee RB (1996) A binding site for SH3 domains targets dynamin to coated pits. J Biol Chem 271: 13–16 [DOI] [PubMed] [Google Scholar]
- Smirnova E, Shurland DL, Newman-Smith ED, Pishvaee B, van der Bliek AM (1999) A model for dynamin self-assembly based on binding between three different protein domains. J Biol Chem 274: 14942–14947 [DOI] [PubMed] [Google Scholar]
- Solomaha E, Palfrey HC (2005) Conformational changes in dynamin on GTP binding and oligomerization reported by intrinsic and extrinsic fluorescence. Biochem J 391: 601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song BD, Leonard M, Schmid SL (2004a) Dynamin GTPase domain mutants that differentially affect GTP binding, GTP hydrolysis, and clathrin-mediated endocytosis. J Biol Chem 279: 40431–40436 [DOI] [PubMed] [Google Scholar]
- Song BD, Yarar D, Schmid SL (2004b) An assembly-incompetent mutant establishes a requirement for dynamin self-assembly in clathrin-mediated endocytosis in vivo. Mol Biol Cell 15: 2243–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag JM, Fykse EM, Ushkaryov Y, Liu JP, Robinson PJ, Sudhof TC (1994) Differential expression and regulation of multiple dynamins. J Biol Chem 269: 4547–4554 [PubMed] [Google Scholar]
- Stowell MH, Marks B, Wigge P, McMahon HT (1999) Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nat Cell Biol 1: 27–32 [DOI] [PubMed] [Google Scholar]
- Susman RD, Quijano-Roy S, Yang N, Webster R, Clarke NF, Dowling J, Kennerson M, Nicholson G, Biancalana V, Ilkovski B, Flanigan KM, Arbuckle S, Malladi C, Robinson P, Vucic S, Mayer M, Romero NB, Urtizberea JA, Garcia-Bragado F, Guicheney P et al. (2010) Expanding the clinical, pathological and MRI phenotype of DNM2-related centronuclear myopathy. Neuromuscul Disord 20: 229–237 [DOI] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst 25: 495–503 [Google Scholar]
- Svergun DI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweitzer SM, Hinshaw JE (1998) Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell 93: 1021–1029 [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P (1995) Tubular membrane invaginations coated by dynamin rings are induced by GTP-gamma S in nerve terminals. Nature 374: 186–190 [DOI] [PubMed] [Google Scholar]
- Thompson HM, Cao H, Chen J, Euteneuer U, McNiven MA (2004) Dynamin 2 binds gamma-tubulin and participates in centrosome cohesion. Nat Cell Biol 6: 335–342 [DOI] [PubMed] [Google Scholar]
- Tuma PL, Stachniak MC, Collins CA (1993) Activation of dynamin GTPase by acidic phospholipids and endogenous rat brain vesicles. J Biol Chem 268: 17240–17246 [PubMed] [Google Scholar]
- van der Bliek AM, Redelmeier TE, Damke H, Tisdale EJ, Meyerowitz EM, Schmid SL (1993) Mutations in human dynamin block an intermediate stage in coated vesicle formation. J Cell Biol 122: 553–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira AV, Lamaze C, Schmid SL (1996) Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 274: 2086–2089 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y (2006) Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci USA 103: 1295–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Baba T, Schmid SL (1997) Ubiquitously expressed dynamin-II has a higher intrinsic GTPase activity and a greater propensity for self-assembly than neuronal dynamin-I. Mol Biol Cell 8: 2553–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock DE, Hinshaw JE, Schmid SL (1996) Dynamin self-assembly stimulates its GTPase activity. J Biol Chem 271: 22310–22314 [DOI] [PubMed] [Google Scholar]
- Wong KB, DeDecker BS, Freund SM, Proctor MR, Bycroft M, Fersht AR (1999) Hot-spot mutants of p53 core domain evince characteristic local structural changes. Proc Natl Acad Sci USA 96: 8438–8442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y, Kinuta M, Abe T, Liang S, Araki K, Cremona O, Di Paolo G, Moriyama Y, Yasuda T, De Camilli P, Takei K (2004) The stimulatory action of amphiphysin on dynamin function is dependent on lipid bilayer curvature. EMBO J 23: 3483–3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA (2001) All phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem 276: 44179–44184 [DOI] [PubMed] [Google Scholar]
- Zhang P, Hinshaw JE (2001) Three-dimensional reconstruction of dynamin in the constricted state. Nat Cell Biol 3: 922–926 [DOI] [PubMed] [Google Scholar]
- Zheng J, Cahill SM, Lemmon MA, Fushman D, Schlessinger J, Cowburn D (1996) Identification of the binding site for acidic phospholipids on the PH domain of dynamin: implications for stimulation of GTPase activity. J Mol Biol 255: 14–21 [DOI] [PubMed] [Google Scholar]
- Zuchner S, Noureddine M, Kennerson M, Verhoeven K, Claeys K, De Jonghe P, Merory J, Oliveira SA, Speer MC, Stenger JE, Walizada G, Zhu D, Pericak-Vance MA, Nicholson G, Timmerman V, Vance JM (2005) Mutations in the pleckstrin homology domain of dynamin 2 cause dominant intermediate Charcot-Marie-Tooth disease. Nat Genet 37: 289–294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.