Regulation of DNA-damage responses and cell-cycle progression by the chromatin remodelling factor CHD4
This paper adds to the emerging concept that chromatin remodelling contributes to DNA-damage responses. CHD4, a known NuRD component, is identified as ATM target, localizes to DNA breaks and contributes to repair while associating with PARPylated factors.
Keywords: cell cycle, CHD4, chromatin remodelling, DNA damage, DNA repair
Abstract
The chromatin remodelling factor chromodomain helicase DNA-binding protein 4 (CHD4) is a catalytic subunit of the NuRD transcriptional repressor complex. Here, we reveal novel functions for CHD4 in the DNA-damage response (DDR) and cell-cycle control. We show that CHD4 mediates rapid poly(ADP-ribose)-dependent recruitment of the NuRD complex to DNA-damage sites, and we identify CHD4 as a phosphorylation target for the apical DDR kinase ataxia-telangiectasia mutated. Functionally, we show that CHD4 promotes repair of DNA double-strand breaks and cell survival after DNA damage. In addition, we show that CHD4 acts as an important regulator of the G1/S cell-cycle transition by controlling p53 deacetylation. These results provide new insights into how the chromatin remodelling complex NuRD contributes to maintaining genome stability.
Introduction
In response to DNA damage, cells initiate a coordinated programme of events, termed the DNA-damage response (DDR), which is critical for maintenance of genome integrity and the prevention of ageing and tumourigenesis (Harper and Elledge, 2007; Jackson and Bartek, 2009). Upon detection of DNA lesions, cells activate local and global DDR events that promote cell-cycle checkpoint signalling and DNA repair. Locally and adjacent to DNA-damage sites, DDR proteins are recruited in an orchestrated manner. For example, in response to DNA double-strand breaks (DSBs), phosphorylation of the histone variant H2AX by DDR protein kinases such as ataxia-telangiectasia mutated (ATM) leads to recruitment of the DDR mediator protein mediator of DNA-damage checkpoint-1 (MDC1; Rogakou et al, 1998; Stucki et al, 2005). This then brings about further chromatin alterations that permit recruitment of additional DDR mediators such as p53-binding protein 1 (53BP1) and breast cancer 1 (BRCA1; Panier and Durocher, 2009). DSBs, DNA single-strand breaks (SSBs) and DNA nicks also activate poly(ADP-ribose) polymerase (PARP) enzymes that modify target proteins with poly(ADP-ribose) chains (PAR chains) at DNA-damage sites, thereby stimulating the recruitment and/or activity of repair factors (Malanga and Althaus, 2005; Hakme et al, 2008; Rouleau et al, 2010). Concomitantly, DNA damage induces a global transcriptional programme that leads to the expression of genes whose products slow down or arrest cell-cycle progression to facilitate DNA repair, or trigger programmed cell death (Harper and Elledge, 2007; Jackson and Bartek, 2009). Central to this transcriptional response is activation of the tumour suppressor p53 through post-translational modifications that include p53 phosphorylation and acetylation (Carter and Vousden, 2009; Kruse and Gu, 2009; Vousden and Prives, 2009).
A critical parameter for initiating the DDR is the accessibility of checkpoint and repair factors to DNA lesions within chromatin (Misteli and Soutoglou, 2009). Indeed, compacted chromatin can be refractory to full DDR activation (Murga et al, 2007), and accumulating evidence suggests pivotal and direct functions for chromatin remodelling factors in relieving such inhibitory effects (Bao and Shen, 2007; Downs et al, 2007; Osley et al, 2007; Clapier and Cairns, 2009; van Attikum and Gasser, 2009). Prominent among these is chromodomain helicase DNA-binding protein 4 (CHD4; also known as Mi-2β), an integral component of the NuRD complex (nucleosome remodelling deacetylase) that is unique in combining chromatin remodelling activity with histone deacetylase and demethylase functions involved in transcriptional repression (Wade et al, 1998; Xue et al, 1998; Zhang et al, 1998; Denslow and Wade, 2007; Ramirez and Hagman, 2009). Notably, CHD4 is a likely target for DDR kinases (Matsuoka et al, 2007; Mu et al, 2007; Stokes et al, 2007), and loss of function of CHD4 or other NuRD components causes accumulation of DNA damage and features of accelerated ageing (Pegoraro et al, 2009). In addition, CHD4-associated NuRD subunits HDAC1 (histone deacetylase 1) and MTA2 (metastasis-associated protein 2) have been implicated in regulating p53 deacetylation and thereby p53 transcriptional activity (Luo et al, 2000). Despite these findings, direct functions for CHD4 in the DDR have not hitherto been described. Here, we show that CHD4 is rapidly recruited to DNA lesions in a PARP-dependent manner and is phosphorylated by ATM in response to DNA damage. We also establish that CHD4 controls cell-cycle progression by regulating p53-mediated G1/S arrest, and promotes DSB repair and cell survival after DNA-damage induction.
Results
CHD4 is recruited to DNA-damage sites as part of the NuRD complex
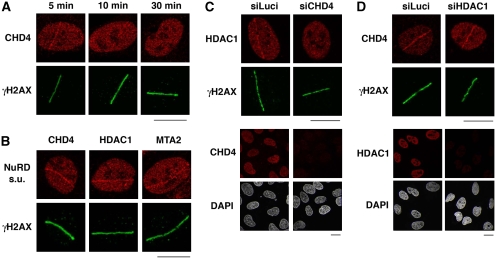
To explore the involvement of CHD4 in the DDR, we first examined whether it was recruited to DNA lesions. When we used laser micro-irradiation to generate localized DNA damage in human U2OS cells, we observed CHD4 accumulation in the damaged regions as revealed by its detection with an anti-CHD4 antibody (Figure 1A; see Supplementary Figure S1A for demonstration of antibody specificity). Notably, this CHD4 re-localization was rapid but transient: CHD4 accumulated at sites of micro-irradiation within a few minutes, but staining intensity then quickly declined and was no longer visible after 30 min (Figure 1A). Similar results were obtained in U2OS cells expressing epitope-tagged CHD4 as well as in other cell types, including BJ primary human fibroblasts, mouse embryonic fibroblasts (Supplementary Figure S1B–D) and HeLa cervical carcinoma cells (data not shown). Although CHD4 did not form detectable foci after cell exposure to ionizing radiation (IR) or genotoxic drugs (data not shown), CHD4 displayed enhanced resistance to detergent extraction very early on after treating cells with the DNA-damaging agent H2O2 (Supplementary Figure S1E), consistent with CHD4 rapid recruitment to damaged chromatin. Interestingly, other components of the NuRD complex—including HDAC1 and MTA2—were also detected at sites of laser-induced damage (Figure 1B). Moreover, CHD4 depletion by short-interfering RNA (siRNA) treatment impaired both HDAC1 and MTA2 recruitment to damaged regions (Figure 1C; Supplementary Figure S2; note that CHD4 depletion did not affect expression levels of HDAC1 or MTA2). In contrast, HDAC1 depletion did not impair CHD4 accrual at sites of DNA lesions (Figure 1D). Collectively, these findings suggested that CHD4 is recruited to DNA-damage sites as part of the NuRD complex and that CHD4 has a leading function in NuRD recruitment.
Figure 1.
CHD4 is recruited to sites of laser-induced DNA damage within the NuRD complex. (A) Immunodetection of CHD4 and γH2AX (damage sites) at the indicated times after laser micro-irradiation in U2OS cells. (B) Recruitment of NuRD complex subunits to sites of laser-induced DNA damage (labelled by γH2AX) 5 min after micro-irradiation in U2OS cells. (C, D) Immunodetection of HDAC1 and CHD4 5 min after laser micro-irradiation in U2OS cells treated with the indicated siRNAs (siLuci: control). Cells were pre-treated with ATM inhibitor to facilitate detection of HDAC1 and CHD4 lines. Lower panels show siRNA efficiency. In laser micro-irradiation experiments, detergent pre-extraction was performed before fixation of the cells for immunostaining. Scale bars, 10 μm.
CHD4 recruitment to DNA-damage sites is PARP dependent
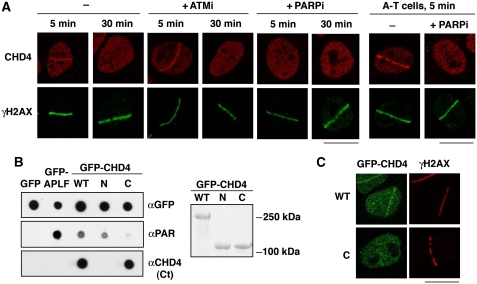
Having discovered the mobilization of CHD4 to DNA lesions, we next sought to determine the mechanism for this. Initially, we focused on the potential function of H2AX phosphorylation (γH2AX) because this histone modification has previously been implicated in the recruitment of certain chromatin remodelling factors to damaged chromatin in yeast (van Attikum and Gasser, 2009). However, CHD4 accumulation at DNA-damage sites was not impaired in H2AX-deficient cells (Supplementary Figure S1D), suggesting a different recruitment mode. We thus tested for the potential contribution of two important enzymes involved in the early steps of the DDR: ATM and PARP. Use of the ATM inhibitor KU-55933 (Hickson et al, 2004) revealed that ATM activity was dispensable for CHD4 recruitment to damage sites; in fact, we consistently observed enhanced CHD4 recruitment upon ATM inhibition (Figure 2A; Supplementary Figure S1B and C; Supplementary Figure S3A shows that CHD4 protein levels were not affected by ATM inhibition). In line with this, CHD4 accumulation at DNA-damage sites was also observed in ATM-deficient A-T fibroblasts (Figure 2A; Supplementary Figure S3B). Similar results were obtained for Seckel syndrome cells (O'Driscoll et al, 2003) deficient for the ATM-related kinase ATR (data not shown).
Figure 2.
PARP-dependent recruitment of CHD4 to sites of DNA damage. (A) Immunodetection of CHD4 and γH2AX (damage sites) at the indicated times after laser micro-irradiation in U2OS (left) and A-T cells (right). Cells were pre-incubated for 1 h with the indicated inhibitors (ATMi, ATM inhibitor; PARPi, PARP inhibitor) before micro-irradiation. (B) PAR-binding assay with immunoprecipitated GFP-CHD4 wild-type (WT: residues 1–1937) or truncated mutants (N: residues 1–758; C: residues 1183–1937). GFP only is used as a negative control and GFP-APLF as a positive control. Right panel: corresponding Ponceau staining of the blot after immunoprecipitation from HEK-293 cells. (C) Recruitment of GFP-CHD4 wild-type (WT) or a truncated mutant (C: residues 1183–1937) to sites of laser-induced damage (γH2AX) 5 min after micro-irradiation in U2OS cells. In all cases, detergent pre-extraction was performed before fixation of the cells for immunostaining. Scale bars, 10 μm.
In striking contrast to the above data, chemical inhibition of the PARP1 and PARP2 enzymes with the compound KU-58948 (Farmer et al, 2005) or PARP1/2 depletion by RNA interference completely abrogated CHD4 accumulation at sites of laser-induced damage (Figure 2A; Supplementary Figures S1B-C and S3C-D; note that PARP inhibition or PARP1/2 depletion did not affect CHD4 levels). As PARP1 and PARP2 are activated by DNA-strand breaks, this PARP dependency of CHD4 recruitment strongly suggested that CHD4 was specifically recruited to such structures. Consistent with this idea, the rapid and transient recruitment of CHD4 to DNA-damage sites mirrored the kinetics of poly(ADP-ribosyl)ation (Supplementary Figure S1F). Moreover, we found that CHD4 directly bound PAR chains (Figure 2B; Supplementary Figure S3E), which concurs with CHD4 having a leading function in recruiting other NuRD subunits to DNA-damage sites.
Although CHD4 does not display canonical PAR-binding domains (Karras et al, 2005; Ahel et al, 2008), sequence analyses revealed the presence of putative PAR-binding motifs in the CHD4 amino- and carboxyl-terminal regions that loosely matched a characterized consensus (Pleschke et al, 2000; Gagne et al, 2008). By carrying out studies with CHD4 deletion derivatives, we found that the amino-terminal, but not carboxyl-terminal, region displayed PAR binding comparable with that exhibited by the full-length protein (Figure 2B), and consistent with this, the CHD4 carboxyl-terminal region displayed defective recruitment to DNA-damage sites (Figure 2C). Taken together, these data showed that CHD4 is rapidly and transiently recruited to damaged chromatin within the NuRD complex. Furthermore, they established that this recruitment occurs in a PARP-dependent manner that likely involves CHD4 binding to PARylated proteins, including PARP1 itself, present at damage sites.
CHD4 is phosphorylated upon DNA damage in an ATM-dependent manner
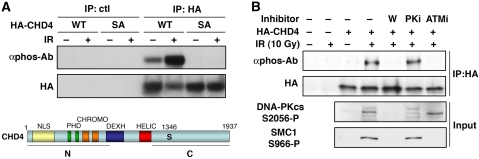
Along with their recruitment to DNA-damage sites, another hallmark of many DDR proteins is their post-translational modification in response to genotoxic stress. Interestingly, proteomic screens in human cells have identified CHD4 as a target for DDR kinases (Matsuoka et al, 2007; Mu et al, 2007; Stokes et al, 2007). More specifically, we noted that one of the putative phosphorylation sites on CHD4 that is conserved in vertebrates (Ser-1346 in isoform 2; Figure 3A) was identified by mass spectrometry (Matsuoka et al, 2007) and by bioinformatic analyses with a high-stringency search mode (http://scansite.mit.edu/). By using a phospho-specific antibody raised against the corresponding motif (GpSQE), we found that transiently expressed HA-CHD4 was indeed phosphorylated in human cells, and that this phosphorylation was markedly increased in response to IR, in a dose- and time-dependent manner (Figure 3A; Supplementary Figure S4A and data not shown). Similarly, we detected phosphorylation of endogenous CHD4 when cells were treated with DNA-damaging agents such as H2O2 and neocarzinostatin (Supplementary Figure S4B and data not shown). Importantly, detection of CHD4 phosphorylation was abrogated when Ser-1346 was mutated to Ala (SA; Figure 3A). Furthermore, we found that this CHD4 phosphorylation was prevented when cells were incubated with Wortmannin (Sarkaria et al, 1998) or the ATM inhibitor KU-55933 (Hickson et al, 2004), but not the DNA-PK inhibitor NU-7441 (Leahy et al, 2004) (see Figure 3B; Supplementary Figure S4B). Collectively, these results showed that CHD4 is phosphorylated after DNA damage on Ser-1346 in an ATM-dependent manner.
Figure 3.
ATM-dependent phosphorylation of CHD4 Ser-1346 in response to DNA damage. (A) Detection of CHD4 phosphorylation (αphos-Ab) 1 h after exposure to 10 Gy ionizing radiation (+ IR) on HA immunoprecipitates from U2OS cells transiently expressing HA-CHD4 wild-type (WT) or S1346A point mutant (SA). The scheme represents CHD4 protein with the amino-acid position of the candidate phospho-serine (S1346); the N and C fragments correspond to CHD4 truncated mutants analysed in Figures 2 and 4. Domain organization of CHD4: NLS (putative nuclear localization signal), PHD (plant homeodomain), CHROMO (chromodomain), DEXH (ATP-binding domain), HELIC (helicase carboxyl-terminal domain). (B) Detection of CHD4 S1346 phosphorylation (αphos-Ab) 30 min after cell exposure to 10 Gy ionizing radiation (IR) on HA immunoprecipitates from U2OS cells transiently expressing HA-CHD4. Cells were pre-treated with the indicated inhibitors (W, Wortmannin; PKi, DNA-PK inhibitor; ATMi, ATM inhibitor). ATM-dependent phosphorylation of SMC1 S966 and DNA-PKcs autophosphorylation on S2056 are used as controls.
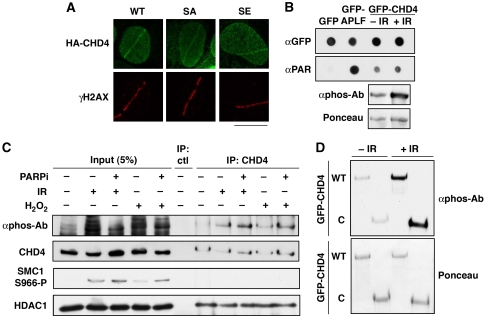
We next investigated possible connections between ATM-dependent phosphorylation of CHD4 and its recruitment to sites of DNA damage along with other NuRD components. Notably, CHD4 phosphorylation did not affect CHD4 interactions with the NuRD subunits MTA2 or HDAC1 (Supplementary Figure S4C and D). In addition, consistent with our finding that ATM activity was dispensable for CHD4 accumulation at DNA-damage sites, mutation of the ATM-target Ser-1346 did not interfere with CHD4 recruitment (Figure 4A). Furthermore, CHD4 phosphorylation after exposing cells to IR did not detectably alter its affinity for PAR (Figure 4B). Reciprocally, preventing CHD4 recruitment to damaged chromatin by PARP inhibition (Figure 4C) or deletion of the CHD4 amino-terminus (Figure 4D) did not impair its phosphorylation. Together, these data established that ATM-dependent CHD4 phosphorylation and PARP-dependent CHD4 recruitment to damaged chromatin are distinct events.
Figure 4.
DNA-damage-induced phosphorylation and recruitment of CHD4 to DNA lesions are distinct events. (A) Recruitment of HA-CHD4 wild-type (WT) and Ser-1346 point mutants (SA, mutated to Ala; SE, mutated to Glu) to sites of laser-induced damage (γH2AX) in transiently transfected U2OS cells 5 min after micro-irradiation. Scale bar, 10 μm. (B) PAR-binding activity of GFP-CHD4 from HEK-293 cells exposed or not to ionizing radiation (IR). GFP only is used as a negative control, GFP-APLF as a positive control. Lower panel shows CHD4 phosphorylation after IR analysed in parallel by western blotting. (C) Detection of CHD4 phosphorylation (αphos-Ab) 30 min after exposure to 10 Gy ionizing radiation (IR) or 500 μM hydrogen peroxide (H2O2) on CHD4 immunoprecipitates from HEK-293 cells treated or not with PARP inhibitor (PARPi). SMC1 phosphorylation is used as a control for DNA damage. The NuRD subunit HDAC1 co-immunoprecipitates with CHD4. (D) Detection of CHD4 phosphorylation (αphos-Ab) 30 min after exposure to 10 Gy ionizing radiation (IR) on GFP immunoprecipitates from H3K293 cells transiently expressing GFP-CHD4 wild-type (WT) or a truncated mutant (C: residues 1183–1937).
CHD4 regulates cellular sensitivity to DNA damage
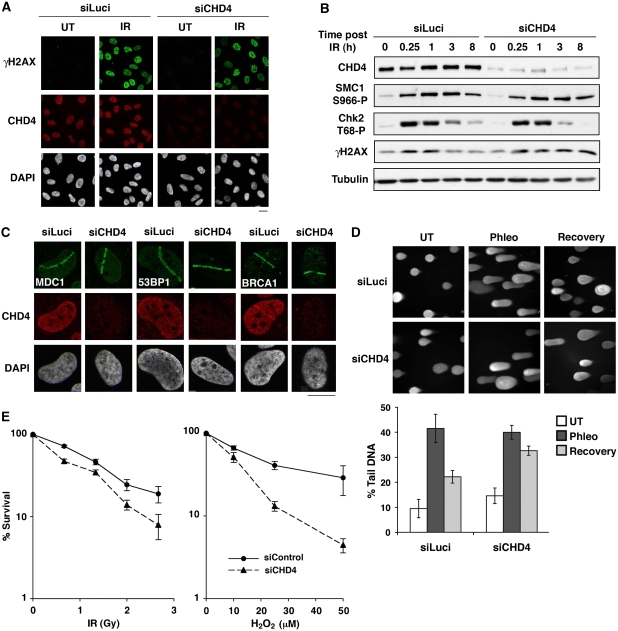
To address the biological significance of CHD4 to the DDR, we analysed the effects of its depletion, focusing initially on PARP-mediated repair of DNA breaks (Caldecott, 2008) and ATM-dependent signalling. Notably, CHD4 depletion did not impair PARP activation in response to DNA damage or PARP recruitment to DNA-damage sites (Supplementary Figure S5A and B), and did not appreciably affect the recruitment or dissociation of the SSB repair factor XRCC1 at sites of laser damage (Supplementary Figure S5C). In line with this, CHD4-depleted cells efficiently repaired DNA breaks arising from H2O2 exposure as measured by alkaline comet assays (Supplementary Figure S5D). In addition, we found that CHD4 depletion did not impair H2AX phosphorylation and focus formation, the phosphorylation/dephosphorylation kinetics of the ATM-targets SMC1 and Chk2 in response to IR (Figure 5A and B), or the recruitment of the ATM-responsive checkpoint mediators MDC1, 53BP1 and BRCA1 to laser-induced and IR-induced DNA damage (Figure 5C and data not shown). Consistent with these findings, CHD4-depleted cells did not display significant defects in G2/M DNA-damage checkpoint activation or checkpoint recovery (Supplementary Figure S6). Importantly, however, we observed persistence of IR-induced H2AX phosphorylation in CHD4-depleted cells (Figure 5B), suggesting that such cells are defective in DSB repair. To directly address this possibility, we performed neutral comet assays. Thus, we found that CHD4 depletion significantly impaired the repair of DSBs produced by the radiomimetic agent phleomycin (Figure 5D). In line with these findings, siRNA-mediated CHD4 depletion also significantly compromised clonogenic cell survival after exposure to IR (Figure 5E). Furthermore, CHD4 depletion led to substantially enhanced cell killing in response to H2O2 treatment both in U2OS and HeLa cells (Figure 5E and data not shown). As discussed further below, these results revealed that, while CHD4 depletion does not overtly impair ATM-dependent signalling and SSB repair—possibly because of partly compensatory chromatin remodelling activities—CHD4 function clearly promotes DSB repair and cell survival after genotoxic challenge.
Figure 5.
CHD4 regulates cell sensitivity to DNA damage. (A) γH2AX foci formation in U2OS cells upon CHD4 depletion (siCHD4) compared with control (siLuci) 1 h after exposure to 10 Gy ionizing radiation (IR). (B) Phosphorylation of DNA-damage checkpoint proteins analysed by western blotting of total extracts prepared from HeLa cells at the indicated times after cell exposure to 10 Gy ionizing radiation (IR) (siCHD4, CHD4 depletion; siLuci, control). (C) Recruitment of MDC1, 53BP1 and BRCA1 to sites of laser-induced damage (γH2AX) 5–10 min after micro-irradiation in HeLa cells upon CHD4 depletion (siCHD4) compared with control (siLuci). Detergent pre-extraction was performed before fixation and immunostaining for BRCA1. Scale bars, 10 μm. (D) Efficiency of DSB repair in control (siLuci) or CHD4-depleted U2OS cells analysed by neutral comet assay after phleomycin treatment (Phleo). Error bars: s.d. from two independent experiments. (E) Clonogenic survival of U2OS cells upon CHD4 knock-down (siCHD4) compared with control (siLuci) in response to ionizing radiation (IR) or hydrogen peroxide (H2O2). H2O2 treatment was for 10 min at the indicated doses. Error bars indicate s.d. from two and three independent experiments, respectively. The results are normalized to plating efficiencies to focus on the effect of CHD4 depletion upon DNA damage. Note that in the absence of DNA-damaging agent, the viability of CHD4-depleted cells is ∼66% of that of control cells.
CHD4 controls the G1/S transition by regulating p53 deacetylation
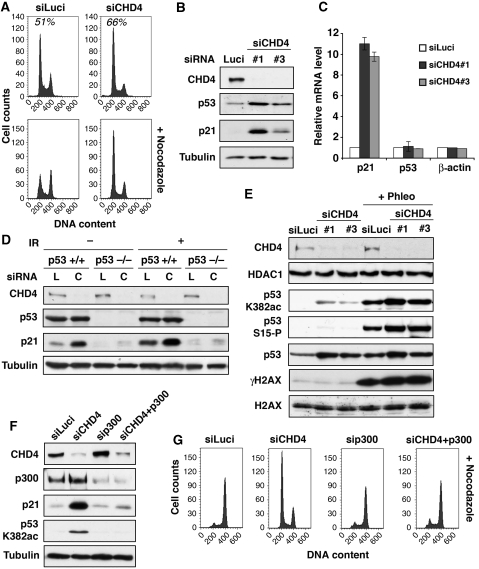
As cell viability in response to DNA damage relies not only on DNA repair but also on proper cell-cycle control, we investigated whether CHD4 affected cell-cycle progression. During the course of these studies, we noticed that in most cell lines—including U2OS cancer cells and non-cancer cells such as retinal pigment epithelial cells—CHD4 depletion resulted in a significant block or delay at the G1/S cell-cycle transition (Figure 6A and data not shown). Given that a master regulator of the passage from G1 to S is the p53-p21 pathway, this prompted us to examine the regulation of this pathway in CHD4-depleted cells. Strikingly, this revealed that CHD4 depletion caused a rapid and substantial accumulation of p21 protein (Figure 6B). The elevation of p21 expression upon CHD4 depletion was also observed at the RNA level (Figure 6C) and was restricted to p53 proficient cells (Figure 6D; Supplementary Figure S7A). Furthermore, we found that p21 induction was generally accompanied by heightened p53 protein levels (Figure 6B and E) and correlated with increased p53 binding to the p21 promoter (Supplementary Figure S7B). Although one potential explanation for these findings was that p53-dependent transcriptional induction of p21 arises as a consequence of endogenous DNA damage that can accumulate in CHD4-depleted cells (Pegoraro et al, 2009; Figure 5D), several lines of evidence argue against this. First, p21 accumulation was observed very early upon CHD4 depletion, before a detectable increase in γH2AX signal (Supplementary Figure S7C), while accumulation of endogenous DNA damage was generally observed only from 72 h siRNA treatment onwards (Figure 5D and data not shown). Moreover, contrary to IR-induced p21 expression, the induction of p21 after CHD4 depletion was not prevented by inhibition of ATM or the other apical DDR kinases ATR and DNA-PK (Supplementary Figure S7D and data not shown).
Figure 6.
CHD4 controls the G1/S cell-cycle transition through p53 deacetylation. (A) Fluorescence-activated cell sorting (FACS) profiles of U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci). Percentages of cells in G1 are indicated. Nocodazole was used to block cell-cycle progression in mitosis (bottom panels). (B) Western-blot analysis of total extracts from U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci) using the indicated antibodies. (C) Quantitative RT–PCR analysis of p21, p53 and β-actin mRNA levels in U2OS cells downregulated for CHD4 (siCHD4) compared with control (siLuci). Error bars indicate s.d. from two independent experiments. (D) Western-blot analysis of p21 induction on total extracts from p53 proficient or deficient HCT116 cells upon CHD4 depletion (C) compared with control siLuciferase (L). + IR: 9 h post-exposure to 10 Gy IR. (E, F) Western-blot analysis of total extracts from U2OS cells treated with the indicated siRNAs. Phleo: 1 h post-exposure to 60 μg/ml Phleomycin. (G) FACS profiles of U2OS cells treated with the indicated siRNAs. Nocodazole was used to block cell-cycle progression in mitosis.
To gain insights into how CHD4 depletion leads to p53-dependent p21 transcription before the induction of detectable DNA damage, we analysed p53 modifications known to be associated with its transcriptional activity. Consistent with CHD4 depletion not rapidly triggering p53 activity through the induction of DNA damage, p53 phosphorylation on Ser-15 was almost undetectable in CHD4-depleted cells before exposure to exogenous damage (Figure 6E). In contrast, we readily detected Lys-382 acetylation on stabilized p53 in these cells, indicating that CHD4 is needed for restraining basal levels of p53 acetylation (Figure 6E). Consistent with this, we found that depletion of the p300 acetyltransferase, which is responsible for p53 Lys-382 acetylation (Carter and Vousden, 2009; Kruse and Gu, 2009), reduced both p53 acetylation and p21 levels, and moreover, rescued cell-cycle progression in CHD4-depleted cells (Figure 6F and G), supporting the importance of acetylation in this cell-cycle arrest. Collectively, these findings established that CHD4 represses p53-dependent p21 transcription and define CHD4 as an important regulator of the G1 to S transition through controlling p53 deacetylation.
Discussion
CHD4 is well characterized as a catalytic subunit of the NuRD transcriptional repressor complex (Wade et al, 1998; Xue et al, 1998; Zhang et al, 1998; Denslow and Wade, 2007; Ramirez and Hagman, 2009). Here, we have uncovered new functions for CHD4 in genome stability and cell-cycle progression. Specifically, we have shown that CHD4 is an important contributor to multiple aspects of the DDR. The recruitment of CHD4 to DNA-damage sites in a PARP-dependent manner and the hyper-sensitivity of CHD4-depleted cells to H2O2 strongly suggest that CHD4 responds to SSBs and possibly oxidative DNA damage, whereas CHD4 phosphorylation by ATM points to additional functions in response to DSBs. Consistent with such a DSB repair function, we have observed that CHD4-depleted cells are hyper-sensitive to IR, display delayed removal of γH2AX after IR exposure and are deficient in repairing DSBs as detected by neutral comet assays. Furthermore, we have established an important function for CHD4 in cell-cycle control at the G1/S transition by regulating p53 deacetylation. It will be interesting to examine whether these functions of CHD4 are distinct, or whether they operate in antagonistic or inter-dependent ways. In particular, CHD4 functions in DSB repair and p53-dependent cell-cycle control are both likely to promote the survival of damaged cells, and their relative contributions need to be further clarified. Notably, we have also observed that ATM inhibition enhances CHD4 accumulation at DNA-damage sites. As ATM phosphorylates CHD4, this result initially suggested to us that phosphorylation might regulate the dissociation kinetics of CHD4 from damage sites. However, we did not observe significant differences in the timing and level of accumulation between wild-type CHD4 and a mutant CHD4 derivative in which the Ser-1346 ATM-target site was mutated. Thus, our interpretation of the effect of ATM inhibitor is that it promotes CHD4 accumulation indirectly by enhancing the PAR signal at DNA-damage sites (shown in Supplementary Figure S1F).
In regards to CHD4 functioning to regulate p53 deacetylation, the mechanism is unlikely to be direct given that CHD4 itself does not display acetyltransferase or deacetylase activities. However, it is noteworthy that, within the NuRD complex, CHD4 associates with HDAC1, an enzyme directly implicated with MTA2 in p53 deacetylation and whose depletion induces p21 expression (Luo et al, 2000; Lagger et al, 2002). Importantly, our data show that HDAC1 and MTA2 levels are unaffected by CHD4 depletion, supporting the idea that CHD4 controls the G1/S cell-cycle transition by promoting the ability of HDAC1 to deacetylate p53. In addition to a function in cell-cycle control, the recruitment of CHD4 and other components of the NuRD complex to DNA breaks indicates that NuRD also has important functions at DNA-damage sites, in particular to ensure efficient repair of DSBs. Although deciphering the underlying mechanism for such a repair function still requires further investigation, it is tempting to speculate that this could be through local remodelling of chromatin to facilitate DNA repair, perhaps in ways similar to those proposed for yeast and other mammalian chromatin remodelling complexes (Park et al, 2006; Ahel et al, 2009; Peng et al, 2009; Sinha et al, 2009). Another possible function for NuRD at DNA-damage sites is to ensure that transcription is locally inhibited, thus preventing transcription from interfering with DNA-damage signalling and/or repair. Interestingly, CHD4 appears to be one of the several, and possibly many, chromatin remodelling factors recruited to DNA breaks in mammalian cells (Murr et al, 2006; Ahel et al, 2009; Gottschalk et al, 2009; Peng et al, 2009; Timinszky et al, 2009). It is, therefore, possible that such factors will display synergistic and/or antagonistic functions in the regulation of chromatin compaction at DNA-damage sites, as happens in the context of transcription (Ramirez-Carrozzi et al, 2006; Clapier and Cairns, 2009). Although such relationships will be of interest to explore in future studies, they may pose technical challenges because defining clear functions for individual chromatin remodelling components will likely require the combined inactivation of functionally overlapping, partially redundant complexes.
Collectively, the data we have provided define CHD4 as a new player in the DDR and in p53-dependent cell-cycle control. These findings thereby help explain recent observations linking NuRD to cellular ageing (Pegoraro et al, 2009) and might shed light on why patients expressing auto-antibodies against CHD4 display higher cancer incidence (Hill et al, 2001). Defining DDR functions for CHD4 and other chromatin remodelling components is also relevant for cancer research in general, given that drugs targeting chromatin-modifying enzymes are being explored as anti-cancer therapies, both for use alone and in combination with DNA-damaging treatments (Ellis et al, 2009).
Materials and methods
Cell culture and transfections
Human U2OS, HEK-293, BJ, HeLa, A-T (from Y Shiloh), p53+/+ and p53−/− HCT116 cells (from B Vogelstein), H2AX+/+ and H2AX−/− mouse embryonic fibroblasts (from A Nussenzweig) were grown in Dulbecco's modified Eagle medium (Invitrogen) supplemented with 10% foetal bovine serum (BioSera), 2 mM L-glutamine, 100 unit/ml penicillin, 100 μg/ml streptomycin and fungizone (Sigma-Aldrich). Cell transfections with plasmid DNA or siRNA duplexes (Supplementary Table S2) were performed by using Lipofectamine 2000 and Lipofectamine RNAiMax (Invitrogen), respectively, following the manufacturer's instructions. Cells were analysed 48–72 h after transfection.
Antibodies
All the antibodies used in this study are commercially available (detailed in Supplementary Table S1) except the phospho-specific antibody used against CHD4, which was provided by Y Shiloh.
Plasmids
An IMAGE clone corresponding to the full-length human CHD4 cDNA (isoform 2, accession number BC038596, clone 5528023 in pCMV-sport6 vector, Geneservice Ltd.) was used to generate plasmids encoding HA-CHD4 and GFP-HA-CHD4 wild-type, truncated mutants (N, C) and point mutants (S1346A, S1346E). The HA tag replaced CHD4 5′UTR region. The GFP sequence was PCR amplified from pEGFP-C1 (Clontech). An NLS sequence was included in CHD4 construct lacking the amino-terminal domain to ensure proper nuclear localization. All constructs were verified by direct sequencing and/or restriction digests. Cloning details and primer sequences (Sigma-Aldrich) are available upon request. GFP-APLF plasmid was described in Ahel et al (2008).
DNA damage and drug treatments
ATM (KU-55933) (Hickson et al, 2004), PARP (KU-58948) (Farmer et al, 2005) and DNA-PK (NU-7441) (Leahy et al, 2004) inhibitors (KuDOS Pharmaceuticals Ltd.) were used at a final concentration of 20, 10 and 2 μM, respectively. Wortmannin (Alexis Biochemicals) was used at 200 μM to inhibit ATM, ATR and DNA-PK. Inhibitors were applied to culture medium 1 h before subsequent treatments. IR was delivered by an X-ray generator (Faxitron X-ray Corporation RX-650, 120 kV, 5 mA, dose rates 10 and 5.3 Gy/min). Treatment with H2O2 (VWR) was for 30 min at 500 μM unless otherwise stated. Treatment with phleomycin (Melford Laboratories) was for 1 h at 60 μg/ml. Nocodazole (Sigma-Aldrich) was used at a final concentration of 40 ng/ml for 20 h.
Laser micro-irradiation
Localized DNA damage was generated by exposure of cells to a UV-A laser (Limoli and Ward, 1993; Lukas et al, 2003). Cells plated on glass-bottom dishes (Willco Wells) were pre-sensitized with 10 μM 5-bromo-2′-deoxyuridine (BrdU, Sigma-Aldrich) in phenol red-free medium (Invitrogen) for 24 h at 37°C. Micro-irradiation was performed with a FluoView 1000 confocal microscope (Olympus) equipped with a 37°C heating stage (Ibidi) and a 405-nm laser diode (6 mW) focused through a 60X UPlanSApo/1.35 oil objective to yield a spot size of 0.5–1 μm. Time of cell exposure to the laser beam was ∼250 ms (fast-scanning mode). Laser settings (0.40 mW output, 50 scans, SIM scanner) were chosen to generate a detectable DDR restricted to the laser path in a pre-sensitization-dependent manner without detectable cytotoxic effects.
Immunofluorescence
Cells on glass coverslips (VWR) or glass-bottom dishes (Willco Wells) were fixed with 2% paraformaldehyde and permeablized with 0.2% Triton-X-100 in PBS. When indicated, permeabilization was carried out before fixation in 10 mM PIPES pH 7.0, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.5% Triton-X-100 for 5 min at room temperature. Samples were then blocked in 5% bovine serum albumin and stained with the appropriate primary (Supplementary Table S1) and secondary antibodies coupled to AlexaFluor 488 or 594 (Molecular Probes). Confocal images were captured on a Nikon Eclipse E800 microscope equipped with Radiance 2100 laser set-up and LaserSharp software (Bio-Rad) or on FluoView1000 Olympus using a × 40 or × 60 oil objective. To avoid bleed-through effects in double-staining experiments, each dye was scanned independently in a multi-tracking mode.
Immunoblotting
Total cell extracts were obtained by scraping cells in Laemmli buffer (0.8% SDS, 4% glycerol, 280 mM β-mercaptoethanol, 25 mM Tris–HCl pH 6.8, 0.005% bromophenol blue). Proteins were resolved by SDS–PAGE, transferred onto nitrocellulose (Protran) and probed using the appropriate primary (Supplementary Table S1) and secondary antibodies coupled to horse-radish peroxidase (Dako, Pierce). Protein detection was performed with ECL reagents (GE Healthcare).
Immunoprecipitation
Cells harvested in PBS were lysed in RIPA buffer (10 mM Tris pH 8, 1% Triton-X-100, 0.1% deoxycholate, 0.1% SDS, 150 mM NaCl) or lysis buffer (20 mM Tris pH 7.5, 40 mM NaCl, 2 mM MgCl2, 0.5% NP-40, 50 U/ml benzonase) supplemented with protease and phosphatase inhibitors and adjusted to 450 mM salt concentration. Lysates were clarified by centrifugation (13 200 r.p.m., 20 min, 4°C) and 500 μg–1 mg proteins were used per immunoprecipitation in RIPA buffer or IP buffer (25 mM Tris pH 7.5, 150 mM NaCl, 1.5 mM DTT, 10% glycerol, 0.5% NP-40) supplemented with protease and phosphatase inhibitors. Proteins were captured with the appropriate antibody and protein A-sepharose Fast-Flow (Sigma) or Dynabeads (Dynal), or directly onto GFP-Trap agarose beads (ChromoTek) for GFP-tagged proteins. Complexes were extensively washed in RIPA or IP buffer. Immunoprecipitation with rabbit serum or from cells that do not express epitope-tagged protein were used as negative controls.
PAR-binding assay
GFP-tagged proteins transiently expressed in human HEK-293 cells were isolated with GFP-Trap beads (ChromoTek) following the above-described immunoprecipitation procedure. Immunoprecipitates were extensively washed in RIPA buffer adjusted to 1 M NaCl to disrupt protein complexes, before incubation for 1 h in Tris-buffered saline—0.1% Tween containing 100 nM purified PAR (Alexis Biochemicals). After extensive washes in TBS-Tween adjusted to 300 mM NaCl, complexes were resuspended in Laemmli buffer, boiled and spotted onto nitrocellulose for immunodetection with anti-GFP and anti-PAR antibodies.
Comet assays
Cells were treated with 150 μM H2O2 in PBS for 10 min on ice or with 60 μg/ml phleomycin for 2 h at 37°C followed by 1 h recovery in culture medium at 37°C. Alkaline and neutral comet assays were as specified in the Comet Assay kit (Trevigen) using GelBond films (Lonza) to support agarose gels. Samples stained with SYBR–Green I were observed under an epifluorescence microscope (Olympus IX71) using a UPlanFLN × 10 objective. Images were analysed with CometScore software (TriTek) by scoring around 100 cells in each case.
Quantitative RT–PCR
RNA extracted with Trizol (Invitrogen) was subject to DNA digestion with Turbo enzyme (Ambion) and reverse transcription with Superscript III RT (Invitrogen). DNA products were quantified by real-time PCR on ABI Prism 7000 sequence detection system (Applied Biosystems) using SYBR–Green mix with the indicated primer pairs (Supplementary Table S3).
Fluorescence-activated cell sorting
Cells were fixed in ice-cold 70% ethanol. DNA was stained with 50 μg/ml propidium iodide (Sigma-Aldrich) in PBS containing 0.1% Triton-X-100 and 0.5 mg/ml DNAse-free RNAse A (Sigma-Aldrich). Samples were processed on an FACSCalibur flow cytometer equipped with CellQuest software (Becton Dickinson). The results were analysed using FlowJo software (TreeStar).
Colony-forming assays
Forty-eight hours after siRNA transfection, cells were replated and exposed to the indicated DNA-damaging agent the following day. After an additional 14-day incubation, colonies were stained with 0.5% crystal violet/20% ethanol and counted. The results were normalized to plating efficiencies.
Supplementary Material
Acknowledgments
We thank Y Shiloh for providing the phospho-specific antibody and A-T cells, A Nussenzweig for H2AX-deficient cells, B Vogelstein for p53-deficient cells, KuDOS Pharmaceuticals Ltd. for DDR-enzyme inhibitors, S Boulton and S West for GFP-APLF construct and M Oren for GST-p53 plasmid. We thank all members of the Jackson laboratory for suggestions and advice, M Blasius for GST-p53 and K Miller and K Dry for critical reading of the manuscript. Research in the Jackson laboratory is supported by grants from Cancer Research UK, the European Union (Integrated Project DNA Repair, LSHG-CT-2005-512113 and Genomic Instability in Cancer and Precancer, HEALTH-F2-2007-201630), the Wellcome Trust and the Biotechnology and Biological Sciences Research Council. This work was also made possible by core infrastructure funding provided by Cancer Research UK and the Wellcome Trust. SEP is funded by Human Frontier Science Program Organization. AK is funded by a Herchel Smith Postdoctoral Research Fellowship. Authors' contributions: All experiments were designed and executed by SEP and AK. LB provided technical assistance. YG raised the phospho-specific antibody used against CHD4. SPJ supervised the project. SEP, AK and SPJ wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ (2009) Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science 325: 1240–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC (2008) Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature 451: 81–85 [DOI] [PubMed] [Google Scholar]
- Bao Y, Shen X (2007) Chromatin remodeling in DNA double-strand break repair. Curr Opin Genet Dev 17: 126–131 [DOI] [PubMed] [Google Scholar]
- Caldecott KW (2008) Single-strand break repair and genetic disease. Nat Rev Genet 9: 619–631 [DOI] [PubMed] [Google Scholar]
- Carter S, Vousden KH (2009) Modifications of p53: competing for the lysines. Curr Opin Genet Dev 19: 18–24 [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR (2009) The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304 [DOI] [PubMed] [Google Scholar]
- Denslow SA, Wade PA (2007) The human Mi-2/NuRD complex and gene regulation. Oncogene 26: 5433–5438 [DOI] [PubMed] [Google Scholar]
- Downs JA, Nussenzweig MC, Nussenzweig A (2007) Chromatin dynamics and the preservation of genetic information. Nature 447: 951–958 [DOI] [PubMed] [Google Scholar]
- Ellis L, Atadja PW, Johnstone RW (2009) Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther 8: 1409–1420 [DOI] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A (2005) Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature 434: 917–921 [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG (2008) Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res 36: 6959–6976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC (2009) Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA 106: 13770–13774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakme A, Wong HK, Dantzer F, Schreiber V (2008) The expanding field of poly(ADP-ribosyl)ation reactions. ‘Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep 9: 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Elledge SJ (2007) The DNA damage response: ten years after. Mol Cell 28: 739–745 [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC (2004) Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res 64: 9152–9159 [DOI] [PubMed] [Google Scholar]
- Hill CL, Zhang Y, Sigurgeirsson B, Pukkala E, Mellemkjaer L, Airio A, Evans SR, Felson DT (2001) Frequency of specific cancer types in dermatomyositis and polymyositis: a population-based study. Lancet 357: 96–100 [DOI] [PubMed] [Google Scholar]
- Jackson SP, Bartek J (2009) The DNA-damage response in human biology and disease. Nature 461: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG (2005) The macro domain is an ADP-ribose binding module. EMBO J 24: 1911–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse JP, Gu W (2009) Modes of p53 regulation. Cell 137: 609–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagger G, O'Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C (2002) Essential function of histone deacetylase 1 in proliferation control and CDK inhibitor repression. EMBO J 21: 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC (2004) Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett 14: 6083–6087 [DOI] [PubMed] [Google Scholar]
- Limoli CL, Ward JF (1993) A new method for introducing double-strand breaks into cellular DNA. Radiat Res 134: 160–169 [PubMed] [Google Scholar]
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J (2003) Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 5: 255–260 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381 [DOI] [PubMed] [Google Scholar]
- Malanga M, Althaus FR (2005) The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol 83: 354–364 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Misteli T, Soutoglou E (2009) The emerging role of nuclear architecture in DNA repair and genome maintenance. Nat Rev Mol Cell Biol 10: 243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, Besusso D, Jung SY, Qin J (2007) A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem 282: 17330–17334 [DOI] [PubMed] [Google Scholar]
- Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, Yang SM, Blasco MA, Skoultchi AI, Fernandez-Capetillo O (2007) Global chromatin compaction limits the strength of the DNA damage response. J Cell Biol 178: 1101–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z (2006) Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol 8: 91–99 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA (2003) A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet 33: 497–501 [DOI] [PubMed] [Google Scholar]
- Osley MA, Tsukuda T, Nickoloff JA (2007) ATP-dependent chromatin remodeling factors and DNA damage repair. Mutat Res 618: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panier S, Durocher D (2009) Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst) 8: 436–443 [DOI] [PubMed] [Google Scholar]
- Park JH, Park EJ, Lee HS, Kim SJ, Hur SK, Imbalzano AN, Kwon J (2006) Mammalian SWI/SNF complexes facilitate DNA double-strand break repair by promoting gamma-H2AX induction. EMBO J 25: 3986–3997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegoraro G, Kubben N, Wickert U, Gohler H, Hoffmann K, Misteli T (2009) Ageing-related chromatin defects through loss of the NURD complex. Nat Cell Biol 11: 1261–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng G, Yim EK, Dai H, Jackson AP, Burgt IV, Pan MR, Hu R, Li K, Lin SY (2009) BRIT1/MCPH1 links chromatin remodelling to DNA damage response. Nat Cell Biol 11: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR (2000) Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem 275: 40974–40980 [DOI] [PubMed] [Google Scholar]
- Ramirez J, Hagman J (2009) The Mi-2/NuRD complex: a critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 4: 532–536 [DOI] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Nazarian AA, Li CC, Gore SL, Sridharan R, Imbalzano AN, Smale ST (2006) Selective and antagonistic functions of SWI/SNF and Mi-2beta nucleosome remodeling complexes during an inflammatory response. Genes Dev 20: 282–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG (2010) PARP inhibition: PARP1 and beyond. Nat Rev Cancer 10: 293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkaria JN, Tibbetts RS, Busby EC, Kennedy AP, Hill DE, Abraham RT (1998) Inhibition of phosphoinositide 3-kinase related kinases by the radiosensitizing agent wortmannin. Cancer Res 58: 4375–4382 [PubMed] [Google Scholar]
- Sinha M, Watanabe S, Johnson A, Moazed D, Peterson CL (2009) Recombinational repair within heterochromatin requires ATP-dependent chromatin remodeling. Cell 138: 1109–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokes MP, Rush J, Macneill J, Ren JM, Sprott K, Nardone J, Yang V, Beausoleil SA, Gygi SP, Livingstone M, Zhang H, Polakiewicz RD, Comb MJ (2007) Profiling of UV-induced ATM/ATR signaling pathways. Proc Natl Acad Sci USA 104: 19855–19860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123: 1213–1226 [DOI] [PubMed] [Google Scholar]
- Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG (2009) A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16: 923–929 [DOI] [PubMed] [Google Scholar]
- van Attikum H, Gasser SM (2009) Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19: 207–217 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137: 413–431 [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP (1998) A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol 8: 843–846 [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W (1998) NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861 [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D (1998) The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 95: 279–289 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.