A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression
Malat1 is a long non-coding RNA that localizes to nuclear speckles, but whose function remains unclear. Here, the Spector and Bessis laboratories show that Malat1 is important for the recruitment of splicing factors to transcription sites, the expression of synaptic genes and as a consequence synaptogenesis.
Keywords: non-coding RNA, nuclear domains, splicing factor, synaptogenesis, transcription
Abstract
A growing number of long nuclear-retained non-coding RNAs (ncRNAs) have recently been described. However, few functions have been elucidated for these ncRNAs. Here, we have characterized the function of one such ncRNA, identified as metastasis-associated lung adenocarcinoma transcript 1 (Malat1). Malat1 RNA is expressed in numerous tissues and is highly abundant in neurons. It is enriched in nuclear speckles only when RNA polymerase II-dependent transcription is active. Knock-down studies revealed that Malat1 modulates the recruitment of SR family pre-mRNA-splicing factors to the transcription site of a transgene array. DNA microarray analysis in Malat1-depleted neuroblastoma cells indicates that Malat1 controls the expression of genes involved not only in nuclear processes, but also in synapse function. In cultured hippocampal neurons, knock-down of Malat1 decreases synaptic density, whereas its over-expression results in a cell-autonomous increase in synaptic density. Our results suggest that Malat1 regulates synapse formation by modulating the expression of genes involved in synapse formation and/or maintenance.
Introduction
A large portion of the eukaryotic genome is transcribed as non-coding RNAs (ncRNAs) of various sizes ranging from ∼20 nucleotides to ∼100 kb (reviewed in Mercer et al, 2009; Wilusz et al, 2009). Despite the increasing number of long ncRNAs (lncRNAs), very few have thus far been assigned a specific function (for review, see Mercer et al, 2009). Whether some of these ncRNAs represent transcriptional noise or are involved in important cellular functions remains a matter of debate (Mattick and Makunin, 2006). However, several observations support the assertion that lncRNAs are likely to have functions in cells (Chamberlain and Brannan, 2001; Willingham et al, 2005; Feng et al, 2006; Mancini-Dinardo et al, 2006; Martianov et al, 2007; Rinn et al, 2007; Yang and Kuroda, 2007; Hirota et al, 2008; Mariner et al, 2008; Nagano et al, 2008; Wang et al, 2008; Yu et al, 2008; Chen and Carmichael, 2009; Clemson et al, 2009; Khalil et al, 2009; Mallik and Lakhotia, 2009; Sasaki et al, 2009; Sunwoo et al, 2009). For instance, large-scale studies have shown that many lncRNAs are conserved, dynamically regulated during differentiation and exhibit tissue-specific expression patterns (Ravasi et al, 2006; Dinger et al, 2008; Mercer et al, 2008; Guttman et al, 2009). In addition, several lncRNAs have been shown to be misregulated in various diseases including cancer and neurological disorders (for reviews, see Costa, 2005; Prasanth and Spector, 2007; Taft et al, 2009; Gupta et al, 2010). In many cases, the subcellular localization of ncRNAs has been determined which has provided significant insight into their functions. For example, Xist/XIST RNA, 15–17 kb in mouse and 19 kb in human, coats the inactive X chromosome from which it is transcribed. This represents part of the mechanism by which transcriptional silencing is achieved (for reviews, see Plath et al, 2002; Heard and Disteche, 2006; Payer and Lee, 2008). Recently, MEN epsilon/beta nuclear-retained ncRNAs (also known as nuclear-enriched autosomal transcript1—NEAT1) were shown to be enriched in nuclear paraspeckles, a novel subnuclear domain that preferentially localized on the periphery of nuclear speckles or interchromatin granule clusters (IGCs). These ncRNAs have a critical function in the establishment and maintenance of paraspeckles (Chen and Carmichael, 2009; Clemson et al, 2009; Sasaki et al, 2009; Sunwoo et al, 2009).
Metastasis-associated lung adenocarcinoma transcript 1 (Malat1) ncRNA was initially characterized as a long polyadenylated ncRNA that is over-expressed in various cancers (Ji et al, 2003; Lin et al, 2006). Recent analysis has shown that the primary transcription products of the Malat1 locus include a 6.7 kb nuclear-retained Malat1 ncRNA, and through processing by the tRNA biogenesis machinery, a cytoplasmic 61-nt tRNA-like ncRNA referred to as mascRNA (Wilusz et al, 2008). Phylogenetic analysis indicates that Malat1 is highly conserved among mammals, up to 90% identity between human and mouse in the last 5 kb of the RNA (data not shown). Such conservation among mammals is indicative of important, yet unknown function(s) of Malat1 ncRNA. The long Malat1 transcript has been localized to nuclear speckles in several cell lines (Hutchinson et al, 2007; Clemson et al, 2009). Nuclear speckles contain a large number of nuclear proteins that are involved in several aspects of mRNP processing, including pre-mRNA splicing and RNA transport (reviewed in Lamond and Spector, 2003). Nuclear speckles are not sites of transcription or pre-mRNA splicing, but represent storage/modification and/or assembly sites of various splicing factors from where pre-mRNA processing factors are recruited to active sites of transcription (reviewed in Lamond and Spector, 2003). A population of poly(A)+ RNA was previously reported to localize to nuclear speckles (Carter et al, 1991; Visa et al, 1993; Huang et al, 1994). Malat1 is the first example of an lncRNA that is specifically enriched in these nuclear domains.
The interesting sub-localization of the abundant Malat1 transcript, as well as its restricted evolutionary conservation among mammals, prompted us to investigate the potential function of Malat1 ncRNA in the mammalian cell nucleus. Here, we provide evidence that the nuclear speckle-enriched Malat1 ncRNA modulates synapse formation in neurons by regulating the expression of genes involved in synaptogenesis.
Results
Malat1 localizes to nuclear speckles in a transcription-dependent manner
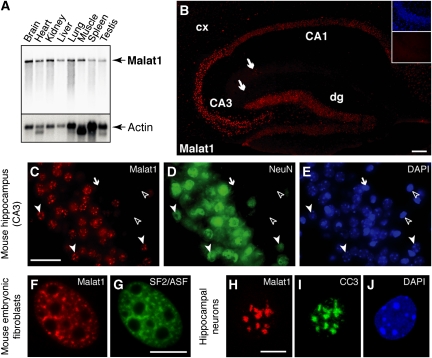
We characterized the expression of the long Malat1 ncRNA in mouse tissues by northern analysis. The ∼6.7 kb Malat1 transcript was detected in all tissue samples examined. We detected the highest levels of Malat1 ncRNA in heart, kidney and brain and a minimum level in spleen and skeletal muscle (Figure 1A; Supplementary Figure 1). As Malat1 shows elevated levels of expression in the brain, we further characterized its expression pattern by RNA fluorescence in situ hybridization (RNA-FISH) to adult mouse brain sections. We used a probe that specifically recognized the nuclear-retained 6.7 kb Malat1 RNA. We found elevated levels of Malat1 transcripts in pyramidal neurons of the hippocampus, Purkinje cells of the cerebellum, neurons of the substantia nigra and motoneurons (Figure 1B; Supplementary Figure 2). Non-neuronal cells in brain sections showed extremely low levels of Malat1 ncRNA (Figure 1C–E; arrow and open arrowheads). Interestingly, in all tissues examined, Malat1 ncRNA RNA-FISH signal displayed a punctate nuclear distribution (Figure 1C), suggesting that Malat1 ncRNA is enriched in a nuclear sub-compartment(s) that appears similar to nuclear speckles (Hutchinson et al, 2007; Clemson et al, 2009). In wild-type mouse embryonic fibroblasts (wt-MEFs), RNA-FISH revealed that Malat1 ncRNA co-localizes with the pre-mRNA-splicing factor SF2/ASF in nuclear speckles (Spector, 2006) (Figure 1F and G). In addition, Malat1 ncRNA could also be detected diffusely in the nucleoplasm (Figure 1F). Similarly, in neurons, Malat1 co-localizes with the CC3 antigen (Figure 1H–J), which is also known to localize to nuclear speckles (Chabot et al, 1995). In contrast to that observed in MEFs, the diffuse nucleoplasmic signal was significantly less in neurons (Figure 1H). The localization of Malat1 ncRNA to nuclear speckles suggests that it may execute its function in relation to gene expression (reviewed in Lamond and Spector, 2003).
Figure 1.
Malat1 is a neuron-enriched nuclear-retained ncRNA that is localized to nuclear speckles. (A) Northern blot analysis of various mouse tissues indicating that Malat1 is detected as a single ∼6.7 kb band with elevated levels in heart, kidney and brain. (B) Malat1 expression is restricted to neurons in the adult mouse hippocampus. RNA-FISH shows the strongest expression in pyramidal neurons of CA1 and CA3 and in granular neurons of the dentate gyrus (dg). Weaker expression can be detected in neurons of the cortex (cx) and in hippocampal interneurons (arrows). Insets show FISH signal with a sense probe (lower) and DAPI staining (upper) in CA3 region. Scale bar, 100 μm. (C–E) Triple labelling of CA3 region showing Malat1 ncRNA by in situ hybridization (C), neuron-specific NeuN immunoreactivity (D) and DAPI labelling of cell nuclei (E). Closed arrowheads indicate cells double positive for Malat1 and NeuN staining. Open arrowheads indicate non-neuronal cells. In a few cases (arrow), weak Malat1 ncRNA signal could be detected in NeuN negative cells. Scale bar, 25 μm. (F–J) Malat1 ncRNA is enriched in nuclear speckles. In wild-type mouse embryonic fibroblasts (wt-MEFs) and in mouse cultured hippocampal neurons, Malat1 ncRNA signal (F, H) co-localizes with SF2/ASF (G) or CC3 immunoreactivity (I), respectively. (J) DAPI staining. Scale bar, 10 μm.
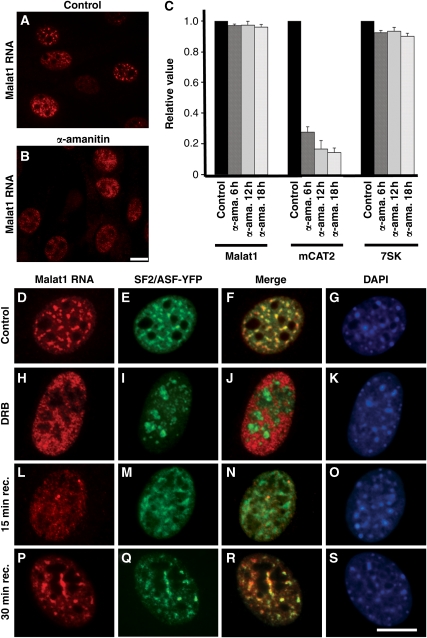
Next, we examined the behaviour of Malat1 ncRNA upon inhibition of RNA polymerase II transcription using α-amanitin (Figure 2B and C) or 5,6-dichloro-1-β-D-ribobenzimidazole (DRB; Figure 2H–K). Both drugs efficiently inhibit RNA pol II-mediated transcription. However, α-amanitin-mediated transcription inhibition is irreversible, whereas transcription in the DRB-treated cells can be reactivated upon removal of the drug from the medium. Both treatments resulted in the re-distribution of Malat1 ncRNA from nuclear speckles to a homogenous nuclear localization. However, real-time RT–PCR showed little to no turnover of Malat1 ncRNA even after prolonged treatment of cells with α-amanitin (Figure 2C) or DRB (data not shown). As previously reported (Bubulya et al, 2004), SF2/ASF localized around the nucleoli in RNA pol II transcription-inhibited cells and relocalized back to the nuclear speckles upon transcription reactivation (Supplementary Figure 3). DRB recovery kinetics revealed that within 15 min of drug removal from the culture medium, SF2/ASF could be detected within nuclear speckles, whereas Malat1 ncRNA continued to show a diffuse nuclear distribution (Figure 2L–O). However, within 30 min of recovery, Malat1 ncRNA was once again enriched in nuclear speckles in which it co-localized with SF2/ASF (Figure 2P–S). Together, these data show that Malat1 ncRNA is a very stable nuclear transcript that associates with nuclear speckles in a transcription-dependent manner.
Figure 2.
Malat1 ncRNA is a stable nuclear RNA that localizes to speckles in a transcription-dependent manner. (A) RNA-FISH to untreated wt-MEFs shows the nuclear speckle localization of Malat1 ncRNA. (B) α-amanitin treatment (50 μg/ml for 6 h) of wt-MEFs results in the re-distribution of Malat1 ncRNA from nuclear speckles to a homogenous nuclear distribution. (C) Malat1 ncRNA levels in untreated and α-amanitin-treated (6, 12 and 18 h) wt-MEFs were assessed by Q–PCR. Malat1 ncRNA showed little to no turnover, similar to the RNA pol III-transcribed ncRNA 7SK. As expected, the protein coding mCAT2 mRNA showed high turnover after α-amanitin treatment. Malat1, mCAT2 and 7SK RNA levels were normalized to β-actin mRNA and were presented relative to RNA levels in untreated (control) cells. The data represents mean and s.d. values of three independent experiments per data point. (D–G) RNA-FISH to wt-MEFs (untreated cells) shows complete co-localization of Malat1 ncRNA with a nuclear speckle marker SF2/ASF. (H–K) Malat1 ncRNA shows homogenous nuclear distribution upon inhibition of RNA pol II transcription by DRB (32 μg/ml for 3 h) and it no longer co-localizes with SF2/ASF. (L–O) Following the removal of DRB from the medium (15 min), Malat1 ncRNA continues to show a homogenous distribution, whereas SF2/ASF relocalizes to nuclear speckles. (P–S) Malat1 ncRNA relocalizes to nuclear speckles within 30 min post-washout of DRB from the medium. RNA-FISH is shown in red (D, H, L, P), YFP-SF2/ASF in green (E, I, M, Q) and DNA is counterstained with DAPI in blue (G, K, O, S). Scale bar, 5 μm.
Malat1 modulates the recruitment of SR proteins to a transcriptionally active locus
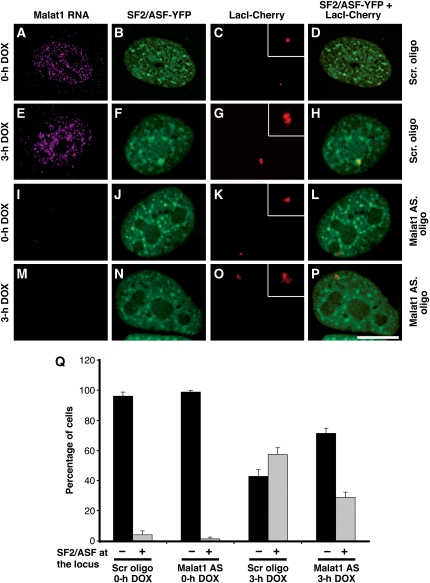
As Malat1 ncRNA is enriched in nuclear speckles, we were interested in addressing whether it has a function in regulating the recruitment of pre-mRNA-splicing factors to transcription sites. To assess such a function to a specific transcription site, we used a previously characterized U2OS cell line in which the transcription site of an inducible transgene array, as well as its mRNA and protein products, can be visualized and quantified (Janicki et al, 2004). As expected from previous experiments using this U2OS cell line (Janicki et al, 2004), the transgene locus, visualized by LacI-mCherry fluorescence, was actively decondensed upon transcriptional induction with doxycycline (compare Figure 3C and G). Upon transcriptional induction of the reporter locus in control cells that were transfected with scrambled oligonucleotides, SF2/ASF was recruited to the actively transcribing gene locus as shown by the increased number of cells displaying SF2/ASF co-localization at the transcription site (Figure 3A–H and Q). Interestingly, only ∼40–45% of the Malat1-depleted cells continued to show a speckled distribution of SF2/ASF. In the rest of the cells, SF2/ASF displayed increased homogenous nuclear distribution (data not shown). Strikingly, in a significant population of Malat1-depleted cells that continue to show speckle localization of SF2/ASF, SF2/ASF recruitment to the induced transcription site was severely compromised (Figure 3I–Q). Importantly, this was not due to a general disruption of the recruitment of the gene expression machinery as the recruitment to the activated transcription site of the rtTa transcriptional activator, CDK9, a component of the pTEF-B kinase complex that phosphorylates the C-terminal domain of RNA pol II, and RNA pol II large subunit was unaffected by knock-down of Malat1 ncRNA (Supplementary Figure 4). To determine whether Malat1 was involved in the recruitment of other splicing factors to the induced transgene, we analysed the recruitment of SC35, another member of the SR family of splicing factors. Similar to that observed for SF2/ASF, the recruitment of SC35 was also affected by the knock-down of Malat1 (Supplementary Figure 5).
Figure 3.
Malat1 ncRNA facilitates the recruitment of SF2/ASF to an active transcription site. (A–D) RNA-FISH to U2OS 2-6-3 cells (Janicki et al, 2004) stably expressing LacI-mCherry and rtTa transactivator (Tet-ON) shows a punctate nuclear localization of Malat1 ncRNA. Note the more homogenous nuclear staining pattern of Malat1 ncRNA in U2OS cells (A, E). In several of cancer cell lines examined, a population of cells (20–25%) showed a nuclear speckle pattern of Malat1 ncRNA unlike primary diploid cell lines and tissues where >70% of the cells exhibited a speckled distribution of Malat1 ncRNA. Cells treated with a scrambled oligonucleotide (A–D) or Malat1-specific oligonucleotide (I–L) in the absence of doxycycline (0 h DOX) do not show SF2/ASF (B, J) at the transcriptionally inactive reporter gene locus (C, K, D, L). (E–H) Upon addition of doxycycline (3 h DOX), the transcriptionally active locus (G) showed enrichment of SF2/ASF (F, H). (I–P) Cells treated with Malat1-specific antisense oligonucleotides showed complete depletion of Malat1 ncRNA (I, M). In the absence of Malat1 ncRNA, upon addition of doxycycline (3 h DOX), a significantly reduced level of SF2/ASF (N, P) was associated with the transcriptionally active locus (O). The inset in figures (C, G, K, O) represents the magnified reporter locus. Scale bar, 5 μm. (Q) The histogram shows the percentage of cells that exhibit recruitment of SF2/ASF to the transcriptionally inactive or active reporter locus in the presence or absence of Malat1 ncRNA. The data represents mean and s.d. values of three independent experiments per data point (n=25 cells/experiment).
Furthermore, Malat1 knock-down did not alter the transcriptional activity of the locus as observed by RT–PCR using primers against the reporter RNA (Supplementary Figure 6). These results show that Malat1 ncRNA modulates the recruitment of SR-type pre-mRNA-splicing factors to/at active transcription sites. Importantly, knock-down of the 6.7 kb Malat1 transcript does not affect the level of mascRNA (Wilusz et al, 2008), indicating that the effect we observed is specifically due to the depletion of the long nuclear-retained Malat1 ncRNA.
Malat1 ncRNA controls the mRNA levels of genes involved in the induction/function of synapses
As Malat1 ncRNA is highly expressed in the brain, we investigated a possible function of Malat1 in neuronal function. We first characterized the cellular processes and components that were most impacted by Malat1 deficiency using DNA microarrays. Malat1 ASO or scrambled oligodeoxynucleotides were transfected into Neuro2A neuroblastoma cells and mRNAs were subsequently hybridized to 44K agilent DNA arrays (G_4122F). We then identified the Gene Ontology (GO) groups that were significantly enriched in the population of genes whose expression were impacted upon Malat1 depletion (Tables I and II; Supplementary Tables I, II and III). The GO groups related to the organization and the function of the nucleus were the most significantly over-represented in Malat1-depleted cells. Interestingly, we also found that the GO groups related to synapse (cellular component; GO:0045202; 1.9-fold enrichment; P=1.40 × 10−2) and dendrite development (biological process; GO: 0016358; 4.3-fold; P=6 × 10−3; see Table I) were also affected upon Malat1 depletion. No other neuron-specific GO group was significantly enriched (P<0.05). Our DNA microarray analysis shows that in neuronal cells, Malat1 preferentially regulates a subset of genes with a significant involvement in dendritic and synapse development.
Table 1. Gene Ontology (GO) analysis of genes down-regulated in Malat1 knock-down Neuro2A cells.
| GO ID | Category | Observed | Expected | Ratio | P (hypergeometric) | |
|---|---|---|---|---|---|---|
| Biological processes | ||||||
| Developmental process | ||||||
| Dendrite development | 0016358 | 54 | 5 | 1.17 | 4.29 | 0.006 |
| Cellular process | ||||||
| Regulation of cell differentiation | 0045595 | 438 | 19 | 9.46 | 2.01 | 0.0032 |
| Cell division | 0051301 | 262 | 15 | 5.66 | 2.65 | 0.0006 |
| Cell proliferation | 0008283 | 819 | 31 | 17.68 | 1.75 | 0.0017 |
| Mitotic cell cycle | 0000278 | 339 | 17 | 7.32 | 2.32 | 0.0011 |
| Mitosis | 0007067 | 220 | 12 | 4.75 | 2.53 | 0.003 |
| Chromosome organization | 0051276 | 445 | 36 | 9.61 | 3.75 | 7.74e−12 |
| Chromatine organization | 0006325 | 347 | 31 | 7.49 | 4.14 | 1.98e−11 |
| Nucleosome assembly | 0006334 | 106 | 16 | 2.29 | 6.99 | 9.63e−10 |
| Localization | ||||||
| mRNA transport | 0051028 | 58 | 5 | 1.25 | 3.99 | 0.037 |
| Metabolic process | ||||||
| Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process | 0006139 | 77 | 6 | 1.66 | 3.61 | 0.0063 |
| Transcription | 0006350 | 2305 | 82 | 49.76 | 1.65 | 1.57e−6 |
| Transcription, DNA dependent | 0006351 | 2107 | 74 | 45.49 | 1.63 | 9.79e−05 |
| Regulation of transcription | 0045449 | 2230 | 78 | 48.14 | 1.62 | 5.96e−06 |
| Transcription from RNA polymerase II promoter | 0006366 | 655 | 25 | 14.14 | 1.77 | 0.0042 |
| DNA metabolic process | 0006259 | 493 | 27 | 10.64 | 2.54 | 9.05e−06 |
| DNA replication | 0006260 | 176 | 18 | 3.8 | 4.74 | 4.84e−08 |
| RNA metabolic process | 0016070 | 2953 | 101 | 63.75 | 1.58 | 3.52e−07 |
| mRNA processing | 0006397 | 259 | 14 | 5.59 | 2.5 | 0.0015 |
| Cellular response to stimulus | 0051716 | 914 | 32 | 19.73 | 1.62 | 0.0047 |
| Response to glucocorticoid stimulus | 0051384 | 88 | 7 | 1.9 | 3.68 | 0.0029 |
| Cellular component | ||||||
| Synapse | 0045202 | 373 | 15 | 7.94 | 1.89 | 0.014 |
| Organelle | 0044427 | 373 | 34 | 7.94 | 4.28 | 7.89e−13 |
| Chromosome | 0005694 | 441 | 35 | 9.38 | 3.73 | 1.89e−11 |
| Chromatin | 0000785 | 220 | 25 | 4.68 | 5.34 | 8.37e−12 |
| Nucleus | 0005634 | 4331 | 177 | 92.14 | 1.92 | 1.24e−23 |
| Nuclear body | 0016604 | 145 | 12 | 3.08 | 3.89 | 6.32e−05 |
| Nucleolus | 0005730 | 361 | 15 | 7.68 | 1.95 | 0.0107 |
| Microtubule cytoskeleton | 0015630 | 476 | 20 | 10.13 | 1.97 | 0.0031 |
| The Gene Ontology groups are indicated for the biological processes and the cellular components. Category, number of reference genes in the category; Observed, number of genes in the category that are down-regulated in Neuro2A cells upon Malat1 knock-down; Expected, expected number in the category; Ratio, ratio of enrichment (R), P-value from hypergeometric test. | ||||||
Table 2. Gene Ontology (GO) analysis of genes up-regulated in Malat1 knock-down Neuro2A cells.
| GO ID | Category | Observed | Expected | Ratio | P (hypergeometric) | |
|---|---|---|---|---|---|---|
| Biological processes | ||||||
| Cellular process | ||||||
| Mitosis | 0007067 | 220 | 17 | 6.29 | 2.70 | 0.0002 |
| Nuclear transport | 0051169 | 248 | 22 | 7.09 | 3.10 | 2.66e−06 |
| Cytoskeleton organization | 0007010 | 427 | 23 | 12.21 | 1.88 | 0.0028 |
| Microtubule-based movement | 0007018 | 104 | 10 | 2.97 | 3.36 | 0.0008 |
| Metabolic process | ||||||
| Lipid metabolic process | 0006629 | 782 | 35 | 22.36 | 3.68 | 0.0058 |
| Glucose metabolic process | 0006006 | 139 | 10 | 3.97 | 2.52 | 0.0067 |
| Ubiquitin-dependent protein catabolic process | 0006511 | 165 | 15 | 4.72 | 3.18 | 7.87e−05 |
| Protein folding | 0006457 | 132 | 12 | 3.77 | 3.18 | 0.0004 |
| Translation | 0006412 | 436 | 24 | 12.47 | 1.92 | 0.0017 |
| Cellular component | ||||||
| Organelle | ||||||
| Endoplasmic reticulum membrane | 0005789 | 491 | 24 | 13.60 | 1.76 | 0.0053 |
| Perinuclear region of cytoplasm | 0048471 | 241 | 15 | 6.68 | 2.25 | 0.003 |
| Mitochondrion | 0005739 | 1018 | 40 | 28.20 | 1.42 | 0.016 |
| Endosome | 0005768 | 270 | 15 | 7.48 | 2.01 | 0.0085 |
| Golgi apparatus | 0005794 | 759 | 33 | 21.03 | 1.57 | 0.0071 |
| Ribosome | 0005840 | 236 | 14 | 6.54 | 2.14 | 0.0062 |
| Vesicle | 0031982 | 593 | 28 | 16.43 | 1.7 | 0.0043 |
| Vacuole | 0005773 | 217 | 16 | 6.01 | 2.66 | 0.0004 |
| Nucleus | 0005634 | 4331 | 148 | 119 | 1.23 | 0.0015 |
| Nucleolus | 0005730 | 361 | 19 | 10 | 2.14 | 0.0058 |
| Cytoskeleton | ||||||
| Spindle | 0005819 | 99 | 10 | 2.74 | 6.02 | 0.0004 |
| Microtubule | 0005874 | 234 | 17 | 6.48 | 2.62 | 0.0003 |
| Centrosome | 0005813 | 176 | 11 | 4.88 | 2.26 | 0.01 |
| The Gene Ontology groups are indicated for the biological processes and the cellular components. Category, number of reference genes in the category; Observed, number of genes in the category that are up-regulated in Neuro2A upon Malat1 knock-down; Expected, expected number in the category; Ratio, ratio of enrichment (R), P-value from hypergeometric test. | ||||||
Malat1 RNA levels in cultured neurons influence synapse formation
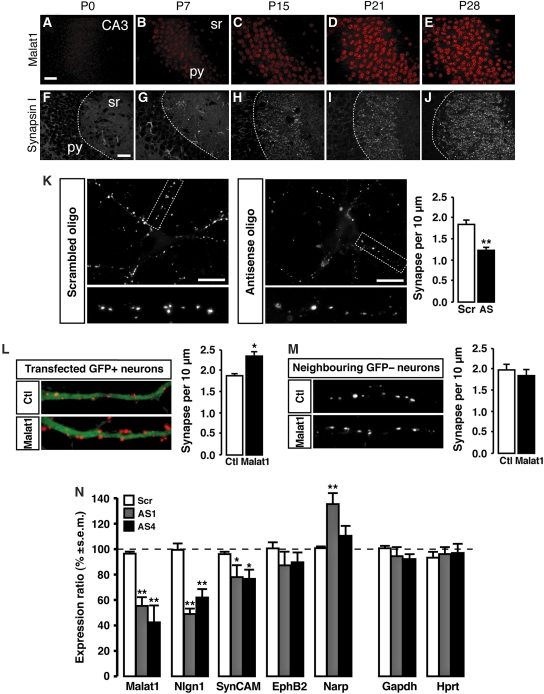
On the basis of the DNA microarray analysis, we further investigated whether Malat1 might be involved in the generation of synapses. We first examined the developmental expression of Malat1 ncRNA. In the hippocampus as well as in Purkinje cells (data not shown), Malat1 is first detected between post-natal day 0 (P0) and P7 and its level increases until P28 (Figure 4A–E). In rodent brain, the first post-natal weeks are known to be periods of intense synaptogenesis on dendrites (Figure 4F–J) (Steward and Falk, 1986, 1991; De Felipe et al, 1997).
Figure 4.
Malat1 regulates genes involved in synaptogenesis in cultured hippocampal neurons. (A–J) Developmental time course of Malat1 RNA-FISH signal (A–E) and synapsin I immunoreactivity (IR) (F–J) in pyramidal neurons (py) of the mouse hippocampus (CA3) between post-natal day 0 (P0) and P28. First punctate signals of synapsin I-IR and first Malat1-FISH signals were detected in the stratum radiatum (sr) at P7 and increases until P28. Scale bar, 25 μm. (K) Synapsin immunoreactivity in cultured hippocampal neurons transfected with Malat1 antisense oligodeoxynucleotides (AS) or control scrambled oligodeoxynucleotide (Scr). Scale bar, 10 μm. Lower panels display higher magnifications of the outlined regions in the upper panels. Histogram shows the quantification of synaptic linear density in three independent experiments. Mean±s.e.m.; **P<0.0001, ANOVA. (L) Synapsin labelling (red) in neurons (green) transfected with a control vector (Ctl) or with Malat1 over-expressing vector (Malat1). Histogram shows the quantification of synaptic linear density in three independent experiments. Mean±s.e.m.; *P<0.001, ANOVA. (M) Synapsin labelling in neurons neighbouring control vector or Malat1 cDNA-transfected neurons. Histogram shows the quantification of synaptic linear density in three independent experiments. Mean±s.e.m.; P=0.49, ANOVA. (N) Quantification by quantitative RT–PCR of mRNA encoding several genes upon transfection of three independent cultures of neurons with Malat1 antisense oligonucleotides (AS1, grey bars; AS4, black bars) or with control scrambled oligodeoxynucleotide (Scr, white bars). Mean±s.e.m. relative to Actin mRNA level **P<0.03; *P<0.05 Mann–Whitney U-test.
We then evaluated the relationship between Malat1 and synapses. Neuro2a cells express genes related to synaptic function, but do not generate synapses per se (McGee, 1980; Spoerri et al, 1980). We, therefore, used well-characterized primary hippocampal neuron cultures for examining the function of Malat1 in synapse formation or development. We examined the effect of Malat1 knock-down or over-expression on synapse density in dendrites of cultured hippocampal neurons (Supplementary Figure 7). We used accumulation of synapsin I immunoreactivity as a marker of synapses because it is expressed from the earliest stages of synapse formation (Ahmari et al, 2000) and, therefore, identifies nascent and mature synapses (Graf et al, 2004). In addition, Malat1 knock-down does not regulate the level of synapsin I mRNA (ratio=−0.026). As shown in Figure 4K, a significant reduction in synaptic density was observed in cultured neurons that were transfected with two independent Malat1 antisense oligonucleotides, as compared with control transfections (−33.5±3.8%; P<0.0001; n=3). Conversely, over-expression of Malat1 resulted in an increased presynaptic bouton density on dendrites (+24.7±5.6% as compared with EGFP vector over-expression; P=0.0008; n=3; Figure 4L). Interestingly, in neurons neighbouring Malat1 over-expressing cells, synaptic density was unchanged (93.2±7.2% as compared with EGFP vector over-expression; P=0.49; n=3; Figure 4M). These data show that the control of synaptogenesis by Malat1 ncRNA is cell autonomous and post-synaptic.
As Malat1 ncRNA controls synapse density in cultured hippocampal neurons, the effect of its knock-down was further investigated on a subset of genes controlling the formation and/or function of synapses (Figure 4N). Neuroligin1 (NLGN1) and synaptic cell adhesion molecule 1 (SynCAM1) post-synaptic proteins are able to drive the recruitment of the presynaptic release machinery through interactions with presynaptic cell adhesion molecules (Chih et al, 2005; Levinson and El-Husseini, 2005; Sara et al, 2005). We measured the effect of decreasing Malat1 ncRNA level on Nlgn1 (Chih et al, 2005) and SynCAM1 (Biederer et al, 2002) mRNAs. Cultured primary mouse hippocampal neurons transfected with two independent Malat1 antisense oligonucleotides showed a significant decrease in the levels of Nlgn1 (−44.3±1.2% of control; P<0.03; n=4) and SynCAM1 (−22.7±1.2%; P<0.05; n=3) transcripts measured by quantitative RT–PCR as compared with cells that were transfected with control oligonucleotides (Figure 4N). In the DNA microarray experiment, a comparable decreased expression of SynCAM1 was observed in the Malat1-depleted cells (−29.8±1.3%). Nlgn1 was not expressed in the Neuro2A cell line. To further evaluate the specificity of Malat1 in cultured hippocampal neurons, we analysed the effect of Malat1 ncRNA knock-down on the mRNA level of the Eph receptor B2 (EphB2) and Neuronal pentraxin-2 (Narp), which control the recruitment of glutamate receptors to synaptic sites (O'Brien et al, 1999; Henderson et al, 2001). We found that knock-down of Malat1 ncRNA did not decrease the transcript levels of EphB2 (88.3±1.4%; P=0.51; n=3). Narp mRNA level was either unchanged or increased depending on the Malat1 antisense oligonucleotide that was used (Figure 4N). Similarly, Malat1 knock-down did not alter the level of transcripts unrelated to synapse function such as Hypoxanthine guanine phosphoribosyl transferase (Hprt) (96.5±1.5%; P=0.44; n=3) or Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) (93.2±0.8% P=0.2; n=4) (Figure 4N). Our results showed that in primary cultured neurons, Malat1 ncRNA regulates transcript levels of Nlgn1 and to a lesser extend SynCAM1. Together, these results suggest that in neurons, Malat1 ncRNA regulates the expression efficiency of a subset of genes controlling synapse formation, which consequently results in the regulation of synaptogenesis.
Discussion
In this study, we have shown that Malat1, a long nuclear-retained ncRNA, modulates the recruitment of SR proteins to an active transcription site of a reporter gene locus. We further showed that in neurons, where it is highly expressed, Malat1 controls the expression of a subset of genes significantly involved in nuclear and synapse function and that it regulates synaptogenesis. Malat1 is the first nuclear-retained lncRNA that has been shown to regulate synapse formation by modulating gene expression.
Within the cell nucleus, Malat1 ncRNA co-localizes with SR-splicing factors in nuclear speckles or IGCs, which are important sub-nuclear domains known to be enriched in factors that regulate transcription and pre-mRNA processing (reviewed in Lamond and Spector, 2003; Hall et al, 2006). We hypothesize that nuclear speckle-enriched Malat1 regulates gene expression by modulating the differential association or activity of SR-splicing factors. SR-splicing factors constitute a family of pre-mRNA-splicing factors that have essential functions in constitutive as well as alternative splicing of pre-mRNAs (reviewed in Long and Caceres, 2009). In general, SR proteins contain one or two RNA-recognition motifs and a serine-arginine dipeptide-rich RS domain (reviewed in Long and Caceres, 2009). In this study, we have shown that Malat1 depletion resulted in decreased recruitment of SR-splicing factors (SF2/ASF, SC35) to a stably integrated transcriptionally active gene locus. We have also recently shown that Malat1-depleted HeLa cells show increased cellular levels of dephosphorylated SF2/ASF (Tripathi and Prasanth, unpublished data). It is known that the phosphorylation of the RS domain influences the recruitment of SR proteins to transcription sites and their binding to pre-mRNA as well as for their function in the recruitment of other splicing factors to form functional spliceosomes (Cao et al, 1997; Misteli et al, 1998; Xiao and Manley, 1998; Stamm, 2008). Our results imply that inefficient recruitment of SR proteins to a transcription site, observed in Malat1-depleted cells, could be due to defects in SR-protein phosphorylation. We have also observed aberrant alternative splicing of a subset of pre-mRNAs including Camk2b kinase, a gene essential for synaptic density and maturation (Shen et al, 1998; Wu and Cline, 1998; Rongo and Kaplan, 1999; Fukunaga and Miyamoto, 2000), in Malat1-depleted HeLa cells (Tripathi, Ellis, Blencowe and Prasanth, unpublished data). These results further confirm the regulatory function of Malat1 in the control of post-transcriptional gene expression. Although Malat1 regulates a basic gene expression event, whole-genome transcription DNA microarray analysis (this study) as well as alternative splicing microarray (Tripathi, Ellis, Blencowe and Prasanth, unpublished data) analyses show that its inhibition affects the level of only a subset of gene transcripts. One hypothesis to explain the selective action of Malat1 is that the association of SR proteins with pre-mRNAs is itself selective. The binding of SR proteins to a particular pre-mRNA is directly dependent on its splice sites and exonic-splicing enhancer sequences (Cartegni et al, 2003). Moreover, different genes can be regulated by different SR proteins (reviewed in Moroy and Heyd, 2007). Therefore, in the absence of Malat1 the binding kinetics of SR proteins would be different in regard to different pre-mRNAs in a tissue-specific manner, thus affecting some transcripts such as Nlgn1 and SynCAM1 in neurons, but not all transcripts. Future analysis will determine which steps of mRNA biogenesis are altered in Malat1-depleted neuronal cells and if the effect on neuronal genes is direct. In addition, as Malat1 ncRNA is also expressed in many other tissues, it is likely to have broader regulatory functions by modulating the expression of specific genes in a tissue-specific manner. Several examples are known of genes that are ubiquitously expressed, but whose altered expression specifically affects neuron-specific functions eventually leading to neurological diseases. The survival of motoneuron protein is expressed in all cells and it is involved in pre-mRNA splicing, but mutations reducing its expression lead to the selective dysfunction of motoneurons and ultimately to spinal muscular atrophies (Burghes and Beattie, 2009). The fragile X mental retardation protein (FMRP) is an RNA-binding protein expressed in many tissues and involved in mRNA trafficking. The absence of FMRP leads to synaptic dysfunction and severe cognitive deficiency (Bassell and Warren, 2008). The superoxide dismutase 1 (SOD1) gene is an ubiquitously expressed metalloprotein, but its mutation specifically induces the death of motoneurons and amyotrophic lateral sclerosis (Pasinelli and Brown, 2006). Therefore, understanding how Malat1 controls the expression of neuron-specific genes will give us insight into the more general question of how the alteration of the expression of a ubiquitously expressed ncRNA functions in a cell-type-specific manner.
A great diversity of long lncRNAs is expressed and regulated in the CNS, where they are thought to have fundamental functions (Mercer et al, 2008). We have identified the involvement of one such lncRNA, Malat1, in regulating genes involved in synapse formation. In the CNS, Malat1 RNA is expressed concomitantly with synaptogenesis. It is known that in neuronal culture, synapse assembly can be induced by Neuroligin1 and SynCAM1 (Chih et al, 2005). We have now shown that in cultured neurons, Malat1 controls the level of Neuroligin1 and SynCAM1 mRNAs and that Malat1 level modulates synapse density in cultured neurons. Our results argue in favour of the growing consensus positioning ncRNAs as essential actors in specific gene regulation during the development and function of the CNS (Mercer et al, 2008).
The number of ncRNAs in eukaryotic genomes has been shown to increase as a function of developmental complexity (Mehler and Mattick, 2006). The biology of the lncRNAs also appears to be far more complex than anticipated (reviewed in Prasanth and Spector, 2007; Wilusz et al, 2009). Given the relative low number of protein coding genes in the mouse and human genomes, it has been suggested that ncRNAs may be part of the mechanism to increase the complexity and fine-tuning of gene expression and allow higher-order functions to develop during evolution (reviewed in Mattick, 2001, 2003; Prasanth and Spector, 2007). In neuronal cells in particular, the restricted expression of ncRNAs especially during development has previously been suggested to have important functions including human brain evolution (Mehler and Mattick, 2006; Pollard et al, 2006). In the brain, the expanding complexity of synaptic networks is one of the hallmarks of evolution. Therefore, one may anticipate that many additional ncRNAs regulating synaptogenesis and other regulatory functions in the nervous system will be identified and characterized (Lein et al, 2007; Mercer et al, 2008).
Materials and methods
Cell culture and drug treatments
The wt-MEFs and U2OS cells were grown in DMEM containing high glucose (Invitrogen, Carlsbad, CA), supplemented with penicillin streptomycin and foetal bovine serum. The U2OS 2-6-3 cell line used in this study has been extensively characterized and described (Janicki et al, 2004). We have now stably expressed the LacI-mCherry and rtTa transactivator (Tet-ON) cDNAs by placing them into a dual promoter expression vector containing the pVitro-2 plasmid (Invivogen). This plasmid was stably integrated into U2OS 2-6-3 cells. U2OS cells were electroporated with 2 μg of plasmid DNA (EYFP-C1-SF2/ASF, EYFP-C1-SC35 or rtTa-EYFP) plus 40 μg of salmon sperm DNA (Amresco, Solon, Ohio) and were seeded onto acid washed coverslips and processed for immunofluorescence localization or RNA-FISH, post-Malat1 antisense oligonucleotide treatment. The SF2/ASF fluorescent protein fusion construct was first generated as an SF2/ASF-GFP fusion protein and characterized previously (Misteli et al, 1997). RNA polymerase II inhibition studies were carried out by incubating cells with either α-amanitin (50 μg/ml; Sigma, St Louis) or DRB (32 μg/ml; Sigma, St Louis) for the indicated times. For DRB recovery kinetic studies, cells were removed from DRB containing medium, washed twice with fresh medium and incubated at 37°C for the indicated times. Neuro2A cell line was grown in DMEM supplemented with 10% foetal calf serum (Biowest, Paris). Neurons were isolated from newborn (post-natal day 0) SWISS mouse hippocampi as described (Goslin et al, 1998) and cultured for 8 days in vitro (DIV) in astrocyte-conditioned medium.
Transfection
Antisense knock-down of Malat1 ncRNA in human U2OS cells was carried out using phosphorothioate-modified DNA oligonucleotides (ISIS Pharmaceuticals): ATGCAAAAACATTAAGTTGG as described elsewhere (Prasanth et al, 2005). In Neuro2A cells, Malat1 depletion was achieved overnight using Lipofectamine 2000 (Invitrogen, Cergy Pontoise) according to manufacturer's instructions. Neurons were transfected at 8 DIV using Lipofectamine 2000 according to manufacturer's instructions. Neurons were incubated in serum-free medium with liposome–DNA complexes for 1 h, rinsed and incubated in astrocyte-conditioned medium for an additional 13 h. For inhibition experiments in neurons and in Neuro2A cells, mixed LNA/DNA oligonucleotides were generated (Proligo, Paris and Eurogentec, Angers) against mouse Malat1 ncRNA (antisense oligonucleotides—AS): AS1 5′-+T+A+AACTGCTCTGGTCA+G+C+C; AS2 5′-+T+T+AATCTACAAGGCCG+A+C+C; AS4 5′-+T+T+TAAGTTCTCTGGTA+T+G+A and scrambled as control: SCR 5′-+A+A+CGTGACACGTTCGG+A+G+A. +N indicates LNA bases. The DNA-based antisense oligonucleotides against Malat1 RNA mediates the RNaseH catalysed degradation of Malat1 RNA as shown for other nuclear-retained RNAs (Lanz et al, 1999). For DNA microarray experiments, Neuro2A cells were transfected with SCR or AS4. For quantitative real-time RT–PCR analysis, neurons were transfected with SCR, AS1 or AS4. For synapse analysis, neurons were transfected with SCR or with AS1+AS2. For the over-expression experiments, the full-length mouse Malat1 cDNA (Genbank accession number AY722410) was cloned into the pCAGG vector (Barthelemy et al, 2004) (kind gift from Dr CE Henderson, CNB, Columbia) and expressed in hippocampal neurons for 14 h.
Northern blot, in situ hybridization, immunochemistry
Northern blot: 2 μg of poly A+ RNA was used in each lane. The blot (Stratagene) was then hybridized with a full-length Malat1 cDNA probe. For histological sections, mice were perfused with 4% formaldehyde in phosphate buffer pH 7.2. Brains were dissected out, post-fixed for 2 h at 4°C in the same fixative, then cryoprotected ON at 4°C in 20% sucrose in phosphate buffer pH 7.2. A total of 40 μm coronal and sagittal sections were made using a cryotome. Neurons were fixed in 4% formaldehyde in phosphate buffer pH 7.2. In situ hybridization (ISH) was adapted fromSchaeren-Wiemers and Gerfin-Moser (1993) and Henrique et al (1995). Sense and antisense digoxigenin-labelled RNA probes (Roche Diagnostics, Meylan) were synthesized from a pGEMT (Promega) vector containing the 720 first nucleotides of Malat1 cDNA. For RNA-FISH, RNA probes were revealed using rhodamine-conjugated anti-digoxigenin antibody (Roche Diagnostics). For RNA-FISH to MEFs, the probes used for ISH were derived from a nick-translated full-length cDNA probe. Immunolocalization studies were performed as described elsewhere (Danglot et al, 2004). Antibodies were mouse monoclonal anti-NeuN antibody (MAB377, Chemicon), mouse monoclonal CC3 antibody (kind gift from Dr O Bensaude, ENS, Paris), mouse monoclonal SF2/ASF antibody (mAb103; Invitrogen), rabbit polyclonal SON antibody (Sharma et al, 2010), mouse monoclonal RNA pol II antibody (H14), rabbit CDK9 antibody (Santa Cruz Biotechnology Inc) and mouse monoclonal anti-synapsinI antibody (Synaptic Systems, Goettingen). RNA-FISH and immunolocalization studies in wt-MEFs and U2OS cells were carried out as described elsewhere (Prasanth et al, 2005).
DNA microarrays
Total RNA was extracted from three independent cultures using the RNAqueous-Micro kit (Ambion). Labelling of the RNA (2–5 μg) with Cy3 or Cy5 dye was carried out by incorporation of amino-allyl dUTP. Hybridization to 44K agilent DNA microarray (G_4122F) was performed according to the protocol developed by the functional genomics platform at the ecole normale supérieure.
Data analysis
Microarray slides were analysed using the GenePix software 6.1 (Axon). Spots flagged by the GenePix software and saturating spots, where the median foreground intensity is >60 000 in one of the two channels, were discarded from the result files. The data were normalized using Goulphar (Lemoine et al, 2006) applying a global Lowess. The complete microarray data and the related protocols are available at the GEO website (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=tbobloyiwcmwmrw&acc=GSE19928). Analysis of the GO groups was performed on genes with a ratio <−0.4 (down-regulated) or >0.4 (up-regulated) using the GO Tree Machine (version 2.0.56) of Vanderbilt University (Zhang et al, 2004) (http://bioinfo.vanderbilt.edu/gotm/) with the following parameters—reference set: the Agilent G_4122F; statistical method: hypergeometric; minimal number of genes: 5.
Quantitative real-time RT–PCR
Total RNA from cultured neurons was extracted using RNeasy Mini kit (QIAgen). RNA (250–500 ng) was reverse transcribed using random hexamers and Superscript II (Invitrogen) according to manufacturer's protocol. Quantitative real-time PCR was performed using QuantiTect SYBR Green PCR kit (QIAgen) and LightCycler 2.0 system (Roche Diagnostics) or as described elsewhere (Prasanth et al, 2005) (primer pairs used for real-time PCR were shown in Supplementary Table IV). Analysis of the data were performed as described (Pfaffl, 2001). Mann–Whitney U-test was used for statistical analysis.
Synaptic density analysis
The modification of Malat1 ncRNA level in transfected neuronal cultured cells was first examined by ISH (Supplementary Figure 7). Analysis of synapsin I puncta density was then performed on the same neurons. Punctae of synapsin immunoreactivity were counted along three primary dendrites for each neuron using Metamorph Software. We chose synapsin I immunoreactivity as this accurately correlates with synapse formation (Fletcher et al, 1991; Zhang and Benson, 2001; Graf et al, 2004). Three independent cultures were examined and 10 neurons per condition and per culture were counted. Analyses were performed blind. Two-way ANOVA was used for statistical analysis.
Supplementary Material
Acknowledgments
We thank O Bensaude, S Marty, X Darzacq, D Hernandez-Verdun, SG Prasanth and members of our laboratories for helpful discussions. We thank Drs Paula Bubulya for SON antibody, Supriya G Prasanth for Q-PCR in non-neuronal cells and C Frank Bennett and Susan M Freier (ISIS Pharmaceuticals, Carlsbad, CA) for Malat1 antisense oligonucleotides used in non-neuronal cells. AB is a fellow of the Centre National de la Recherche Scientifique. Research was funded by the Institut pour la Recherche sur la Moelle épinière et l'Encéphale and the Association Française contre les Myopathies and the 7th framework program Moodinflame (222963) (AB) and NIH/NIGMS 42694 (DLS) and NIH/NCI 5PO1CA013106-38 (DLS).
Footnotes
The authors declare that they have no conflict of interest.
References
- Ahmari SE, Buchanan J, Smith SJ (2000) Assembly of presynaptic active zones from cytoplasmic transport packets. Nat Neurosci 3: 445–451 [DOI] [PubMed] [Google Scholar]
- Barthelemy C, Henderson CE, Pettmann B (2004) Foxo3a induces motoneuron death through the Fas pathway in cooperation with JNK. BMC Neurosci 5: 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassell GJ, Warren ST (2008) Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron 60: 201–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Sudhof TC (2002) SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297: 1525–1531 [DOI] [PubMed] [Google Scholar]
- Bubulya PA, Prasanth KV, Deerinck TJ, Gerlich D, Beaudouin J, Ellisman MH, Ellenberg J, Spector DL (2004) Hypophosphorylated SR splicing factors transiently localize around active nucleolar organizing regions in telophase daughter nuclei. J Cell Biol 167: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao W, Jamison SF, Garcia-Blanco MA (1997) Both phosphorylation and dephosphorylation of ASF/SF2 are required for pre-mRNA splicing in vitro. RNA 3: 1456–1467 [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Wang J, Zhu Z, Zhang MQ, Krainer AR (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31: 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter KC, Taneja KL, Lawrence JB (1991) Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol 115: 1191–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabot B, Bisotto S, Vincent M (1995) The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res 23: 3206–3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain SJ, Brannan CI (2001) The Prader-Willi syndrome imprinting center activates the paternally expressed murine Ube3a antisense transcript but represses paternal Ube3a. Genomics 73: 316–322 [DOI] [PubMed] [Google Scholar]
- Chen L-L, Carmichael GG (2009) Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35: 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chih B, Engelman H, Scheiffele P (2005) Control of excitatory and inhibitory synapse formation by neuroligins. Science 307: 1324–1328 [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF (2005) Non-coding RNAs: new players in eukaryotic biology. Gene 357: 83–94 [DOI] [PubMed] [Google Scholar]
- Danglot L, Rostaing P, Triller A, Bessis A (2004) Morphologically identified glycinergic synapses in the hippocampus. Mol Cell Neurosci 27: 394–403 [DOI] [PubMed] [Google Scholar]
- De Felipe J, Marco P, Fairen A, Jones EG (1997) Inhibitory synaptogenesis in mouse somatosensory cortex. Cereb Cortex 7: 619–634 [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, Sunkin SM, Crowe ML, Grimmond SM, Perkins AC, Mattick JS (2008) Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 18: 1433–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD (2006) The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 20: 1470–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher TL, Cameron P, De Camilli P, Banker GA (1991) The distribution of synapsin I and synaptophysin in hippocampal neurons developing in culture. J Neurosci 11: 1617–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga K, Miyamoto E (2000) A working model of CaM kinase II activity in hippocampal long-term potentiation and memory. Neurosci Res 38: 3–17 [DOI] [PubMed] [Google Scholar]
- Goslin K, Asmussen H, Banker G (1998) Rat hippocampal neurons in low-density culture. In Culturing Nerve Cells, Banker G, Goslin K (eds), pp 339–370. Cambridge [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM (2004) Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119: 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LL, Smith KP, Byron M, Lawrence JB (2006) Molecular anatomy of a speckle. Anat Rec A Discov Mol Cell Evol Biol 288: 664–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E, Disteche CM (2006) Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev 20: 1848–1867 [DOI] [PubMed] [Google Scholar]
- Henderson JT, Georgiou J, Jia Z, Robertson J, Elowe S, Roder JC, Pawson T (2001) The receptor tyrosine kinase EphB2 regulates NMDA-dependent synaptic function. Neuron 32: 1041–1056 [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D (1995) Expression of a Delta homologue in prospective neurons in the chick. Nature 375: 787–790 [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K (2008) Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456: 130–134 [DOI] [PubMed] [Google Scholar]
- Huang S, Deerinck TJ, Ellisman MH, Spector DL (1994) In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol 126: 877–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A (2007) A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL (2004) From silencing to gene expression: real-time analysis in single cells. Cell 116: 683–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, Thomas M, Berdel WE, Serve H, Muller-Tidow C (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. PNAS 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL (2003) Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol 4: 605–612 [DOI] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW (1999) A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97: 17–27 [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176 [DOI] [PubMed] [Google Scholar]
- Lemoine S, Combes F, Servant N, Le Crom S (2006) Goulphar: rapid access and expertise for standard two-color microarray normalization methods. BMC Bioinformatics 7: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson JN, El-Husseini A (2005) Building excitatory and inhibitory synapses: balancing neuroligin partnerships. Neuron 48: 171–174 [DOI] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS (2006) A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene 26: 851–858 [DOI] [PubMed] [Google Scholar]
- Long JC, Caceres JF (2009) The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417: 15–27 [DOI] [PubMed] [Google Scholar]
- Mallik M, Lakhotia S (2009) The developmentally active and stress-inducible non-coding hsr{omega} gene is a novel regulator of apoptosis in Drosophila. Genetics 183: 831–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini-Dinardo D, Steele SJS, Levorse JM, Ingram RS, Tilghman SM (2006) Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 20: 1268–1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA (2008) Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell 29: 499–509 [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A (2007) Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445: 666–670 [DOI] [PubMed] [Google Scholar]
- Mattick JS (2001) Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep 2: 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS (2003) Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25: 930–939 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15 (Spec No 1): R17–R29 [DOI] [PubMed] [Google Scholar]
- McGee R (1980) Regulation of presynaptic cellular function. Biochemical studies using clonal neuronal cells. Mol Cell Biochem 33: 121–133 [DOI] [PubMed] [Google Scholar]
- Mehler MF, Mattick JS (2006) Non-coding RNAs in the nervous system. J Physiol 575 (Pt 2): 333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS (2009) Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105: 716–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Cáceres JF, Clement JQ, Krainer AR, Wilkinson MF, Spector DL (1998) Serine phosphorylation of SR proteins is required for their recruitment to sites of transcription in vivo. J Cell Biol 143: 297–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T, Caceres JF, Spector DL (1997) The dynamics of a pre-mRNA splicing factor in living cells. Nature 387: 523–527 [DOI] [PubMed] [Google Scholar]
- Moroy T, Heyd F (2007) The impact of alternative splicing in vivo: mouse models show the way. RNA 13: 1155–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P (2008) The air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720 [DOI] [PubMed] [Google Scholar]
- O'Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P (1999) Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron 23: 309–323 [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH (2006) Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci 7: 710–723 [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT (2008) X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 42: 733–772 [DOI] [PubMed] [Google Scholar]
- Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29: e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Mlynarczyk-Evans S, Nusinow DA, Panning B (2002) Xist RNA and the mechanism of X chromosome inactivation. Annu Rev Genet 36: 233–278 [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, Kern AD, Dehay C, Igel H, Ares M Jr, Vanderhaeghen P, Haussler D (2006) An RNA gene expressed during cortical development evolved rapidly in humans. Nature 443: 167–172 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL (2005) Regulating gene expression through RNA nuclear retention. Cell 123: 249–263 [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL (2007) Eukaryotic regulatory RNAs: an answer to the ‘genome complexity' conundrum. Genes Dev 21: 11–42 [DOI] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, Grimmond SM, Hume DA, Hayashizaki Y, Mattick JS (2006) Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res 16: 11–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, Chang HY (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongo C, Kaplan JM (1999) CaMKII regulates the density of central glutamatergic synapses in vivo. Nature 402: 195–199 [DOI] [PubMed] [Google Scholar]
- Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, Sudhof TC, Kavalali ET (2005) Selective capability of SynCAM and neuroligin for functional synapse assembly. J Neurosci 25: 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, Hirose T (2009) MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. PNAS 106: 2525–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A (1993) A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry 100: 431–440 [DOI] [PubMed] [Google Scholar]
- Sharma A, Takata H, Shibahara K, Bubulya A, Bubulya PA (2010) Son is essential for nuclear speckle organization and cell cycle progression. Mol Biol Cell 21: 650–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen K, Teruel MN, Subramanian K, Meyer T (1998) CaMKIIbeta functions as an F-actin targeting module that localizes CaMKIIalpha/beta heterooligomers to dendritic spines. Neuron 21: 593–606 [DOI] [PubMed] [Google Scholar]
- Spector DL (2006) SnapShot: cellular bodies. Cell 127: 1071. [DOI] [PubMed] [Google Scholar]
- Spoerri PE, Glees P, Dresp W (1980) The time course of synapse formation of mouse neuroblastoma cells in monolayer cultures. Cell Tissue Res 205: 411–421 [DOI] [PubMed] [Google Scholar]
- Stamm S (2008) Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem 283: 1223–1227 [DOI] [PubMed] [Google Scholar]
- Steward O, Falk PM (1986) Protein-synthetic machinery at postsynaptic sites during synaptogenesis: a quantitative study of the association between polyribosomes and developing synapses. J Neurosci 6: 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Falk PM (1991) Selective localization of polyribosomes beneath developing synapses: a quantitative analysis of the relationships between polyribosomes and developing synapses in the hippocampus and dentate gyrus. J Comp Neurol 314: 545–557 [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL (2009) MEN {varepsilon}/{beta} nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft R, Pang K, Mercer T, Dinger M, Mattick J (2009) Non-coding RNAs: regulators of disease. J Pathol 220: 126–139 [DOI] [PubMed] [Google Scholar]
- Visa N, Puvion-Dutilleul F, Harper F, Bachellerie JP, Puvion E (1993) Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res 208: 19–34 [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Rei, Du K, Pascual G, Tempst P, Rosenfeld M, Glass C, Kurokawa R (2008) Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature 454: 126–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG (2005) A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL (2008) 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Cline HT (1998) Stabilization of dendritic arbor structure in vivo by CaMKII. Science 279: 222–226 [DOI] [PubMed] [Google Scholar]
- Xiao SH, Manley JL (1998) Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 17: 6359–6367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Kuroda MI (2007) Noncoding RNAs and intranuclear positioning in monoallelic gene expression. Cell 128: 777–786 [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H (2008) Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 451: 202–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Schmoyer D, Kirov S, Snoddy J (2004) GOTree machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Benson DL (2001) Stages of synapse development defined by dependence on F-actin. J Neurosci 21: 5169–5181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.