ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery
By dissecting the so far loosely defined roles of the Werner syndrome helicase at stalled replication forks, this study shows that two highly related kinases can differentially regulate specific WRN phosphorylation sites and distinct WRN functions at various stages after replication arrest.
Keywords: genome instability, RecQ helicases, replication fork arrest, Werner syndrome
Abstract
Accurate response to replication arrest is crucial to preserve genome stability and requires both the ATR and ATM functions. The Werner syndrome protein (WRN) is implicated in the recovery of stalled replication forks, and although an ATR/ATM-dependent phosphorylation of WRN was observed after replication arrest, the function of such modifications during the response to perturbed replication is not yet appreciated. Here, we report that WRN is directly phosphorylated by ATR at multiple C-terminal S/TQ residues. Suppression of ATR-mediated phosphorylation of WRN prevents proper accumulation of WRN in nuclear foci, co-localisation with RPA and causes breakage of stalled forks. On the other hand, inhibition of ATM kinase activity or expression of an ATM-unphosphorylable WRN allele leads to retention of WRN in nuclear foci and impaired recruitment of RAD51 recombinase resulting in reduced viability after fork collapse. Altogether, our findings indicate that ATR and ATM promote recovery from perturbed replication by differently regulating WRN at defined moments of the response to replication fork arrest.
Introduction
An accurate response to stressed replication is crucial for maintaining genome stability. If perturbed replication forks are not properly handled, cells may accumulate chromosomal rearrangements as frequently observed in cancers and a subset of genetic diseases collectively referred to as chromosome fragility syndromes. The Werner syndrome (WS) is a chromosome fragility and cancer-prone disease caused by mutations in the human RecQ helicase WRN (Muftuoglu et al, 2008). Several pieces of evidence suggest that the primary function of WRN is related to recovery of stalled replication forks. At the biochemical level, WRN shows a remarkable preference towards substrates that mimic structures associated with stalled replication forks (Brosh et al, 2002; Choudhary et al, 2004; Machwe et al, 2006) and, at the cellular level, loss of WRN causes S-phase defects, hypersensitivity to agents interfering with replication and fragility of chromosomal regions that are considered natural hotspots of replication fork stalling (Poot et al, 1999, 2001; Pichierri et al, 2001; Pirzio et al, 2008; Sidorova et al, 2008). Consistently, replication fork arrest triggers extensive subnuclear re-localisation of WRN and co-localisation with RPA and replication foci (Constantinou et al, 2000; Rodriguez-Lopez et al, 2003; Franchitto and Pichierri, 2004). The exact contribution of WRN to replication fork recovery is not fully appreciated, but it has been suggested that it may facilitate replication restart by either promoting recombination or by processing intermediates at stalled forks in a way that counteracts unscheduled recombination (Pichierri, 2007; Sidorova, 2008). Recent data from our group indicate that loss of WRN determines activation of an alternative pathway of fork recovery resulting in DSB accumulation that is subsequently repaired through recombination (Franchitto et al, 2008), supporting the hypothesis that, upon replication fork stalling, WRN acts to limit fork collapse.
Recovery of stalled replication forks involves the co-ordinated action of several proteins under the control of the replication checkpoint kinase ATR (Cimprich and Cortez, 2008). Loss of ATR function determines hypersensitivity to replication fork arrest and accumulation of chromosomal breakage in response to stressed replication (Cliby et al, 1998; Casper et al, 2002; Cortez, 2003), which are phenotypes reminiscent of those associated with WS cells. Indeed, WRN is phosphorylated after replication arrest in an ATR-dependent manner and both ATR and WRN act in a common pathway to stabilise common fragile sites (Pichierri et al, 2003; Pirzio et al, 2008). However, it is currently unknown if WRN is a substrate of ATR and which are the functional consequences of this phosphorylation on the recovery of stalled forks.
We examined the phosphorylation of WRN by ATR and evidenced that WRN can be phosphorylated at multiple sites located at its C-terminal region. We found that phosphorylation of WRN is functionally related to the prevention of fork collapse and DSB accumulation carried out by ATR upon replication arrest. Interestingly, we observed that ATR and ATM-dependent phosphorylation of WRN influence different steps of the replication fork recovery reaction. Although phosphorylation by ATR regulates WRN subnuclear re-localisation and interaction with RPA, preventing DSB formation at stalled forks, ATM-dependent phosphorylation becomes essential during recovery of collapsed forks influencing the ability of RAD51 to form nuclear foci.
Results
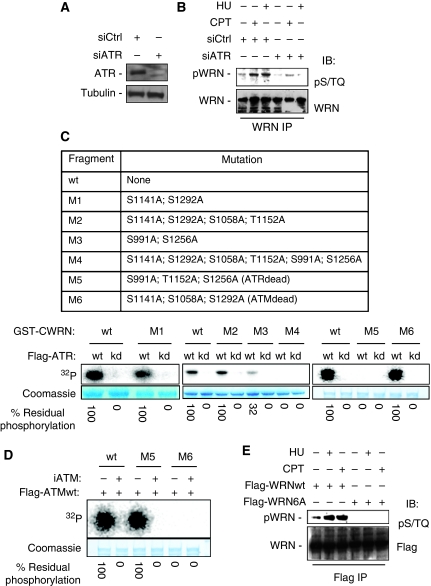
ATR phosphorylates the WRN protein at multiple sites on its C-terminal region
The minimal consensus sequence of ATR or ATM kinase is the Ser/Thr-Gln (S/TQ) motif and the human WRN protein contains six S/TQ motifs clustered at the C-terminal region, suggesting that it could be a direct substrate of ATR and/or ATM kinase activity. As ATR, but not ATM, is the primary kinase involved in the response to replication stress, we first evaluated whether the WRN S/TQ sites were phosphorylated by ATR. To this aim, HeLa cells, in which ATR was depleted (Figure 1A), were treated with campthotecin (CPT) or hydroxyurea (HU) to induce replication arrest and cell lysates were subjected to WRN immunoprecipitation. Using an antibody that specifically recognises phospho-S/TQ motifs (pS/TQ), we found that phosphorylation of WRN was barely detectable in untreated conditions and increased greatly after treatments (Figure 1B). Interestingly, depletion of ATR resulted in reduced WRN immunoreactivity to the anti-pS/TQ antibody, suggesting that most, if not all, of the phosphorylation is ATR related under perturbed replication (Figure 1B). To analyse which of the six S/TQ sites of WRN (S991, S1058, S1141, T1152, S1256 and S1292) were the candidate targets of ATR, we mutated several S/TQ sites in WRN simultaneously (Figure 1C). To exclude any phosphorylation by a co-precipitating kinase, we first verified by ATR immunocomplex kinase assay that the wild-type (wt) fragment of WRN was efficiently phosphorylated by ATR wt, but not by the kinase-dead form of the enzyme (ATRkd) (Figure 1C). Similarly, the analysis using the M1 fragment containing the two Ala substitutions at previously identified ATM substrates (Kim et al, 1999; Matsuoka et al, 2007) as well as the M2, in which two additional residues were mutated, were efficiently phosphorylated by ATR (Figure 1C), clearly suggesting that ATR and ATM do not target the same residues. However, phosphorimaging analysis showed that the M3 fragment still presented a residual level of phosphorylation (about 32% of wt, Figure 1C), whereas the M4, containing all the six potential S/TQ sites changed into Ala, was not phosphorylated at all (Figure 1C), excluding the presence of non-canonical ATR phosphorylation sites. To identify the actual ATR phosphorylation sites, we generated the M5 fragment in which S991, T1152 and S1256 were mutated into Ala and the M6 fragment containing Ala substitutions at the ATM putative substrates (Figure 1C). The M5 fragment was not phosphorylated by ATR, whereas M6 showed a level of phosphorylation similar to that of the wt fragment (Figure 1C). To confirm the observed non-overlapping phosphorylation by ATR and ATM, we performed ATM kinase assays using the M5 (ATRdead) and M6 (ATMdead) fragments as substrates. The results presented in Figure 1D confirmed that the S/TQ residues phosphorylated by ATR are not substrate for ATM and vice versa. Interestingly, addition of the ATM inhibitor KU55933 to the assays prevented phosphorylation of the M5 fragment, excluding phosphorylation by other kinases (Figure 1D).
Figure 1.
WRN is phosphorylated by ATR at multiple sites of the C-terminal region. (A) Depletion of ATR. HeLa cells were transfected with control siRNAs or siRNAs directed against ATR, and 48 h later, cell lysates were subjected to immunoblotting with anti-ATR antibody. Tubulin was used as loading control. (B) Evaluation of WRN phosphorylation in ATR-depleted cells. After siRNA transfection, HeLa cells were treated with 2 mM HU or 10 μM CPT for 6 h. Cell extracts were immunoprecipitated (IP) using anti-WRN antibody followed by immunoblotting with an anti-pST/Q antibody. Total WRN was used to evaluate the amount of WRN immunoprecipitated. (C) Identification of ATR phosphorylation sites on the C-terminal region of WRN by immunocomplex kinase assay. Top, schematic representation of the GST-tagged C-terminal wild-type or mutant forms of WRN. Locations of multiple Ala substitutions are indicated. Below, in vitro kinase assay. GST-C-terminal wild-type (wt) or mutants WRN (M1, M2, M3, M4, M5 and M6) were incubated with the wild-type or inactive form of ATR immunopurified from HeLa cells in the presence of 32P-ATP (for details see ‘Materials and methods'). After separation by SDS–PAGE, the presence of phosphorylation was assessed by phosphorimaging. The amount of C-terminal WRN fragments used is shown by Coomassie staining. Phosphorylation levels were expressed as the percentage of residual phosphorylation of each mutant fragment compared with the wild type. (D) Analysis of in vitro phosphorylation of C-terminal WRN fragment by ATM. The wild type (wt) and both the M5 and M6 mutant fragments containing, respectively, Ala changes at the ATR or at the ATM phosphorylation sites were incubated with immunopurified Flag-ATM with or without 10 μM KU55933 (iATM). (E) Analysis of mutant WRN phosphorylation. HEK293T cells were transfected with plasmids expressing a Flag-tagged full-length WRN wild type or carrying Ala substitutions at all the six S/TQ sites (6A) and 48 h later treated with 2 mM HU or 10 μM CPT for 6 h. Cell extracts were immunoprecipitated (IP) using anti-WRN antibody following immunoblotting with an anti-pST/Q antibody. Anti-Flag tag antibody was used to evaluate the amount of wild-type or mutant form WRN immunoprecipitated.
Altogether, in vitro kinase assays indicate that S991, T1152 and S1256 are ATR substrates, suggesting that the C-terminal fragment of the protein can undergo multiple phosphorylation events also in vivo.
To confirm that the C-terminal WRN region might be phosphorylated in multiple residues, we transiently expressed in HEK293T cells an N-terminal-truncated fragment of WRN (WRNΔN) and analysed the presence of phosphorylated species of the fragment. The immunoblotting analyses revealed that Myc-WRNΔN migrated as a doublet consisting of two species, designed as β and γ, in both untreated and treated extracts (Supplementary Figure 1). Treatment of extracts from either control or CPT-treated cells with λ-phosphatase eliminated both the β and γ forms of Myc-WRNΔN and resulted in the appearance of a single faster-migrating species that we named α form, confirming that the β and γ forms represented phosphorylated species (Supplementary Figure 1).
To exclude that WRN could be phosphorylated at additional S/TQ sites after replication arrest, we transfected HEK293T cells with constructs expressing the wt full-length Flag-tagged WRN protein (FlagWRNwt) or a mutant in which all the six C-terminal S/TQ sites were changed into Ala (Flag-WRN6A). Immunoblotting analysis of cells immunoprecipitated using anti-Flag antibody showed that, with or without treatment, mutation of the six C-terminal S/TQ sites completely abrogated WRN phosphorylation, as assessed by anti-pS/TQ antibody (Figure 1E).
Altogether, these results show that WRN is an ATR substrate both in vitro and in vivo, evidencing that multiple C-terminal phosphorylation sites can be targeted in response to replication arrest.
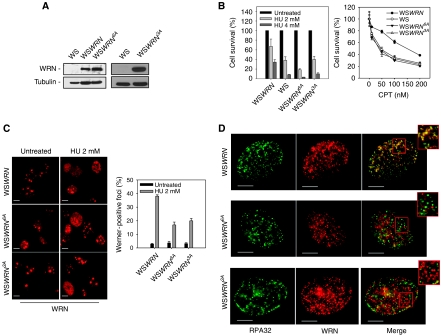
Efficient re-localisation of WRN in nuclear foci and co-localisation with RPA require ATR phosphorylation
Having shown that ATR and ATM do not target the same WRN residues and that in response to perturbed replication WRN is phosphorylated by ATR, the WRN6A protein can be considered as an ATR-unphosphorylable mutant under these experimental conditions. To assess whether phosphorylation by ATR could influence the nuclear dynamics of WRN after replication stress, we generated cell lines stably expressing a Flag-tagged wt WRN (WSWRN) or the WRN6A mutant (WSWRN6A). Even though WSWRN6A cells expressed levels of the WRN protein similar to that of WS cells (Figure 2A), hypersensitivity to replication stressing agents was comparable with that seen in WS cells (Figure 2B). Moreover, immunofluorescence analysis showed that formation of WRN foci in WSWRN6A cells was greatly reduced after HU-induced replication fork stalling as compared with cells expressing wt WRN (Figure 2C). Consistently with previous reported data (Constantinou et al, 2000), in WSWRN cells, HU treatment led to re-localisation of WRN in nuclear foci co-localising with RPA at sites of replication fork stalling in the majority of the nuclei (Figure 2D). In contrast, only a minor fraction of the nuclei positive for WRN foci revealed co-localisation with RPA in WSWRN6A cells (Figure 2D). In addition, nuclear foci formed by the mutant form of WRN preferentially co-localised with the DSB-marker γ-H2AX (Supplementary Figure 2), suggesting that they might pinpoint spontaneous DSBs or collapsed replication forks. Interestingly, cells expressing the WRN3A mutant, containing Ala substitutions only at the identified ATR phosphorylation sites (S991, T1152 and S1256; WSWRN3A), showed a defect in WRN subnuclear re-localisation and co-localisation with RPA similar to that observed in the WRN6A mutant (Figure 2C and D).
Figure 2.
WRN re-localisation and co-localisation with RPA upon replication arrest depend on its phosphorylation by ATR. (A) Western blotting on extracts from WS cells stably expressing the Flag-tagged wild-type WRN (WSWRN), the 6A mutant (WSWRN6A) or the ATR-unphosphorylable form of WRN (WSWRN3A) showing levels of WRN using an anti-WRN antibody. WS cells were used as negative control and tubulin as loading control. (B) Cells were treated with different doses of HU or CPT for 16 h and allowed to grow in drug-free medium for 2 weeks before analysis of clonogenic survival. Survival is expressed as percentage of the untreated cultures. Data are presented as mean±s.e. from three independent experiments. (C) Analysis of WRN re-localisation to nuclear foci after replication arrest. Images show WRN nuclear distribution with or without 8 h treatment. The graph shows the percentage of WRN-positive nuclei. Data are presented as means of three independent experiments. Error bars represent standard errors. (D) Analysis of WRN and RPA co-localisation after replication arrest. Cells were treated with 2 mM HU for 8 h and subjected to immunofluorescence using anti-WRN and anti-RPA32 antibodies. Representative images from cells treated with HU for 8 h are presented. Insets show an enlarged portion of the nuclei for a better evaluation of the co-localisation status of WRN with RPA32 foci. Scale bars, 10 μm.
These studies indicate that phosphorylation by ATR is essential for correct WRN subnuclear re-localisation at sites of replication fork stalling after HU-induced replication stress.
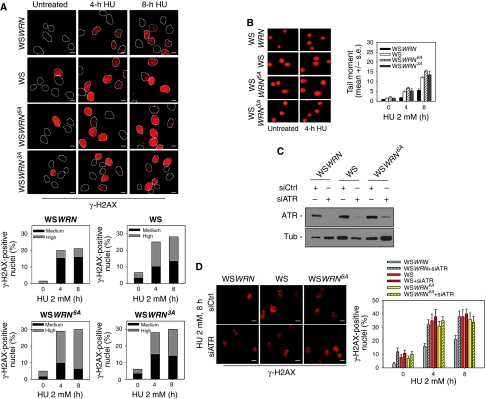
ATR phosphorylation of WRN prevents the appearance of DSBs upon replication fork stalling
In the absence of WRN, stalled forks undergo collapse and DSBs are formed (Franchitto et al, 2008). To assess whether ATR phosphorylation was required for the protective function of WRN at stalled forks, WS, WSWRN, WSWRN6A and the WSWRN3A cells were analysed for their ability to accumulate DSBs. In WS cells, HU-induced replication fork stalling resulted in a large number of nuclei with high γ-H2AX fluorescence (Figure 3A), which appeared reduced in percentage and intensity by expressing the wt form of WRN (Figure 3A). In contrast, expression of either the completely unphosphorylable form of WRN (WRN6A) or the ATR-unphosphorylable mutant (WRN3A) induced an accumulation of γ-H2AX-positive nuclei as the absence of WRN (Figure 3A). Noteworthy, expression of the WRN6A mutant led to enhanced number of γ-H2AX-positive nuclei with highly fluorescence respect to that observed in parental WS cells (Figure 3A).
Figure 3.
WRN phosphorylation by ATR is required to prevent accumulation of DNA breakage after replication arrest. (A) Analysis of DNA breakage using γ-H2AX immunostaining. Cells were exposed to HU for the indicated times and stained with an antibody against γ-H2AX. In the panel, representative images from each experimental point are shown. Graphs show the percentage of γ-H2AX-positive nuclei with medium or high intensity of γ-H2AX fluorescence. (B) DNA breakage as detected using a neutral comet assay. Cells were treated with HU for the indicated times and then subjected to comet assay. In the panel, representative images are shown. Data are presented as mean tail moment and as means of three independent experiments. Error bars represent standard errors. Where not depicted, standard errors were <15% of the mean. (C) Depletion of ATR. WS, WSWRN or WSWRN6A cells were transfected with control siRNAs or siRNAs directed against ATR and cell lysates subjected to immunoblotting with anti-ATR antibody. Tubulin was used as loading control. (D) Analysis of DNA breakage by γ-H2AX in WS, WSWRN or WSWRN6A cells in which ATR was depleted using RNAi. After transfection, cells were treated with 2 mM HU for the indicated times and stained with an antibody against γ-H2AX. In the panel, representative images are shown. Graphs show the percentage of γ-H2AX-positive nuclei. Data are presented as means of three independent experiments and error bars represent standard errors. Scale bars, 10 μm.
As γ-H2AX immunostaining does not necessarily mark only DSBs upon replication arrest, we used neutral comet assay to verify accumulation of these lesions at stalled forks in the WRN phosphorylation mutants. Comet assay confirmed that HU treatment determined the formation of DSBs in WS cells, which could be prevented by expressing the wt form of WRN (Figure 3B) and showed that WSWRN6A cells, consistently with the γ-H2AX data, showed levels of breakage even more abundant than those observed in the absence of WRN or in the presence of the WRN3A mutant (Figure 3B).
It has been previously reported that ATR functions in preserving fork stability after replication stress (Cortez, 2003; Chanoux et al, 2009). As we observed that the expression of the unphosphorylable mutant form of WRN leads to accumulation of DSBs after replication arrest, we asked whether ATR-mediated avoidance of DSBs at stalled forks might directly involve phosphorylation of WRN. Thus, we depleted ATR function by RNAi in WSWRN, WS and WSWRN6A cells (Figure 3C) to analyse the level of DSB accumulation after HU treatment. As shown in Figure 3D, abrogation of ATR in wt cells (WSWRN+siATR) treated with HU led to a two-fold increase in the percentage of nuclei with γ-H2AX foci respect to the WSWRN cells. However, the absence of ATR in WRN-deficient (WS+siATR) cells or expressing the unphosphorylable form of WRN (WSWRN6A+siATR) did not enhance the effect of HU-induced accumulation of DSBs (Figure 3D). Similar results were obtained from comet assay (data not shown). To confirm that ATR phosphorylation of WRN is specifically related to the function of ATR in the stability of stalled forks, we analysed activation of the S-M checkpoint in cells expressing the WRN6A mutant. Progression into mitosis in the presence of HU was impaired only in ATR RNAi-treated cells (Supplementary Figure 3), suggesting that the function of ATR in promoting cell cycle arrest is unrelated to phosphorylation of WRN.
Our data are consistent with the hypothesis that phosphorylation of WRN may mediate the ATR-dependent function of replication fork stability preservation, avoiding degeneration of forks into DSBs.
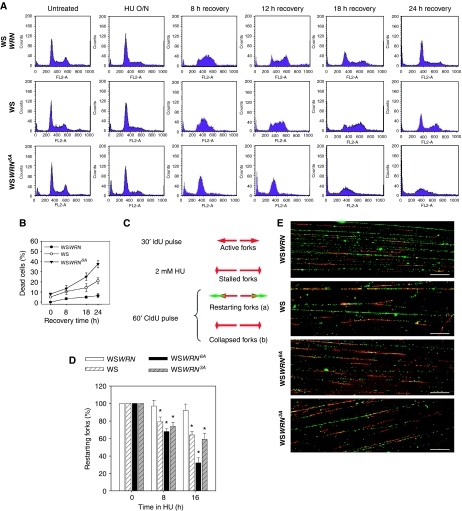
WRN phosphorylation is required for the recovery from perturbed replication
Given the function of WRN phosphorylation in the correct handling of stalled forks and the observation that expression of WRN6A leads to a higher sensitivity to HU than the absence of the WRN protein, we evaluated the possibility that loss of S/TQ phosphorylation of WRN could affect the ability of the cells to recover cell cycle progression after replication stress. Flow cytometry analyses showed that WSWRN cells readily resumed cell cycle progression after release from the HU-induced block and completely recovered from the arrest within 18–24 h (Figure 4A). Similarly, WS cells arrested in response to HU exposure recovered normally cell cycle progression, even if with a delayed kinetic (24 h) if compared with the wt counterpart (Figure 4A). In contrast, at 12 h from release, the majority of WSWRN6A cells were still accumulated in S phase (Figure 4A), and even though they restarted cell cycle progression at later times, they also underwent extensive cell death (Figure 4A and B).
Figure 4.
WRN phosphorylation is necessary for the cellular recovery from prolonged replication arrest and for replication fork restart. (A) Analysis of cell cycle progression after replication arrest. Cells were treated overnight with 2 mM HU, samples collected at the indicated recovery times and subjected to FACS analysis. (B) Evaluation of cell viability by LIVE/DEAD assay. WS, WSWRN or WSWRN6A cells were treated with 2 mM HU for the indicated times. Cell viability was evaluated as described in ‘Materials and methods'. Data are presented as per cent of dead cells and as means of three independent experiments. Error bars represent standard errors. (C) Schematic representation of experimental design used to measure replication fork recovery and examples of replication track labelling after recovery, labelled replication tracks showing stalled forks recovering upon HU removal (a) or forks collapsed after HU treatment (b). (D) Graph shows quantification of restarting forks evaluated by dividing the number of restarting forks (i.e. ‘a' tracks) by the total number of forks (i.e. a+b). WS, WSWRN, WSWRN6A or WSWRN3A cells were treated with 2 mM HU for the indicated times and recovered for 1 h in IdU-containing medium before DNA fibre assay. Data are presented as means±s.e. from three independent experiments. *Statistically significant (P<0.01) by a Student's t-test. (E) Images of DNA fibres visualised by immunofluorescence detection. Images derived from samples treated with HU for 16 h. Scale bars, 20 μm.
To test whether cell death observed in WSWRN6A cells depended on defective restart of stalled/collapsed forks after DSB formation, we analysed the ability of stalled forks to recover DNA synthesis by the DNA fibre assay as previously described (Merrick et al, 2004; Franchitto et al, 2008). Using this protocol, stalled forks, which resume DNA synthesis once HU is removed, show both IdU and CldU labelling, whereas forks that have been inactivated present only IdU labelling (Figure 4C). After release from 8 h HU treatment, almost the totality of stalled forks recovered in WSWRN cells, whereas WS and WSWRN6A cells showed a reduced number of restarting forks, as shown by poor CldU labelling (Figure 4D). However, and in contrast with WS cells, prolonged replication arrest (16 h) resulted in a further reduction of restarting forks in WSWRN6A cells (Figure 4D and E). On the other hand, expression of WRN3A, which has mutated only the ATR phosphorylation sites, did not result in a defect in fork restart greater than that observed in the absence of WRN (Figure 4D and E).
Taken together, these results indicate that expression of the ATR and ATM-unphosphorylable WRN6A mutant leads to a more severe phenotype than that observed in WS cells or in cells expressing the WRN3A mutant, especially after DSB formation and fork collapse, affecting the ability of cells to resume replication under replication stress condition.
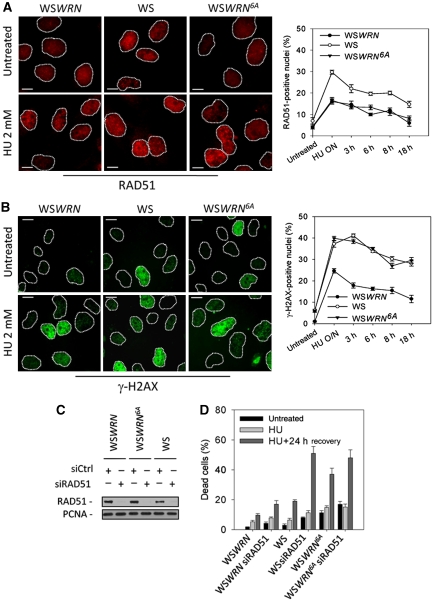
Loss of phosphorylation of WRN affects the RAD51-dependent recovery of stalled forks
The reduced ability of WSWRN6A cells to resume from S-phase arrest suggests that the completely unphosphorylable WRN protein may act in a dominant-negative manner, affecting the other pathways involved in the recovery from replication blockage in the absence of an active WRN. As WS cells use RAD51-dependent recombination to recover from replication arrest (Franchitto et al, 2008), we investigated whether loss of ATR and ATM phosphorylation of WRN could interfere with engagement of recombination. In WS cells, increased RAD51 foci formation correlates with enhancement of recombination (Franchitto et al, 2008), thus, we used RAD51 foci as readout of its involvement in replication recovery. As expected, in WS cells, HU treatment increased formation of RAD51 foci, which is reverted by expressing wt WRN (Figure 5A). However, WSWRN6A cells showed levels of RAD51-positive nuclei comparable with those observed in WSWRN cells (Figure 5A), but the amount of DSBs was equivalent to that exhibited by the parental WS cells (Figure 5B). Furthermore, treatment with etoposide, an agent inducing DSBs irrespective of DNA replication, resulted in similar percentages of RAD51-positive nuclei in all the cell lines, suggesting that defective accumulation of RAD51 foci seen in WSWRN6A cells was specific for DSBs created at collapsed forks (Supplementary Figure 4). Then, we determined whether the elevated HU sensitivity of the WSWRN6A cells could be attributable to concomitant impairment of WRN- and RAD51-dependent replication fork recovery mechanisms. Given that viability of WS cells after prolonged replication arrest is affected by RAD51 depletion (Franchitto et al, unpublished), we compared survival of WS and WSWRN6A cells, in which RAD51 was depleted by RNAi (Figure 5C). We found that, after RAD51 depletion, cell death was about 20% in wt cells and >50% in WS cells (Figure 5D). In contrast, despite the WSWRN6A cells were more sensitive than WS cells to HU, cell death was only slightly enhanced by RAD51 depletion reaching values similar to those of WS siRAD51 cells (Figure 5D).
Figure 5.
Analysis of the activation of RAD51-dependent pathway after replication arrest. (A) Analysis of RAD51 re-localisation in foci. Cells were treated with HU for 18 h, released into drug-free medium for the indicated times and stained with an antibody against RAD51. Images show cells with RAD51 re-localisation in foci at 18 h of HU. Graph shows the percentage of RAD51-positive nuclei for each experimental condition. (B) Evaluation of DNA breakage accumulation by γ-H2AX immunofluorescence. Cells were treated as in (A). Representative images from the 18 h HU treatment. Graph shows the percentage of γ-H2AX-positive nuclei for each experimental condition. (C) Depletion of RAD51. WSWRN, WSWRN6A and WS cells were transfected with control siRNAs or siRNAs directed against RAD51 and cell lysates subjected to immunoblotting with anti-RAD51 antibody. PCNA was used as loading control. (D) Evaluation of cell viability by LIVE/DEAD assay. After siRNAs transfection, WSWRN, WS and WSWRN6A cells were treated overnight with 2 mM HU and recovered in drug-free medium for 24 h. Data are presented as per cent of dead cells and as means of three independent experiments. Error bars represent standard errors. Scale bars, 10 μm.
Altogether, our results indicate that expression of the unphosphorylable form of WRN leads to elevated levels of replication-associated DSBs at stalled forks, but impaired activation of the RAD51-dependent fork recovery response.
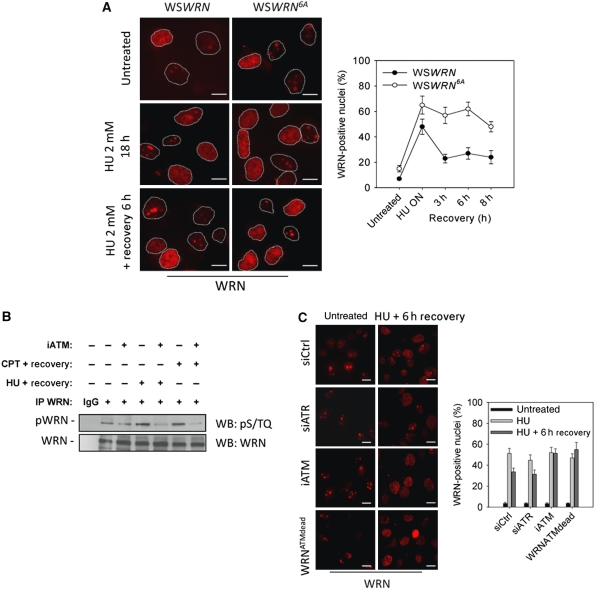
Phosphorylation of WRN by ATM influences WRN subnuclear dynamics after recovery from stalled or collapsed forks and allows RAD51 accumulation in foci
Having shown that expression of the WRN6A mutant interferes with RAD51-dependent fork recovery, we investigated how loss of WRN phosphorylation could produce such effect. Correct execution of recombination requires timely and coordinate interchange of proteins at the lesion and, given that accumulation of RAD51 foci is elevated after prolonged replication inhibition and early after recovery (Petermann et al, 2010; Franchitto et al, unpublished), we analysed whether inability of WSWRN6A cells to form RAD51 foci could correlate with altered subnuclear dynamics of the WRN mutant at later time of treatment and after recovery. Although WSWRN6A cells showed reduce re-localisation of WRN in nuclear foci early after replication arrest (see Figure 2B), upon prolonged HU treatment, WRN6A was re-localised as efficiently as the wt protein (Figure 6A). Most importantly, although the percentage of WRN foci in WSWRN cells decreased after recovery, it remained sustained in WSWRN6A cells (Figure 6A) and largely (>80%) co-localised with γ-H2AX (Supplementary Figure 2).
Figure 6.
Loss of ATM-dependent phosphorylation impairs WRN de-localisation during the recovery from prolonged replication arrest. (A) Analysis of WRN re-localisation in foci. WSWRN or WSWRN6A cells were treated with 2 mM HU for 18 h (HU O/N), recovered for the indicated times and stained with an antibody against WRN. Representative images are shown. Scale bars, 10 μm. Graph shows the percentage of WRN-positive nuclei for each experimental point. (B) Analysis of WRN phosphorylation during recovery from replication fork stalling. WSWRN cells were treated with 10 μM CPT or 2 mM HU for 8 or 18 h, respectively, and recovered for 3 h before preparation of whole cell extracts for anti-WRN IP. When indicated, cells were also treated from 1 h before inducing replication arrest to the sampling time with 10 μM of the KU55933, an ATM inhibitor (iATM). Anti-WRN IP were separated by SDS–PAGE and subjected to WB using an anti-pS/TQ antibody. One-third of IPs was subjected to WB using total anti-WRN antibody. IgG represents Ctrl IP. (C) Analysis of WRN re-localisation in foci in the absence of ATM-dependent phosphorylation. WSWRN or WSWRNATMdead cells were treated with 2 mM HU for 18 h (HU O/N), with or without 8 h of recovery. In some cases, WSWRN cells were transfected 48 h before treatment with Ctrl siRNAs or ATR siRNA, whereas exposure to 10 μM KU55933 during treatments and recovery was used to inhibit ATM (iATM). Representative images are shown. Scale bars, 10 μm. Graph shows the percentage of WRN-positive nuclei for each experimental point. Data are presented as means of three independent experiments and error bars represent standard errors.
Both ATR and ATM are required for recovery from replication-dependent DSBs and to regulate RAD51 foci formation (Yuan et al, 2003; Sorensen et al, 2005; Trenz et al, 2006). Our results suggest that ATR regulates WRN subnuclear dynamic early after HU treatment, thus we tested the possibility that ATM phosphorylation could influence WRN de-localisation at later times once DSBs were formed at stalled forks, as reported upon prolonged replication inhibition (Hanada et al, 2007). As the WRN6A is a combined ATR and ATM phosphorylation mutant, we first analysed whether ATM-dependent phosphorylation of WRN took place during recovery from prolonged replication inhibition, when an evident de-localisation of WRN from nuclear foci was observed (see Figure 6A). To this end, we treated WSWRN cells with an ATM inhibitor (KU55933) and induced replication arrest with HU or CPT, an agent able to produce DSBs at stalled forks. As shown in Figure 6B, after recovery from replication arrest, WRN was phosphorylated in S/TQ residues and such phosphorylation was largely prevented by treatment with the ATM inhibitor, suggesting that ATM phosphorylation of WRN and its de-localisation from nuclear foci were functionally related. To verify this hypothesis, we took advantage from our kinase assays data (see Figure 1C and D) and previous results published by others (Kim et al, 1999; Matsuoka et al, 2007), indicating that ATR and ATM target different WRN C-terminal residues. Thus, we generated a WS-derived cell line stably expressing a mutant form of WRN that contained Ala substitutions at S1058, S1141 and S1292 (WSWRNATMdead), which can be phosphorylated by ATR, but not by ATM. Interestingly, expression of this ATM-unphosphorylable WRN protein was able to revert the characteristic accumulation of DSBs observed in WSWRN6A cells after replication arrest (WSWRNATMdead; Supplementary Figure 5A and B). Using WSWRNATMdead cells, we analysed WRN subnuclear dynamics after recovery from prolonged HU treatment, and we obtained that ATM inhibition or expression of the WRNATMdead mutant similarly prevented WRN de-localisation from nuclear foci (Figure 6C). In contrast, depletion of ATR by RNAi did not affect de-localisation of WRN upon recovery from the arrest (Figure 6C), in agreement with robust ATM activation observed in these cells (Supplementary Figure 5C).
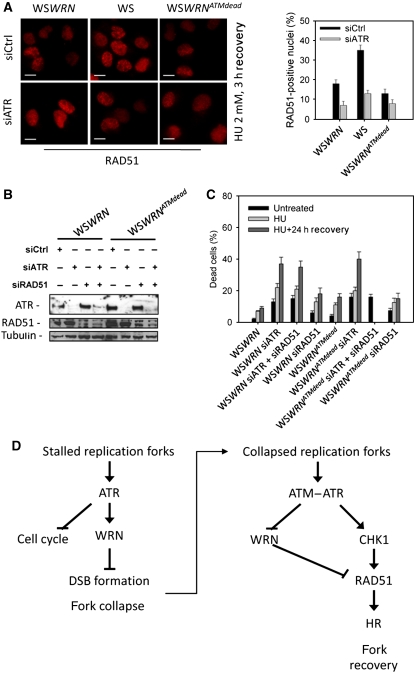
To analyse whether ATM-dependent WRN de-localisation from stalled/collapsed forks was essential for subsequent formation of RAD51 foci under conditions causing replication-dependent DSBs, we treated WSWRNATMdead, WSWRN and the parental WS cells with HU, and examined RAD51 foci assembly after recovery. We also down-regulated ATR by RNAi to experimentally increase the number of DSBs formed at stalled replication forks (see Figure 3D). As expected, we detected a more elevated percentage of RAD51-positive nuclei in WS cells and, in agreement with an ATR-dependent control over RAD51 localisation (Sorensen et al, 2005), abrogation of ATR by RNAi reduced RAD51 assembly in foci in WSWRN and WS cells (Figure 7A). In contrast, in cells lacking ATM-dependent phosphorylation of WRN (WSWRNATMdead), HU did not induce accumulation of RAD51 in foci and this result was not significantly affected by ATR depletion (Figure 7A). Worth noting, chemical inhibition of ATM in wt cells or expression of WRNATMdead similarly affected formation of RAD51 foci during the recovery from CPT treatment, which directly generates replication-dependent DSBs (Supplementary Figure 6A). Moreover, inhibition of ATM was able to reduce formation of RAD51 foci also in siATR-treated cells (Supplementary Figure 6B).
Figure 7.
Formation of RAD51 foci and recovery from prolonged replication arrest requires WRN de-localisation from nuclear foci. (A) Analysis of RAD51 re-localisation in foci in the absence of ATM-dependent WRN phosphorylation. WSWRN, WS or WSWRNATMdead cells were transfected with control siRNAs or siRNAs directed against ATR and treated with 2 mM HU for 18 h followed by a 3 h recovery period before being analysed for the presence of RAD51 foci. Scale bars, 10 μm. (B) Concomitant depletion of ATR and RAD51. WSWRN or WSWRNATMdead cells were transfected with control siRNAs or siRNAs directed against ATR and/or RAD51 and cell lysates subjected to immunoblotting with anti-ATR or anti-RAD51 antibody. Tubulin was used as loading control. (C) Evaluation of cell viability by LIVE/DEAD assay. After concomitant depletion of ATR and RAD51, WSWRN and WSWRNATMdead cells were treated for 18 h with HU before recovery in drug-free medium. Cell viability was evaluated as described in ‘Materials and methods'. Data are presented as per cent of dead cells and as means of three independent experiments. Error bars represent standard errors. (D) Schematic model of the dual function of ATR and ATM in ensuring correct recovery from stalled and collapsed forks after HU treatment.
Our data suggest that, after formation of DSBs at stalled forks, cellular recovery requires the recruitment of RAD51 in foci, which is dependent on ATM-mediated WRN de-localisation and such a mechanism probably cooperates with direct control of RAD51 by ATR/CHK1 (Sorensen et al, 2005). Thus, we hypothesised that deregulation of each of these two parallel control mechanisms would have determined a loss of viability upon prolonged replication stress, which was comparable with that caused by loss of RAD51. To test this hypothesis, we evaluated viability after prolonged HU treatment with or without recovery in cells expressing either the wt or WRNATMdead protein in which ATR or RAD51 function was down-regulated by siRNAs. As shown in Figure 7B, RNAi efficiently depleted both ATR and RAD51 in the two isogenic cell lines. As expected, depletion of ATR sensitised wt cells to cell death induced by HU, and concomitant depletion of RAD51 and ATR did not further increase the number of dead cells (Figure 7C). Interestingly, either down-regulation of RAD51 in wt cells or expression of the WRNATMdead protein gave a similar enhancement of lethality induced by HU treatment (Figure 7C). Similarly to what was observed in wt cells, depletion of ATR alone or in combination with RAD51 determined comparable levels of cell death also in the WSWRNATMdead cells (Figure 7C).
Collectively, our findings imply that ATM-dependent regulation of WRN de-localisation from replication fork stalling sites and ATR-dependent control of RAD51 both contribute to ensure cellular recovery after DSB formation at stalled forks.
Discussion
To deal with replication stress, eukaryotic cells have evolved an accurate checkpoint response under the control of the ATR kinase (Cimprich and Cortez, 2008; Friedel et al, 2009), which is devoted to avoidance of chromosome rearrangements accumulation (Myung et al, 2001; Glover et al, 2005; Arlt et al, 2006) and suppression of tumour development (Halazonetis et al, 2008). In this study, we report that ATR- and ATM-dependent phosphorylation of WRN is instrumental in prevention of DSB formation at stalled forks and in ensuring efficient fork recovery upon the occurrence of the replication-dependent DSB.
Previous data from our and other groups have envisaged a possible cross-talk between WRN and ATR (Pichierri et al, 2003; Otterlei et al, 2006). Our in vitro data indicate that the C-terminal region of WRN is directly targeted by ATR kinase activity at S991, T1152 and S1256, and our in vivo data provide the first evidence of a functional correlation between ATR and WRN in response to replication stress. Interestingly, functional cross-talks between the related human RecQ helicase BLM and both ATR and ATM have been extensively reported after perturbed replication or DNA damage (Ababou et al, 2000; Beamish et al, 2002; Davies et al, 2004).
One of the prominent functions of the ATR-dependent checkpoint is to preserve the integrity of the stalled replisome, counteracting fork collapse and DSB accumulation (Cimprich and Cortez, 2008). Indeed, loss of ATR function results in enhanced formation of DSBs and chromosome breakage (Cliby et al, 1998; Liu et al, 2000; Casper et al, 2002; Zachos et al, 2005; Chanoux et al, 2009), but this is also a feature of cells having lost WRN function (Pichierri et al, 2001; Franchitto et al, 2008). We observe that WRN phosphorylation by ATR is essential to prevent DSB formation after replication stress. Most importantly, loss of ATR function in WSWRN6A cells does not enhance DSB accumulation after HU treatment, indicating that ATR performs its protective function against DSB formation at stalled fork through the phosphorylation of WRN. Early after induced replication fork stalling, WRN re-localises at sites of arrested replication and co-localises with RPA, which is loaded onto ssDNA accumulating at arrested forks (Constantinou et al, 2000; Franchitto and Pichierri, 2004). Phosphorylation by ATR seems to affect the ability of WRN to re-localise at sites of replication fork stalling, as both the WSWRN6A and the WSWRN3A cells show a reduced percentage of WRN foci and a decreased fraction of WRN co-localisation with RPA. However, in case of prolonged replication arrest, the phosphorylation mutant regains the ability to form foci and, interestingly, these late WRN foci show co-localisation with the γ-H2AX foci, implying that stalled replication forks are probably collapsed into DSBs.
These findings suggest that early after replication stress, phosphorylation by ATR is not absolutely required for WRN translocation in foci, but rather to stabilise the protein at the sites of stalled forks. Consistently, lower salt concentrations are sufficient to release the WRN6A mutant from chromatin after HU treatment (unpublished). As the C-terminal region of WRN is involved in mediating protein–protein association, WRN phosphorylation could promote the interaction with other proteins making WRN able to stay at stalling sites (Bohr, 2008). Alternatively, phosphorylation could regulate directly the association with DNA through the two C-terminal DNA-binding sites (von Kobbe et al, 2003). Interestingly, interaction of the BLM RecQ helicase with some of its protein partners has been reported to be regulated through ATR or ATM-dependent phosphorylation (Rao et al, 2005; Tripathi et al, 2008).
After prolonged replication arrest induced by HU, cells expressing WRN6A mutant show reduced ability to recover stalled forks and restart cell cycle progression from the S-phase arrest, undergoing extensive cell death. This phenotype is even more severe than that associated with the complete loss of WRN (Franchitto et al, 2008) or with inactivation of just the three ATR phosphorylation sites (WRN3A). As WRN6A is not phosphorylable by both ATR and ATM, our data suggest that loss of phosphorylation by ATM may confer to the WRN6A mutant the ability to act in a dominant-negative way on other pathways activated to ensure recovery from replication inhibition, especially when DSBs are formed. Replication-associated DSBs are mainly repaired by RAD51 (Arnaudeau et al, 2001), and in WS cells, RAD51 foci are up-regulated (Pichierri et al, 2001). Moreover, survival of WRN-deficient cells after replication arrest requires recombination enzymes (Franchitto et al, 2008). We find that expression of WRN6A mutant interferes with up-regulation of RAD51 foci and that cell death observed in WSWRN6A cells, after recovery from prolonged HU treatment, is comparable with that induced by down-regulating RAD51 in the parental WS cells. Most importantly, loss of RAD51 does not affect cell death in WSWRN6A cells, indicating that the WRN6A mutant interferes with RAD51 function. Consistently with our data, it has been recently reported that RAD51 performs an important function late in the recovery from replication stress, when, after prolonged replication inhibition, stalled replication forks collapse into DSBs (Petermann et al, 2010). Interestingly, the WRN6A mutant shows a persistent localisation in nuclear foci during the recovery from prolonged replication inhibition, which is exactly the opposite occurring to RAD51 foci (Franchitto et al, unpublished observations) (Petermann et al, 2010). It has been reported that formation of RAD51 foci after replication inhibition is mediated by ATR through CHK1-dependent phosphorylation of the RAD51 protein (Sorensen et al, 2005), whereas, after IR or treatment with radiomimetic drugs, RAD51 foci are under the control of ATM (Morrison et al, 2000; Yuan et al, 2003). In addition, also recovery from DSBs associated to perturbed replication requires ATM (Trenz et al, 2006). Thus, ATM and ATR cooperate whenever DSBs are introduced at the forks to stimulate recombination-mediated restart. Our experiments show that phosphorylation of WRN early upon replication inhibition is almost exclusively ATR dependent, whereas phosphorylation of WRN during recovery after prolonged replication stress induced by HU or CPT treatment leading to DSB formation is essentially ATM dependent. Our data indicate that once DSBs are formed and RAD51 is required to support replication fork restart and viability, WRN has to move away from fork stalling sites and this de-localisation is mediated by ATM. Thus, it seems that even though ATM and ATR share the same consensus sequence, they differently regulate WRN during well-defined moments of the response to replication fork arrest. Consistently, our biochemical experiments indicate that the three different residues (S1058, S1141 and S1292), previously identified as potential ATM substrates (Kim et al, 1999; Matsuoka et al, 2007), are not phosphorylated at all by ATR. Furthermore, expression of the WRNATMdead protein is able to revert the accumulation of DSBs typically detected in WS cells, likely because ATR phosphorylation sites are intact. Indeed, cells expressing the ATR-unphosphorylable WRN3A mutant show an accumulation of DSBs that is comparable with that observed in the absence of the protein. However, once DSBs are formed after prolonged replication arrest, inhibition of ATM or the expression of the WRNATMdead allele leads to defective RAD51 focus formation even in an ATR wt background, suggesting that the absence of the ATM kinase activity is correlated with the inability to promote fork recovery.
Collectively, our findings led us to propose a model (Figure 7D) whereby phosphorylation by ATR is instrumental for the early function of WRN, avoiding DSB formation at stalled forks, whereas ATM promotes de-localisation of WRN from collapsed forks to prepare the way for RAD51-mediated replication recovery. In view of this, expression of WRN6A is detrimental as it not only determines a large fraction of inactivated stalled replication forks, but also interferes with activation of the alternative RAD51-dependent DSB salvage pathway. Following DSB formation, replication recovery and viability depend on proper activation of RAD51 function based on at least two levels of regulation: displacement of WRN from the forks and licensing of RAD51 foci assembly (Figure 7D, left arm).
Thus, these data indicate that phosphorylation of WRN has a vital function in the S-phase checkpoint response, promoting the failsafe recovery from replication arrest and the engagement of recombination upon fork collapse.
Materials and methods
Cell cultures
The SV40-transformed WS fibroblasts (AG11395) were obtained from Coriell Cell Repositories. The AG11395 cell line carries an Arg368 stop mutation that gives rise to a truncated protein lacking the NLS and that undergoes degradation.
WSWRN, WSWRN6A, WSWRNATMdead and WSWRN3A were generated by transfection of the AG11395 (WS) with the pCMV-FLAGWRN2, the pCMV-FLAGWRN6A, the pCMV-FLAGWRN3A1058−1141−1292 or the pCMVFLAGWRN3A991−1152−1256 plasmid followed by selection of stable transfectants using G418. HeLa and HEK293T cells were obtained from ATCC. All the cell lines were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Boehringer Manheim) and incubated at 37°C in an humified 5% CO2 atmosphere.
Site-directed mutagenesis and cloning
Site-directed mutagenesis of the WRN cDNA was performed on the pCMV-FLAGWRN2 plasmid that contains the wt ORF sequence of WRN. The indicated S/T>A mutations were introduced sequentially by the Quick-change XL kit (Stratagene) using mutagenic primer pairs designed according to the manufacturer's directions. Each mutated plasmid was verified by full sequencing of the WRN ORF. For the in vitro kinase assays, the 940–1432 C-terminal fragment of WRN (CWRN) was amplified by PCR using the pCMV-FLAGWRN2 or its derivatives containing the indicated S/T>A mutations as template. The resulting C-terminal fragment (wt, M1, M2, M3, M4, M5 or M6) was cloned into the pGEX4T-1 vector (Stratagene).
Production of recombinant CWRN fragments and in vitro immunocomplex kinase assay
Expression of the wt and the mutant GST-CWRN fragments was induced in the BL21 pLys bacterial strain by adding 1 mM IPTG for 2 h at 37°C. The different GST-CWRN fragments were affinity purified using GSH-magnetic beads (Promega) and eluted using 30 mM glutathione in 50 mM Tris/Cl pH 8. For immunocomplex kinase assay, wt ATR (ATRwt) or its kinase-dead form (ATRkd) was immunopurified from HeLa cells transiently transfected with the pFLAG-ATRwt or pFLAG-ATRkd constructs (kindly provided by Dr Cimprich, Stanford University, Stanford, USA) using anti-Flag-conjugated-magnetic beads (Dynal). Similarly, FLAG-ATM was immunopurified from HEK293T cells transiently transfected with the pcDNA3.1-Flag-His-ATM plasmid (kindly provided by Dr Kastan, St Jude Children's Research Hospital, Memphis, USA). Immunopurified ATRwt, ATRkd or ATM was incubated with 200 ng of the indicated GST-CWRN fragment for 30 min at 37°C in the presence of 10 μCi γ32P-ATP and 5 μM ATP. After the incubation, the GST-CWRN fragments were separated from the Flag-ATR beads and phosphorylation levels assessed by SDS–PAGE followed by Coomassie staining and densitometric analysis by phosphorimaging.
Treatments, RNAi and transfection
HU, CPT and etoposide (Sigma-Aldrich) were added to culture medium at the indicated concentrations from stock solutions prepared in PBS (HU) or DMSO (CPT and etoposide) to induce DNA replication arrest or DNA damage. To inhibit ATM activity, we used the KU55933 compound (Calbiochem) at a 10 μM concentration.
ATR or RAD51 expression was knocked down by transfection with siRNAs directed against the coding sequence of the gene (SmartPool; Thermo Fisher Scientific). Transfection was performed using Interferin (Polyplus transfection) at 10 nM, according to the manufacturer's instruction. Protein down-regulation was confirmed by western blotting 48 h after transfection. GFP-targeting siRNAs (Thermo Fisher Scientific) were used as a control.
DNA expression plasmids were transfected using DreamFect (OZ Bioscience) according to the manufacture's instruction. Protein expression levels were analysed by western blotting 48 h post-transfection.
Clonogenic survival assay
For clonogenic survival, cells were treated with HU or CPT for 16 h at the indicated doses. The next day, cells were re-plated at low density for colony formation. After 14–21 days, colonies were fixed in 70% ethanol and stained with 1 mg/ml of crystal violet in water.
The number of colonies for each experimental point was expressed as a percentage of the untreated control.
Immunofluorescence
Immunofluorescence was performed as described (Franchitto et al, 2008). For WRN analysis, cells were subjected to a modified in situ fractionation protocol (Franchitto and Pichierri, 2004) before fixing in 4% PFA/PBS. Images were acquired as greyscale files using Metaview software (MDS Analytical Technologies) and processed using Adobe Photoshop CS3 (Adobe). For each time point, at least 200 nuclei were examined by two independent investigators and foci were scored at × 60. Only nuclei showing more than five bright foci were counted as positive and co-localisation scored as positive if >70% of the WRN and RPA foci overlapped.
Immunoprecipitation and western blotting
For IP experiments, 2.5 × 106 cells were used. Lysates were prepared using Co-IP buffer (1% Triton X-100, 0.5% Na-dehoxycolate, 150 mM NaCl, 2.5 mM MgCl2, 1 mM EGTA, 1 mM EDTA, 20 mM Tris/Cl pH 8) supplemented with phosphatase and protease inhibitors. One-milligram of lysate was incubated overnight at 4°C with 20 μl of Dynabeads M-280- Tosylactivated (Invitrogen) conjugated with 5 μg of anti-Flag or 5 μg of anti-WRN antibody according to the manufacturer. After extensive washing in RIPA buffer, proteins were eluted in 2 × electrophoresis buffer and subjected to SDS–PAGE and western blotting. For the analysis of whole protein content, cells were washed with PBS and lysed in standard RIPA buffer. Twenty micrograms of total protein were resolved by SDS–PAGE and transferred to nitrocellulose (PROTRAN, Schleicher and Schuell). Where indicated, lysates were incubated in the presence or absence of 400U λ-phosphatase (NEB) for 1 h at 30°C. Detection was carried using ECL plus according to the manufacturer's instructions (Amersham) and, where needed, protein amount evaluated by densitometry after normalisation using the PCNA or tubulin signal. The following antibodies were used: rabbit anti-WRN (Santa Cruz Biotechnologies), mouse anti-FLAG (Sigma-Aldrich), rabbit anti-pS/TQ (Cell Signaling Technologies), mouse anti-RAD51 (GenTex), rabbit anti-ATR (Calbiochem), mouse anti-pS1981ATM (Rockland), mouse anti-PCNA (Santa Cruz Biotechnology) and mouse anti-β-tubulin (Sigma-Aldrich).
Cell cycle analysis by flow cytometry
Cells were processed for flow cytometry as described (Franchitto et al, 2008) and data analysed with CellQuest software.
DNA fibre assay
The efficiency of replication recovery was measured using the DNA fibre assay. First, DNA replication sites were labelled with 25 μM IdU (30 min) and then cells were treated with 2 mM HU for 6 or 16 h to induce replication fork stalling. After treatment, cells were washed twice with PBS and incubated for 60 min in fresh medium with 100 μM CldU. DNA fibres were prepared and analysed as described (Hanada et al, 2007; Franchitto et al, 2008).
Comet assay
DNA breakage induction was evaluated by Comet assay (single cell gel electrophoresis) in non-denaturing conditions as described (Olive et al, 1991; Pichierri et al, 2001). A minimum of 200 cells was analysed for each experimental point. Apoptotic cells (smaller comet head and extremely larger comet tail) were excluded from the analysis to avoid artificial enhancement of the tail moment.
LIVE/DEAD staining
Cell viability was evaluated by the LIVE/DEAD assay (Sigma-Aldrich) according to the manufacturer. Cell number was counted in randomly chosen fields and expressed as per cent of dead cells (number of red nuclear stained cells/total cell number). For each time point, at least 200 cells were counted.
Supplementary Material
Acknowledgments
We are grateful to Dr Karlene Cimprich for providing the Flag-ATR wt and kinase-dead expression constructs and Dr Michael Kastan for providing the Flag-ATM expression construct. We thank Dr Orazio Sapora for assistance in flow cytometry experiments and helpful discussion. This work was supported by Fondazione Telethon to PP (Grant no. GGP04094) and, in part, by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC) to PP, AF and MB.
Authors' contribution: FA, LMP, performed experiments and analysed data; AF and PP designed experiments, analysed data and wrote the paper; MB discussed data and gave conceptual advice.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M (2000) ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene 19: 5955–5963 [DOI] [PubMed] [Google Scholar]
- Arlt MF, Durkin SG, Ragland RL, Glover TW (2006) Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst) 5: 1126–1135 [DOI] [PubMed] [Google Scholar]
- Arnaudeau C, Lundin C, Helleday T (2001) DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol 307: 1235–1245 [DOI] [PubMed] [Google Scholar]
- Beamish H, Kedar P, Kaneko H, Chen P, Fukao T, Peng C, Beresten S, Gueven N, Purdie D, Lees-Miller S, Ellis N, Kondo N, Lavin MF (2002) Functional link between BLM defective in Bloom's syndrome and the ataxia-telangiectasia-mutated protein, ATM. J Biol Chem 277: 30515–30523 [DOI] [PubMed] [Google Scholar]
- Bohr VA (2008) Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci 33: 609–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh RM Jr, Waheed J, Sommers JA (2002) Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J Biol Chem 277: 23236–23245 [DOI] [PubMed] [Google Scholar]
- Casper AM, Nghiem P, Arlt MF, Glover TW (2002) ATR regulates fragile site stability. Cell 111: 779–789 [DOI] [PubMed] [Google Scholar]
- Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ (2009) ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem 284: 5994–6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary S, Sommers JA, Brosh RM Jr (2004) Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of Werner syndrome protein. J Biol Chem 279: 34603–34613 [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D (2008) ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol 9: 616–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH (1998) Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J 17: 159–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC (2000) Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep 1: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez D (2003) Caffeine inhibits checkpoint responses without inhibiting the ataxia-telangiectasia-mutated (ATM) and ATM- and Rad3-related (ATR) protein kinases. J Biol Chem 278: 37139–37145 [DOI] [PubMed] [Google Scholar]
- Davies SL, North PS, Dart A, Lakin ND, Hickson ID (2004) Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol Cell Biol 24: 1279–1291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franchitto A, Pichierri P (2004) Werner syndrome protein and the MRE11 complex are involved in a common pathway of replication fork recovery. Cell Cycle 3: 1331–1339 [DOI] [PubMed] [Google Scholar]
- Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P (2008) Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J Cell Biol 183: 241–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedel AM, Pike BL, Gasser SM (2009) ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol 21: 237–244 [DOI] [PubMed] [Google Scholar]
- Glover TW, Arlt MF, Casper AM, Durkin SG (2005) Mechanisms of common fragile site instability. Hum Mol Genet 14 (Spec No. 2): R197–R205 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J (2008) An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355 [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R (2007) The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104 [DOI] [PubMed] [Google Scholar]
- Kim ST, Lim DS, Canman CE, Kastan MB (1999) Substrate specificities and identification of putative substrates of ATM kinase family members. J Biol Chem 274: 37538–37543 [DOI] [PubMed] [Google Scholar]
- Liu Q, Guntuku S, Cui XS, Matsuoka S, Cortez D, Tamai K, Luo G, Carattini-Rivera S, DeMayo F, Bradley A, Donehower LA, Elledge SJ (2000) Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev 14: 1448–1459 [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK (2006) The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45: 13939–13946 [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER III, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y, Shiloh Y, Gygi SP, Elledge SJ (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316: 1160–1166 [DOI] [PubMed] [Google Scholar]
- Merrick CJ, Jackson D, Diffley JF (2004) Visualization of altered replication dynamics after DNA damage in human cells. J Biol Chem 279: 20067–20075 [DOI] [PubMed] [Google Scholar]
- Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S (2000) The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J 19: 463–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muftuoglu M, Oshima J, von Kobbe C, Cheng WH, Leistritz DF, Bohr VA (2008) The clinical characteristics of Werner syndrome: molecular and biochemical diagnosis. Hum Genet 124: 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD (2001) Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell 104: 397–408 [DOI] [PubMed] [Google Scholar]
- Olive PL, Wlodek D, Banath JP (1991) DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res 51: 4671–4676 [PubMed] [Google Scholar]
- Otterlei M, Bruheim P, Ahn B, Bussen W, Karmakar P, Baynton K, Bohr VA (2006) Werner syndrome protein participates in a complex with RAD51, RAD54, RAD54B and ATR in response to ICL-induced replication arrest. J Cell Sci 119: 5137–5146 [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T (2010) Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P (2007) Interplay between wrn and the checkpoint in s-phase. Ital J Biochem 56: 130–140 [PubMed] [Google Scholar]
- Pichierri P, Franchitto A, Mosesso P, Palitti F (2001) Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol Biol Cell 12: 2412–2421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichierri P, Rosselli F, Franchitto A (2003) Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene 22: 1491–1500 [DOI] [PubMed] [Google Scholar]
- Pirzio LM, Pichierri P, Bignami M, Franchitto A (2008) Werner syndrome helicase activity is essential in maintaining fragile site stability. J Cell Biol 180: 305–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Gollahon KA, Rabinovitch PS (1999) Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum Genet 104: 10–14 [DOI] [PubMed] [Google Scholar]
- Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS (2001) Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J 15: 1224–1226 [DOI] [PubMed] [Google Scholar]
- Rao VA, Fan AM, Meng L, Doe CF, North PS, Hickson ID, Pommier Y (2005) Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage. Mol Cell Biol 25: 8925–8937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS (2003) Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech Ageing Dev 124: 167–174 [DOI] [PubMed] [Google Scholar]
- Sidorova JM (2008) Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 7: 1776–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorova JM, Li N, Folch A, Monnat RJ Jr (2008) The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7: 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T (2005) The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat Cell Biol 7: 195–201 [DOI] [PubMed] [Google Scholar]
- Trenz K, Smith E, Smith S, Costanzo V (2006) ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 25: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Kaur S, Sengupta S (2008) Phosphorylation-dependent interactions of BLM and 53BP1 are required for their anti-recombinogenic roles during homologous recombination. Carcinogenesis 29: 52–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C, Thoma NH, Czyzewski BK, Pavletich NP, Bohr VA (2003) Werner syndrome protein contains three structure-specific DNA binding domains. J Biol Chem 278: 52997–53006 [DOI] [PubMed] [Google Scholar]
- Yuan SS, Chang HL, Lee EY (2003) Ionizing radiation-induced Rad51 nuclear focus formation is cell cycle-regulated and defective in both ATM(−/−) and c-Abl(−/−) cells. Mutat Res 525: 85–92 [DOI] [PubMed] [Google Scholar]
- Zachos G, Rainey MD, Gillespie DA (2005) Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Mol Cell Biol 25: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.