A polarity factor takes the lead in chromosome segregation
Several recent studies shed light on how bacteria achieve rapid and accurate chromosome segregation through an interplay of Par-type partitioning systems, cytokinesis regulators and a polarity determinant.
EMBO J 29 18, 3068–3081 (2010); published online August272010
Little is known about how rapid and accurate chromosome segregation, a vital step that must be completed before cell division, is achieved in bacterial cells. Several studies, including one by Schofield et al in this issue of The EMBO Journal, now shed light on this process by elucidating novel interactions between the chromosome partitioning system ParAB-parS, the polarity determinant TipN and the cytokinesis regulators FtsZ/MipZ, exploring also the underlying biochemistry and the committing event for the poleward motion during segregation.
Early models viewed bacterial cells as simple ‘bags' of proteins and DNA, in which all biological processes were mediated by diffusion. However, the observation that the Escherichia coli chromosome was 700–900 μm in length (Cairns, 1963), with the cell itself only measuring 2–3 μm (Cullum and Vicente, 1978), suggested a requirement for active mechanisms to unpack, replicate, segregate and condense the chromosome. Subsequent investigations demonstrated that homologs of the Par type 1 system, best studied in the context of E. coli plasmid partitioning, were involved in chromosome segregation in other bacteria (Gerdes et al, 2010). This system possesses three components, the ParA ATPase, the ParB site-specific DNA-binding protein and the chromosomal parS site, located near the origin of replication (ori), which is bound by ParB. ParA provides or regulates the motor force that drives the parS/ParB complex of the newly replicated chromosome to the new pole, although until now the mechanism of force generation has not been well understood. In the model organism Caulobacter crescentus (hereafter referred to as Caulobacter), a mechanistic link between the Par system and regulation of cell division was discovered by Thanbichler and Shapiro (2006): The ParA-like ATPase MipZ inhibits polymerization of the FtsZ cytokinetic tubulin homolog into the Z-ring and binds to newly replicated parS/ParB complexes, which become rapidly segregated to opposite poles. This leads to a gradient of MipZ, peaking at the poles and dipping where the Z-ring forms.
Caulobacter division is asymmetric and influenced by polarity determinants, producing daughters that differ in size and polar features: a smaller motile dispersal (swarmer) cell possessing a polar flagellum and pili, and a larger reproductive (stalked) cell with a polar stalk that carries an adhesive holdfast at its tip. The stalked end always gives rise to a new stalked cell, and the other end, closer to the division plane, always spawns a swarmer cell. Polarity is thus hardwired into Caulobacter division, and the correct interpretation of polarity cues relies on landmarks such as the coiled-coil protein TipN: it is first sequestered to the newborn pole where the flagellum is built, and absent from the old pole where the stalk elaborates. This polarity axis of TipN allows downstream effectors, like flagellar assembly factors, to localize to the correct pole (Huitema et al, 2006; Lam et al, 2006). In cells lacking TipN (TipN−), flagellar structural proteins and regulators frequently mislocalize (Huitema et al, 2006; Lam et al, 2006) and flagella assemble at erroneous cellular sites (Huitema et al, 2006; Lam et al, 2006). In addition, the bias of the division plane is reversed (towards the stalked pole), resulting in a smaller stalked and a larger swarmer daughter cell (Lam et al, 2006). This unexplored TipN− phenotype provided the starting point for the current study by Schofield et al (2010) in this issue.
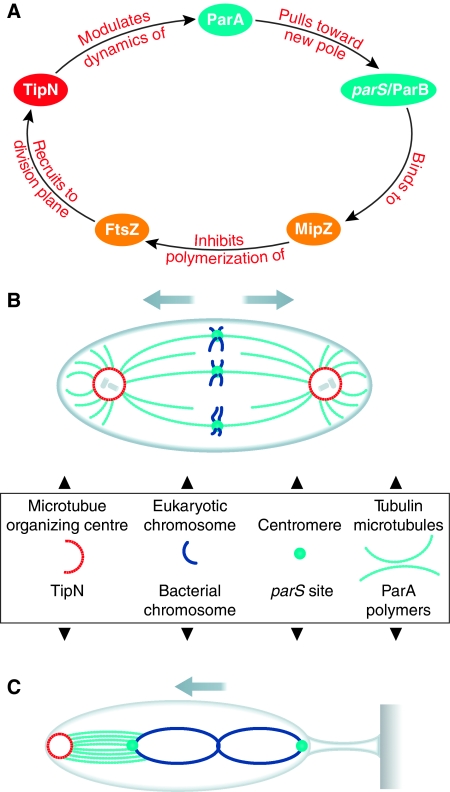
First, real-time kymographs revealed that the timing of FtsZ and MipZ dynamics was altered in the TipN− mutant. The Z-ring formed much later during the cell cycle and at an aberrant position compared with wild-type (WT) cells. Moreover, although MipZ travels to the newborn (TipN marked) pole in a rapid, continuous and unidirectional manner in WT, TipN− cells exhibited slow, intermittent and erratic movement of MipZ. Considering that MipZ co-migrates with the parS/ParB complex (Thanbichler and Shapiro, 2006), and that ParA has been implicated in energizing or regulating the translocation of the parS locus, the authors examined the localization of ParA. ParA formed a ‘cloud' over the nucleoid, which promptly retracted towards the newborn pole. In the TipN− mutant, this retraction was compromised, indicating that TipN influences the dynamics of ParA. Subsequent pull-down and FRET experiments corroborated the hypothesis that TipN and ParA interact at the pole. These data yield a model in which TipN at the new pole binds and sequesters ParA when it is released from the DNA-bound ‘cloud', thus preventing ParA from returning behind the parS/ParB/MipZ ternary complex and from pulling the ori back towards the old pole. In the absence of TipN, this function is lost, resulting in an erratic and delayed translocation of MipZ that correlates with the delayed and mispositioned Z-ring and the apparent stalked-pole proximal constriction bias. Intriguingly, this dependency also unearths an underlying feedback loop (Figure 1A) in which polar TipN regulates the correct positioning of the Z-ring via MipZ, whereas the incorporation of TipN at the new poles during division is mediated by FtsZ (or interactions with the downstream factors FtsN, TolA and/or Pal (Moll et al, 2010; Yeh et al, 2010)).
Figure 1.
(A) Feedback loop mediated by TipN through the chromosome segregation machinery (the Par system) and cytokinesis regulators (MipZ and FtsZ). (B) Schematic diagram of chromosome segregation in a eukaryotic cell. (C) Schematic diagram of chromosome segregation in a Caulobacter cell. Arrows indicate the direction of chromosome movement.
How ParA energizes the translocation of the parS locus to the new pole was investigated by Ptacin et al (2010), who used super-resolution single molecule fluorescence microscopy in live Caulobacter to resolve the ParA ‘cloud' to linear filaments of polymerized ParA. In vitro, polymerization of ParA occurs upon non-specific binding of DNA, with further addition of ParB inducing depolymerization, presumably reflecting the retraction of the parS/ParB complex to the new pole in vivo. Importantly, these authors showed that the C-terminal domain of TipN directly interacts with ParA, accounting for the observed TipN-dependent poleward retraction of ParA in vivo. Taken together, this suggests striking parallels to eukaryotic chromosome segregation, with ParA proposed to act in a similar way to spindle microtubules that depolymerize to pull chromosomes into the two daughter cells, and TipN serving a function analogous to that of the spindle-organizing centrosome, promoting poleward movement through polymer retraction (Figure 1B and C).
In a third recent study, Shebelut et al (2010) used real-time imaging at high temporal resolution to delineate the order of events during translocation, showing parS segregation locus to be a multistep process comprising four major stages: (1) polar release, (2) polar retraction, (3) early translocation from pole to midcell and (4) late translocation from midcell to pole. Importantly, the Par system was only required for late translocation, which exhibited significantly higher velocity than early translocation. Steps 2 and 3 appear to occur by a non-specific bulk separation mechanism(s), with this initial separation sufficing to enable the Par system to recognize and rapidly translocate the ori destined for the new pole.
Together, these articles demonstrate that prokaryotic chromosome segregation in Caulobacter involves spatio-temporally concerted action of a polarity determinant and a dedicated segregation machinery, at a level of complexity resembling that of eukaryotes.
Footnotes
The authors declare that they have no conflict of interest.
References
- Cairns J (1963) The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol 6: 208–213 [DOI] [PubMed] [Google Scholar]
- Cullum J, Vicente M (1978) Cell growth and length distribution in Escherichia coli. J Bacteriol 134: 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Howard M, Szardenings F (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141: 927–942 [DOI] [PubMed] [Google Scholar]
- Huitema E, Pritchard S, Matteson D, Radhakrishnan SK, Viollier PH (2006) Bacterial birth scar proteins mark future flagellum assembly site. Cell 124: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Lam H, Schofield WB, Jacobs-Wagner C (2006) A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124: 1011–1023 [DOI] [PubMed] [Google Scholar]
- Moll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M (2010) DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol Microbiol 77: 90–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacin JL, Lee SF, Garner EC, Toro E, Eckart M, Comolli LR, Moerner WE, Shapiro L (2010) A spindle-like apparatus guides bacterial chromosome segregation. Nat Cell Biol 12: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield WB, Lim HC, Jacobs-Wagner C (2010) Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J 29: 3068–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shebelut CW, Guberman JM, van Teeffelen S, Yakhnina AA, Gitai Z (2010) Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci USA 107: 14194–14198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Yeh Y-C, Comolli LR, Downing KH, Shapiro L, McAdams H (2010) The Caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol (in press; doi:10.1128/JB.00607-10) [DOI] [PMC free article] [PubMed] [Google Scholar]