Abstract
Background
The phytopathogenic fungus Diplodia seriata, whose genome remains unsequenced, produces severe infections in fruit trees (fruit blight) and grapevines. In this crop is recognized as one of the most prominent pathogens involved in grapevine trunk disease (or grapevine decline). This pathology can result in the death of adult plants and therefore it produces severe economical losses all around the world. To date no genes or proteins have been characterized in D. seriata that are involved in the pathogenicity process. In an effort to help identify potential gene products associated with pathogenicity and to gain a better understanding of the biology of D. seriata, we initiated a proteome-level study of the fungal mycelia and secretome.
Results
Intracellular and secreted proteins from D. seriata collected from liquid cultures were separated using two-dimensional gel electrophoresis. About 550 cytoplasmic proteins were reproducibly present in 3 independent extractions, being 53 identified by peptide mass fingerprinting and tandem mass spectrometry. The secretome analysis showed 75 secreted proteins reproducibly present in 3 biological replicates, being 16 identified. Several of the proteins had been previously identified as virulence factors in other fungal strains, although their contribution to pathogenicity in D. seriata remained to be analyzed. When D. seriata was grown in a medium supplemented with carboxymethylcellulose, 3 proteins were up-regulated and 30 down-regulated. Within the up-regulated proteins, two were identified as alcohol dehydrogenase and mitochondrial peroxyrredoxin-1, suggesting that they could play a significant role in the pathogenicity process. As for the 30 down-regulated proteins, 9 were identified being several of them involved in carbohydrate metabolism.
Conclusions
This study is the first report on proteomics on D. seriata. The proteomic data obtained will be important to understand the pathogenicity process. In fact, several of the identified proteins have been reported as pathogenicity factors in other phytopathogenic fungi. Moreover, this proteomic analysis supposes a useful basis for deepening into D. seriata knowledge and will contribute to the development of the molecular biology of this fungal strain as it has been demonstrated by cloning the gene Prx1 encoding mitochondrial peroxiredoxin-1 of D. seriata (the first gene to be cloned in this microorganism; data not shown).
Background
The fungal family Botryosphaeriaceae (Botryosphaeriales, Ascomycetes) is a complex family that, according to a recent broad-based phylogenetic study, includes ten genera [1]. The majority of these genera comprise members described as fungal endophytes, present in virtually all woody host examined (both gymnosperm and angiosperm species) [1,2], and being the dominating endophyte communities in Eucalyptus and Pinus in some environments [3]. One of the most important members of this family is Diplodia (which includes teleomorphs distributed in several genera). Diplodia seriata (before named as Botryosphaeria obtusa) is an important phytopathogenic fungus that has been isolated from at least 34 different hosts, causing severe infections in fruit trees (mainly pomes and stone fruits) [4]. Also recently, D. seriata has been described as an important pathogen of grapevines (Vitis vinifera), involved in grapevine trunk disease causing the death of adult plants [5]. Grapevine decline, or grapevine trunk disease, is currently recognized as the main threat to vineyards all around the world since no effective treatment to fight this fungal pathology is presently available [6,7]. D. seriata can be isolated from grapevines exhibiting dieback of shoots, spurs and branches, as well as severe internal wood necrosis symptoms [8]. It is also a cosmopolitan grapevine pathogen which has been detected all around the world [9]. Its contribution to grapevine decline is of major importance since it has been the most frequently isolated pathogen in an extensive survey on affected grapevine plants carried out in Spain. In fact, 63% of the total Botryosphaeria-like species isolated from young plants and 28% of the pathogenic isolates from adult plants were identified as D. seriata, which represent around 36% of the total isolated pathogen fungi associated to grapevine decline [10]. Despite the importance of D. seriata as phytopathogen, little is known about its biological life cycle, its basic molecular aspects and the mechanisms involved in the pathogenicity of this fungal species. Botryosphaeriaceae species are known to produce phytotoxic compounds like exopolysaccharides and other toxic organic compounds which induce phytotoxic effects on plants [11,12], but unfortunately no D. seriata pathogenicity-related genes has been deposited yet in databases.
Proteomic studies are one of the most powerful methods to evaluate the final result of gene expression. In the present case two-dimensional gel electrophoresis (2-DE) has been the technique of choice to obtain a referential global picture of the cytoplasmic and extracellular protein maps. Up to now, this technique has been successfully applied to analyze not only the proteome and/or secretome of many different fungal strains [13,14], but also plant-pathogen interactions, including analyses of several phytopathogenic fungi [15-17]. Regarding phytopathogenicity, the phylum Ascomycota comprises highly appreciated species in food and pharmaceutical industries, but also others dreaded as phytopathogens due to their secreted proteins [18]. However, the understanding of the molecular basis of the secretion process in these microorganisms is still very limited. As reviewed above, a considerable number of proteomic analyses have been carried out in fungal species in the last years, mainly concerning strains of medical or agricultural interest. Nevertheless, to our knowledge, no proteomic data have been reported yet for D. seriata.
Thus, the main goal of this manuscript is the achievement of an initial approach to define the D. seriata proteome and secretome. Protocols for protein extraction from mycelium, protein separation by 2-DE and mass spectrometry (MS) analyses have been optimized. Furthermore, proteome differences have also been tested by growing the strain in the presence of carboxymethylcellulose (CMC). Since cellulose is one of the major components of the plant cell wall, proteins overexpressed during growth on CMC could be expected to play a significant role in the pathogenicity process. In fact, this strategy has been used to identify putative pathogenicity factors in the phytopathogenic fungus Botrytis cinerea [19,20]. We decided to try this strategy since D. seriata is an endophytic fungi that exerts its pathogenicity by developing inside the vascular system of grapevine, and unfortunately to date no experimental approach is available that mimics the natural conditions in which D. seriata is developing its pathogenicity. In this way, this manuscript contributes to the knowledge of biological basic aspects of this important phytopathogenic fungus, and, therefore, means a first step in the development of the molecular biology of this fungal strain. In fact, the gene encoding mithocondrial peroxyrredoxin-1 was cloned, sequenced and characterized (data not shown) based on the amino acid sequence of several internal peptides identified by tandem mass spectrometry (MS/MS).
Results and discussion
Analysis of fungal growth in the experimental conditions tested
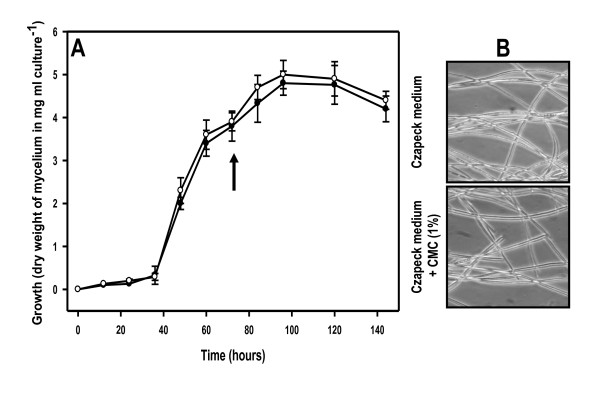
Fig. 1A shows the growth curves of D. seriata in liquid cultures developed in Czapeck medium and the same medium supplemented with CMC (1%). They were obtained from experiments carried out in the same experimental conditions to perform the proteome analysis. As it can be seen in Fig. 1, both curves are very similar, which obviously indicated that CMC did not have any significant effect over the growth rates. These data clearly indicated that most CMC could not be assimilated by D. seriata, as occurs in many others fungi. As it can be deduced from the graphic, the fungal growth in both experimental conditions increased rapidly beyond 36 h, reaching their maximum around 96 h. Samples for proteome analysis were taken at 72 h of growth corresponding approximately to the middle of the exponential phase of growth. The microscopic observation of mycelia samples collected at that time showed (Fig. 1B) a normal mycelium structure with no symptoms of cellular lysis.
Figure 1.
(A).- Growth curve of D. seriata liquid cultures performed in Czapeck liquid medium (●) and the same medium supplemented with CMC (1%) (○). The sample collection points (72 h for every culture) are indicated by an arrow. (B).- Microscopic observation (400 ×) of 72 h old mycelia samples (corresponding to the sample collection points) showing that the mycelium exhibited a normal structure and integrity (no cellular lysis symptoms were observed at this time).
Visualization and analysis of the secreted proteins
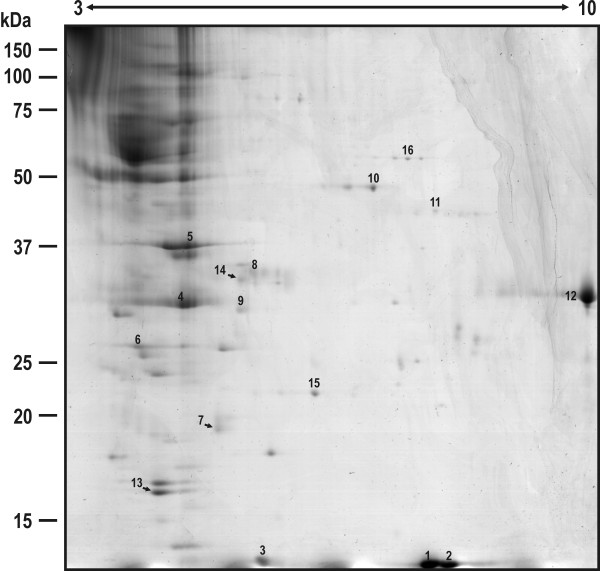
The secreted proteins from D. seriata analyzed by 2-DE are showed in Fig. 2. All the secreted proteins that were subsequently identified by PMF and MS/MS analyses are numbered and listed in Table 1. Seventy-five extracellular proteins were reproducibly visualized in three independent experiments and sixteen could be identified by MS/MS. This low proportion of identified secreted proteins against visualized secreted proteins is not uncommon when the proteome of fungal species is analyzed, specially as in the case of D. seriata the genome remains unsequenced, which obviously makes difficult the identification of many proteins. In fact, when the extracellular proteomes of other fungal strains such as Aspergillus flavus [21], Trichoderma reesei [22], Trichoderma harzianum [23], or the phytopathogen Sclerotinia sclerotiorum [24] have been analyzed, a low ratio of identified proteins was also obtained. In the case of D. seriata, this fact could reflect that many of the non-identified secreted proteins are specific to this fungal species, or perhaps specific to endophytic fungi. The lack of proteomic analyses and of a significant number of characterized genes in endophytic fungi undoubtedly represents a limiting factor in the identification of proteins in these phytopathogenic fungi. In fact, up to our knowledge, no data regarding extense proteome and/or secretome analyses of other endophytic fungi are available (with the exception of a proteome analysis of Neotyphodium lolii, an endophytic fungus from the perennial ryegrass Lolium perenne) [25] and, in particular, there are no available data of other endophytic ascomycetes taxonomically close to Botryosphaeriales.
Figure 2.
2-DE of CBB-stained 12.5% SDS-PAGE of the secreted proteins of D. seriata.
Table 1.
Summary of the proteins secreted by D. seriata.
| Spot | Acc. number | Protein identitya | PM/Seq cov./scoreb | Best peptides (PRO-BLAST) | Identified by | Experimental MW/pI | Theoretical MW/pI | Organism | Signal P |
|---|---|---|---|---|---|---|---|---|---|
| S1 | gi|256724272 | Hypothetical protein | 2/20%/83 | MASCOT | 13.2/7.6 | 13.5/9.6 | Nectria haematococca | NNc | |
| S2 | gi|255935271 | Pc13g02210 | 2/14%/79 | KIIYPAYTDK ARIKGDVVKPDKSYAPKAIP |
PRO-BLAST | 13.2/7.8 | 14.5/3.7 | Penicillium chrysogenum | 0.999 |
| S3 | gi|256724272 | Hypothetical protein | 2/20%/82 | MASCOT | 13.4/5.6 | 13.5/9.6 | Nectria haematococca | NN | |
| S4 | gi|27924024 | glucosidase | 4/11%/97 | GFNIGATNADGSC*Kd EGGHVSISAVSRSEDLYRGDSDASK |
PRO-BLAST | 30.2/4.7 | 33.4/9.3 | Fusarium sporotrichioides | 0.999 |
| S5 | gi|256732553 | Predicted protein (necrosis and ethylene inducing peptide 2 precursor) | 2/7%/97 | GELHNDAFALFYAWYFAPG AVLAHDAVVGFAETVPFVGVWHAHK |
PRO-BLAST | 35.7/4.7 | 24.69/7.75 | Nectria haematococca | 0.999 |
| S6 | gi|239931549 | Secreted protein | 2/9%/105 | SGASSWTAAR SSSFPHWLTLSGSC*NTR |
PRO-BLAST | 26.3/4.2 | 23.03/8.35 | Streptomyces ghanaensis | 0.989 |
| S7 | gi|189192036 | Hypothetical protein | 3/18%/94 | MASCOT | 19.4/5.1 | 22.5/7.6 | Pyrenophora tritici-repentis | 0.999 | |
| S8 | gi|256726602 | Hypothetical protein (necrosis-and ethylene-inducing protein 1) | 3/8%/77 | FGLFYAWYFPK ALSPHDSLTKDSYVR |
PRO-BLAST | 33.8/5.5 | 26.99/9.2 | Nectria haematococca | 1.000 |
| S9 | gi|156061869 | Hypothetical protein (necrosis-and ethylene-inducing protein 1) | 6/21%/77 | MASCOT | 26.0/5.2 | 26.9/6.6 | Sclerotinia sclerotiorum | 0.994 | |
| S10 | gi|189196154 | NADP-specific glutamate DH | 9/31%/302 | MASCOT | 48.0/6.9 | 49.1/5.9 | Pyrenophora tritici-repentis | No | |
| S11 | gi|171677093 | Unnamed protein product (aspartate aminotransferase) | 5/19%/80 | MASCOT | 43.4/7.6 | 43.0/8.3 | Podospora anserina | No | |
| S12 | gi|237868680 | Peptidase 1 | 4/8%/109 | KTSGNFIASVASMSLGFESR KC*FPTEEDRRSGVDVYVVDTGLYTAHSEF |
PRO-BLAST | 30.9/9.4 | 38.7/7.9 | Pleurotus ostreatus | 1.000 |
| S13 | gi|255946596 | Pc22g00190 (cell wall protein PhiA) | 2/9%/81 | KGIGYTTGAEPTPK ASTPKKLYVDR |
PRO-BLAST | 16.2/4.4 | 18.9/5.6 | Penicillium chrysogenum | 0.996 |
| S14 | gi|182412796 | Hypothetical protein | 2/2%/81 | FGHWSPNASLATAAK YGPLSFVKGK |
PRO-BLAST | 33.1/5.4 | 74.5/6.8 | Opitutus terrae | 0.990 |
| S15 | gi|261355012 | Superoxide dismutase | 5/18%/111 | GHGLYHDHSLFWENLAC*NLWPK SYIPPLPYAYDAIEPAISR |
PRO-BLAST | 22.0/6.3 | 26.8/10.3 | Verticillium albo-atrum | No |
| S16 | gi|189196326 | Glutathione reductase | 2/4%/90 | SFDPIIKDTITKEYER VAPHYLVLNGSGGLASGR |
PRO-BLAST | 58.1/7.3 | 55.8/6.1 | Pyrenophora tritici-repentis | No |
a The assignment of the biological or enzymatic function to the hypothetical proteins (based on BlastP analyses) is shown between parentheses and in italic characters.
b Number of peptides matched/Sequence coverage (%)/Score.
c NN: Non-classically secreted proteins.
d C*: Cys as S carbamidomethyl derivative
A clear biological or enzymatic function could be solely attributed to ten out of the sixteen secreted proteins annotated (spots S4, S5, S8, S9, S10, S11, S12, S13, S15, and S16), whereas the rest (spots S1-S3, S6, S7, and S14) were annotated as hypothetical, putative uncharacterized protein or secreted proteins. In order to achieve a better identification of these proteins, their biological or enzymatic functions were determined. For this, the amino acid sequence of the hypothetical protein was used to perform a BlastP search against the non-redundant protein sequence database at NCBI (in these cases the putative biological functions are indicated between parentheses and in italics in Tables 1 and 2). In this way, spots S8 and S9 were determined to correspond to a necrosis and ethylene-inducing protein 1 (Nep1), whereas the spot 5 was determined to correspond to a necrosis and ethylene-inducing protein 2 precursor (Nep2). By the same approach, S11 was identified as an aspartate aminotransferase and S13 as a protein similar to the cell wall protein PhiA.
Table 2.
List of cytoplamic proteins of D. seriata.
| Spot | Acc. number | Protein identitya | PM/Seq cov./scoreb | Identified by | Total Ion scorec | Experimental MW/pI | Theoretical MW/pI | Organism |
|---|---|---|---|---|---|---|---|---|
| 1 | gi|238498522 | Phosphoglycerate kinase | 12/30%/251 | MASCOT | 44.0/8.3 | 44.4/7.7 | Aspergillus flavus | |
| 2 | gi|67903236 | Hypothetical protein (peptidyl-prolyl cis-trans isomerase or cyclophilin 1) | 5/47%/219 | MASCOT | 18.2/8.5 | 17.8/8.9 | Aspergillus nidulans | |
| 3 | gi|169601412 | Hypothetical protein (enolase) | 11/30%/595 | MASCOT | 48.1/6.2 | 47.4/5.2 | Phaeosphaeria nodorum | |
| 4 | gi|198425409 | Predicted Zn-finger U1-like | 12/54%/88 | MASCOT | 12.3/4.9 | 22.6/9.5 | Ciona intestinalis | |
| 5 | gi|156053161 | Hypothetical protein (aldehyde reductase) | 2/5%/82 | MASCOT | 77 | 36.2/7.8 | 37.9/8.8 | Sclerotinia sclerotiorum |
| 6 | gi|169625443 | Hypothetical protein (triosephosphate isomerase) | 7/23%/257 | MASCOT | 25.0/6.9 | 27.1/6.1 | Phaeosphaeria nodorum | |
| 7 | gi|46581334 | Hypothetical protein | 6/23%/112 | MASCOT | 30.2/7.4 | 16.8/6.3 | Desulfovibrio vulgaris | |
| 8 | gi|189210148 | Signal transduction protein (CipC-like antibiotic response protein) | 3/19%/117 | MASCOT | 13.1/4.8 | 14.9/4.9 | Pyrenophora tritici-repentis | |
| 9 | gi|115400267 | Transaldolase | 6/18%/165 | MASCOT | 35.2/5.2 | 35.2/6.2 | Aspergillus terreus | |
| 10 | gi|85107722 | Hypothetical protein (phosphoglycerate kinase) | 10/22%/217 | MASCOT | 45.2/7.9 | 45.3/6.2 | Neurospora crassa | |
| 11 | gi|120691 | Glyceraldehyde-3-phosphate dehydrogenase | 8/23%/334 | MASCOT | 38.3/8.2 | 36.6/6.7 | Cochliobolus lunatus | |
| 12 | gi|189204622 | Fructose-bisphosphate aldolase | 4/9%/156 | MASCOT | 140 | 38.4/6.7 | 40.2/5.5 | Pyrenophora tritici-repentis |
| 13 | gi|189197705 | Glycolipid transfer protein | 5/25%/98 | MASCOT | 22.6/6.9 | 24.0/5.8 | Pyrenophora tritici-repentis | |
| 14 | gi|126139808 | Hypothetical protein (thiamine biosynthesis enzyme) | 5/20%/124 | MASCOT | 39.1/6.5 | 38.1/5.4 | Pichia stipitis | |
| 15 | gi|212542889 | Glycerol dehydrogenase Gcy1 putative | 4/17%/306 | MASCOT | 33.6/8.4 | 33.7/6.0 | Aspergillus fumigatus | |
| 16 | gi|67903236 | Hypothetical protein (peptidyl-prolyl cis-trans isomerase or cyclophilin 1) | 6/50%/214 | MASCOT | 18.5/7.9 | 17.8/8.9 | Aspergillus nidulans | |
| 17 | gi|70985178 | Thiamine biosynthesis protein | 8/25%/325 | MASCOT | 37.2/6.0 | 38.6/6.0 | Aspergillus fumigatus | |
| 18 | gi|255943905 | Malate dehydrogenase | 9/26%/471 | MASCOT | 34.6/7.9 | 35.8/8.4 | Penicillium chrysogenum | |
| 19 | gi|254583736 | Hypothetical protein (fumarate reductase) | 5/9%/161 | MASCOT | 144 | 56.9/6.7 | 54.0/8.6 | Zygosaccharomyces rouxii |
| 20 | gi|154285406 | Nucleoside-diphosphate kinase | 6/38%/234 | MASCOT | 13.3/9.4 | 16.9/7.8 | Ajellomyces capsulatus | |
| 21 | gi|164429080 | Adenosine kinase | 5/12%/252 | MASCOT | 236 | 38.1/5.3 | 49.4/6.0 | Neurospora crassa |
| 22 | gi|218722748 | Conserved hypothetical protein | 4/19%/157 | MASCOT | 51.4/4.9 | 42.8/5.8 | Talaromyces stipitatus | |
| 23 | gi|189196154 | NADP-specific glutamate dehydrogenase | 10/22%/504 | MASCOT | 46.4/7.5 | 49.1/5.9 | Pyrenophora tritici-repentis | |
| 24 | gi|58265982 | Eukaryotic translation initiation factor 5A-1 | 5/30%/391 | MASCOT | 20.0/4.9 | 17.9/5.3 | Cryptococcus neoformans | |
| 25 | gi|145613040 | Conserved hypothetical protein (mitochondrial peroxiredoxin PRX1) | 4/21%/306 | MASCOT | 25.4/6.6 | 25.2/5.7 | Magnaporthe grisea | |
| 26 | gi|225877962 | Putative saccharopine dehydrogenase | 4/11%/146 | MASCOT | 131 | 45.2/5.6 | 43.5/5.2 | Gibberella fujikuroi |
| 27 | gi|194669270 | Endonuclease reverse transcriptase | 23/24%/88 | MASCOT | 37.8/6.8 | 150.3/9.7 | Bos taurus | |
| 28 | gi|169625443 | Hypothetical protein (triosephosphate isomerase) | 5/19%/178 | MASCOT | 25.4/6.4 | 27.1/6.1 | Phaeosphaeria nodorum | |
| 29 | gi|255711238 | KLTH0B02596p | 9/70%/73 | MASCOT | 39.5/6.0 | 16.3/5.4 | Lachancea thermotolerans | |
| 30 | gi|154318339 | Hypothetical protein (Elongation factor 1β) | 2/20%/87 | PRO-BLASTd | 35.2/4.1 | 17.7/4.7 | Botryotinia fuckeliana | |
| 31 | gi|67903236 | Hypothetical protein (peptidyl-prolyl cis-trans isomerase or cyclophilin 1) | 5/35%/168 | MASCOT | 19.0/8.0 | 17.8/8.9 | Aspergillus nidulans | |
| 32 | gi|189196154 | NADP-specific glutamate dehydrogenase | 11/29%/541 | MASCOT | 46.4/7.2 | 49.1/5.9 | Pyrenophora tritici-repentis | |
| 33 | gi|145613040 | Conserved hypothetical protein (mitochondrial peroxiredoxin PRX1) | 4/21%/305 | MASCOT | 26.0/6.5 | 25.2/5.7 | Magnaporthe grisea | |
| 34 | gi|156057169 | ATP sulfurylase | 11/22%/98 | MASCOT | 74.6/4.4 | 64.2/6.6 | Sclerotinia sclerotiorum | |
| 35 | gi|169596136 | Hypothetical protein (piruvate kinase) | 12/28%/271 | MASCOT | 72.8/6.8 | 58.4/5.7 | Phaeosphaeria nodorum | |
| 36 | gi|154315126 | S-Adenosylmethionine synthetase | 5/18%/136 | MASCOT | 43.9/5.8 | 58.4/5.7 | Botryotinia fuckeliana | |
| 37 | gi|148361511 | Vacuolar serine protease | 7/20/115 | MASCOT | 15.7/6.1 | 41.0/5.8 | Cladosporium cladosporioides | |
| 38 | gi|85094513 | Hypothetical protein (phosphoglyceromutase) | 9/17/222 | MASCOT | 64.5/6.1 | 57.0/5.4 | Neurospora crassa | |
| 39 | gi|145232889 | Hypothetical protein (pyruvate decarboxylase) | 6/18/123 | MASCOT | 75.1/5.3 | 63.0/6.3 | Aspergillus niger | |
| 40 | gi|116196014 | Hypothetical protein (Heat shock 70 kDa protein, mitochondrial) | 12/20/369 | MASCOT | 84.8/5.4 | 68.2/5.6 | Chaetomium globosum | |
| 41 | gi|258575103 | 3'(2'),5'-Bisphosphate nucleotidase | 10/25/335 | MASCOT | 41.5/5.6 | 38.1/5.0 | Uncinocarpus reesii | |
| 42 | gi|258577135 | Rab GPD dissociation inhibitor alpha | 9/25/396 | MASCOT | 61.8/5.5 | 52.0/5.7 | Uncinocarpus reesii | |
| 43 | gi|255936655 | Pc13g09300 (pyruvate decarboxylase) | 7/19/107 | MASCOT | 75.4/5.4 | 63.3/5.7 | Penicillium chrysogenum | |
| 44 | gi|114321348 | Peptidoglycan synthetase FtsI | 11/17/93 | MASCOT | 30.6/6.9 | 62.7/8.0 | Alkalilimnicola ehrlichii | |
| 45 | gi|39945784 | Hypothetical protein (UDP-glucose 4-epimerase Gal10) | 5/15/344 | MASCOT | 323 | 39.8/6.6 | 40.9/5.9 | Magnaporthe grisea |
| 46 | gi|6647556 | 3-Isopropylmalate dehydrogenase A | 5/16/198 | MASCOT | 41.4/5.8 | 38.8/5.7 | Aspergillus niger | |
| 47 | gi|74627960 | Mannitol-1-phosphate 5-dehydrogenase | 7/17/191 | MASCOT | 45.7/5.7 | 43.7/6.0 | Alternaria alternata | |
| 48 | gi|255942351 | Hypothetical protein (UDP-galactoyranose mutase) | 9/21/250 | MASCOT | 60.1/7.0 | 57.2/5.6 | Penicillium chrysogenum | |
| 49 | gi|145232889 | Hypothetical protein (pyruvate decarboxylase) | 6/18/118 | MASCOT | 74.0/5.9 | 63.0/6.3 | Aspergillus niger | |
| 50 | gi|576627 | Alcohol dehydrogenase | 4/16/240 | MASCOT | 36.0/8.2 | 37.6/6.4 | Candida albicans | |
| 51 | gi|255933774 | Pc12g14620 (flavohemoprotein) | 04/08/1972 | PRO-BLASTe | 42.5/7.5 | 46.0/6.0 | Penicillium chrysogenum | |
| 52 | gi|261359248 | Aldose reductase | 02/05/1991 | PRO-BLASTf | 37.0/7.5 | 33.6/5.4 | Verticillium albo-atrum | |
| 53 | gi|242812056 | Alcohol dehydrogenase | 04/06/1999 | PRO-BLASTg | 36.1/8.1 | 38.9/6.0 | Talaromyces stipitatus |
a The assignment of the biological or enzymatic function to the hypothetical proteins (based on BlastP analyses) is shown between parentheses and in italic characters.
b Number of peptides matched/Sequence coverage (%)/Score
c Total and best ion scores are shown for proteins showing a sequence coverage value less than 15%
d Two best peptides (PRO-BLAST): SYLVGYASPKADV/APDAAKFPHAAR
e Two best peptides (PRO-BLAST): RDPIIAEHGNTITTR/SMAPHINHIAATRNPVR
f Two best peptides (PRO-BLAST): DIGTDYVDLYLIHDAKKTGGR/ALGLSNFSR
g Two best peptides (PRO-BLAST): EFPANEDHIAFAAR/KIVHPPKVR
Twelve of the proteins visualized in the secretome were predicted to contain a Signal P motif (proteins S2, S4-S9, S12-S14) or to contain a non-classical protein secretion motif (proteins S1 and S3) indicating their extracellular location. Four intracellular proteins (lacking of Signal P motif) were present in the secretome of D. seriata: NADP-specific glutamate dehydrogenase (spot S10), aspartate aminotransferase (spot S11), superoxide dismutase (spot S15) and glutathione reductase (spot S16). Their presence might indicate that some cell lysis may have occurred during fungal growth (although the microscopy observation of mycelia at 72 h of growth did not let detect any sign of cellular lysis in the liquid cultures. See Fig. 1B), and/or preparation of the secretion protein samples, resulting in the release of some cytoplasmic proteins, even though care was taken to minimize this possibility. This problem has also been encountered by other authors analyzing the secretome of phytopathogenic microorganisms, concluding that the release of cytoplasmic proteins is likely an unavoidable consequence of sample preparation [24,26]. Alternatively, these intracellular proteins could be secretion-signal-less proteins that in some conditions are located at the cell surface, or detected in culture supernatants, like it has been reported by other fungi. These secretion-signal-less proteins are secreted by some unknown mechanism. Many of these proteins are glycolytic enzymes, chaperones, translation factors and others suggesting that they could be 'moonlighting' (multifunctional) proteins. In fact, a glutathione reductase (spot S16) has been identified as a secretion-signal-less protein in S. cerevisiae [27]. Also, Murad and colleagues have described a extracellular aminotransferase secreted by the entomopathogenic fungi Metarhizium anisopliae [28].
As it might be expected, some of the secreted proteins identified are proteins or enzymes that could be involved in pathogenesis. Thus, an enzyme like glucosidase (spot S4), responsible for the degradation of the polysaccharydes found in the plant cell wall, would allow the invading phytopathogenic fungus to penetrate more easily the plant cells. Other proteins involved in peptide and/or protein degradation such as peptidase 1 (spot S12) could be responsible for the degradation of cell wall proteins and the inactivation or inhibition of some plant defense response proteins, whose expression may be induced by pathogen challenge. This possibility has been suggested for some secreted proteases from the phytopathogenic ascomycete Sclerotinia sclerotiorum [24]. Spots S5, S8 and S9 were identified as necrosis and ethylene-inducing proteins (Nep1 and Nep2). Nep proteins represent a new class of necrotic elicitors. Indeed, the extracellular Nep1 protein, initially identified from the culture filtrate of Fusarium oxysporum, has the ability to induce different kind of injuries in susceptible plants, such as necrosis in cacao or cell death and necrotic spots in Arabidopsis thaliana [29,30]. Plant responses to Nep proteins include induction of pathogen related (PR) genes, accumulation of ROS and ethylene, changes in K+ and H+ channel fluxes, callose apposition, altered cell respiration, induction of the hypersensitive response and localized cell death [29]. Finally, spot S13 could correspond to a cell wall protein PhiA. In Aspergillus nidulans, PhiA protein is highly up-regulated by exposition to several antifungal compounds [31] and it is involved in phialide and conidium normal development [32]. These data suggest that this protein could play a significant role as a defense mechanism against some antifungal responses, as when the plant produces toxic metabolites and, at the same time, the synthesis of PhiA initiates to ensure fungi survival by producing conidia [32].
Although we also made several attempts in order to characterize the secretome of D. seriata when growing in the presence of CMC the gels obtained were always of very poor quality due to irresoluble technical problems. The capability of D. seriata to metabolize CMC is limited and therefore high amounts of CMC remained in the supernatant cultures that interfered with the protein precipitation protocols. We tried to remove CMC from supernatants by several methods, including extensive dialysis and precipitation of CMC by using ammonium sulfate (80%), or PEG-CaCl2, but in all cases no satisfactory results were obtained.
Visualization and analysis of cytoplasmic proteins
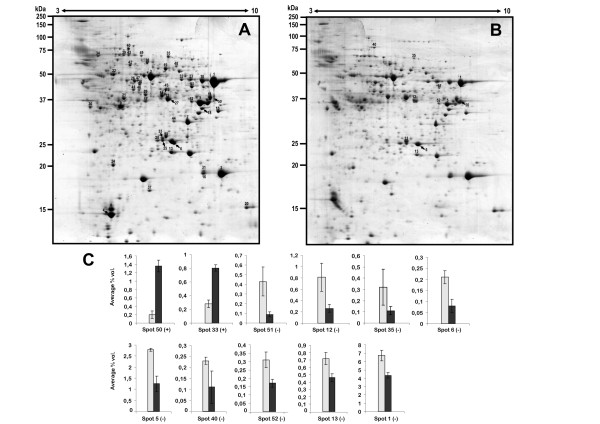
The proteins extracted from the whole mycelia of D. seriata and separated by 2-DE are showed in Fig. 3. Of the 550 mycelial proteins reproducibly visualized in the 2-DE gels from three independent protein extractions, 53 proteins were identified by PMF and MS/MS as shown in Table 2. However, 25 out of these 53 proteins were annotated in databases as hypothetical or putative uncharacterized proteins. As indicated above for the secreted proteins, a BlastP search against the non-redundant protein sequence database (NCBI) was performed by using their amino acid sequences (their putative biological functions are indicated between parentheses and in italics in Table 2) in order to achieve a better identification of the biological or enzymatic functions of these hypothetical proteins. This strategy let us assign a clear biological or enzymatic function to 22 out of the 25 hypothetical proteins.
Figure 3.
2-DE CBB-stained 12.5% SDS-PAGE gels of D. seriata cytoplasmic proteins corresponding to a liquid culture carried out in normal conditions (A), and to a liquid cultured developed in the presence of CMC. (C).- Density plots graphically representing the (%) volumen average corresponding to the proteins up-regulated (+) and down-regulated (-) are shown. (%) Vol. corresponds to the volume of each spot divided by the total volume of all the spots in the gel. Differentially expressed proteins were considered when the ratio of the (%) Vol. average (3 gels) between conditions was higher than 1.5 and the p-value deduced from the t-test was <0.05. The density plots corresponding to each category (up- and down-regulated proteins) are ordered from higher to lower ratios (as showed in Table 3).
The 53 annotated proteins represented 44.7% of the total spot volume (%Vol), whereas it is remarkable that the first 12 spots identified (numbered 1 to 12 in Table 2) corresponded to 29.75% of the total spot volume. These values obviously indicated that these proteins are highly abundant in the cytoplasm of D. seriata and probably suggest a relevant cellular function. In fact, 6 out of the 12 more abundant proteins were glycolytic enzymes like phosphoglycerate kinase (spots 1 and 10), triose phosphate isomerase (spot 6), enolase or phosphopyruvate dehydratase (spot 3), glyceraldehyde 3-phosphate dehydrogenase or GAPDH (spot 11) and fructose bisphosphate aldolase (spot 12). This can be explained biologically if we take into account that the glycolytic pathway plays a central role in the cellular energetic metabolism, and obviously glycolytic enzymes are found among the most expressed proteins in many different organisms.
As D. seriata is a phytopathogenic fungus, the annotation of proteins of importance in the pathogenesis process is of special interest. In this way, the case of GAPDH is significant since it is a very complex enzyme which possesses, besides its role in the glycolytic cycle, up to other fifteen different functions in mammalian cells and microbial pathogens [33], including a role as virulence factor for a vast variety of microbial pathogens [34,35]. Curiously, the second most abundant cytoplasmic protein (spot 2; 4.33%Vol) was identified as a peptidyl prolyl cis-trans isomerase (also spots 16 and 31) or cyclophilin. Cyclophilins have peptidyl prolyl cis-trans isomerase activity that performs protein folding by catalyzing the isomerization of peptide bonds preceding proline residues [36]. More interestingly, cyclophilins have been reported as an important virulence factor in the plant infection of rice by the ascomycete fungus Magnaporthe grisea [37], in the yeast Cryptococcus neoformans [38] and in the later infection stages or plant colonization by Botrytis cinerea [39].
Another abundant protein (spot 8) was identified as a signal transduction protein. A BlastP search let us confirm that this protein showed high amino acid identity (58%) to a CipC-like antibiotic response protein from Aspergillus fumigatus (gi|70997948). CipC-like antibiotic response protein (or concanamycin-induced protein) has been identified as an up-regulated protein in several fungal strains exposed to antibiotics, e.g. A. nidulans. In addition, in S. sclerotiorum, seven ESTs corresponding to CipC were identified during interactions with Brassica napus and classified as pathogenesis-associated genes [40]. In the case of the phytopathogenic soilborne fungus Verticillium dahlia, CipC protein has been suggested to help to its survival in soil by inactivating antibiotics produced by other soil microorganisms [41]. Spot 18 was identified as malate dehydrogenase (MDH). This enzyme catalyzes the reversible conversion of oxalacetate and malate. Oxalacetate is an oxalic acid precursor which has been described as a pathogenicity factor in B. cinerea [42], since the secretion of this acid is supposed to create an acidic environment that makes easier the pathogenic activity of the fungus. A role of MDH in pathogenesis has also been suggested for B. cinerea since this enzyme is produced exclusively or at higher levels in a more virulent B. cinerea 2100 strain [43]. Apart from the above-mentioned proteins, we could also identify (spot 40) a putative mitochondrial heat shock 70 kDa protein (HSP70) which is a molecular chaperone essential for tolerance to protein denaturing stresses as well as for protein biogenesis. HSP70 seems to be involved in fungal pathogenesis since it has been reported to facilitate the infection of coffee leaves by the fungus Colletotrichum gloeosporioides [44].
Some of the in-gel visualized proteins that we tried to identify could not be annotated, probably due to the fact that homologous proteins are not present in databases, which obviously could indicate that most of them might be novel or uncharacterized. In fact, proteomic analysis of many fungal species, particularly filamentous fungi, is difficult due to the lack of publicly available genome sequence data and the problems associated with cross-species comparisons [15].
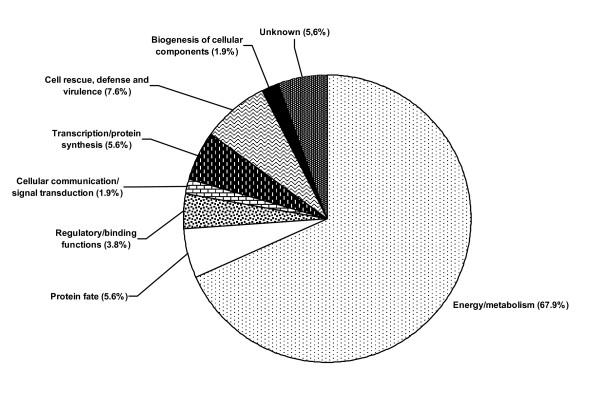
Functional classification of cytoplasmic proteins
Fig. 4 shows a graphical representation of the distribution of the identified cytoplasmic proteins based on a functional classification carried out according to the MIPS Functional Catalogue (FunCat description). Most of the identified proteins (67.9%) were metabolic enzymes or proteins involved in energy production. The proteins involved in energy production could be categorized into two groups. First, those involved in the glycolytic pathway like phosphoglycerate kinase (spots 1 and 10), enolase (spot 3), triosephosphate isomerase (spots 6 and 28), glyceraldehyde-3-phosphate dehydrogenase (spot 11), aldolase (spot 12), phosphoglyceromutase (spot 38) and pyruvate kinase (spot 35). Second, those that are components of the tricarboxylic acid pathway and mithocondrial proteins: pyruvate decarboxylase (spots 39, 43, 49) and malate dehydrogenase (spot 18). Several of the proteins identified are involved in carbohydrate metabolism such as UDP-glucose 4-epimerase (spot 45), mannitol-1-phophate-5-dehydrogenase (spot 47), UDP-galactopyranoside mutase (spot 48), alcohol dehydrogenase (spots 50 and 53) and aldose reductase (spot 52); whereas others such as NADP-specific glutamate dehydrogenase (spots 23 and 32), saccharopine dehydrogenase (spot 26) and S-adenosylmethionine synthetase (spot 36) are engaged in amino acid metabolism. Besides, several proteins involved in nucleic acid metabolism were annotated such as a nucleoside-diphosphate kinase (spot 20), adenosine kinase (spot 21), biphosphate nucleotidase (spot 41) and two proteins related to thiamine biosynthesis (spots 14 and 17).
Figure 4.
Functional classification of identified cytoplasmic proteins of D. seriata according to the MIPS Functional Catalogue (FunCat description). The listed percentage corresponds solely to the proportion of the identified proteins and does not take into account the proteins that were not annotated.
There were also proteins (7.6%) implicated in cell rescue, defense and virulence like mitochondrial peroxiredoxin 1 (spots 25 and 33), a heat shock 70 kDa protein (spot 40) and a flavohemoprotein (spot 51).
The remaining functional categories included a lower number of proteins. Thus 5.6% were classified as unknown proteins. The same percentage (5.6%) was estimated to be proteins involved in protein fate, like peptidyl-prolyl cis-trans isomerase (spots 2, 16, and 31). Proteins with regulatory and binding functions, like a predicted Zn-finger U1-like protein (spot 4) and a Rab GDP dissociation inhibitor alpha (spot 42) represented 3.8%; whereas proteins with a role in the transcription/translation processes, like an eukaryotic translation initiation factor 5A-1 (spot 24), an endonuclease reverse transcriptase (spot 27) and an elongation factor 1β (spot 30) represented 5.6% of the characterized proteins. As for spot 44 (biogenesis of cellular components, 1.9%), it was identified as a putative peptidoglycan synthetase FtsI, which obviously does not make any sense in a fungal strain, indicating that the primary role of this protein is still to be determined. Finally, solely one protein (spot 8; 1.9%) was classified into the cellular communication/signal transduction category.
Differentially expressed cytoplasmic proteins in the presence of CMC
Since cellulose is one of the major components of the plant cell wall, those proteins up-regulated when a phytopathogenic fungal strain grows in its presence are supposed to have a significant role in the pathogenicity process. This approach has been recently postulated as useful for the identification of putative pathogenicity factors in Botrytis cinerea [19,20]. As reported above, the analysis of the corresponding sets of 2-DE gels revealed an average of 550 protein spots in the proteome of D. seriata. Three proteins were overexpressed when the fungus grew in the presence of CMC [CMC is a cellulose derivative with carboxymethyl groups bound to the hydroxyl groups of the glucose unit. There can be up to three substituents on each glucose unit, thus the maximal degree of substitution is 3]. Two of them were identified as mitochondrial peroxiredoxin-1 and alcohol dehydrogenase (Table 3), suggesting a putative role in the pathogenicity process. The third protein could not be identified despite the good quality of the PMF spectra, probably due to the absence of similar proteins in protein databases.
Table 3.
Identified proteins of D. seriata up-regulated (+) and down-regulated (-) in the presence of CMCa
| Spot no. | Expression level | Ratio | p-value | Protein identityb |
|---|---|---|---|---|
| 50 | + | 6.72 | 0.0001 | Alcohol dehydrogenase |
| 33 | + | 2.82 | 0.0001 | Conserved hypothetical protein (Mitochondrial peroxiredoxin PRX1) |
| 51 | - | 5.00 | 0.0082 | Pc12g14620 (flavohemoprotein) |
| 12 | - | 3.16 | 0.0102 | Fructose-bisphosphate aldolase |
| 35 | - | 2.96 | 0.0444 | Hypothetical protein (piruvate kinase) |
| 6 | - | 2.77 | 0.0025 | Hypothetical protein (triosephosphate isomerase) |
| 5 | - | 2.24 | 0.0008 | Hypothetical protein (aldo/keto reductase) |
| 40 | - | 2.07 | 0.0254 | Hypothetical protein (heat shock 70 kDa protein, mitochondrial) |
| 52 | - | 1.87 | 0.0045 | Aldose reductase |
| 13 | - | 1.56 | 0.0053 | Glycolipid transfer protein |
| 1 | - | 1.54 | 0.0022 | Phosphoglycerate kinase |
a Differentially expressed proteins were considered when the ratio of the %Vol average (3 gels) between strains was higher than 1.5 and the p-value deduced from the t-test was <0.05.
b The assignment of the biological or enzymatic function to the hypothetical proteins (based on BlastP analyses) is shown between parentheses and in italic characters.
Peroxiredoxins (PRXs) are an ubiquitous family of thiol-specific antioxidant proteins that help to control intracellular peroxide levels. They also control cytokine-induced peroxide levels which mediate signal transduction in mammalian cells. Their broad distribution and high levels of expression suggest that they are both an ancient and important enzyme family [45]. On the other hand, as a ROS, hydrogen peroxide seems to play a significant role in plants under severe environmental conditions, including biotic stresses produced by phytopathogen infection. In fact, hydrogen peroxide participates in many resistance mechanisms, including reinforcement of the plant cell wall, phytoalexin production, and enhancement of resistance to various stresses [46]. Taking together all these evidences, we might speculate that an increase in the H2O2 levels in grapevine, as a result of fungal infection, could be counteracted by an increase in the peroxiredoxin levels in the fungal cells. In fact, peroxiredoxins have been proved to be important not only for resistance to H2O2 stress but also for virulence in the dimorphic fungus Cryptococcus neoformans, since they contribute to virulence in a mouse model [47]. Also it has been reported that peroxiredoxin Tsa1 is highly induced during 37°C hyphal growth in Candida albicans (the pathogenic form of this dimorphic yeast), and that the thioredoxin system genes are induced after peroxide exposure [48]. All these data support that an increase in the levels of peroxiredoxins in fungal strains could significantly contribute to the development of the pathological response in phytopathogenic fungi.
Alcohol dehydrogenases (ADH) are complex enzymes. Up to 7 different ADHs (ADH1 to ADH7) have been identified in Sacharomyces cerevisiae [49]. Although the primary role of ADH (ADH1) is to catalyze the production of ethanol from acetaldehyde in cells grown anaerobically or in the presence of a glucose excess, they can also achieve other cellular functions such as the catalyzation of the opposite reaction (ADH2 and ADH5 can convert ethanol accumulated during aerobic growth to acetaldehyde) or the participation in aldehyde reduction in oxidative conditions (ADH6), allowing the use of 2,3-butanediol as carbon and energy source [50]. The putative meaning of increased levels of ADH observed in the presence of CMC (and its putative role in virulence) seems more difficult to decipher. In fact, in the pathogen C. albicans, it has been reported that Adh1p restricts its ability to form biofilms on catheter surfaces through an ethanol-based mechanism and that the disruption of the ADH1 gene does not affect its ability to form the pathogenic hyphal form of this fungus [51]. We might speculate that the higher levels of ADH detected in the presence of CMC could be related to a tendency of the fungus growth to perform an anaerobic metabolism under this condition. Perhaps, this could reflect some difficulty to access oxygen when D. seriata grows inside the trunk of grapevines (in the vascular system), where availability of oxygen may be reduced. In these conditions, a higher ADH activity could be beneficial in order to obtain energy. However, we must not forget that in the presence of CMC up to four glycolytic enzymes are down-regulated (Table 3): fructose-bisphosphate aldolase, triose phosphate isomerare, phosphoglycerate kinase and pyruvate kinase, which obviously implicated a slower rate of efficiency of the glycolytic pathway, and therefore lower intracellular pyruvate levels available for ethanol production. This fact could be compensated by the nitrate reduction (remember that nitrate is the sole nitrogen source in the Czapeck media used) to nitrite, and the subsequent reduction to ammonia, coupled with the catabolic oxidation of ethanol to acetate and substrate-level phosphorylation that supports growth under anaerobic conditions [52].
Out of the 30 proteins down-regulated in the presence of CMC, solely 9 could be identified (Table 3). Most of these proteins are glycolytic enzymes (spots 12, 6, 35 and 1) or proteins involved in carbohydrate metabolism (aldose reductase, spot 52), which is difficult to understand if we bear in mind that the fungal strain is growing in a culture media supplemented with CMC and containing sucrose. Regarding this point, we could speculate that the metabolism of CMC could have some negative/inhibitory effect on some key enzymes of the glycolytic pathway. It should be noted that CMC can be degraded at different rates depending on the effectiveness of the different types of cellulases produced by the fungal strain to attack CMC. Different sugars (glucose, cellobiose, cellotriose, cellotetraose and cellopentaose) can be generated as degradation products [53]. The identification of these CMC-degradation products suggests that the carboxy-methyl groups of CMC could be released as acetate, which might exert a repressive effect on the levels of different glycolytic enzymes, as it has been shown for the bacteria Escherichia coli [54]. In this way, the CMC metabolism could be the cause in the observed diminishing of the levels of several glycolytic enzymes. Alternatively, it could be also possible that the reduced amounts of some glycolytic enzymes could be due by an increased carbon flow through the pentose phosphate pathway.
Finally, we could emphasize that although, as indicated above, the growth of phytopathogenic fungi in the presence of CMC has been postulated as a promising tool for the identification of putative proteins important for pathogenicity, our results indicated that, at least in the case of D. seriata, no significant differences could be detected in the proteome of this fungus grown in a defined medium and in the same medium supplemented with CMC, since a limited number of proteins were up- or down-regulated. Unfortunately, Fernández-Acero and colleagues [20] did not compare the growth of Botrytis cinerea proteome under normal conditions and cellulose-degradation conditions, in order to establish differences in the proteomes.
Conclusion
The main aim of this study was to perform a proteome analysis of the significant phytopathogenic and endophytic fungus D. seriata in order to understand better its lifecycle as well as its ability to infect grapevine plants producing trunk diseases. This is the first approach to study the proteome of this fungal strain which contributes to cause severe economic losses to wine industry all around the world. Notoriously, a high proportion of the identified proteins in the secretome could have a significant role in the pathogenicity process, including a glucosidase and a peptidase that could be involved in the degradation of the vegetable cell wall. More interestingly, three necrosis and induced proteins (Nep) were identified in the secretome of D. seriata. This is particularly intriguing since Nep proteins share a high degree of sequence similarity and they are able to induce an hypersensitive-like death response in a variety of plants, also inducing necrotic responses in plant tissues [55]. The putative role of D. seriata Nep proteins in the initiation or progression of the grapevine infection process needs to be further investigated in order to determine their significance for pathogenicity. Moreover, the detection of a putative secreted PhiA protein needs to be highlighted, since a high production of this protein has been suggested to ensure fungi survival by inducing a higher conidia production rate in response to the presence of toxic metabolites.
Regarding cytoplasmic proteins and in addition to the annotated proteins involved in basic cellular functions such as energy and metabolism, protein fate or transcription and protein synthesis, several proteins identified as virulence factor in other fungal strains were annotated. Among them, we can cite GAPDH, 3 peptidyl prolyl cis-trans isomerases (or cyclophilins), a CipC-like antibiotic response protein, malate dehydrogenase and a mitochondrial heat shock 70 kDa protein.
The differences observed in the proteome by growing the fungi in the presence of CMC were minor than expected, since solely three proteins were up-regulated and thirty down-regulated. These data could mean that this strategy might not be very effective to identify putative pathogenicity factors in phytopathogenic fungi. Finally, we would like to add that we hope that this study means a first step to advance in the necessary molecular biology studies of D. seriata that can contribute to deciphering the role of these putative pathogenicity factors in the development of grapevine decline diseases.
Finally, we hope that the proteomic data obtained will let us in a future develop further studies. In fact, and based on the peptide sequences obtained in this work, we have cloned and sequenced the D. seriata Prx1 gene (data not shown). This is the first gene of this important phytopathogen to be cloned, which supposes a significant contribution for the developing of the Molecular Biology in this microorganism. In fact, a transformation methodology for D. seriata is currently being developed to try to interrupt or attenuate the expression of Prx1 gene in order to check its putative role in the virulence process. The development of this methodology will let us in a future to check the involvement of other putative virulence proteins identified in this study in the pathogenicity process.
Materials and methods
Culture conditions
The fungal pathogen Diplodia seriata (isolated from an affected Vitis vinifera plant) was routinely maintained on potato dextrose agar (PDA) (Scharlau Chemie S.A., Barcelona, Spain) plates at 4°C. Liquid cultures were performed through the following protocol: a total of 12 Czapeck agar plates (sucrose, 3%; NaNO3, 0.4%; K2HPO4, 0.1%; MgSO4·7H20, 0.1%; FeSO4·7H20, 0.001%; and agar, 2%) were surface-covered with cellophane sheets. Plates were inoculated in the center with an agar plug (0.5 cm) removed from the outer edge of a 6-day-old fungal colony grown on Czapeck agar plates and incubated at 25°C for 5 days. The mycelia from two Czapeck plates were removed from the cellophane and disrupted with a beater. Next, mycelia were transferred to flasks containing 150 ml of Czapeck liquid medium (control conditions), or Czapeck liquid medium supplemented with 1% carboxymethylcellulose (CMC) (Sigma, St. Louis, MO, USA). The cultures were incubated in an orbital shaker at 120 rpm and 25°C for up to 7 days. The whole fungal proteome and secretome of D. seriata were prepared respectively from fungal mycelia and culture supernatants from 3-day-old liquid cultures. Three independent replicates were analyzed. Culture supernatants were collected by filtration and secreted proteins were immediately precipitated. Mycelia were harvested by filtration, washed with sterile water and stored at -20°C until use. Fungal growth was estimated as dry-weight of mycelia by filtering 5 ml of liquid cultures through a glass fiber filter (0.5 μm pore size). Mycelia were washed 3 times with deionized water and dried at 80°C for 48 h before weighing.
Protein extraction
Cytoplasmic proteins were isolated by using a protocol based on that of Fernández-Acero and colleagues [56], which was modified as follows. Frozen mycelia were ground to a fine powder with a pre-cooled mortar and pestle using liquid nitrogen. Two grams of powder were stirred (4°C for 2 h) in 10 ml of 10 mM potassium-phosphate buffer (K2HPO4-KH2PO4) (pH 7.4) containing 0.07% (w/v) 1,4-DTT and supplemented with Complete™ protease inhibitor cocktail (Roche, Mannheim, Germany) (one tablet per 10 ml of buffer). The cellular extracts were clarified twice by centrifugation (5 min, 15 000 × g). Proteins were precipitated for at least 1 h at -20°C after the addition of 1 volume of 20% trichloroacetic acid (TCA) in acetone containing 0.14% (w/v) DTT. The protein pellet was washed twice with acetone followed by an additional washing step with 80% acetone. The proteins were finally resuspended in 500 μl of sample buffer: 8 M urea, 2% CHAPS, 0.5% IPG Buffer pH 3-10 NL (GE Healthcare, Uppsala, Sweden), 20 mM DTT, and 0.002% bromophenol blue. The insoluble fraction was discarded by centrifugation at 15 000 × g for 5 min. The supernatant was collected and protein concentration was determined using the Bio-Rad Protein Reagent (Bio-Rad, Hercules, CA, USA) and bovine serum albumin as standard.
To prepare the secreted proteins for the secretome analysis the proteins from culture supernatants (100 ml) were precipitated for at least 1 h at -20°C after the addition of 1 volume of 20% TCA in acetone containing 0.14% (w/v) DTT. The precipitated proteins were harvested by centrifugation at 15 000 × g for 20 min at 4°C. The protein pellet was washed twice with acetone followed by an extra washing step with 80% acetone, and then resuspended in 500 μl of sample buffer. The insoluble fraction was discarded by centrifugation (5 min, 15 000 × g). Protein concentration was determined as indicated above.
2-D gel electrophoresis
For the analysis of the cytoplasmic and extracellular proteomes 18-cm IPG strips [pH 3-10 NL (non-linear), GE Healthcare] were loaded with 350 μg of soluble proteins in the sample buffer (see above). Proteins were focused using an Ettan IPGphor II (GE Healthcare) at 20°C according to the following program: 1 h, 0 V; and 12 h, 30 V (rehydratation); 2 h, 60 V; 1 h, 500 V; 1 h, 1 000 V; 30 min gradient up to 8000 V; and finally 7 h at 8,000 V until 50 kVh. Focused IPG gels were equilibrated twice for 15 min in a buffer containing 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS, 0.002% bromophenol blue and 1% DTT. For the second equilibration step, DTT was replaced by 4.0% iodoacetamide. The second dimension was run on a 12.5% SDS-PAGE gel in an Ettan Dalt Six apparatus (GE Healthcare) for 45 min at 3 W per gel, and then for 3.5 h at 18 W per gel. Precision Plus Protein Standards (Bio-Rad) were used as protein markers. Gels were stained following the "Blue Silver" staining method [57] using 0.12% CBB G-250 (Sigma), 10% ammonium sulfate, 10% phosphoric acid and 20% methanol.
Gel analysis
2-D images were captured by scanning stained gels using a ImageScanner II (GE Healthcare) previously calibrated with a greyscale marker (Kodak, Rochester, NY, USA), digitalized with Labscan 5.00 (v 1.0.8) software (GE Healthcare) and analyzed with the ImageMaster™ 2D Platinum v 5.0 software (GE Healthcare). Three gels of each strain obtained from three independent cultures (biological replicates) were analyzed to guarantee representative results. After automated spot detection, spots were checked manually to eliminate any possible artifacts, such as streaks or background noise. Biological replicates showed less than 10% and 5% of variability in the number of protein spots detected for intracellular and extracellular proteomes, respectively. The patterns of each sample were overlapped and matched, using landmark features, to detect potential differentially expressed proteins. The gel presenting the highest spot number was selected as Master gel and the coefficient of determination (r2) for each gel pair was calculated in order to ensure the analyses reliability (pair average r2: 0.82). Spot normalization, as an internal calibration to make data independent of experimental variations among gels, was made using relative volumes (%Vol) to quantify and compare the gel spots. %Vol corresponds to the volume of each spot divided by the total volume of all the spots in the gel. Differentially expressed proteins were considered when the ratio of the %Vol average (3 gels) between conditions was higher than 1.5 and the p-value deduced from the t-test was <0.05.
Protein Identification by MALDI-TOF/TOF MS and MS/MS
The protein spots of interest were manually excised from CBB-stained gels by biopsy punches, placed in a microcentrifuge tube, and washed twice with ddH2O. The proteins were digested following the method of Havlis et al. [58]: samples were dehydrated with ACN (Carlo Erba, Barcelona, Spain) for 5 min, reduced with 10 mM DTT (GE Healthcare) in 25 mM ammonium bicarbonate (Fluka, Buchs, Switzerland) for 30 min at 56°C and subsequently alkylated with 55 mM iodoacetamide (MS grade; Sigma) in 25 mM ammonium bicarbonate for 15 min in the dark. Finally, samples were digested overnight at 37°C with 10.0 ng/μl of sequencing-grade modified porcine trypsin (Promega, Madison, WI, USA) in 25 mM ammonium bicarbonate (pH 8.5). After digestion, the supernatant was collected, speed-vacuum dried and resuspended in 4 μl of 50% ACN, 0.1% TFA in ddH2O. One μl was spotted on a MALDI target plate and air dried for 10 min at room temperature. Subsequently, 0.5 μl of matrix [3 mg/ml of α-cyano-4-hydroxy-trans-cinnamic acid (LaserBio Labs, Sophia-Antipolis, France)] diluted in 0.1% TFA-ACN/H2O (1:1, v/v) was added to the dried peptide digestion and air dried for another 5 min at room temperature. The samples were analyzed with a 4800 Proteomics Analyzer MALDI-TOF/TOF mass spectrometer (Applied Biosystems, Foster City, CA, USA). A 4700 proteomics analyzer calibration mixture (Cal Mix 5; Applied Biosystems) was used as external calibration. All MS spectra were internally calibrated using peptides from the trypsin digestion. The peptides observed (up to 65 peptides per spot) were collected and represented as a list of monoisotopic molecular weights with a S/N greater than 20 using the 4000 Series Explorer v3.5.3 software (Applied Biosystems). All known contaminant ions (such as trypsin- and keratin-derived peptides) were excluded from further consideration in protein database search and later MS/MS analysis. Hence, from each MS spectrum, the fifteen most intensive precursors with a S/N greater than 20 were selected for MS/MS analyses with CID (atmospheric gas was used) in 2-kV ion reflector mode and precursor mass windows of ±7 Da. The default calibration was optimized for the MS/MS spectra processing.
For protein identification, Mascot Generic Files combining MS and MS/MS spectra were automatically created and used to interrogate a non-redundant protein database using a local license of Mascot v 2.2 from Matrix Science through the Global Protein Server (GPS) v 3.6 (Applied Biosystems). The search parameters for the PMF and MS/MS spectra were set as follows: i) NCBInr (2009.11.03) sequence databases were used; ii) taxonomy: All entries (9993394 sequences), Fungi (644746 sequences); iii) fixed and variable modifications were considered (Cys as S carbamidomethyl derivative and Met as oxidized methionine); iv) one missed cleavage site was allowed; v) precursor tolerance was set at 100 ppm and MS/MS fragment tolerance at 0.3 Da; vi) peptide charge was 1+; vii) the algorithm was set to use trypsin as the enzyme. Protein candidates provided by this combined PMF and MS/MS search were considered as valid when the global Mascot score was greater than 83 (total database) or 71 (fungi database) with a significance level of p < 0.05. Further additional criteria were taken into account for confident identification. First, the protein match should have at least 15% sequence coverage. Second, only those proteins with a Mascot ions score (see Table 2: Total Ion Score) above 77 in the MS/MS analysis (with a significance level of p < 0.05) should be considered as valid. Due to the almost absolute absence of protein or DNA sequences deposited in databases for D. seriata, those non-confirmed proteins after Mascot database search were identified by de novo sequencing and BLAST similarity searching following the procedure described by Liska and Shevchenko [59]. All MS/MS spectra for a sample were sequenced de novo using ProBLAST software (Applied Biosystems). The best fragmented ions were manually selected and the top ten candidate peptide sequences for each MS/MS were combined into a single text-format search string by means of the DeNovo Explorer software (Version 3.6) included in GPS Explorer Software (Applied Biosystems). This search string was used for similarity searches using the BLASTP algorithm [version 1.4.0] against an in-house protein sequence database containing the NCBInr sequence database (date: 2010.01.28; 10385566 sequences; 3543119913 residues) under the following parameters: i) fixed modifications: Cys as S carbamidomethyl derivative; ii) mass tolerance: 0.3 Da iii) E-value threshold: 20; iv) scoring matrix: PAM30MS; v) algorithm enzyme: trypsin. Protein identification significance was judged using the BLASTP scoring algorithm. Only those proteins matched by a minimum of two peptide sequences with a score higher than 52 according to DeNovo Explorer software were included in the results list, providing more stringent criteria for the identification. Information about the peptides identified can be found in the additional file 1 (Cytoplasmic PRO-BLAST peptides), additional file 2 (Secretome PRO-BLAST peptides), and additional file 3 (MASCOT Search Results).
Protein functional annotation
For proteins with no assigned function, homology searches were performed using the BlastP program against all non-redundant protein sequences deposited in the NCBI database http://blast.ncbi.nlm.nih.gov/Blast.cgi. Protein alignments were considered to be significant when their e-value was below 10-30. The secretion mechanism was predicted using SignalP (for classical secretion signal motifs) and SecretomeP (for non-classical signal motifs) software [60,61]. The identified proteins were distributed into categories according to their involvement in the biological process based on the MIPS Functional Catalogue (FunCat: http://mips.helmholtz-muenchen.de/proj/funcatDB/search_main_frame.html) [62].
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
RC carried out the D. seriata cultures and protein isolation, interpreted data and performed the gene cloning. CB performed 2DGE and acquired of MS/MS data and de novo sequencing. RMM helped with the design of experiments, the construction of the genomic library and cloning of Prx1 gene. JJRC carried out the design of experiments and the coordination of the study, helped with the data analysis and drafted the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Cytoplasmic proteins identified by means of PRO-BLAST search. Fragmentation spectra obtained from cytoplasmic proteins after de novo sequencing using ProBLAST software and properly identified by BLAST similarity searching are showed.
Extracellular proteins identified by means of PRO-BLAST search. Fragmentation spectra obtained from secreted proteins after de novo sequencing by means of ProBLAST software and suitably identified by BLAST similarity searching are showed.
MASCOT Search Results. Data (observed, expected and calculated mases, ppm and misscleavages) corresponding to those proteins identified by combined PMF and MS/MS search using MASCOT are indicated.
Contributor Information
Rebeca Cobos, Email: rebeca.cobos@unileon.es.
Carlos Barreiro, Email: c.barreiro@unileon.es.
Rosa María Mateos, Email: rosa.mateos@unileon.es.
Juan-José R Coque, Email: jjrubc@unileon.es.
Acknowledgements
This work has been financed with the benefits of a Research Contract sponsored by Bodegas Vega Sicilia S.A.U. (Valbuena de Duero, Valladolid, Spain). R. Cobos and R.M. Mateos were supported by a Research Contract financed by Bodegas Vega Sicilia. C. Barreiro was supported by the Program 'Personal Técnico de Apoyo' of the Ministry of Science and Innovation of Spain and the European Social Fund (ESF) (PTA-2003-01-00689). The technical support of M.D. Gutiérrez (Universidad Complutense de Madrid, Spain) is acknowledged.
References
- Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO, Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ. Phylogenetic lineages in the Botryosphaeriaceae. Stud Mycol. 2006;55:235–253. doi: 10.3114/sim.55.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slippers B, Wingfield MJ. Botryosphaeriaceae as endophytes and latent pathogens of woody plants: diversity, ecology and impact. Fungal Biol Rev. 2007;21:90–106. doi: 10.1016/j.fbr.2007.06.002. [DOI] [Google Scholar]
- Burgess TI, Sakalidis M, Hardy GES. Gene flow of the canker pathogen Botryosphaeria australis between Eucalyptus globulus plantations and native eucalypt forests in Western Australia. Mycol Res. 2006;31:559–566. [Google Scholar]
- Slippers B, Smit WA, Crous PW, Coutinho TA, Wingfield BD, Wingfield MJ. Taxonomy, phylogeny and identification of Botryosphaeriaceae associated with pome and stone fruit tress in South Africa and other regions of the world. Plant Pathol. 2007;56:128–139. doi: 10.1111/j.1365-3059.2006.01486.x. [DOI] [Google Scholar]
- Van Niekerk JM, Fourie PH, Halleen F, Crous PW. Botryosphaeria spp. as grapevine trunk disease pathogens. Phytopathol Mediterr. 2006;45:S43–S54. [Google Scholar]
- Mugnai L, Graniti A, Surico G. Esca (black measles) and brown wood-streaking: two old and elusive diseases of grapevines. Plant Dis. 1999;83:404–418. doi: 10.1094/PDIS.1999.83.5.404. [DOI] [PubMed] [Google Scholar]
- Fussler L, Kobes N, Bertrand F, Maumy M, Grosman J, Savary S. A Characterization of grapevine trunk diseases in France from data generated by the National Grapevine Wood Diseases Survey. Phytopathol. 2008;98:571–579. doi: 10.1094/PHYTO-98-5-0571. [DOI] [PubMed] [Google Scholar]
- Auger J, Esterio M, Ricke G, Pérez I. Black dead arm and basal canker of Vitis vinifera vc. Red Globe caused by Botryosphaeria obtusa in Chile. Phytopathol Mediterr. 2005;44:93–99. doi: 10.1094/PDIS.2004.88.11.1286A. [DOI] [PubMed] [Google Scholar]
- Taylor A1, GEStJ Hardy, Wood P, Burgess T. Identification and pathogenicity of Botryosphaeria species associated with grapevine decline in Western Australia. Austral Plant Pathol. 2005;34:187–195. doi: 10.1071/AP05018. [DOI] [Google Scholar]
- Martín MT, Cobos R. Identification of fungi associated with grapevine decline in Castilla y León (Spain) Phytopathol Mediterr. 2007;46:18–25. [Google Scholar]
- Martos S, Andolfi A, Luque J, Mugnai L, Surico G, Evidente A. Production of phytotoxic metabolites by five species of Botryosphaeriaceae causing decline on grapevines, with special interest in the species Neofusicoccum luteum and N. parvum. Eur J Plant Pathol. 2008;121:451–461. doi: 10.1007/s10658-007-9263-0. [DOI] [Google Scholar]
- Barbosa AM, Steluti RM, Dekker RF, Cardoso MS, Da Silva C. Structural characterization of Botryosphaeran: a (1→3; 1→6)-β-D-glucan produced by the ascomyceteous fungus, Botryosphaeria sp. Carbohydrate Res. 2003;338:1691–1698. doi: 10.1016/S0008-6215(03)00240-4. [DOI] [PubMed] [Google Scholar]
- Shah P, Atwood JA, Orlando R, El Mubarek H, Podila GK, Davis MR. Comparative proteomic analysis of Botrytis cinerea secretome. J Proteome Res. 2009;8:1123–1130. doi: 10.1021/pr8003002. [DOI] [PubMed] [Google Scholar]
- Jami MS, Barreiro C, García-Estrada C, Martín JF. Proteome analysis of the penicillin producer Penicillium chrysogenum: Characterization of protein changes during the industrial strain improvement. Mol Cell Proteomics. 2010;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nandakumar MP, Marten MR. Proteomics of filamentous fungi. Trends Biotech. 2007;25:395–400. doi: 10.1016/j.tibtech.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Fernández-Acero FJ, Carbú M, Garrido C, Vallejo I, Cantoral JM. Proteomic advances in phytopathogenic fungi. Curr Proteom. 2007;4:79–88. doi: 10.2174/157016407782194620. [DOI] [Google Scholar]
- Mehta A, Brasileiro ACM, Souza DSL, Romano E, Campos MA, Grossi-de-Sá MF, Silva MS, Franco OL, Fragoso RR, Bevitori R, Rocha TL. Plant-pathogen interactions: what is proteomics telling us? FEBS J. 2008;275:3731–3746. doi: 10.1111/j.1742-4658.2008.06528.x. [DOI] [PubMed] [Google Scholar]
- Bouws H, Wattenberg A, Zorn H. Fungal secretomes-nature's toolbox for white biotechnology. Appl Microbiol Biotechno. 2008;80:381–388. doi: 10.1007/s00253-008-1572-5. [DOI] [PubMed] [Google Scholar]
- Brito N, Noda J, González C. Botrytis cinerea endo-β-1,4-glucanase Cel5A is expressed during infection but is not required for pathogenesis. Physiol Mol Plant Pathol. 2005;66:213–221. doi: 10.1016/j.pmpp.2005.06.005. [DOI] [Google Scholar]
- Fernández-Acero FJ, Colby T, Harzen A, Cantoral JM, Schmidt J. Proteomic analysis of the phytopathogenic fungus Botrytis cinerea during cellulose degradation. Proteomics. 2009;9:2892–2902. doi: 10.1002/pmic.200800540. [DOI] [PubMed] [Google Scholar]
- Medina ML, Kiernan UA, Francisco WA. Proteomic analysis of rutin-induced secreted proteins from Aspergillus flavus. Fungal Genet Biol. 2004;41:327–335. doi: 10.1016/j.fgb.2003.11.014. [DOI] [PubMed] [Google Scholar]
- Vinzant TB, Adney WS, Decker SR, Baker JO, Kinter MT, Sherman NE, Fox JW, Himmel ME. Fingerprinting Trichoderma reesei hydrolases in a commercial cellulase preparation. Appl Bioche Biotechno. 2001;91:99–107. doi: 10.1385/ABAB:91-93:1-9:99. [DOI] [PubMed] [Google Scholar]
- Suárez MB, Sanz L, Chamorro MI, Rey M, González FJ, Llobel A, Monte E. Proteomic analysis of secreted proteins from Trichoderma harzianum. Identification of a fungal cell wallinduced aspartic protease. Fungal Genet Biol. 2005;42:924–934. doi: 10.1016/j.fgb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Yajima W, Kav NNV. The proteome of the phytopathogenic fungus Sclerotinia sclerotiorum. Proteomics. 2006;6:5995–6007. doi: 10.1002/pmic.200600424. [DOI] [PubMed] [Google Scholar]
- Basset SA, Bond JJ, Kwan FYS, McCulloch AF, Haynes PA, Johnson RD, Bryan GT, Jordan TW. Proteomic analysis of a filamentous fungal endophyte using EST datasets. Proteomics. 2009;9:2295–2300. doi: 10.1002/pmic.200800585. [DOI] [PubMed] [Google Scholar]
- Kazemi-Pour N, Condemine G, Hugouvieux-Cotte-Pattat N. The secretome of the plant pathogenic bacterium Erwinia chrysanthemi. Proteomics. 2004;4:3177–3186. doi: 10.1002/pmic.200300814. [DOI] [PubMed] [Google Scholar]
- Nombela C, Gil C, Chaffin WL. Non-conventional protein secretion in yeast. Trends Microbiol. 2006;14:15–21. doi: 10.1016/j.tim.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Murad AM, Noronha EF, Miller RNG, Costa FT, Pereira CD, Mehta A, Caldas RA, Franco OL. Proteomic analysis of Metarhizium anisopliae secretion in the presence of the insect pest Callosobruchus maculatus. Microbiol. 2008;154:3766–3774. doi: 10.1099/mic.0.2008/022913-0. [DOI] [PubMed] [Google Scholar]
- Bae H, Kim MS, Sicher RC, Bae HJ, Bailey BA. Necrosis- and ethylene-inducing peptide from Fusarium oxysporum induces a complex cascade of transcripts associated with signal transduction and cell death in Arabidopsis. Plant Physiol. 2006;141:1056–1067. doi: 10.1104/pp.106.076869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García O, Macedo JAN, Tiburcio R, Rincones J, Bittencourt LMC, Ceita GO, Micheli F, Gesteira A, Mariano AC, Schiavinato MA, Medrano FJ, Meinhardt LW, Pereira GAG, Cascardo JC. Characterization of necrosis and ethylene-inducing proteins (NEP) in the basidiomycete Moniliophthora perniciosa, the causal agent of witches' broom in Theobroma cacao. Mycol Res. 2007;111:443–455. doi: 10.1016/j.mycres.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Meyer V, Damveld RA, Arentshorst M, Stahl U, van den Hondel CAMJJ, Ram AFJ. Survival in the presence of antifungals. Genome-wide expression profiling of Aspergillus niger in response to sublethal concentrations of caspofungin and fenpropimorph. J Biol Chem. 2007;282:32935–32948. doi: 10.1074/jbc.M705856200. [DOI] [PubMed] [Google Scholar]
- Melin P, Schnürer J, Wagner GH. Characterization of phiA, a gene essential for phialide development in Aspergillus nidulans. Fungal Genet Biol. 2003;40:234–241. doi: 10.1016/S1087-1845(03)00108-7. [DOI] [PubMed] [Google Scholar]
- Deveze-Alvarez M, García-Soto J, Martinez-Cadena G. Glyceraldehyde-3-phosphate dehydrogenase is negatively regulated by ADP-ribosylation in the fungus Phycomyces blakesleeanus. Microbiol. 2001;147:2579–2584. doi: 10.1099/00221287-147-9-2579. [DOI] [PubMed] [Google Scholar]
- Pancholi V, Chhatwal GS. Housekeeping enzymes as virulence factors for pathogens. Int J Med Microbiol. 2003;293:391–401. doi: 10.1078/1438-4221-00283. [DOI] [PubMed] [Google Scholar]
- Alderete JF, Millsap KW, Lehker MW, Benchimol M. Enzymes on microbial pathogens and Trichomonas vaginalis: molecular mimicry and functional diversity. Cell Microbiol. 2001;3:359–70. doi: 10.1046/j.1462-5822.2001.00126.x. [DOI] [PubMed] [Google Scholar]
- Wang P, Heitman J. The cyclophilins. Genome Biol. 2005;6:226–231. doi: 10.1186/gb-2005-6-7-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud MC, Balhadere PV, Talbot NJ. A Magnaporthe grisea cyclophilin acts as a virulence determinant during plant infection. Plant Cell. 2002;14:917–930. doi: 10.1105/tpc.010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Cardenas ME, Cox GM, Perfect JR, Heitman J. Two cyclophilin A homologs with shared and distinct functions important for growth and virulence of Cryptococcus neoformans. EMBO Rep. 2001;2:511–518. doi: 10.1093/embo-reports/kve135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viaud MC, Brunet-Simon A, Brygoo Y, Pradier JM, Levis C. Cyclophilin A and calcineurin functions investigated by gene inactivation, cyclosporin A inhibition and cDNA arrays approaches in the phytopathogenic fungus Botrytis cinerea. Mol Microbiol. 2003;50:1451–1465. doi: 10.1046/j.1365-2958.2003.03798.x. [DOI] [PubMed] [Google Scholar]
- Li R, Rimmer R, Buchwaldt L, Sharpe AG, Séguin-Swartz G, Coutu C, Hegedus DD. Interaction of Sclerotinia sclerotiorum with a resistant Brassica napus cultivar: expressed sequence tag analysis identifies genes associated with fungal pathogenesis. Fungal Genet Biol. 2004;41:735–753. doi: 10.1016/j.fgb.2004.03.001. [DOI] [PubMed] [Google Scholar]
- El-Bebany AF, Rampitsch C, Daayf F. Proteomic analysis of the phytopathogenic soilborne fungus Verticillium dahliae reveals differential protein expression in isolates that differ in aggressiveness. Proteomics. 2010;10:289–303. doi: 10.1002/pmic.200900426. [DOI] [PubMed] [Google Scholar]
- Lyon GD, Goodman BA, Willianson B. In: Botrytis biology, pathology, and control. Elad Y, Willianson B, Tudzynski P, Delen N, editor. Dordrecht: Kluwer; 2004. pp. 119–142. [Google Scholar]
- Fernández-Acero FJ, Jorge I, Calvo E, Vallejo I, Carbú M, Camafeita E, Garrido C, López JA, Jorrin J, Cantoral JM. Proteomic analysis of phytopathogenic fungus Botrytis cinerea as a potentia tool for identifying pathogenicity factors, therapeutic targets and for basic research. Arch Microbiol. 2007;187:207–215. doi: 10.1007/s00203-006-0188-3. [DOI] [PubMed] [Google Scholar]
- Chena ZJ, Ribeiroa A, Silvaa MC, Santosa P, Guerra-Guimarães L, Gouveia M, Fernandez D, Rodrigues CJ Jr. Heat shock-induced susceptibility of green coffee leaves and berries to Colletotrichum gloeosporioides and its association to PR and hsp70 gene expression. Physiol Mol Plant Pathol. 2003;63:181–190. doi: 10.1016/j.pmpp.2003.11.004. [DOI] [Google Scholar]
- Wood ZA, Schröder E, Harris JR, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/S0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Quan LJ, Zhang B, Shi WW, Li HY. Hydrogen peroxide in plants: a versatile molecule of the reactive oxygen species network. J Int Plant Biol. 2008;50:2–18. doi: 10.1111/j.1744-7909.2007.00599.x. [DOI] [PubMed] [Google Scholar]
- Missall TA, Lodge JK, McEwen JE. Mechanisms of resistance to oxidative and nitrosative stress: implications for fungal survival in mammalian hosts. Euk Cell. 2004;3:835–846. doi: 10.1128/EC.3.4.835-846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W, Yoo Y, Kim JM, Shin D, Jeon HB. Identification of proteins highly expressed in the hyphae of C. albicans by two-dimensional electrophoresis. Yeast. 2003;20:1053–1060. doi: 10.1002/yea.1022. [DOI] [PubMed] [Google Scholar]
- de Smidt O, du Preez JC, Albertyn J. The alcohol dehydrogenases of Saccharomyces cerevisiae: a comprehensive review. FEMS Yeast Res. 2008;8:967–978. doi: 10.1111/j.1567-1364.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- Larroy C, Fernandez MR, Gonzalez E, Pares X, Biosca JA. Characterization of the Saccharomyces cerevisiae YMR318C (ADH6) gene product as a broad specificity NADPH-dependent alcohol dehydrogenase: relevance in aldehyde reduction. Biochem J. 2002;36:163–172. doi: 10.1042/0264-6021:3610163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee PK, Mohamed S, Chandra J, Kuhn D, Liu S, Antar OS, Munyon R, Mitchell AP, Andes D, Chance MR, Rouabhia M, Ghannoum MA. Alcohol dehydrogenase restricts the ability of the pathogen Candida albicans to form a biofilm on catheter surfaces through an ethanol-based mechanism. Infect Immun. 2006;74:3804–3816. doi: 10.1128/IAI.00161-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Takaya N, Sakairi M, Shoun H. Oxygen requirement for the denitrification by the filamentous fungus Fusarium oxysporum. Arch Microbiol. 2001;175:19–25. doi: 10.1007/s002030000231. [DOI] [PubMed] [Google Scholar]
- Karlsson J, Momcilovic D, Wittgren B, Schülein M, Tjerneld F, Brinkmalm G. Enzymatic degradation of carboxymethyl cellulose hydrolyzed by the endoglucanases Cel5A, Cel7B, and Cel45A from Humicola insolens and Cel7B, Cel12A and Cel45Acore from Trichoderma reesei. Biopolymers. 2002;63:32–40. doi: 10.1002/bip.1060. [DOI] [PubMed] [Google Scholar]
- Oh MK, Rohlin L, Kao KC, Liao JC. Global expression profiling of acetate-grown Escherichia coli. J Biol Chem. 2002;277:13175–13183. doi: 10.1074/jbc.M110809200. [DOI] [PubMed] [Google Scholar]
- Pemberton CL, Salmond GPC. The Nep1-like proteins--a growing family of microbial elicitors of plant necrosis. Mol Plant Pathol. 2004;5:353–359. doi: 10.1111/j.1364-3703.2004.00235.x. [DOI] [PubMed] [Google Scholar]
- Fernández-Acero FJ, Jorge I, Calvo E, Vallejo I, Carbú M, Camafeita E, López JA, Cantoral JM, Jorrín J. Two-dimensional electrophoresis protein profile of the phytopathogenic fungus Botrytis cinerea. Proteomics. 2006;6:S88–S96. doi: 10.1002/pmic.200500436. [DOI] [PubMed] [Google Scholar]
- Candiano G, Bruschi M, Musante L, Santucci L, Ghiggeri GM, Carnemolla B, Orecchia P, Zardi L, Righetti PG. Blue silver: a very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis. 2004;25:1327–1333. doi: 10.1002/elps.200305844. [DOI] [PubMed] [Google Scholar]
- Havlis J, Thomas H, Sebela M, Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- Liska AJ, Shevchenko A. Expanding the organismal scope of proteomics: cross-species protein identification by mass spectrometry and its implications. Proteomics. 2003;3:19–28. doi: 10.1002/pmic.200390004. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunakm S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Jensen LJ, Blom N, von Heijne G, Brunak S. Feature based prediction of non-classical and leaderless protein secretion. Protein Eng Des Sel. 2004;17:349–356. doi: 10.1093/protein/gzh037. [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cytoplasmic proteins identified by means of PRO-BLAST search. Fragmentation spectra obtained from cytoplasmic proteins after de novo sequencing using ProBLAST software and properly identified by BLAST similarity searching are showed.
Extracellular proteins identified by means of PRO-BLAST search. Fragmentation spectra obtained from secreted proteins after de novo sequencing by means of ProBLAST software and suitably identified by BLAST similarity searching are showed.
MASCOT Search Results. Data (observed, expected and calculated mases, ppm and misscleavages) corresponding to those proteins identified by combined PMF and MS/MS search using MASCOT are indicated.