Abstract
Summary: Domain mapping of disease mutations (DMDM) is a database in which each disease mutation can be displayed by its gene, protein or domain location. DMDM provides a unique domain-level view where all human coding mutations are mapped on the protein domain. To build DMDM, all human proteins were aligned to a database of conserved protein domains using a Hidden Markov Model-based sequence alignment tool (HMMer). The resulting protein-domain alignments were used to provide a domain location for all available human disease mutations and polymorphisms. The number of disease mutations and polymorphisms in each domain position are displayed alongside other relevant functional information (e.g. the binding and catalytic activity of the site and the conservation of that domain location). DMDM's protein domain view highlights molecular relationships among mutations from different diseases that might not be clearly observed with traditional gene-centric visualization tools.
Availability: Freely available at http://bioinf.umbc.edu/dmdm
Contact: mkann@umbc.edu
1 INTRODUCTION
The domain mapping of disease mutations (DMDM) database provides an aggregated view of all human coding disease-related mutations and SNPs for each protein domain. Domains are the structural, functional and evolutionary units of proteins (Holm and Sander, 1996; Murzin et al., 1995; Orengo et al., 1997). Most proteins contain multiple domains in a variety of domain combinations; and different domain combinations are associated with different protein functions (Bornberg-Bauer et al., 2005; Doolittle, 1995; Nikitin and Lisacek, 2003). Thus, the aggregated view of all human mutations at the domain level is an extremely useful tool for visualizing the molecular events that lead to diseased and healthy states in organisms. DMDM will also aid scientists to generate new hypotheses concerning the role of protein domains in key complex biological systems.
2 METHODS
HMMer's semi-global implementation (Eddy, 1996) was used to search for complete domains in human proteins from the RefSeq (Pruitt et al., 2007) and SWISS-PROT (Boeckmann et al., 2003) databases. Hidden Markov models for protein domains from SMART (Letunic et al., 2006), COG (Tatusov et al., 2003), CDD (Marchler-Bauer et al., 2007) and Pfam (Finn et al., 2008) were built using multiple sequence alignments from CDD with the hmmerbuild tool. The human disease mutations and SNPs mapped onto these domains were extracted from the OMIM (McKusick, 2007), SWISS-PROT and the dbSNP (Sherry et al., 2001) databases.
3 DMDM NAVIGATION LAYERS
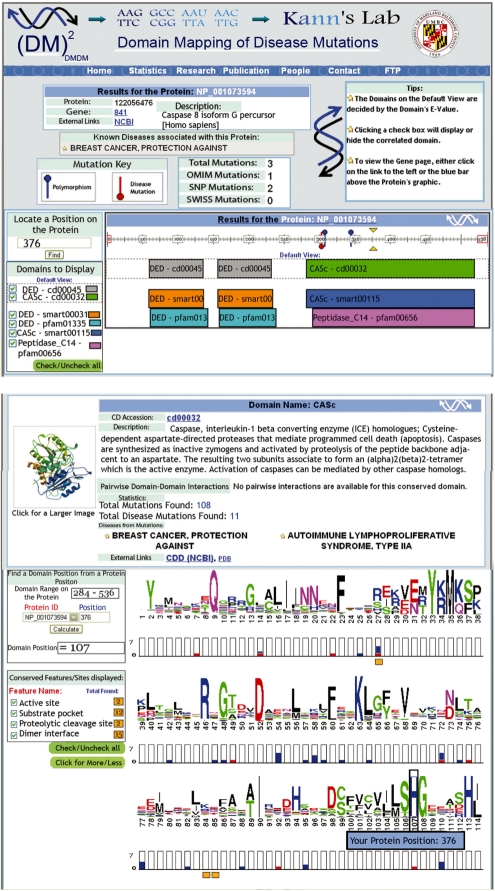
The data in DMDM can be visualized at three levels: gene, protein and protein domain. A search within DMDM can be performed at any of these three levels, or by disease name, using multiple search options. For instance, users may search by description, which is useful when only a keyword about the molecular entity is known, or by any gene or protein identifier. The results of the search in any of the layers consist of a summary of the information at the top of the page that includes a description, identifiers and external links to the gene, protein or domain. The summary is followed by either a graphical display of the information and/or tables of domain and mutational information, with key identifiers and relevant links. Proteins, both in the gene and protein layers, are depicted as a scaled bar that indicates the amino acid positions; the corresponding domains are shown below the bar. By selecting a region of the protein, information and links about the subset of mutations found around that region are displayed on a separate page along with a graphical display of each mutation.
The domain layer, an example of which is illustrated in Figure 1, displays three levels of information: sequence logos, mutational data and conserved functional features/sites for each domain position. Multiple sequence alignment information, obtained from CDD for each conserved domain model, is displayed using sequence logos [WebLogo software (Crooks et al., 2004)]. Mutational data for each human protein with one or more domains is represented by histograms under each position on the sequence logo. The third level, which was extracted from the CDD manual annotation and which is displayed below the histogram bars, provides the functional information for each domain position. The height of the histogram's bars represents the number of mutations found at individual domain positions for all human proteins that match that domain. Polymorphisms are represented in blue and disease mutations in red.
Fig. 1.
Screen shots of two DMDM queries using: (a) the NP_001073594 protein (GI:122056476) and (b) the CASc (cd00032) domain. The above display only shows a subset of the actual results available to users on the DMDM site.
Redundant mutations that share location, amino acid types and gene, but that are in different proteins are counted only once. When selecting a position on the histogram bar, a list of all mutations in that position, including redundancies, are displayed on a separate page. The upper left boxes in domain pages can be used to locate a particular protein position in the display. The check boxes provide control over the display of functional features shown for each domain position. The histogram bars provide a unique display of all the available information regarding human mutations, polymorphisms and disease mutations that were mapped to a particular domain position.
4 CONCLUSIONS AND FUTURE DEVELOPMENTS
The DMDM database is an online resource created for displaying human mutations with other relevant functional information at the protein domain level. This domain-centric database provides an ideal framework for studying biological processes relevant to human health, as well as for the integration of other molecular events, such as protein translational modifications and alternative splicing events affecting protein domains. Updates to DMDM will be performed every six months. In the future, DMDM will be expanded to map and integrate additional experimental data from the next generation of sequence, gene expression and proteomic experiments.
ACKNOWLEDGEMENTS
The authors wish to thank Russ B. Altman, Donna Maglott, Aron Marchler-Bauer and Mileidy W. Gonzalez for their helpful comments on the manuscript and feedback on the webpage design.
Funding: National Institutes of Health (NIH) [1K22CA143148 to M.G.K. (PI); R01LM009722 to M.G.K. (collaborator)].
Conflict of Interest: none declared.
REFERENCES
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E, et al. The evolution of domain arrangements in proteins and interaction networks. Cell. Mol. Life Sci. 2005;62:435–445. doi: 10.1007/s00018-004-4416-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF. The multiplicity of domains in proteins. Annu. Rev. Biochem. 1995;64:287–314. doi: 10.1146/annurev.bi.64.070195.001443. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Hidden Markov models. Curr. Opin. Struct. Biol. 1996;6:361–365. doi: 10.1016/s0959-440x(96)80056-x. [DOI] [PubMed] [Google Scholar]
- Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C. Mapping the protein universe. Science. 1996;273:595–603. doi: 10.1126/science.273.5275.595. [DOI] [PubMed] [Google Scholar]
- Letunic I, et al. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA. Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 2007;80:588–604. doi: 10.1086/514346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin AG, et al. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Nikitin F, Lisacek F. Investigating protein domain combinations in complete proteomes. Comput. Biol. Chem. 2003;27:481–495. doi: 10.1016/j.compbiolchem.2003.09.003. [DOI] [PubMed] [Google Scholar]
- Orengo CA, et al. CATH—a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]