Abstract
Motivation: Experiments such as ChIP-chip, ChIP-seq, ChIP-PET and DamID (the four methods referred herein as ChIP-X) are used to profile the binding of transcription factors to DNA at a genome-wide scale. Such experiments provide hundreds to thousands of potential binding sites for a given transcription factor in proximity to gene coding regions.
Results: In order to integrate data from such studies and utilize it for further biological discovery, we collected interactions from such experiments to construct a mammalian ChIP-X database. The database contains 189 933 interactions, manually extracted from 87 publications, describing the binding of 92 transcription factors to 31 932 target genes. We used the database to analyze mRNA expression data where we perform gene-list enrichment analysis using the ChIP-X database as the prior biological knowledge gene-list library. The system is delivered as a web-based interactive application called ChIP Enrichment Analysis (ChEA). With ChEA, users can input lists of mammalian gene symbols for which the program computes over-representation of transcription factor targets from the ChIP-X database. The ChEA database allowed us to reconstruct an initial network of transcription factors connected based on shared overlapping targets and binding site proximity. To demonstrate the utility of ChEA we present three case studies. We show how by combining the Connectivity Map (CMAP) with ChEA, we can rank pairs of compounds to be used to target specific transcription factor activity in cancer cells.
Availability: The ChEA software and ChIP-X database is freely available online at: http://amp.pharm.mssm.edu/lib/chea.jsp
Contact: avi.maayan@mssm.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Gene expression profiling using microarrays, or now RNA-seq, are popular methods to measure the level of mRNA in mammalian cells at a genome-wide scale. Yet, since mRNA levels only weakly correlate with protein level, data collected from mRNA profiling provides little clues on how cells are regulated by the activity of transcription factors, co-regulator complexes and cell signaling pathways. Many methods have been applied to study transcriptional regulation both experimentally and computationally. Recently, large-scale experimental methods that profile the binding of transcription factors to DNA at the genome-wide level have emerged. These methods include ChIP-chip (Iyer et al., 2001), ChIP-seq (Johnson et al., 2007), ChIP-PET (Wei et al., 2006) and DamID (Vogel et al., 2007) (these four methods are referred to as ChIP-X for shorthand hereinafter). Results from such experiments report the binding of specific transcription factors to DNA in proximity of target gene loci, commonly listing hundreds to thousands of potential regulatory interactions. Such interactions are often reported in Excel spreadsheets or PDF tables as Supplementary Materials to research articles, or as raw data files provided as short-sequence-reads in FASTA format, making such data difficult for reuse. So far, information from genome-wide ChIP-X studies, as well as low-throughput transcription-factor/DNA interaction studies, is utilized to develop binding-site sequence-motifs. For example, JASPAR (Sandelin et al., 2004) and TRANSFAC (Matys et al., 2003) are two popular databases that collect information about potential binding sites into logo-motifs also known as binding-site matrices. These databases contain collections of transcription factors with information on regulatory-motif elements. With binding-site matrices, it is straight forward to map potential binding sites across an entire genome. Alternatively, conserved sequences near gene-coding regions across and within species can suggest transcription-factor binding sites and be used to improve predictions of functional transcription-factor/DNA interactions using multiple alignments combined with logo-motif search. The obvious emerging alternative method is to utilize ChIP-X data for linking transcription factors to gene expression changes by computing binding site over-representation. By compiling ChIP-X experiments into a gene-list library database, we can rank transcription factors most likely responsible for the observed changes in gene expression based on statistical enrichment analysis. We show that this method is powerful, capable of capturing interesting underlying biology with clear signals. Such an approach likely works well because it considers the chromatin structure of a specific cellular state, in a specific experiment; and as such, it is a more direct way to infer transcription factor regulation compared to sequence-based methods.
2 IMPLEMENTATION
We manually collected data from ChIP-X experiments into a database of gene lists by extracting lists from the supporting materials of publications. In this ChIP-X database, each record contains a list of genes potentially regulated by a specific transcription factor under a specific condition. We only included publications that describe ChIP-X experiments applied to profile human or mouse cells. Besides manually extracting the gene lists reported by the authors from the publications' supporting materials, when it was possible and available, we also generated gene lists directly from the raw data files that belong to each publication. We implemented our own method for indexing, peak calling and gene matching to process the raw ChIP-seq and ChIP-chip data using a standard process (see Supplementary data for detail). The manually curated portion of the database as of July 26, 2010 contains 189 933 extracted interactions, from 84 publications, describing the binding of 92 transcription factors to 31 932 target genes. Several publications reported target genes for more than one factor, and several factors were profiled by different groups using different conditions and cell types. The automatically generated portion of the database contains 19 ChIP-seq and 10 ChIP-chip publications with 203 ChIP-seq and 22 ChIP-chip individual experiments. Both parts of this ChIP-X database are expected to continually grow. Additionally, since peak height varies significantly across experiments, we also implemented an interactive visualization tool that gives users the control to dynamically set the peak height cut-off for a specific experiment using a slider (Fig. 1).
Fig. 1.
Screen-shot from the ChIP-X database web application. Users can interactively adjust the normalized peak height threshold to determine the genes that are regulated by the transcription factor.
For comparing input gene lists across species, human and mouse gene IDs were merged using homologene. However, species are separated in the database and the user can perform the analysis on each species separately. The automatically generated lists displayed more variability in total gene calling per experiment per transcription factor when using a fixed peak height, in combination with the same indexing and peak calling methods. This suggests that there is intrinsic variability in average peak height and number of identified peaks across different ChIP-seq and ChIP-chip experimensts. The ChIP-X database is utilized to create a web-based interactive software application called ChIP Enrichment Analysis (ChEA) (Fig. 2). With ChEA, users can cut and paste input lists of mammalian gene symbols, typically gene lists that significantly changed in expression level from genome-wide gene expression profiling studies. Then, the software computes over-representation for targets of transcription factors from the ChIP-X database. To compute statistical enrichment, we implemented the Fisher exact test with the Bonferroni's correction, where the proportions for the test are the number of genes in the input list, the number of genes identified in the ChIP-X experiment, the genes that are shared among the two lists and the number of overall targets in the ChIP-X database (∼30 000). The program reports a ranked list of ChIP-X experiments that show statistically significant overlap with the input list. Indentified genes from the input list, potentially regulated by a specific transcription factor, are also connected and visualized as a network using known protein–protein interactions. To construct the protein–protein interaction network we used the networks we consolidated for the program Genes2Networks (Berger et al., 2007), as well as all mammalian interactions downloaded from the KEGG pathway database (Kanehisa et al., 2008). The ChEA software application and the interactive features of the ChIP-X database are implemented with JavaScript, AJAX, Java Server Pages, with a back-end MySQL database, as well as an interactive Adobe Flash-based network and peak-calling viewer developed using ActionScript 3.0. The system can also be downloaded and installed as a standalone Java desktop application, or as a command-line tool. The tool also provides search capabilities for finding transcription factor regulators for specific genes, or for finding experiments for specific transcription factors, including the ability to find which genes are regulated by more than one transcription factor (Supplementary Fig. S1). Users can also browse the content of the database online. The database containing all interactions extracted manually from the ChIP-X experimental data, as well as the indexed files created from the raw data analysis can be downloaded from our web site.
Fig. 2.
Screen-shot from the ChEA program web application. Users can cut and paste input lists of genes in the text box on the left. The system reports a ranked list of transcription factors/experiments (concatenated string that includes the transcription factor and the PubMed ID linking the factor to a specific study) based on over-representation of transcription factor putative targets in the input list.
3 RESULTS
3.1 Overlapping target genes among transcription factors
The collection of gene lists for each transcription factor from the ChIP-X studies allows us to reconstruct initial distance matrices that connect transcription factors based on different measures of binding site similarity. The first approach is based on shared target genes. If two factors share many targets, it is likely that they regulate genes together and potentially physically interact. We used the Jaccard Coefficient and hierarchical clustering to visualize such overlap (Fig. 3; Supplementary Figs S2–S5). Such analysis unravels known clusters linking the polycomb group members near the self-renewal embryonic stem-cell members, and clustering cancer related oncogenic transcription factors such as Myc and Cyclin-D1, as well as less obvious relations. Similarity between pairs of transcription factors profiled using ChIP-X studies can be further examined by looking at the proximity of binding peaks. Hence, the second type of distance matrix counts the number of proximal peaks for pair-wise factors (Supplementary Figs S6 and S7 and Supplementary Material). Given an arbitrary threshold, such distance matrices can be transformed into networks (Supplementary Figs S8–S10). Such networks can be used to further visualize potential transcription factor complexes and help in placing new ChIP-X results within the context of nuclear transcription regulators from prior studies based on target similarity, binding proximity and protein–protein interactions.
Fig. 3.
Transcription factor/target gene similarity distance table based on hierarchical clustering on 122 experiments from the manually extracted ChIP-X database. Transcription factors are considered similar if target genes reported from a ChIP-X study implicate significant overlap (Jaccard's Coefficient). Detailed labels of all experiments are shown in the Supplementary Figures.
3.2 Case studies: gene expression analyses with ChEA
To demonstrate the usefulness of ChEA we used the ChIP-X database for the analysis of mRNA expression data. We demonstrate the utility of the system for three different applications: The first case study is the re-analysis of gene lists that were extracted as biomarker features from two studies that developed a classifier for breast cancer metastasis using tissue from patients profiled with mRNA expression microarrays. The second application is the re-analysis of microarray data collected after over-expression of key transcription factors in embryonic stem cells. Whereas the last example cross references the ChIP-X database with the connectivity map (CMAP), a database of over 6000 chemical perturbations followed by results from genome-wide gene-expression microarrays (Lamb et al., 2006). Joining the ChIP-X and CMAP databases provides predictions for combinations of small molecule drugs that could be used to potentially down-regulate Myc activity in different cancers.
3.3 Case study 1: linking breast cancer signature genes to transcription factors
The first case study demonstrates how the ChEA system can be used for comparative analysis of two reports that identified a biomarker set for invasive breast cancer inferred from microarray studies. In two independent publications, the authors used two independently collected compendiums of mRNA expression microarrays from patients with breast cancer tumors to find a signature set of genes that can differentiate between benign and malignant breast cancers (van't Veer et al., 2002; Wang et al., 2005). Both studies produced lists of genes considered as selected feature ‘biomarkers’. Surprisingly, the two lists, containing 162 and 73 genes, share very little overlap (two genes) which is statistically insignificant. However, when both gene lists are used as input for ChEA separately, they show enrichment for SMAD2/3 gene targets. The P-value of 2.2E-05 for the list of 73 genes (the third-most significant out of all other experiments in the ChIP-X database) and also 9.5E-05 for the list of the 162 genes (fourth most significant ChIP-X experiment). When the lists are combined, SMAD2/3 is at the top of the list of enriched factors with an improved P-value of 9.3E-10 (Table 1; Supplementary Tables S1–S4).
Table 1.
Overlap summary among two prior reports that extracted biomarker sets from microarray mRNA profiling of breast cancer tissue from patients, and the top-ranked ChIP-X study identified by ChEA
| Experiment | Targets | Targets | P-values | |
|---|---|---|---|---|
| input | (database) | |||
| Van't Veer et al. (2002) | E2F1-18555785 | 35 | 4172 | 2.56E-07 |
| CREB1-15753290 | 15 | 957 | 1.15E-06 | |
| CUX1-19635798 | 27 | 3052 | 3.54E-06 | |
| SMAD2/3-18955504 | 20 | 1936 | 9.50E-06 | |
| Wang et al. (2005) | ZFP281-18757296 | 16 | 2004 | 7.72E-06 |
| HNF4A-19822575 | 29 | 6083 | 2.20E-05 | |
| SMAD2/3-18955504 | 15 | 1936 | 2.23E-05 | |
| Combined | SMAD2/3-18955504 | 35 | 1936 | 9.30E-10 |
| ZFP281-18757296 | 32 | 2004 | 9.58E-08 | |
| E2F1-18555785 | 50 | 4172 | 1.17E-07 |
Complete tables are provided at Supplementary Tables S1–S4.
Careful examination of the 35 genes that were identified as overlapping among the Smad2/3 targets from the two studies point to several genes that have been previously reported to play a role in breast cancer metastasis (Supplementary Table S4). In particular, MMP9 and CD44 are both highly implicated in breast cancer metastasis. MMP9 and CD44 are listed in GeneRifs for 23 and 17 articles returned by the query search ‘breast cancer’, respectively. MMP9 is a metalo-protease that digests the extra-cellular matrix during invasion, whereas changes in CD44 expression likely play a role in evading the host immune response. The results from the ChEA analysis clearly implicate that TGF-β/SMAD2/3 signaling plays a dominant role in breast cancer metastasis and can be used to further explain the origins of the discrepancy between the original two studies. It has been well-established that TGF-β/SMAD2/3 signaling is playing an important role in breast cancer metastasis (Koumoundourou D, 2007; Liapis et al., 2007; Xie et al., 2002). However, the ChEA analysis combined with the microarray profiling provides unbiased global additional support for such hypothesis. Our results also complement a network analysis approach applied to the same data using protein interactions. Chuang et al. (2007) ‘connected’ the breast cancer biomarkers identified by the two independent studies using known protein-protein interactions to find that a SMAD2/3 sub-network, among other sub-networks, is up-regulated in metastasized tumors. Here we linked such results to transcriptional regulation evidence from ChIP-X studies. Gene-expression profiling from different cancers, collected from patients or cell types, can now be linked to a transcription factor regulatory signature using ChEA. Such signature may hint, in a direct way, to the molecular regulatory mechanisms altered in any specific cancer subtype.
3.4 Case study 2: re-analysis of gene over-expression followed by mRNA profiling of mESCs
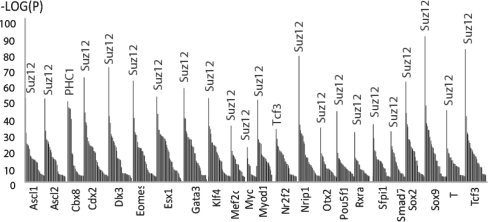
For the second case study, we re-analyzed results from a report where the authors over-expressed 50 transcription factors, one-by-one, in mouse embryonic stem cells (mESCs) and then measured the effect of such perturbations on gene-expression response using mRNA microarrays (Nishiyama et al., 2009). Among the 50 transcription factors used, all the well-known mESCs regulators are included, i.e. Oct4, Nanog and Sox2. The study identified Cdx2 as the transcription factor with the most dramatic effect when over-expressed, and as such it was selected for conducting a ChIP-seq experiment. We re-analyzed the results from the Nishiyama et al. study by inputting the top 500 genes that changed mostly as compared to the control for each of the 50 perturbations using ChEA. Surprisingly, the two studies that reported ChIP-X results for Suz12 binding appeared as the most statistically enriched for binding sites for almost all of the perturbations (Fig. 4; Supplementary Table S5). The P-values for overlap with Suz12 targets were very significant, reaching for example, 1.65E-89 for the up-regulation of Sox9. Additional confidence is added due to the fact that the ChEA database contains two independent Suz12 ChIP-X experiments that do not fully overlap, and both studies appeared at the top for almost all the gain-of-function experiments. Suz12 is a member of the polycomb group (PcG) complex responsible for methylation of lysine 9 and 27 of histone 3. Such methylation is known to cause transcriptional suppression of differentiation genes (Ru and Yi, 2004). Hence, the fact that all the changes in gene expression observed in this study are strongly associated with Suz12 targets, regardless of the perturbation applied to mESCs, may implicate that almost all perturbations cause differentiation. This suggests that the quantitative level of many components of the self-renewal machinery must be critically balanced to maintain the pluripotency state. It seems that the type of perturbation in itself was less critical as any perturbation induce similar global changes in chromatin rearrangements.
Fig. 4.
ChEA analysis of the top 500 genes that changed in their mRNA expression after 50 over-expression experiments of single transcription factors in mESCs. The P-value rankings from ChEA for each transcription factor over-expression perturbation are inverse log transformed. The top-ranked transcription factors reported by ChEA are labeled (peaks in the bar graph). Out of the 50 perturbations, only those factors that reached a low P-value of 1.0E-22 are labeled for clarity. Full results are available in Supplementary Table S5.
3.5 Case study 3: cross-referencing ChEA with CMAP for designing multiple drug treatments for cancer
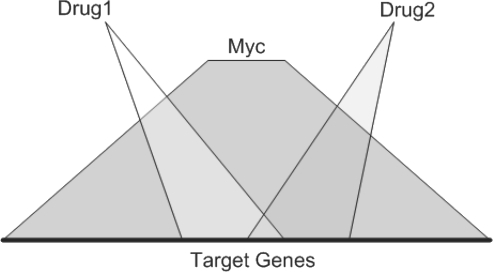
Another opportunity offered by the ChIP-X database, and the ChEA gene-list enrichment analysis software, is to combine ChEA with the Connectivity Map (CMAP) (Lamb, 2006, 2007). Such combination of databases can be used for identifying and ranking small molecules that can potentially be used for controlling the activity of specific transcription factors. CMAP is a dataset of mRNA microarray expression profiling of drug-stimulated mammalian cancer cells. CMAP contains ∼6000 perturbations with ∼1300 single drugs, sometimes in different concentrations, cell types or other variable experimental conditions. Examining the genes that increased or decreased significantly after a perturbation, we can use ChEA to rank the transcription factors that most likely regulate (statistically over represented transcription factors) the genes that increase or decrease in expression due to the drug perturbation. This ranking can be used to design combinations of drugs that can potentially counteract the activity of specific transcription factors in a specific cellular context (Fig. 5).
Fig. 5.
Illustration of the concept of using pair-wise drug perturbations to target the Myc target gene space.
Algorithmically, we can define two families of sets: one describing the relationship between drugs and the mRNAs they affect based on CMAP, and the other family of sets describing transcription factors and their target genes based on entries from the ChIP-X database. The drug-mRNA family of sets DR contains the top 500 genes that increased or decreased in expression given drug i and perturbation j from CMAP, and where j is used as the set label. Hence, the cardinality of all sets DRj is always 500. Elements in DRj sets are gene symbols. The second family of sets TR is made of target genes from all the ChIP-X experiments. Hence, TRi contains gene symbols reported to be targets of transcription factor i in experiment j where j is the set label. Now we can operate on these families of sets to find the best pair-wise combinations of drugs that cover the target space for transcription factor i. First, we can compute the union of DR with itself to create a new family of sets DP describing how pairs of drugs may affect gene expression in an additive manner. For all DRi and for all DRj where i not equal to j
The additive assumption was chosen for simplicity, however there is some evidence that it might be reasonable (Geva-Zatorsky et al., 2010). Hence DP contains a family of (n2/2)−n sets, where n is the number of drug perturbation experiments in CMAP. Note that the family of sets DP contains also genes symbols as the elements of the sets. We are now ready to score how drug perturbation pairs may affect the activity of transcription factor k in experiment j. Such score is simply the intersection between DP and TR,
The scoring scheme can be used to suggest, for example, how we can use small molecules to induce the activity of specific transcription factors such as Oct4 for iPS reprogramming, or for blocking uncontrolled cell proliferation by targeting Myc. In this case study, we devise mechanisms to down-regulate the activity of the transcription factor Myc and hence potentially block the proliferation of cancer cells. When we examined the results of inputting all lists of the mostly down-regulated 500 genes as reported in the ranked lists from CMAP entered in a batch mode into ChEA, we noticed that Myc appears often as the top-ranked transcription factor for binding in proximity to genes that decreased in expression after many drug perturbations. This is expected since Myc is a known oncogene, and all cell-lines in CMAP are human cancer cells, and many of the drugs that were used to create CMAP are anti-cancer drugs. For illustration, we ranked pairs of drugs based on their combined coverage of Myc targets (Fig. 5). Our strategy optimizes selection of drug-pairs that do not have similar effects on Myc targets. Table 2 provides the resulting top 10 pair-wise entries (a Perl script with an input table is provided as supporting materials, Script S1, Supplementary Table S6). The top 10-ranked list of pair-wise drugs suggests combinations of drug treatments for further maximally reducing Myc transcriptional regulatory activity. The combinations we identified include known cancer drugs as well as other drugs. For example, monastrol is a known cancer drug that targets kinesin-5, a motor protein important in mitosis (Mayer et al., 1999), whereas clonidine is a alpha-adrenergic agonist (Andén et al., 1970) that is an anti-hypertensive used to aid sleeping and treat ADHD. Hence, it is likely acting through a different pathway to regulate a subset of Myc-regulated target genes. Our initial approach of combining and ranking pairs of drugs to regulate the activity of specific transcription factors can be further improved in many ways. One possibility is to compute the likelihood that a combination of more than two drugs will cover a specific transcription factor target space. This can be achieved, for example, with algorithms such as the probabilistic generative model for GO enrichment analysis (Lu et al., 2008). Our initial formulation can also be extended by using quantitative values instead of sets, and include statistical randomization as control. In summary, the approach presented in this third case study provides a step forward toward rationale combinatorial application of drugs to treat specific cancers with a transcription-factor anchoring. Such an approach is amenable also for improving iPS reprogramming strategies by, for example, designing combination of drugs that would activate Oct4 or other key stem-cell self-renewal factors for reprogramming somatic cells into iPS cells. Many other similar applications to attempt to control cell fate are possible. The strategy should also work for other types of gene expression microarray datasets in other contexts.
Table 2.
Top-ranked pairs of drug perturbations from CMAP experiments that cover the gene target space of Myc as determined by several ChIP-X experiments independently
| ExpID |
|||
|---|---|---|---|
| Drug1 | Drug2 | Targets for | Total |
| D1/D2/overlap | targets | ||
| monastrol-614 | clonidine-1555 | 170/215/8 | 377/500 |
| monastrol-614 | colchicine-1598 | 170/210/8 | 372/500 |
| monastrol-614 | tolazoline-2000 | 170/208/7 | 371/500 |
| nocodazole-1393 | laudanosine-1741 | 178/191/6 | 363/500 |
| nocodazole-1393 | valproic acid-1047 | 178/192/7 | 363/500 |
| monastrol-614 | dihydroergocristine-1745 | 170/201/8 | 363/500 |
| nocodazole-1393 | tolazoline-2000 | 178/208/24 | 362/500 |
| monastrol-614 | methylergometrine-1607 | 170/203/11 | 362/500 |
| monastrol-614 | bromocriptine-2007 | 170/203/11 | 362/500 |
| monastrol-614 | tretinoin-1548 | 170/202/10 | 362/500 |
D1, drug1; D2, drug2; ExpID, experiment ID from CMAP.
4 DISCUSSION
One of the reasons high-throughput genome-wide ChIP-X studies are expected to be more useful and accurate than computational sequence-based methods is because the sequence-based approaches do not take into consideration the chromatin state of the cell under a specific experimental condition, cell type or organism. Hence, ChIP-X databases and tools such as ChEA are expected to perform more accurately when combined with data from mRNA gene expression studies as compared to computational sequence based methods. A match between a transcription factor and changes in gene expression will be found not only by linking the changes in expression to transcription factor binding sites, but also linking such binding to a specific prior experiment which has been conducted under similar condition. Combining different types of ChIP-X experiments from different papers, cell-types and experimental conditions, using different statistical cut-offs and experimental techniques is challenging. We chose to either use the criteria applied by the authors of each study, or apply our own standard method for finding peaks and calling target genes. Both approaches are simple and relatively unbiased. The two approaches complement each other in regards to coverage. The raw data route excludes many of the studies currently in the database since there are many ChIP-X publications that only provide the target list without the raw data. There are also many ChIP-X raw data files available in the public domain without a publication that contains an author extracted gene list. Regardless, we expect that the database will rapidly continue to grow. Moreover, multiple entries for the same transcription factor can increase the confidence for functional binding sites (Wu and Ji, 2010). Our initial analysis shows that overlap among different ChIP experiments using the same factor increases functional gene predictability. For example, we examined the overlap among independent Oct4 ChIP-X studies and compared the consensus overlapped genes with an Oct4 knock-down followed by a microarray study. Initial results demonstrate that functional genes prediction improves when multiple independent studies are combined, but this should be further investigated in future studies. Since we keep track in our database on information such as the cell type, organism, experimental method, distance to start site and peak height, we implemented filters that can be used by users to exclude the analysis from including specific organism, cell-type or experimental method; as well as calibrate the gene calling threshold for peak height and distance to start site. For future studies we plan to integrate ChIP-X data with lost-of-function/gain-of-function microarray studies as well as include more histone modification ChIP-X studies. Many of the studies that report global transcription factor binding to DNA using whole genome-wide ChIP-X experiments also often conduct global mRNA experiments after knock-down or over-expression of the transcription factors that were used in the ChIP-X studies, as well as profiling specific histone modifications or polymerase binding using ChIP-X technologies. By combining mRNA microarrays of RNA-seq together with ChIP-X transcription factor, polymerase binding and histone modification studies, we can determine which binding sites are functional, as well as which functional sites are activation or inhibition sites. By combining expression data with ChIP-X we should be able to obtain a signed and directed network which is desired for understanding pathways, improving enrichment analyses and performing dynamical simulations. The ChIP-X database and ChEA web-based software tool was generated utilizing code from our previous work of developing a kinase-substrate database and software system for kinase enrichment analysis (KEA) (Lachmann and Ma'ayan, 2009). These two software systems can potentially be combined. Since we know the group of transcription factors that regulate genes based on changes at the mRNA level under a certain experimental condition or in a specific disease based on tissue expression profiling, we can use known protein–protein interactions to build a sub-network to connect these transcription factors. Then, we can link this sub-network, as input for KEA, to obtain the protein kinases and pathways that most likely regulate the transcription factor centered sub-network (Bromberg et al., 2008). Such an approach can be used to understand cell regulation at the cell signaling network level given mRNA expression profiling data and suggest kinase inhibitors as drug-targets (Ma'ayan and He, 2010).
5 CONCLUSIONS
In summary, the ChIP-X database and the ChEA software provide an alternative way for researchers to analyze mRNA expression data in context of genome-wide transcription-factor ChIP-X experiments collected and organized into a prior knowledge database and an interactive web-based software system. As more transcription factors are profiled, under different experimental conditions, the database is expected to grow and improve in accuracy and coverage. Using other tools and databases in combination with ChEA, integrative creative new applications in systems biology can be made possible.
Funding: National Institutes of Health (grants P50GM071558-01A27398, R01DK088541, R01GM054508, R01DA15446 and KL2RR029885-0109).
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Andén NE, et al. Evidence for a central noradrenaline receptor stimulation by clonidine. Life Science. 1970;9:513–523. doi: 10.1016/0024-3205(70)90207-9. [DOI] [PubMed] [Google Scholar]
- Berger S, et al. Genes2Networks: connecting lists of gene symbols using mammalian protein interactions databases. BMC Bioinformatics. 2007;8:372. doi: 10.1186/1471-2105-8-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg KD, et al. Design logic of a cannabinoid receptor signaling network that triggers neurite outgrowth. Science. 2008;320:903–909. doi: 10.1126/science.1152662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang H.-Y, et al. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3 doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geva-Zatorsky N, et al. Protein dynamics in drug combinations: a linear superposition of individual-drug responses. Cell. 2010;140:643–651. doi: 10.1016/j.cell.2010.02.011. [DOI] [PubMed] [Google Scholar]
- Iyer VR, et al. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Johnson DS, et al. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koumoundourou D, et al. Prognostic significance of TGFbeta-1 and pSmad2/3 in breast cancer patients with T1-2,N0 tumours. Anticancer Res. 2007;27:2613–2620. [PubMed] [Google Scholar]
- Lachmann A, Ma'ayan A. KEA: kinase enrichment analysis. Bioinformatics. 2009;25:684–686. doi: 10.1093/bioinformatics/btp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J, et al. The connectivity map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Lamb J. The connectivity map: a new tool for biomedical research. Nat. Rev. Cancer. 2007;7:54–60. doi: 10.1038/nrc2044. [DOI] [PubMed] [Google Scholar]
- Liapis G, et al. Effect of the different phosphorylated Smad2 protein localizations on the invasive breast carcinoma phenotype. Apmis. 2007;115:104–114. doi: 10.1111/j.1600-0463.2007.apm_517.x. [DOI] [PubMed] [Google Scholar]
- Lu Y, et al. A probabilistic generative model for GO enrichment analysis. Nucleic Acids Res. 2008;36:e109. doi: 10.1093/nar/gkn434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma'ayan A, He JC. Protein kinase target discovery from genome-wide mRNA expression profiling. Mount Sinai J. Med. 2010;77:345–349. doi: 10.1002/msj.20192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer TU, et al. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- Nishiyama A, et al. Uncovering early response of gene regulatory networks in ESCs by systematic induction of transcription factors. Cell Stem. Cell. 2009;5:420–433. doi: 10.1016/j.stem.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru C, Yi Z. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Sandelin A, et al. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Veer LJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- Vogel MJ, et al. Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat. Protoc. 2007;2:1467–1478. doi: 10.1038/nprot.2007.148. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. The Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- Wei CL, et al. A global map of p53 transcription-factor binding sites in the human genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- Wu H, Ji H. JAMIE: joint analysis of multiple ChIP-chip experiments. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq314. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, et al. Alterations of Smad signaling in human breast carcinoma are associated with poor outcome: a tissue microarray study. Cancer Res. 2002;62:497–505. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.