Abstract
Chemokine receptor 5 (CCR5) is a cell surface protein required for HIV-1 infection. It is important to detect the amount and observe the spatial distribution of the CCR5 receptors on the cell surfaces. In this report, we describes the metal nanoparticles which were specially designed as molecular fluorescent probes for imaging of CCR5 receptors on the T-lymphocytic PM1 cell surfaces. These CCR5 monoclonal antibodies (mAbs) metal complexes were prepared by labeling mAbs with Alexa Fluor 680 followed by covalent binding the labeled mAbs on the 20 nm silver nanoparticles. Compared with the labeled mAbs without metal, the mAb-metal complexes were found to display enhanced emission intensity and shortened lifetime due to interactions between fluorophores and metal. The mAb-metal complexes were incubated with the PM1 cell lines. The confocal fluorescent intensity and lifetime cell images were recorded on single cells. It was observed that the mAb-metal complexes could be clearly distinguished from the cellular autofluorescence. By analyzing a pool of cell images, we observed that most CCR5 receptors appeared as clusters on the cell surfaces. The fluorophore-metal complexes developed in this report are generally useful for detection of cell surface receptors and provide a new class of probe to study the interaction between the CCR5 receptors with viral gp120 during HIV infections.
Keywords: Chemokine receptor 5 (CCR5), monoclonal antibody (mAb), Alexa Fluor 680, mAb-metal complex, T-lymphocytic PM1 cell, fluorescence cell image
Introduction
The chemokine receptor CCR5 is expressed as a co-receptor on the cell surfaces of T-lymphocytes [1,2]. It is recognized by the R5 virus during the initial stage of infection [3,4]. Therefore, the detection of the CCR5 receptors, including amount and distribution on the cell surfaces, may provide insight into understanding the molecular mechanism of infection. Fluorescence microscope has been widely applied to detect the various target molecules on the cell surfaces [5–7]. Cell imaging is also expected to be useful for elucidating cellular signal transductions and for medical diagnostics. In the present report, we describe the use of cell imaging to observe the CCR5 receptors on the surfaces of T-lymphocytic PM1 cells.
The information available from fluorescent cell imaging depends on the biochemical and optical properties of the imaging agents. These agents typically consist of fluorophores conjugated to the antibodies that may provide binding specificity [8–10]. However, there are several limitations with the use of organic fluorophores. The optical brightness or photon emission rate of single fluorophore is limited, and it thus difficult to depth single fluorophores in the presence of cellular autofluorescence in cell imaging. Additionally, single fluorophores often display photobleaching and/or blinking. And finally, most organic fluorophores have emission lifetimes close to that of cellular autofluorescence (2 – 10 ns), which makes it difficult to distinguish the probe emission from cellular autofluorescence. Hence, it is a need for novel imaging agents with improved intrinsic optical properties.
During the past 10 years, there has been a great interest in the use of metal nanoparticles as optical probes. Such metal nanoparticles are known to display strong interactions with light due to collective oscillation of electrons on the metal surfaces [11–13]. These oscillations are known as plasmons. Fluorophores can interact with these plasmons in a manner that improves the optical properties of the fluorophores. These improvements include an increase in intensity, quantum yield, and photostability, and a decrease in blinking and lifetime [14,15]. We used this phenomenon to develop fluorophore-metal nanoparticle complexes and applied the complexes for cell imaging to detect the membrane-bound receptors [16,17].
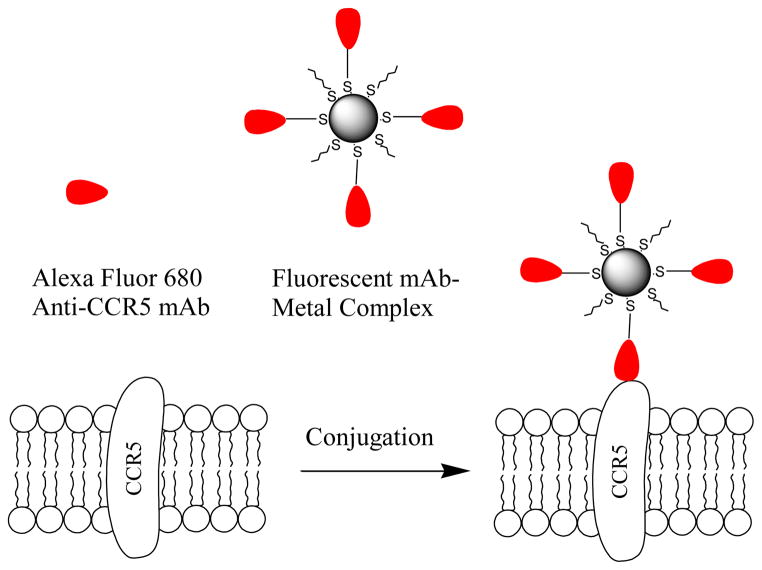
In the present paper, these probes were used to detect the CCR5 receptors on the cell surfaces of T-lymphocytes. The mAb-metal complexes were synthesized by binding the fluorescently labeled mAb molecules on the 20 nm silver nanoparticles. The mAb-metal complexes were then incubated with T-lymphocytic PM1 cells (Figure 1). We selected a long wavelength fluorophore Alexa Fluor 680 to minimize the contribution of cellular autofluorescence in cell imaging [18]. The average number of the CCR5 receptors per T-lymphocytes and the distribution on the cell surfaces were determined in the research.
Figure 1.
Alexa Fluor 680 labeled anti-CCR5 mAb or mAb-metal complex were immuno-interacted with target CCR5 receptor on T-lymphocytic PM1 cell.
Materials and Methods
All reagents and spectroscopic grade solvents were purchased from Sigma-Aldrich. Alexa Fluor 680 succinimidyl ester was purchased from Invitrogen. The dialysis membrane (MWCO 4,000) was purchased from Spectrum Laboratories, Inc. The Nanopure water (>18.0 MΩ.cm) from a Millipore Milli-Q was used in all experiments. (2-mercapto-propionylamino) acetic acid 2,5-dioxo-pyrrolidin-1-ylester was synthesized as we have described previously [19]. PM1 cells were from Dr. R. R. Redfield’s laboratory at the Institute of Human Virology. Monoclonal CCR5 antibody (45531) was obtained from the NIH AIDS Research and Reference Reagent Program.
Fluorescently labeling anti-CCR5 mAb
Anti-CCR5 mAb was labeled with Alexa Fluor 680 succinimidyl ester via a reactive ester reaction [20]. Typically, anti-CCR5 mAb was dissolved in 10 mM PBS buffer that was adjusted to pH = 8.0 with sodium hydrogencarbonate. Alexa Fluor 680 succinimidyl ester was then added to the solution at a molar ratio of dye/mAb = 5/1. The solution was stirred for 2 h at 4°C. The unbound fluorophore was removed by gel filtration column chromatography on Sephadex G-150, followed by dialysis against 10 mM PBS buffer at pH = 7.2.
Preparation of mAb-metal complex
Silver nanoparticles with an average diameter of 20 nm were synthesized in a modified Brust reaction as reported previously [17]. The metal nanoparticles were coated with organic monolayers of N-(2-mercaptopropionyl) glycine (tiopronin) [16,17] and then partially substituted by (2-mercapto-propionylamino) acetic acid 2,5-dioxo-pyrrolidin-1-ylester via ligand exchange [21]. The substitution was carried out in a mixture solution of N,N′-dimethylformamide (DMF) and water (v/v = 1/1) with the silver nanoparticle concentration of 4×10−8 M, as estimated from the extinction coefficient, and succinimidyl ester concentration of 8×10−7 M. The reaction solution was stirred for 24 h at room temperature. The suspension was removed by centrifugation at 6,000 rpm, and the residue was washed with ethanol to remove the non-reacted organic components. The purified metal nanoparticles were dispersed in 10 mM PBS buffer at pH = 7.2.
The labeled anti-CCR5 mAbs were covalently bound on the silver nanoparticles via condensation [22]. Typically, the succinimidylated metal nanoparticles (2×10−8 M) and labeled mAb (4×10−7 M) were co-dissolved in 10 mM PBS buffer at pH = 8.0. The reaction solution was continuously stirred for 2 h at 4°C. The suspension was first centrifuged at 2,000 rpm for 2 min. to remove the aggregated metal nanoparticles, and then at 8,000 rpm to recover the metal nanoparticles. The residue was redispersed in 10 mM PBS buffer at pH = 7.2. One drop of diluted ammonium hydroxide was added into solution to eliminate any non-reacted succinimidyl moieties on the metal nanoparticles. The sample was then centrifuged at 8,000 rpm and washed with 10 mM pH = 7.2 PBS buffer. The residue was dispersed in 10 mM PBS buffer and stored at 4°C.
Cell culture and immuno-reaction on the cell surface
T-lymphocytic PM1 cells were grown in the RPMI-1640 culture medium (Sigma) supplemented with 10% (v/v) heat-inactivated fetal bovine serum (Atlanta Biologicals Inc. GA) that contained 200 units/ml penicillin, 200 units/ml streptomycin (Invitrogene) and recombinant human interleukin (100 U/ml) (Roche, Indianapolis, Indiana, USA). The cells were grown for 6 days prior to the conjugation experiments [23]. The number of cells was counted to be ca. 5 × 106 cells/ml. PM1 cells in 500 L aliquots were incubated with the mAb-metal complex in serial concentrations of 200, 100, 40, 15, 3, 0.3 × 10−12 M for 2 h at room temperature. The labeled cell lines were washed with PBS-Mg solution and then re-suspended in 500 L PBS buffer. 20 L cell-suspended solutions were taken and cast on the cleaned glass coverslips and dried in air for cell imaging.
Cell imaging, TEM and spectral measurements
Absorption spectra were monitored with a Hewlett Packard 8453 spectrophotometer. Ensemble fluorescence spectra were recorded with a Cary Eclipse Fluorescence Spectrophotometer. Cell images were obtained with a time-resolved confocal microscope (MicroTime 200, PicoQuant) [21], which consists of an inverted confocal microscope coupled to high-sensitivity detection optics. A single mode pulsed laser diode (635 nm, 100 ps, 40 MHz) (PDL800, PicoQuant) was used as the excitation source. An oil immersion objective (Olympus, 100×, 1.3NA) was used for both focusing laser light onto sample and collecting the emission. The emission signal was passed through a dichroic mirror, focused onto a 75 μm pinhole for spatial filtering to reject out-of-focus light, and detected with a single photon avalanche diode (SPAD) (SPCM-AQR-14, Perkin Elmer Inc). Bandpass filters were used to eliminate the scattering incident light and minimize the spectral crosstalk. The photon counts were collected with a TimeHarp 200 board and stored in Time-Tagged Time-Resolved Mode (TTTR), which allows every detected photon to be accepted with a specific excitation pulse and spatial location.
Transmission electron micrographs (TEM) were taken with a side-entry Philips electron microscope at 120 keV. Samples were cast from water solutions onto standard carbon-coated (200–300 Å) Formvar films on copper grids (200 mesh) by placing a droplet of a 1 mg/mL aqueous sample solution on the grids. The size distribution of the metal cores was analyzed with Scion Image Beta Release 2 using data from at least 200 nanoparticles.
Results and Discussion
Prior to cell imaging, we characterized the spectral properties of the mAb-metal complexes. From the absorption spectrum (Figure S1) and extinction coefficient of Alexa Fluor 680 [20], we determined that each anti-CCR5 mAb molecule contained an average of 3.4 fluorophores. We found this number of fluorophore to provide adequate brightness without interfere with the immune reactivity [20]. Upon excitation at 650 nm, the labeled mAbs exhibited an emission maximum of 701 nm (Figure S1), close to the free Alexa Fluor 680. On the TEM images, the metal nanoparticles showed homogeneous sizes with 80% in a range of 20±10 nm. Larger nanoparticles are known to be able to enhance the fluorescence more efficiently [17]. However, we selected the smaller size particles because there were less steric hindrances to bind with the target molecules on the cell surfaces [22].
For use with cell imaging, it is important for the nanoparticles to be chemically stable and not aggregate in the culture medium. In order to improve the stability of metal nanoparticles in buffer, the metal nanoparticles were coated with organic monolayers. Covalent binding sites for anti-CCR5 mAbs on the metal nanoparticles were obtained by partial substitution of the surface-bound ligands with (2-mercapto-propionylamino) acetic acid 2,5-dioxo-pyrrolidin-1-ylester. Such a surface exchange is known to occur at the molar ratio of 1:1 [21], so we expect that there are approx. 20 succinimidyl ligands substituted per metal nanoparticle. The labeled mAb molecules were then added in a 20-fold excess relative to the silver nanoparticles to reduce possible aggregation of the metal nanoparticles. The absorbance spectrum of the mAb-metal complexes (Figure S2) showed an identical plasmon resonance to that of individual silver nanoparticles suggesting that the nanoparticles were not significantly aggregated. The metal nanoparticles were observed as clear single particles on the TEM image with an average 20 nm diameter (inset of Figure S2) furthermore supporting the fact the metal nanoparticles did not aggregate. Upon excitation at 650 nm the mAb-metal complexes exhibited an emission maximum of 705 nm (Figure S3), which is a slight red-shift from the labeled mAbs without metal.
In order to determine the number of mAb on a single metal nanoparticle, we removed the metal core of silver nanoparticle by adding several drops of 0.1 N NaCN [24]. The plasmon resonance was found to progressively disappear as the metal was removed by NaCN. As a result, the bound mAb molecules were released into solution. The concentration of dissolved metal nanoparticle in solution was estimated from the intensity of plasmon resonance before the treatment; and the concentration of released mAb was estimated from the emission intensity after the treatment. The data indicated that there was an average of 8.3 mAb molecules per silver nanoparticle. We also noticed that the fluorescence intensity decreased upon the dissolution of metal (Figure S2), which was due to the loss of the metal - fluorophore interaction [17,21].
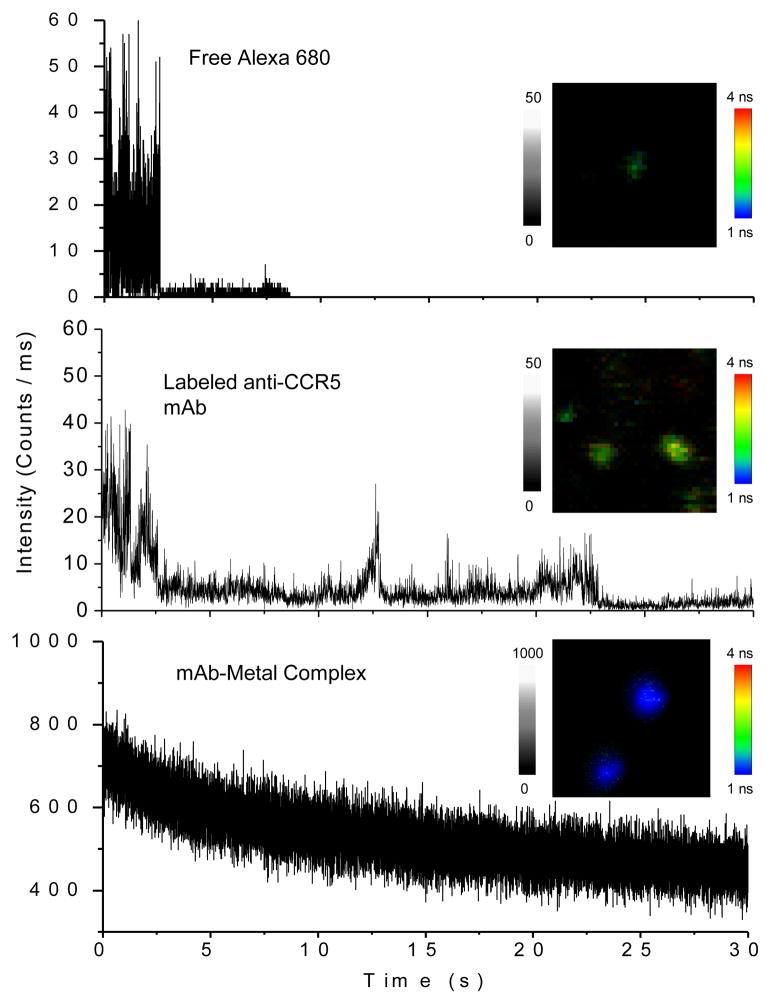
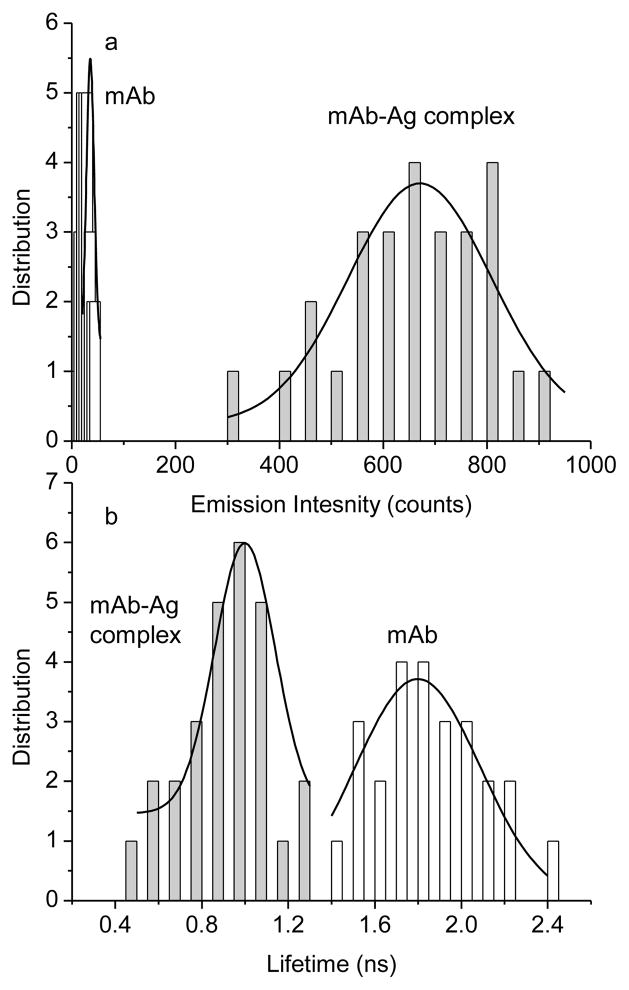
The optical properties of single mAb-metal complexes were evaluated with the time-resolved confocal microscope. Individual round spots were observed in the images (Figure 2) that appeared to be due to single nanoparticles. The time traces from apparent single mAb-metal complexes decayed exponentially with the observation time due to the continuous photobleaching of multiple fluorophores on each particle. In contrast, the time traces from single labeled mAb molecules without metal displayed multi-step photobleaching indicating that mAb molecule had several fluorophores. The emission rates of these mAb molecules were lower than those of the mAb-metal complexes. The time traces of single Alexa Fluor 680 molecules displayed typical one-step photobleaching. For each sample, at least 50 emission spots were collected for the analysis of the emission intensity and lifetime (Figure 3). The results showed that a labeled mAb molecule exhibited a 3-fold increase of emission intensity comparing with a free Alexa Fluor 680 molecule. This is consistent with our estimation that each mAb molecule is labeled by 3.4 fluorophores. The lifetime was almost unchanged. The mAb-metal complex displayed a 20-fold increase of emission intensity relative to the labeled mAb without metal (Figure 3a) that we believe was due to a combination of increased rates of excitation and emission. Simultaneously, the lifetime was dramatically shortened from 1.8 ns to 0.9 ns (Figure 3b), which was due to the increase of radiative rates of fluorophores when coupled with metal nanoparticle [15]. We found that the lifetime of the mAb-metal complex is significantly shorter than that of the cellular autofluorescence (2 – 10 ns). There was a range of intensity or lifetime for the single mAb-metal complexes, which was probably due to the loading number of fluorophores per mAb molecule, metal nanoparticle size, and emission distribution. However, these ranges were ±50% proximately acceptable for the current experiments. Relative to the labeled mAbs without metal, the mAb-metal complexes display enhanced emission intensities and shortened lifetimes, so we anticipate these emission signals can be isolated from the cellular autofluorescence in cell imaging.
Figure 2.
Representative time traces of single Alexa Fluor 680, fluorescent anti-CCR5 mAb, and mAb-metal complex upon excitation at 635 nm. The insets represent the respective typical single molecule or nanoparticle fluorescence images recorded in either emission intensity or lifetime. The scales of diagrams were 5 × 5 μm and the resolutions were 100 × 100 pixel with an integration of 0.6 ms/pixel.
Figure 3.
Histograms of emission intensity and lifetime collected from the fluorescence images of labeled mAbs in the absence of metal and mAb-metal complexes.
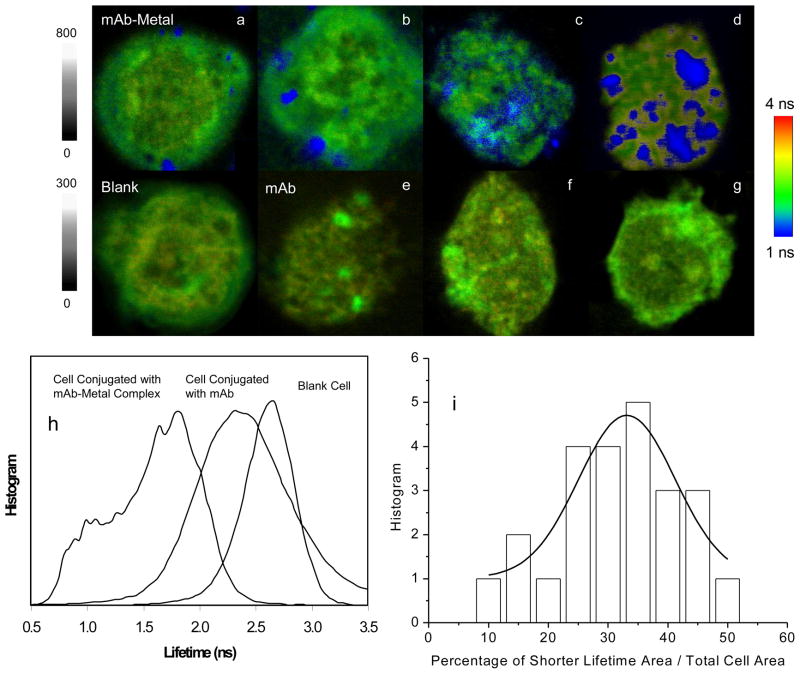
To determine the optimal condition for binding the mAb-metal complexes with the CCR5 receptors on the cell surfaces, the cells were incubated with increasing concentration of mAb-metal complex. The incubated cells were then washed with buffer and cast on the coverslips for cell imaging. As a control, the cells were also incubated under the same conditions with the labeled mAbs but without metal nanoparticles. Representative single cell images are presented in Figure 4. As expected, the emission spots from the mAb-metal complexes were clearly distinguished on the intensity and lifetime cell images due to stronger emission intensities and shorter lifetimes. In the controls, the emission signals from the labeled mAbs without metal were dim and blurry. W also noticed that when the cells were incubated with a low concentration of mAb-metal complex, individual round emission spots were obtained from individual probes complexes, which presumably corresponded with single CCR5 molecules or small clusters on the cell surfaces. With increasing probe concentration, the emission spots became dense and continuous throughout the cell images, suggesting that the mAb-metal complexes were aggregated in the large clusters.
Figure 4.
Upper panel: representative single PM1 cell images that were collected from the cells under the incubation with different concentrations of mAb-metal complex (a–d) or free mAb (bottom panel). The cell lines were incubated with increasing concentration of mAb-metal complex at (a) 0.3; (b) 3; (c) 15, and (d) 40 pM; or with increasing concentration of mAb at (e) 0; (f) 10; (g) 55, and (h) 120 pM, respectively. The images were recorded either in emission intensity or lifetime. The scales of diagrams are 15 × 15 μm and the resolutions are 400 × 400 pixel with an integration of 0.6 ms/pixel. Bottom panel: (h) lifetime distributions over the entire images of blank cell and cell by the mAb-metal complex or labeled mAb in the absence of metal and (i) percentage distribution of occupation area by the mAb-metal complex throughout the entire cell image.
The emission intensity and lifetime cell images were processed with PicoQuant analysis software. At least 20 single cell images were analyzed for each probe concentration to obtain the average emission intensity and lifetime over the entire cell. These average values were plotted against the incubation concentration (Figure S3), showing that the emission intensity increased but the lifetime decreased with the concentration. Both curves reached plateaus at 40 pM, suggesting that the immuno-reaction on the cell surface was saturated. We obtained the similar curves from the cell images with the labeled mAbs, showing similar tendencies but much smaller scales (Figure S3). It is noticed that the error bars in Figure S3 are relatively large, which is probably due to the heterogeneous distributions of the CCR5 receptors on the T-lymphocytic cell surfaces [25].
In order to test the specificity of the mAb-metal complex binding with the CCR5 receptor, we performed similar experiment using the complex that was not specific for CCR5. In the experiment, Cy5-avidin conjugates were covalently bound on the 20 nm silver nanoparticles followed by incubation with the PM1 cells. The collected cell images displayed only few or no emission spots, implying that without the specific anti-CCR5 antibodies, the avidin-metal complexes could not be efficiently conjugated on the cell surfaces. Thus, the cell images with the mAb-metal complexes reflect the specific interactions of the complexes with the CCR5 receptors.
The use of fluorescence lifetime, in stead of intensity, may provide an additional mean to detect the target molecules in a cell image [10]. When the lifetime distributions throughout the entire cell surfaces were studied with the different treatments, clear differences were obtained. The blank cell lifetime images have their most signals above 2.0 ns. In contrast, the cell lifetime images with the mAb-metal complexes have their most signals below 2 ns. The cell lifetime images with the labeled mAbs have their most signals between these two extremes. Thus, the lifetime distribution curves from the blank cells and the cells with the labeled mAbs have a significant overlap whereas those from the blank cells and the cells with the mAb-metal complexes have almost no overlap. These differences in the lifetime allowed us to resolve the signals of the mAb-metal complexes from those of mAbs alone or from cellular autofluorescence.
The cell images with the mAb-metal complexes (Figure 3d) revealed the clusters of fluorescence throughout the cell images. Hence, we suggest most of the CCR5 receptors appear as clusters on the cell surfaces. As a result, it is difficult to count the CCR5 receptors by counting the emission spots on the cell images. Thus, we utilized the occupation percentage to represent the presence of CCR5 receptors. Using the OriginPro-7 software, the data on the single cell lifetime image was totally converted to one followed by integration. This value represents the area of the entire cell. Then, the lifetime data over 1.5 ns on the single cell lifetime image, which may mostly come from the cellular autofluorescence, were removed, and the left data were subsequently converted to one followed by integration. This value represents the area of CCR5 receptors on the cell. The ratio of two integrated values was regarded as the occupation percentage of the CCR5 receptors. By analyzing a population of at least 20 cell lifetime images with the mAb-metal complexes, we can give the histogram of the occupation percentage in Figure 4i. We observed a broad distribution range of 10 – 50% and a maximum value of 30%. The result from flow cytometry measurement demonstrates that there is an average of 4,000 CCR5 receptors on the PM1 cells [23]. The current results reveal that most of the CCR5 receptors appear as clusters on about 1/3 area of the PM1 cells as well as few as individual receptors. This observation is important in understanding the HIV virtual infection and disease progress in future works.
Conclusion
In this paper, we report that the emission signals from the mAb-metal complexes can be distinguished clearly from the cellular autofluorescence on the cell images. This unequivocal separation allows us to detect the amount of CCR5 molecule and observe the spatial distribution on the cell surface with the minimum interference of cellular autofluorescence. The use of the mAb-metal complex as molecular imaging probe can provide a new way to visualize the interaction between the CCR5 receptor with the HIV-1 viral gp120 on the cell surface that is an important step during HIV infection.
Supplementary Material
Acknowledgments
This research was supported by grants from NIH (EB009509, HG-002655, HG005090, EB006521, and CA134386).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Dragic T, Litwin V, Allaway GP, Martin SR, Huang YX, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson DAD, Cormier CE, Dragic T. CCR5 and CXCR4 Usage by Non-Clade B Human Immunodeficiency Virus Type 1 Primary Isolates. J Virol. 2002;76:3059–3064. doi: 10.1128/JVI.76.6.3059-3064.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lusso P. HIV and the chemokine system: 10 years later. Embo J. 2006;25:447–456. doi: 10.1038/sj.emboj.7600947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Y, Paxton WA, Wolinsky SM, Neumann AU, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau NR, Phair J, Ho DD, Koup RA. The role of a mutant CCR 5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 5.Ntziachristos V, Chance B. Breast imaging technology: Probing physiology and molecular function using optical imaging - applications to breast cancer. Breast Cancer Res. 2001;3:41–46. doi: 10.1186/bcr269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hawrysz DJ, Sevick-Muraca EM. Imaging disease with near-infrared fluorescent probes. Neoplasia. 2000;2:388–417. doi: 10.1038/sj.neo.7900118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weissleder R, Tung CH, Mahmood U, Bogdanov A. Uses of fluorescent proteins to visualize cancer in vivo: ex vivo imaging using fluorescent proteins. Nature Biotechnol. 1999;17:375–378. doi: 10.1038/7933. [DOI] [PubMed] [Google Scholar]
- 8.Smiley RD, Hammes GG. Single molecule studies of enzyme mechanisms. Chem Rev. 2006;106:3080–3094. doi: 10.1021/cr0502955. [DOI] [PubMed] [Google Scholar]
- 9.Michalet X, Weiss S, Jager M. Single-molecule fluorescence studies of protein folding and conformational dynamics. Chem Rev. 2006;106:1785–1813. doi: 10.1021/cr0404343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lakowicz JR. Principles of Fluorescence Spectroscopy. 3. Springer Published; New York: 2006. [Google Scholar]
- 11.Sokolov K, Chumanov G, Cotton TM. Enhancement of molecular fluorescence near the surface of colloidal metal films. Anal Chem. 1998;70:3898–3905. doi: 10.1021/ac9712310. [DOI] [PubMed] [Google Scholar]
- 12.van Dijk MA, Lippitz M, Orrit M. Far-field optical microscopy of single metal nanoparticles. Acc Chem Res. 2005;38:594–601. doi: 10.1021/ar0401303. [DOI] [PubMed] [Google Scholar]
- 13.Hao E, Li S, Bailey RC, Zou S, Schatz GC, Hupp JT. Optical properties of metal nanoshells. J Phys Chem B. 2004;108:1224–1229. [Google Scholar]
- 14.Lakowicz JR. Radiative Decay Engineering: Biophysical and Biomedical Applications Anal. Biochem. 2001;298:1–24. doi: 10.1006/abio.2001.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakowicz JR. Radiative decay engineering 5: metal-enhanced fluorescence and plasmon emission Anal. Biochem. 2005;337:171–194. doi: 10.1016/j.ab.2004.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Fu Y, Liang D, Zhao RY, Lakowicz JR. Fluorescent avidin-bound silver particle: a strategy for single target molecule detection on a cell membrane. Anal Chem. 2009;81:883–889. doi: 10.1021/ac801932m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Fu Y, Chowdry MH, Lakowicz JR. Single molecule studies on fluorescently labeled silver particles: effects of particle size. J Phys Chem C. 2008;112:18–26. doi: 10.1021/jp074938r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lal S, Clare SE, Halas NJ. Nanoshell-enabled photothermal cancer therapy: impending clinical impact. Acc Chem Res. 2008;41:1842–1851. doi: 10.1021/ar800150g. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Roll D, Geddes CD, Lakowicz JR. Aggregation of silver nanoparticle-dextran adducts with concanavalin A and competitive complexation with glucose. J Phys Chem B. 2004;108:12210–12214. doi: 10.1021/jp037772c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brinkley MA. brief survey of methods for preparing protein conjugates with dyes, haptens and crosslinking reagents. Bioconj Chem. 1992;3:2–13. doi: 10.1021/bc00013a001. [DOI] [PubMed] [Google Scholar]
- 21.Templeton AC, Wuelfing WP, Murray RW. Monolayer-protected cluster molecules. Acc Chem Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, Fu Y, Liang D, Nowaczyk K, Zhao RY, Lakowicz JR. Single cell fluorescence imaging using metal plasmon-coupled probe 2: lifetime image and single molecule counting. Nano Lett. 2008;8:1179–1186. doi: 10.1021/nl080093z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heredia A, Gilliam B, DeVico A, Le N, Bamba D, Flinko R, Lewis G, Gallo RC, Redfield RR. CCR5 density levels on primary CD4 T cells impact the replication and Enfuvirtide susceptibility of R5 HIV-1. AIDS. 2007;21:1317–1322. doi: 10.1097/QAD.0b013e32815278ea. [DOI] [PubMed] [Google Scholar]
- 24.Rosi NL, Mirkin CA. Nanostructures in biodiagnostics. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]
- 25.Jacques R, Pierre P, Michel S, Vincent B, Pascal A, Odile A, Marie-Christine P, Jacques C, Jean-François E, Pierre C. CD4 T cell surface CCR5 density as a host factor in HIV-1 disease progression. Aids. 2001;15:1627–1634. doi: 10.1097/00002030-200109070-00004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.