Abstract
The history of plant biology is inexorably intertwined with the conception and discovery of auxin, followed by the many decades of research to comprehend its action during growth and development. Growth responses to auxin are complex and require the coordination of auxin production, transport, and perception. In this overview of past auxin research, we limit our discourse to the mechanism of auxin action. We attempt to trace the almost epic voyage from the birth of the hormonal concept in plants to the recent crystallographic studies that resolved the TIR1-auxin receptor complex, the first structural model of a plant hormone receptor. The century-long endeavor is a beautiful illustration of the power of scientific reasoning and human intuition, but it also brings to light the fact that decisive progress is made when new technologies emerge and disciplines unite.
The history of research on auxin—from its discovery almost a century ago to the recent determination of the structure of its receptor—illustrates the power of emerging technology and interdisciplinary approaches.
The simple hormone related to tryptophan, indole-3-acetic acid (IAA or auxin), has probably been the most intensely studied molecule in plants as it impacts virtually every facet during their life cycle. In fact, a total failure in IAA production has not been reported for any plant alive. Thus, it is not surprising that auxin biology is one of the oldest fields of experimental plant research and that the underlying mechanisms of its action have captivated many generations of scientists.
The regulation of growth and development by IAA is largely executed via the coordination of a triumvirate of complex processes: auxin metabolism, auxin translocation, and auxin response. The intricate maze of metabolic reactions related to IAA, encompassing its spatio-temporal patterns of biosynthesis, reversible conjugation, and degradation, is still unfolding. For example, significant progress has been made charting the biosynthetic pathways by a combination of genetic and biochemical approaches, which revealed the operation of at least five different routes to IAA. Our current understanding of the redundant metabolic processes that determine auxin supply has recently been reviewed (Woodward and Bartel 2005; Delker et al. 2008; Chandler 2009). The delivery of auxin from its biosynthetic sources to its sites of perception follows two major, unrelated modes of transportation: rapid long-distance movement via the phloem sap, and slower cell-to-cell distribution over shorter distances. The latter process is unique among plant hormones and signaling molecules because of its tissue-dependent directionality, which is established by the asymmetric subcellular localization of auxin influx and efflux carrier proteins. The chemiosmotic hypothesis of polar auxin transport proposed a mechanistic framework and made astonishingly visionary predictions (Rubery and Sheldrake 1974; Raven 1975). The interference of local, cell-specific auxin biosynthesis and directional intercellular auxin transport causes a differential distribution of auxin in a given tissue, which, in its extreme, can give rise to distinct and steep auxin maxima or minima, or establish more graded differences in auxin concentration between its cells. Such auxin gradients are often influenced by diverse internal and external cues and have been implicated in the regulation of numerous auxin-mediated processes relevant to the adaptation of plant form and function. Research on polar auxin distribution enjoyed a renaissance during the past decade and the remarkable progress made has been documented in various excellent reviews (see Benjamins and Scheres 2008; Petrášek and Friml 2009; Vanneste and Friml 2009).
The positional information encrypted in the landscape of differential auxin distribution in a field of cells is believed to determine individual cell responses to the hormone (Vanneste and Friml 2009), which brings into focus the mechanism of auxin perception and ensuing action. In this article, we revisit the long-sought and sometimes hard-fought quest to understand these fundamental processes. After more than seven decades of incremental progress, occasionally disrupted by stagnation or distraction, today it is well established that auxin primarily acts by reprogramming gene expression to influence plant growth (Chapman and Estelle 2009). However, it was only recently that the initial mechanism of auxin perception was uncovered, which revealed a surprisingly short path to the execution of transcriptional response (Tan et al. 2007), envisioned more than two decades ago (Theologis 1986). In this brief overview, we highlight the almost epic voyage from the emergence of the hormone concept in plants, followed by the discovery of IAA and the struggle to understand its mode of action, to the first structural model of a plant hormone receptor, the Rosetta stone in auxin biology.
BIRTH OF “AUXANO”
The concept of hormone action is deeply rooted in the history of botany and can be traced back for more than 250 years. The phenomenon of correlations, that is the influence exerted by one plant organ on another, or in today’s words, functional relationships between distant parts of a plant, was first formulated in 1758 by Henri Lois Duhamel du Monceau. His studies on growth and strength of wood led him to conclude that two streams of sap moving in opposite directions are responsible for establishing such correlations. A typical experiment would show that interruption of the downward moving sap by clamps or ring wounds caused swellings that often formed callus tissue and gave rise to root formation above the site of injury. For about a century, the idea of physiological correlations lost its appeal to the study of plant anatomy, however, only until the discovery of the sieve elements together with their associated role in sap conductance. The concept regained momentum in the 1880s when Julius von Sachs unified known facts on correlation phenomena with advances in morphology. He proposed a first coherent framework of root-forming, flower-inducing, and other special substances, which move in different directions through the plant to control its growth and development (see Went and Thimann 1937).
Directional growth responses such as phototropism and geotropism were soon regarded as a special kind of correlation phenomenon and their study directly led to the discovery of auxin. Charles Darwin, who became interested in plant tropisms, showed that light and gravity are perceived by the tips of shoots and roots, and that the effect of an asymmetric stimulus is transmitted to the lower region beneath the tip, which then responds with differential growth, causing curvature. For the phototropic response, he performed simple but insightful experiments on coleoptiles of etiolated canary grass seedlings by shielding either the tip or the elongating base from unilateral illumination. He also noted that when the tip was removed, no curvature occurred in the lower part of the coleoptile. The essence of these observations is encapsulated in his famous and widely cited phrase “we must, therefore, conclude that when seedlings are freely exposed to a lateral light, some influence is transmitted from the upper to the lower part, causing the latter to bend”, published in his book The Power of Movement in Plants (1880). Thus, Darwin is credited for conceiving the idea of a transmittable correlation factor regulating growth at a distant site. However, the nature of this factor remained elusive for more than 30 years. Building on Darwin’s observations, Peter Boysen-Jensen showed that the phototropic stimulus is transmitted from an excised tip across a gelatin barrier into the lower part of an Avena coleoptile where it still elicits the curvature response. Arpad Paál further developed this line of experimentation, which supported the diffusible nature of the correlation carrier. His crucial contribution was to show that an excised and off-center reattached tip could induce curvature of the base even in darkness. He was the first to conclude that the tip must produce and release a chemical substance that travels toward the coleoptile base to promote growth and, in addition, that unilateral light causes an asymmetric transmission of this substance, which later became a cornerstone of the Cholodny-Went hypothesis. Thus, Paál’s conclusion meets the original definition of hormones by Bayliss and Starling (1904), who wrote: “the peculiarity of these substances (hormones) is that they are produced in one organ and carried by the blood current to another organ, on which their effect is manifested” (see Went and Thimann 1937).
The implication of Paál’s work sparked attempts for a direct demonstration of the postulated growth-promoting substance. P. Stark developed a method to replace excised tips with asymmetrically attached agar blocks containing various tissue extracts, but bending of the decapitated coleoptiles could not be induced because of low abundance and instability of the growth factor, as we know today. A breakthrough was achieved in 1926–1928 by Frits Went who adapted Stark’s agar block method and advanced Paál’s line of reasoning to make the definitive discovery of the hormone, named auxin soon after its structural identification in 1934/35 (a term derived from the Greek verb auxano, which means “to grow or to expand”). He placed excised Avena coleoptile tips on agar blocks that received the growth-promoting substance by diffusion and could then serve as an artificial but potent stimulus source for inducing curvature of decapitated coleoptiles. Went developed the Avena coleoptile curvature test to a quantitative bioassay, which he applied to determine some of the physicochemical properties of the hormone. Whereas Frits Went succeeded in capturing the substance by a simple diffusion technique, its isolation and the determination of its chemical identity followed a meandering path. Because of insufficient analytical methods to track minute amounts of the hormone, its purification was carried out with human urine and culture filtrates of several fungi, both of which are rich sources of substances active in the Avena bioassay. After a false start leading to auxin mimics in urine (incorrectly termed auxin a and auxin b), the subsequent analysis of a third component (ironically named heteroauxin) identified the structure of auxin as indole-3-acetic acid, which was also found in fungal cultures (Kögl et al. 1934; Thimann and Koepfli 1935). It was not until a decade later that IAA was eventually discovered (Haagen-Smit et al. 1946) in a plant (Zea mays), and it soon became clear that IAA is the principal auxin in all plant species.
SETTING SAILS FOR EXPLORATION
After the landmark experiments of that early period, research on auxin diversified and followed several major trajectories that are intensely explored to this day. The recognition of polar auxin transport, which is essential for establishing asymmetries during cell growth and plant development, was closely connected to the discovery of auxin. The classical experiments by Frits Went, who showed movement of auxin through excised coleoptile segments in an orientation-dependent fashion, were systematically continued by van der Weij’s thesis research in 1932. He showed that directional auxin transport in coleoptile segments (apically to basally) is independent of auxin gradients established by donor and receiver agar blocks, and that the transport rate is far greater than the diffusion rate of the hormone. The availability of [14C]-carbon (a by-product of the Manhattan project in the 1940s that proved essential for the elucidation of carbon fixation by the Calvin cycle) made possible the synthesis of radiolabeled IAA. Assays with [14C]-IAA revealed tissue uptake by a minor diffusion component followed by polar transport, which is readily saturated and energy-dependent. Transport studies with short pulses of the tracer led to the concept of “bound” auxin (immobilized or conjugated IAA) and opened the way to the study of auxin homeostasis and metabolism (see Thimann 1977).
Soon after the chemical identification of IAA, a number of structurally related compounds with auxin activity were reported. These findings sparked systematic studies of structure-activity relationships for several decades, which often used auxin-dependent curvature of split pea stems as a more robust bioassay. The large body of experimental data led to predictions of structural requirements for auxin activity and spatial features of a hypothetical receptor site (see Thimann 1969). The recent elucidation of crystal structures for the TIR1 complex in association with an Aux/IAA degron peptide together with one of three different auxin compounds (including the natural IAA and two synthetic auxins) confirmed most of the early structure–function predictions and can be viewed as a historical milestone of this long line of investigation (Tan et al. 2007). The search for auxin receptors by a biochemical strategy (isolation of auxin-binding proteins) resulted in the identification of ABP1 (Hertel et al. 1972). Although reverse genetic studies support its function as an extracellular auxin receptor (Jones et al. 1998; Chen et al. 2001; Braun et al. 2008), additional components of a putative ABP1 pathway remain to be discovered.
An influential direction of research was initiated by James Bonner (1933), who showed in “straight growth” assays that auxin induces rapid growth (classically defined as an irreversible increase in volume) of isolated Avena coleoptile segments by cell enlargement. These simpler bioassays were soon extended to excised stems of numerous other plants and provided the experimental system for investigating the biochemical basis of auxin action. Whereas auxin activity was initially defined as the hormone promoting cell enlargement of coleoptiles and stems, it soon became clear that auxin also plays seemingly unrelated and more long-term roles during the life cycle of a plant. The first surprise was the demonstration that auxin stimulates cambial activation and cell division in sunflower (Snow 1935). The description of other, now well-known auxin effects soon followed suit, such as root-growth inhibition, which was extensively used in structure-activity studies and later in screens for auxin-insensitive mutants, or the inhibition of axillary bud growth, which explained the phenomenon of apical dominance. Other reported auxin effects include inhibition of leaf and fruit abscission, stimulation of adventitious and lateral root formation, induction of vascular differentiation, or elevation of ethylene production (see Thimann 1977). With the discovery and identification of additional plant hormones since the 1950s, considerable effort was placed to explore the interactions of auxin with other plant hormones, most notably with cytokinin, ethylene, brassinoisteroids, and jasmonic acid.
THE LONG JOURNEY TO THE SHORT PATH OF AUXIN ACTION
The very earliest observation leading to the discovery of auxin rested on the fact that IAA promotes cell enlargement. As determined in “straight growth” assays of stem sections, stimulation of cell elongation is among the fastest hormonal responses known with a lag period of 10–25 minutes (Brummell and Hall 1987). Thus, it is not surprising that this classic and simple growth response, which is not complicated by cell division, made the phenomenon of accelerated cell elongation an attractive experimental system for investigating the primary mechanism of auxin action.
Effector of Enzyme Activity?
Before the discovery of nucleic acids as carriers of genetic information in the 1940s and 1950s, research followed the emerging paradigm of biochemistry and attempted to establish a connection between the activity of enzymes essential for growth and auxin, with the hormone presumed to act as a coenzyme. Bonner (1936) recognized the dependence on oxygen availability of auxin-induced growth, which supported this proposition. In the following years through the 1950s, Bonner, Thimann, and their colleagues showed that a wide spectrum of compounds, which inactivate enzymes of the Krebs cycle and respiration, also inhibit auxin-induced growth. However, a much hoped-for specific effect of auxin on a metabolic reaction never materialized. A related line of research began soon after the discovery of auxin with the proposal that the hormone stimulates growth by increasing the plasticity of the cell wall (see Thimann 1977). Both biochemical concepts of that early period survived to this day. ATP-dependent proton pumping across the plasma membrane, directly regulated by auxin, is a principal tenet of the acid growth hypothesis of auxin-induced cell growth (see the following discussion), which is still controversially discussed among plant physiologists (see Kutschera vs. Grebe 2006).
The Gene Activation Hypothesis
Independent work in Folke Skoog’s laboratory in the early 1950s showed that the ratio of auxin to cytokinin as well as their concentrations in the growth medium profoundly altered not only the growth of tobacco pith tissue but also the relative levels of nucleic acids in the tissue (Silberger and Skoog 1953). These observations led to the proposal that the mechanism of plant hormone action involves changes in nucleic acid metabolism (Skoog 1954). A series of follow-up studies provided much evidence that supported and expanded Skoog’s original observation to other plant species and organs, which were shown to accumulate RNA in response to both naturally occurring and synthetic auxins (Trewavas 1968, Key 1969). Research in the 1960s showed enhanced incorporation of radioactive precursors into RNA and proteins after prolonged treatment (several hours) of excised stem sections with auxin. In addition, the use of rather specific inhibitors of RNA and protein biosynthesis such as actinomycin D and cycloheximide clearly revealed that auxin-induced cell elongation requires continued RNA and protein synthesis. Collectively, these correlative observations led to the concept of “growth-limiting” RNAs and proteins and provided the basis for the gene activation hypothesis (Key 1969), which postulated that auxin regulates the synthesis of specific RNAs necessary for cell growth.
The Acid Growth Hypothesis
The hypothesis that gene activation reflects the primary event in auxin action was seriously challenged in the 1970s. The principal argument against the gene-centered view focused on the discrepancy between the timing of auxin-induced gene expression and the rapid kinetics of auxin-stimulated cell elongation. Although the experimental methods at that time remained of insufficient resolving power to show concurrent synthesis of specific RNAs and proteins in response to auxin, a refined technique for measuring growth of excised oat coleoptiles with high resolution determined the lag phase for auxin-dependent growth by cell enlargement at ∼10 minutes (Evans and Ray 1969). Thus, considering known transcription and translation rates in animal systems, it appeared increasingly unlikely that auxin-induced gene expression could establish a comprehensive growth response within this short time frame. The impasse gave way to the acid growth hypothesis, which built on the early idea of direct auxin action on the cell wall (Rayle and Cleland 1970; Hager et al. 1971). According to this view, auxin induces acidification of the apoplast via activation of a proton-secreting plasma membrane ATPase, which subsequently causes a relaxation of the load-bearing cell wall elements by activating wall-loosening proteins such as expansins (Cosgrove 2005). However, no experimental evidence convincingly showed a direct effect of auxin on a transport system such as an H+-ATPase. Before the discovery of rapid gene regulation by auxin, Vanderhoef and Dute (1981) proposed two parallel modes of auxin action in an attempt to merge the two contradictory hypotheses. Their “dual site” hypothesis is based on the observation that auxin-induced growth displays a biphasic pattern and that the two phases are experimentally separable (Vanderhoef and Stahl 1975). The initial but transient increase of elongation rate was proposed to be a consequence of auxin-induced proton secretion, whereas the second phase of maximal elongation rate was thought to be mediated and sustained by auxin-dependent gene expression.
New Technologies Deliver a Fresh Breeze
With no technical advance in sight, the once forceful gene activation hypothesis lost its appeal to many plant physiologists during the 1970s who embraced the acid growth hypothesis as a refreshing thought about a long-standing question. Nonetheless, a few unimpressed researchers withstood the general trend and continued their studies to explore the effect of auxin on gene expression. At the beginning of the next decade, the powerful tools of molecular biology became available and were eagerly adopted by those laboratories, which then rigorously showed that auxin does indeed rapidly alter the expression of specific genes. Although the development of two-dimensional protein agarose gel electrophoresis (2D-PAGE) allowed for higher resolution of complex protein mixtures, rapid auxin-induced changes in the overall pattern of cellular proteins were still difficult to detect because long in vivo labeling periods were required (>1 h) to achieve sufficient specific radioactivity for detecting newly synthesized polypeptides. Labile or low-abundant proteins would also be missed by applying this approach alone. However, the combination of 2D-PAGE with in vitro translation of purified total or poly(A) RNA provided the much needed method for monitoring the spectrum of polypeptides that are synthesized at discrete and very short time points after the application of the hormone. Analysis of in vitro translation products of mRNAs isolated from auxin-treated soybean and pea tissues by 2D-PAGE showed that exposure to the hormone alters the abundance of specific mRNAs in a progressive manner (Zurfluh and Guilfoyle 1982; Theologis and Ray 1982). Interestingly, the earliest increases in the amounts of a few translatable RNAs occurred within 15–20 minutes after auxin application, i.e., concomitantly with or possibly earlier than the initiation of cell elongation and proton secretion, and were gradually followed by changes in mRNA abundance at later time points (>2 h). Whether auxin alters mRNA abundance by increasing transcription rates or influencing posttranscriptional events could only be answered after employing recombinant DNA technology to generate and identify the complementary DNA molecules followed by RNA blot hybridization and in vitro nuclear run-off assays. This powerful new kit of molecular tools enabled the isolation of cDNA probes necessary for testing the gene activation hypothesis. In the early 1980s, three research groups reported the selection of cDNA clones for auxin-responsive mRNAs, which were subsequently used to unambiguously show that auxin rapidly activates transcription (5–15 min) of a select set of genes (Walker and Key 1982; Hagen and Guilfoyle 1985; Theologis et al. 1985). It is noteworthy that the rapid kinetics of auxin-mediated gene induction in pea epicotyls (Koshiba et al. 1995) clearly precede (by 10–15 min) the onset of apoplastic acidification and cell elongation measured in the same experimental system (Jacobs and Ray 1976). Together with the abrogation of auxin-induced proton secretion and cell elongation by inhibitors of RNA and protein synthesis (Theologis et al. 1985), the most likely scenario of auxin-regulated cell enlargement reflects a sequence of events in which auxin only indirectly promotes cell wall acidification via de novo gene expression (Theologis 1986).
The Compass of Early Genes
The strong experimental support for the gene activation hypothesis and the prospect that rapid change of specific gene expression constitutes a primary mechanism of auxin action electrified the field, which soon followed the tracks of the early gene paradigm. The study of immediate early genes or primary response genes, whose expression is independent of de novo protein synthesis by definition, played a significant role for the understanding of how growth factors act in animal cells (Herschman 1991). The stimulus-responsive promoter elements of primary genes represent a bridgehead from which to explore the short and direct signaling pathway in reverse, whereas the products of early genes often function as regulators of the stimulus-specific response. Several classes of primary auxin-responsive genes have been identified (Aux/IAAs, GH3-like, SAURs) and much has been learned from the study of the Aux/IAA gene family, which directly links auxin perception to the control of nuclear gene expression (Abel and Theologis 1996; Mockaitis and Estelle 2008). Many of its members are rapidly induced (5–60 min) by a variety of auxin compounds at physiologically relevant concentrations (Abel et al. 1995). Interestingly, inhibitors of protein synthesis, which are useful to distinguish between primary and secondary gene regulation, induce several Aux/IAA mRNAs even in the absence of the hormone. This intriguing observation was studied in great detail in pea epicotyls and the evidence supported the interpretation that Aux/IAA genes are under control of a short-lived transcriptional repressor (Koshiba et al. 1995). These experiments provided a first hint to the importance of proteolysis in auxin signaling, which was further supported by the finding that at least some Aux/IAA genes encode extremely short-lived proteins of low abundance (Abel et al. 1994). A detailed promoter analysis of representative members of each primary gene class (Ballas et al. 1993; Liu et al. 1994; Li et al. 1994) identified a common auxin-responsive element (AuxRE), which is often shared among early auxin genes. Subsequently, Ulmasov et al. (1997) used an AuxRE derivative as bait in a yeast one-hybrid screen and identified the associated auxin-response factor (ARF1), the founding member of the ARF family of auxin-related transcriptional regulators. Most of the 29 Aux/IAA polypeptides encoded by the Arabidopsis genome are characterized by the presence of four conserved domains (I–IV). The carboxy-terminal domains III and IV mediate homo- and heterodimerization of Aux/IAA polypeptides, as well as the interaction between Aux/IAA and ARF proteins (Remington et al. 2004; Overvoorde et al. 2005). Almost all ARF proteins, encoded by 23 genes in Arabidopsis, share two similar domains at their carboxyl terminus and recognize AuxREs via a conserved, plant-specific DNA binding domain (B3) on their amino-terminal half (Remington et al. 2004; Okushima et al. 2005). Depending on the amino-acid composition of their variable internal region, the largely constitutively expressed ARF proteins activate or repress gene transcription (Ulmasov et al. 1999). Thus, given that the amino-terminal domain I of Aux/IAA proteins functions as a potent repressor domain (Tiwari et al. 2004) that is able to recruit a transcriptional corepressor (Szemenyei et al. 2008), the physical association of Aux/IAA and ARF proteins establishes a negative feedback loop in primary gene regulation, which often shapes a transient auxin response (Abel et al. 1995). The observation that ARF activity can be modulated by additional transcription factors such as MYB77 (Shin et al. 2007), and that chromatin remodeling factors are required for Aux/IAA function (Fukaki et al. 2006), suggest a greater complexity of auxin-dependent transcriptional regulation. Conserved domain II confers instability to Aux/IAA proteins and comprises a transferable degron peptide with a characteristic GWPPV amino-acid motif at its core. Remarkably, elevating auxin level rapidly accelerates (<2 min) proteasome-dependent Aux/IAA protein destruction, indicating that derepression of primary genes by auxin-stimulated proteolysis is an immediate-early and pivotal event in auxin signal transduction (Gray et al. 2001; Ramos et al. 2001; Zenser et al. 2001).
Arabidopsis Genetics Take the Lead
Since the establishment of Arabidopsis thaliana as the model organism for studying the fundamentals of plant development and physiology in the late 1980s (Meyerowitz and Pruitt 1985; Meyerowitz 1987), genetic approaches have been developed to dissect hormone action in this reference species (Klee and Estelle 1991; Estelle 1992). A forward genetic strategy based on root growth inhibition by auxin was taken in parallel to the pursuit of the early gene concept and validated the importance of the ARF-Aux/IAA circuit for auxin-regulated transcription and directly guided the way into the realm of auxin perception. One group of auxin-resistant mutants in Arabidopsis provided overwhelming support for the critical role of Aux/IAA protein abundance. Gain-of-function mutations in a number of Aux/IAA genes were identified that change a conserved amino-acid residue within the degron peptide of domain II (Rouse et al. 1998; Mockaitis and Estelle 2008). As a consequence, the altered Aux/IAA proteins are stabilized and repress ARF function, which often results in dramatic developmental defects because of decreased auxin sensitivity. It is thought that specific responses to auxin are mediated by pairs of interacting Aux/IAA and ARF proteins that are coexpressed in planta. For such established combinations, recessive arf mutations confer similar phenotypes as dominant aux/iaa mutations, which underscore the biological significance of negative feedback regulation in auxin-responsive gene expression (Tatematsu et al. 2004, Weijers et al. 2005).
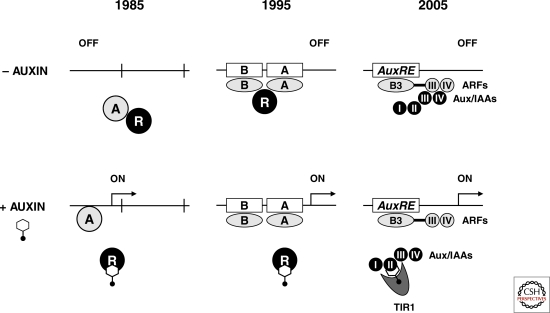
A second group of mutations conferring resistance to auxin or its transport inhibitors stabilize Aux/IAA proteins by disabling components of the SCFTIR1 complex or its associated regulatory proteins (Leyser et al. 1993; Mockaitis and Estelle 2008). SCF (Skp1/Cullin/F-box) complexes are the largest class of E3 ubiquitin protein ligases in plants and catalyze the ubiquitinylation of protein substrates as a prelude to their regulated degradation by the 26S proteasome. Target proteins are recruited to the SCF complex via a specificity-lending F-box protein subunit that is tethered to its scaffold by an adaptor protein. Mutations in TIR1 (TRANSPORT INHIBITOR RESPONSE 1), which encodes a leucine-rich-repeat (LRR) containing F-box protein, confer reduced auxin response. The TIR1 protein is localized to the cell nucleus and interacts with core SCF subunits, thus establishing the SCFTIR1 complex as a central regulator of auxin signaling (Gray et al. 1999; Ruegger et al. 1998). A series of meticulous biochemical studies showed that the SCFTIR1 complex physically interacts with Aux/IAA proteins via their degron peptide in an auxin-dependent manner (Gray et al. 2001; Dharmasiri et al. 2003; Kepinski et al. 2004). Surprisingly, auxin just alone, and not as initially thought auxin-dependent Aux/IAA substrate modification, promotes TIR1:Aux/IAA association by binding directly to the TIR1 subunit (Dharmasiri et al. 2005; Kepinski and Leyser 2005). This unexpected result strongly suggests that nuclear TIR1 and Aux/IAA targets are sufficient for auxin sensing by the SCFTIR1 pathway, which activates early genes by Aux/IAA removal. Thus, auxin signaling is simple and direct, as suspected from the rapid kinetics of primary gene activation (Theologis 1986; Ballas et al. 1995; Koshiba et al. 1995) (Fig. 1).
Figure 1.
The progression of models for the regulation of auxin-inducible genes by derepression. The model on the left was proposed by Theologis (1986) based on published data (Theologis et al. 1985) and suggests control of primary genes by a short-lived protein repressor (R) that inhibits a transcriptional activator (A). Ten years later, this model was refined (center panel) after analyzing the auxin-responsive region of the PS-IAA4/5 promoter by linker scanning mutations, which identified two domains (A and B). Domain A contains typical auxin-responsive DNA elements (AuxRE) and both domains interact with positive transcription factors (Ballas et al. 1995), presumably with auxin-response factors (ARF) (Ulmasov et al. 1997). The current simplified model (right panel) reflects the progress made during the past 15 years by combining molecular, biochemical, and foremost genetic approaches taken by several laboratories (see text), which culminated in the identification of TIR1 as an auxin receptor (Dharmasiri et al. 2005; Kepinski and Leyser 2005) and in the structural model of a TIR1-auxin complex in association with a degron peptide of Aux/IAA domain II (Tan et al. 2007).
Landfall and Homecoming: “Seeing is Believing”
How does auxin enhance TIR1 affinity for its targets? Crucial insight into this pressing question was provided by structural biology. Tan et al. (2007) reported the crystal structures of the complex formed by TIR1 and its ASK1 (Arabidopsis SKP1) adapter, as well as of the TIR1-ASK1 complex in association with an Aux/IAA degron peptide together with one of three different auxins (IAA, 1-NAA, and 2,4-D). The F-box motif of TIR1 interacts with ASK1 to form a stem-like structure that is capped by the solenoid fold of the TIR1-LRR domain. A single pocket on the top surface of the LRR domain binds to auxin and the degron peptide. Interestingly, auxin binding does not induce an allosteric switch or significant conformational change, but the planar ring system of each auxin examined covers up the polar bottom of the binding pocket to form a continuous hydrophobic interaction surface for accommodating the core GWPPV motif of the degron peptide. This regulatory mechanism is consistent with the weak affinity of SCFTIR1 to its Aux/IAA substrates observed in the absence of auxin and with their short basal half-lives. It also explains why several natural and synthetic compounds that only share a planar unsaturated ring structure and a side chain with a carboxyl group, the latter providing anchorage via salt bridges to an internal inositol hexakisphosphate (IP6) cofactor, display “auxin activity” in many bioassays. As long as these diverse compounds can be anchored to the bottom of the TIR1-LRR pocket, are small enough to be accommodated by the auxin-binding cavity, and provide sufficient hydrophobic contact surface for GWPPV adhesion, Aux/IAA proteins will be marked for degradation. The structural model of the TIR1-Aux/IAA coreceptor complex is a remarkable achievement in the long quest to understand an entire auxin signaling pathway and establishes a novel mechanism for sensing small molecules.
CONCLUDING REMARKS
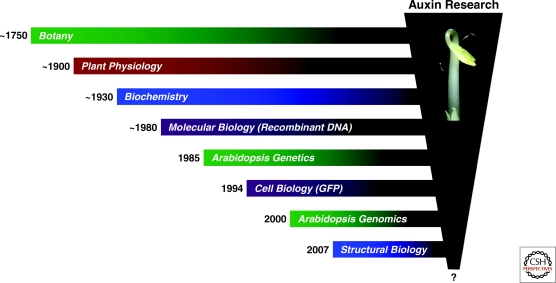
The century-long endeavor from the conception of auxin to the understanding of its perception is a beautiful illustration of the power of scientific reasoning and human intuition, but it also brings to light the fact that decisive progress is made when new technologies emerge and disciplines unite (Fig. 2). The model of auxin action, as we see it today, is largely a sharper image of early ideas and visionary hypotheses, which were difficult to test with the methods at their time. To exemplify this point, Kenneth V. Thimann, the patriarch of auxin biology, wrote in 1969: “ … the molecular structure which an auxin requires for its activity leads to the concept of a specific charge distribution on a surface, at which the auxin molecule becomes oriented. In some way the placement of the auxin molecule there activates the surface, or … may activate another part of the surface which thus acquires enzymatic activity” (p. 29). Thus, Thimann unwittingly and quite correctly sketched out the mechanism of auxin perception by the TIR1 ubiquitin ligase some 40 years ago! Since then, the field witnessed the rise of molecular biology, which amplified the strength of plant physiology and biochemistry. Another turning point was the decision to establish Arabidopsis as the model and reference species for fundamental plant research, which accelerated the pace of progress by harnessing the power of genetics into a tractable experimental system. The leading laboratories studying auxin response in pea, soybean, and mung bean and the like soon converted to the little mustard weed. Cell biology and its advances in noninvasive imaging were the next additions to the arsenal of tools, which proved invaluable for the study of polar auxin transport and its underlying mechanisms. Recently, structural biology entered the arena and the relentless cross fire delivered together with forward genetics and modern biochemistry forced auxin to give up one of its most coveted secrets. What might trigger the next wave of insight? New biophysical tools will further push the limits of single cell analysis in planta and provide much needed techniques for directly sensing the levels and monitoring the fluxes of auxin and its numerous metabolic precursors and derivatives in a single cell or subcellular compartment. The imaging of single macromolecules and of their assemblies and cellular functions in real time will deepen our mechanistic comprehension of auxin action. On the other hand, advances in genome and information sciences will illuminate the role of auxin signaling at the systems level during plant-environment interactions as well as in an evolutionary context. Between the reductionist and holistic approaches of future research, we are left to wonder: What’s next, ‘Auxano’?
Figure 2.
Advances in auxin research. A reductionist approach to the understanding of auxin action (indicated by the vertical trapeze) was facilitated (1) by the development of new technologies, such as adopting recombinant DNA technology, or engineering GFP (green fluorescent protein) to a noninvasive intracellular reporter system (Chalfie et al. 1994), (2) by the introduction of Arabidopsis thaliana as a model and reference organism for research in plant biology and genomics (Meyerowitz and Pruitt 1985; Arabidopsis Genome Initiative 2000), and (3) by the synergism of merging different disciplines. The inset shows two classic biological systems for studying auxin response, dark-grown pea and Arabidopsis seedlings.
Footnotes
Editors: Mark Estelle, Dolf Weijers, Karin Ljung, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Abel S, Theologis A 1996. Early genes and auxin action. Plant Physiol 111: 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Oeller PW, Theologis A 1994. Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci 91: 326–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel S, Nguyen D, Theologis A 1995. The PS-IAA4/5-like family of early auxin-inducible mRNAs in Arabidopsis thaliana. J Mol Biol 251: 533–549 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Ke M, Theologis A 1993. Identification of the auxin-responsive element, AuxRE, in the primary indoleacetic acid-inducible gene, PS-IAA4/5, of pea (Pisum sativum). J Mol Biol 233: 580–596 [DOI] [PubMed] [Google Scholar]
- Ballas N, Wong LM, Ke M, Theologis A 1995. Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc Natl Acad Sci 92: 3483–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss WM, Starling E 1904. The chemical regulation of the secretory process. Proc Roy Soc B 73: 310–322 [Google Scholar]
- Benjamins R, Scheres B 2008. Auxin: The looping star in plant development. Annu Rev Plant Biol 59: 443–465 [DOI] [PubMed] [Google Scholar]
- Bonner J 1933. The action of the plant growth hormone. J Gen Physiol 17: 63–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner J 1936. The growth and respiration of the Avena coleoptiles. J Gen Physiol 20: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun N, Wyrzykowska J, Muller P, David K, Couch D, Perrot-Reichenmann C, Fleming AJ 2008. Conditional repression of auxin binding protein 1 reveals that it coordinates cell division and cell expansion during postembryonic shoot development in Arabidopsis and tobacco. Plant Cell 20: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummell DA, Hall JL 1987. Rapid cellular responses to auxin and the regulation of growth. Plant Cell Environ 10: 523–543 [Google Scholar]
- Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC 1994. Green fluorescent protein as a marker for gene expression. Science 263: 802–805 [DOI] [PubMed] [Google Scholar]
- Chandler JW 2009. Local auxin production: A small contribution to a big field. BioEssays 31: 60–70 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M 2009. Mechanism of auxin-regulated gene expression in plants. Ann Rev Genet 43: 265–285 [DOI] [PubMed] [Google Scholar]
- Chen J-G, Ullah H, Young JC, Sussman MR, Jones AM 2001. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev 15: 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove DJ 2005. Growth of the plant cell wall. Nature 6: 850–861 [DOI] [PubMed] [Google Scholar]
- Darwin C 1880. The power of movement in plants. John Murray, London [Google Scholar]
- Delker C, Raschke A, Quint M 2008. Auxin dynamics: The dazzling complexity of a small molecule’s message. Planta 227: 929–941 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M 2005. The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Jones AM, Estelle M 2003. Auxin action in a cell-free system. Curr Biol 13: 1418–1422 [DOI] [PubMed] [Google Scholar]
- Estelle M 1992. The plant hormone auxin: Insight in sight. BioEssays 14: 439–443 [DOI] [PubMed] [Google Scholar]
- Evans ML, Ray PM 1969. Timing of the auxin response in coleoptiles and its implications regarding auxin action. J Gen Physiol 53: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaki H, Taniguchi N, Tasaka M 2006. PICKLE is required for SOLITARY-ROOT/IAA14-mediated repression of ARF17 and ARF19 activity during Arabidopsis lateral root initiation. Plant J 29: 153–168 [DOI] [PubMed] [Google Scholar]
- Gray WM, del Pozo JC, Walker L, Hobbie L, Risseeuw E, Banks T, Crosby WL, Yang M, Ma H, Estelle M 1999. Identification of an SCF ubiquitin-ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev 13: 1678–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M 2001. Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle TJ 1985. Rapid induction of selective transcription by auxins. Mol Cell Biol 5: 1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haagen-Smit AJ, Dandliker WB, Wittwer SH, Murneek AE 1946. Isolation of 3-indoleacetic acid from immature corn kernels. Am J Bot 33: 118–120 [Google Scholar]
- Hager A, Menzel H, Krauss A 1971. Versuche und Hypothese zur Primärwirkung des Auxins beim Streckungswachstum. Planta 100: 47–75 [DOI] [PubMed] [Google Scholar]
- Herschman HR 1991. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem 60: 281–319 [DOI] [PubMed] [Google Scholar]
- Hertel R, Thompson K-S, Russo VEA 1972. In-vitro auxin binding to particulate cell fractions from corn coleoptiles. Planta 107: 325–340 [DOI] [PubMed] [Google Scholar]
- Jacobs M, Ray PM 1976. Rapid auxin-induced decrease in free space pH and its relationship to auxin-induced growth in maize and pea. Plant Physiol 58: 203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im K-H, Savka MA, Wu M-J, Dewitt NG, Shillito R, Binns AN 1998. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science 282: 1114–1117 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2004. Auxin-induced SCFTIR1-Aux/IAA interaction involves stable modification of the SCFTIR1 complex. Proc Natl Acad Sci 101: 12381–12386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kepinski S, Leyser O 2005. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Key JK 1969. Hormones and nucleic acid metabolism. Ann Rev Plant Physiol 20: 449 [Google Scholar]
- Klee H, Estelle M 1991. Molecular genetic approaches to plant hormone biology. Annu Rev Plant Physiol Plant Mol Biol 42: 529–551 [Google Scholar]
- Kögl F, Erxleben H, Haagen-Smit AJ 1934. Über die Isolierung der Auxine a und b aus pflanzlichen Materialien. IX. Mitteilung. Physiol Chem 243: 209–226 [Google Scholar]
- Koshiba T, Ballas N, Wong LM, Theologis A 1995. Transcriptional regulation of PS-IAA4/5 and PS-IAA6 early gene expression by indoleacetic acid and protein synthesis inhibitors in pea (Pisum sativum). J Mol Biol 253: 396–413 [DOI] [PubMed] [Google Scholar]
- Kutschera U, Grebe M 2006. Acid growth and plant development. Science 311: 952–954 [DOI] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M 1993. Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Li Y, Lui ZB, Shi X, Hagen G, Guilfoyle TJ 1994. An auxin-inducible element in soybean SAUR promoters. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle TJ 1994. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6: 645–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz EM 1987. Arabidopsis thaliana. Ann Rev Genet 21: 93–111 [DOI] [PubMed] [Google Scholar]
- Meyerowitz EM, Pruitt RE 1985. Arabidopsis thaliana and plant molecular genetics. Science 229: 1214–1218 [DOI] [PubMed] [Google Scholar]
- Mockaitis K, Estelle M 2008. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol 24: 55–80 [DOI] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D, et al. 2005. Functional genomic analysis of the auxin response factor gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 17: 444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Onodera C, Quach H, et al. 2005. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17: 3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrášek J, Friml J 2009. Auxin transport routes in plant development. Development 136: 2675–2688 [DOI] [PubMed] [Google Scholar]
- Ramos JA, Zenser N, Leyser O, Callis J 2001. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell 13: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA 1975. Transport of indoleacetic acid in plant cells in relation to pH and electrical potential gradients, and its significance for polar IAA transport. New Phytol 74: 163–172 [Google Scholar]
- Rayle DL, Cleland RE 1970. Enhancement of wall loosening and elongation by acid solution. Plant Physiol 46: 250–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remington DL, Vision TJ, Guilfoyle TJ, Reed JW 2004. Contrasting modes of diversification in the Aux/IAA and ARF gene families. Plant Physiol 135: 1738–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stimberg P, Estelle M, Leyser O 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279: 1371–1373 [DOI] [PubMed] [Google Scholar]
- Rubery PH, Sheldrake AR 1974. Carrier-mediated auxin transport. Planta 118: 101–121 [DOI] [PubMed] [Google Scholar]
- Ruegger M, Dewey E, Gray WM, Hobbie L, Turner J, Estelle M 1998. The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast Grr1p. Genes Dev 12: 198–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19: 2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberger J, Skoog F 1953. Changes induced by indole-acetic acid in nucleic acid content and growth of tobacco pith tissue. Science 118: 443. [DOI] [PubMed] [Google Scholar]
- Skoog F 1954. Substances involved in normal growth and differentiation of plants. Brookhaven Symposium in Biology 6: 1. [PubMed] [Google Scholar]
- Snow R 1935. Activation of cambial growth by pure hormones. New Phytol 34: 347–360 [Google Scholar]
- Szemenyei H, Hannon M, Long JA 2008. Topless mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319: 1384–1386 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LIA, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446: 640–645 [DOI] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, et al. 2004. MASSUGU2 encodes aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16: 379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Ray PM 1982. Early auxin-regulated polyadenylated mRNA sequences in pea stem tissue. Proc Natl Acad Sci 79: 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theologis A, Huynh TV, Davis RW 1985. Rapid induction of specific mRNAs by auxin in pea epicotyl tissue. J Mol Biol 183: 53–68 [DOI] [PubMed] [Google Scholar]
- Theologis A 1986. Rapid gene regulation by auxin. Ann Rev Plant Physiol 37: 407–438 [Google Scholar]
- Thimann KV 1969. The auxins. The Physiology of Plant Growth and Development, New York [Google Scholar]
- Thimann KV 1977. Hormones action in the whole life of plants. University of Massachussetts Press, Amherst [Google Scholar]
- Thimann KV, Koepfli JB 1935. Identity oft he growth-promoting and root-forming substances of plants. Nature 135: 101 [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle TJ 2004. Aux/IAA proteins contain a potent transcriptional repression domain. Plant Cell 16: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas A 1968. Relationship between plant growth hormones and nucleic acid metabolism. Prog Phytochem 1: 113 [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1997. ARF1, a transcription factor that binds auxin response elements. Science 276: 1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1999. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci 96: 5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhoef LN, Dute RR 1981. Auxin-regulated wall loosening and sustained growth in elongation. Plant Physiol 67: 146–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderhoef LN, Stahl CA 1975. Separation of two responses to auxin by means of cytokinin inhibition. Proc Nat Acad Sci 72: 1822–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J 2009. Auxin: A trigger for change in plant development. Cell 136: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Walker JC, Key JL 1982. Isolation of cloned cDNAs to auxin-responsive poly(A)+ RNAs of elongating soybean hypocotyl. Proc Natl Acad Sci 79: 7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jäger KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jürgens G 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24: 1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Went F, Thimann KV 1937. Phytohormones. MacMillan Company, New York [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: Regulation, action and interaction. Ann Bot 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenser N, Ellsmore A, Leasure C, Callis J 2001. Auxin modulates the degradation rate of Aux/IAA proteins. Proc Natl Acad Sci 98: 11795–11800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurfluh LL, Guilfoyle TJ 1982. Auxin-induced changes in the population of translatable messanger RNA in elongating sections of soybean hypocotyls. Plant Physiol 69: 338. [DOI] [PMC free article] [PubMed] [Google Scholar]