Abstract
Internal membrane bound structures sequester all genetic material in eukaryotic cells. The most prominent of these structures is the nucleus, which is bounded by a double membrane termed the nuclear envelope (NE). Though this NE separates the nucleoplasm and genetic material within the nucleus from the surrounding cytoplasm, it is studded throughout with portals called nuclear pore complexes (NPCs). The NPC is a highly selective, bidirectional transporter for a tremendous range of protein and ribonucleoprotein cargoes. All the while the NPC must prevent the passage of nonspecific macromolecules, yet allow the free diffusion of water, sugars, and ions. These many types of nuclear transport are regulated at multiple stages, and the NPC carries binding sites for many of the proteins that modulate and modify the cargoes as they pass across the NE. Assembly, maintenance, and repair of the NPC must somehow occur while maintaining the integrity of the NE. Finally, the NPC appears to be an anchor for localization of many nuclear processes, including gene activation and cell cycle regulation. All these requirements demonstrate the complex design of the NPC and the integral role it plays in key cellular processes.
Nuclear pores mediate selective transport into and out of the nucleus, but also function as the tethering sites for proteins involved in numerous other processes, including RNA processing and gene regulation.
Taxonomically speaking, all life on earth falls into one of two fundamental groups, the prokaryotes and the eukaryotes. The prokaryotes, the first group to evolve, are single cell organisms bounded by a single membrane. About 1.5 billion years later, a series of evolutionary innovations led to the emergence of eukaryotes. Eukaryotes have multiple inner membrane structures that allow for compartmentalization within the cell, and therefore differentiation of the cell and regulation within it. Ultimately, the greater cellular complexity of eukaryotes allowed them to adopt a multicellular lifestyle, as seen in the plants, fungi and animals of today (reviewed in Field and Dacks 2009).
Internal membrane bound structures sequester all genetic material in eukaryotic cells. The most prominent of these structures, which gives the eukaryotes their Greek-rooted name, is the nucleus—the central “kernel” (gr. “karyo-”) of the cell. The nucleus is bounded by a double membrane termed the nuclear envelope (NE), which separates the nucleoplasm and genetic material from the surrounding cytoplasm. However the genetic material in the nucleus is not totally isolated from the rest of the cell. Studded throughout the NE are portals called nuclear pore complexes (NPCs). The NPC is a highly selective, bidirectional transporter for a tremendous range of cargoes. Going into the nucleus, these cargoes include inner nuclear membrane proteins and all the proteins in the nucleoplasm. Going out are RNA-associated proteins that are assembled into ribosomal subunits or messenger ribonucleoproteins (mRNPs). Once transported, the NPC must ensure these cargos are retained in their respective nuclear and cytoplasmic compartments. All the while the NPC must prevent the passage of nonspecific macromolecules, yet allow the free diffusion of water, sugars, and ions. These many types of nuclear transport are regulated at multiple stages, providing a powerful extra level of cellular control that is not necessary in prokaryotes. Assembly, maintenance, and repair of the NPC must somehow occur while maintaining the integrity of the NE. Finally, the NPC appears to be an anchor for localization of many nuclear processes, including gene activation and cell cycle regulation (reviewed in Ahmed and Brickner 2007; Hetzer and Wente 2009). All these requirements demonstrate the complex design of the NPC and the integral role it plays in key cellular processes.
STRUCTURE OF THE NPC: SET UP OF THE MACHINE
The specifications of the NPC’s transport machinery represent a huge engineering challenge for evolution. No transitional forms of this elaborate transport system have yet been found in modern day organisms to reveal how it evolved. However, recent clues show that the NPC itself retains in its core a fossil of its ancient origins, indicating that the same mechanism that generated the internal membranes of eukaryotes might also have been responsible for the NPCs and the transport machinery.
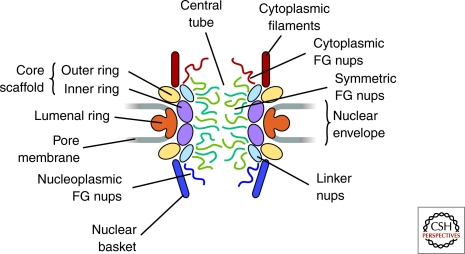
In the electron microscope, the NPC appears as a complex cylindrical structure with strong octagonal symmetry, measuring some 100–150 nm in diameter and 50–70 nm in thickness depending on the organism (reviewed in Wente 2000; Lim et al. 2008). This overall appearance seems broadly conserved throughout all eukaryotes. The two membranes of the NE, the outer and inner membranes, join only in a specialized, sharply curved piece of “pore membrane” that forms a grommet in the NE within which the NPC sits. Within each NPC is a core structure containing eight spokes surrounding a central tube. This central hole (∼30 nm diameter and ∼50 nm long) is where the nucleoplasm connects to the cytoplasm and where macromolecular exchange occurs. Peripheral filaments are attached to the core, filling the central hole as well as emanating into the nucleoplasm and cytoplasm. These filaments form a basket-like structure on the nuclear side of the NPC (Fig. 1).
Figure 1.
Major structural features of the NPC (based on the architectural map of Alber et al. (2007b); see Table 1 and main text for details).
One can envision the NPC as being comprised of layers of interacting proteins, starting with the core structure, moving outwards through its peripheral filaments, and then to associating clouds of soluble transport factors and peripherally associating protein complexes in the nucleus and cytoplasm (Rout and Aitchison 2001). These protein interactions can occur on radically different time scales. Some proteins form relatively permanent associations with the core structure, and so are termed nuclear pore complex components or “nucleoporins” (“Nups”). Other proteins associate transiently with the NPC, either constantly cycling on and off or attaching only at particular times in the cell’s life cycle. The NPC is covered in binding sites for these transiently associating proteins. Because the NPC is neither a motor nor an enzyme, the interactions provided by its binding sites wholly define the function of the NPC.
Recent work, mainly in the yeast Saccharomyces cerevisiae and in vertebrates, has begun to elucidate the molecular architecture of the NPC (Rout et al. 2000; Cronshaw et al. 2002; Alber et al. 2007b). Given its large size, the main body of the NPC comprises a surprisingly small number of ∼30 unique proteins (Table 1). However, because of the NPC’s eightfold symmetry, these Nups are each present in multiple copies (usually 16 per NPC) resulting in around 400 polypeptides for each NPC in every eukaryote (Rout et al. 2000; Cronshaw et al. 2002; DeGrasse et al. 2009). Further redundancy is evident from the recent mapping of the yeast NPC. Indeed, the NPC’s structure is modular, consisting of a few highly repetitive protein fold types (Devos et al. 2006; Alber et al. 2007b; DeGrasse et al. 2009). This suggests that the bulk of the NPC’s structure has evolved through multiple duplications of a small precursor set of genes encoding just a handful of progenitor Nups.
Table 1.
Nucleoporin homologs of yeast and vertebrates
| NPC substructure | Yeast components | Vertebrate components |
|---|---|---|
| Outer Ring | Nup84 subcomplex (Nup84, Nup85, Nup120, Nup133, Nup145C, Sec13, Seh1) | Nup107-160 complex (Nup160, Nup133, Nup107, Nup96, Nup75, Seh1, Sec13, Aladin, Nup43, Nup37) |
| Inner Ring | Nup170 subcomplex (Nup170, Nup157, Nup188, Nup192, Nup59, Nup53) | Nup155 subcomplex (Nup155, Nup205, Nup188, Nup35) |
| Cytoplasmic FG Nups and Filaments | Nup159, Nup42 | Nup358, Nup214, Nlp1 |
| Lumenal Ring | Ndc1, Pom152, Pom34 | Gp210, Ndc1, Pom121 |
| Symmetric FG Nups | Nsp1, Nup57, Nup49, Nup145N, Nup116, Nup100 | Nup62, Nup58/45, Nup54, Nup98 |
| Linker Nups | Nup82, Nic96 | Nup88, Nup93 |
| Nucleoplasmic FG Nups and Filaments | Nup1, Nup60, Mlp1, Mlp2 | Nup153, Tpr |
To understand its evolutionary origins, the NPC of the highly divergent Trypanosoma was recently characterized (DeGrasse et al. 2009). Despite significant divergence in primary structure, the Trypanosome NPC consists mainly of the motifs and domains found in vertebrate and yeast NPCs, indicating on a molecular level that the basic structural components of the NPC are conserved across all eukaryotes. Importantly, this also strongly implies that the last common eukaryotic ancestor had many features in common with contemporary NPCs, and perhaps provided a key adaptive advantage for this organism that has been retained, little changed, ever since.
The structural proteins making up the bulk of the spokes and rings give the NPC its shape and strength (Fig. 1). These core proteins of the NPC also maintain the stability of the nuclear envelope and facilitate the bending of the pore membrane into the inner and outer NE membranes. The most equatorial rings, termed the inner rings, are comprised of the Nup170 complex (yeast) or Nup155 complex (vertebrates) (Aitchison et al. 1995; Grandi et al. 1997; Miller et al. 2000) (Fig. 1). The inner rings are sandwiched between the outer rings, which are comprised of the Nup84 complex (yeast) or Nup107 complex (vertebrates) (Table 1) (Siniossoglou et al. 1996; Fontoura et al. 1999; Siniossoglou et al. 2000; Belgareh et al. 2001; Vasu et al. 2001). Together, these Nup complexes form a scaffold that hugs the curved surface of the pore membrane and helps form the central tube through which macromolecular exchange occurs (Alber et al. 2007a; Alber et al. 2007b).
Nups in the core scaffold represent roughly half the mass of the whole NPC and are composed almost entirely of either β-propeller folds, α-solenoid folds, or a distinct arrangement of both in an amino-terminal β-propeller followed by a carboxy-terminal α-solenoid fold. The core scaffold of all eukaryotes appears to retain this basic fold composition (Devos et al. 2004; Devos et al. 2006; DeGrasse et al. 2009). Strikingly, there are similarities between the structures of the core NPC scaffold curving around the pore membrane and other membrane-associated complexes such as clathrin/adaptin, COPI, and COPII (Fig. 1) (Devos et al. 2004; Devos et al. 2006). Clathrin/adaptin is involved in coat-mediated endocytosis at the plasma membrane, and COPI and COPII are responsible for coat-mediated vesicular transport between the plasma membrane and endomembrane systems such as the Golgi and ER. Indeed, the similarities between core scaffold Nups and coating complexes have been borne out in numerous crystallographic studies (Berke et al. 2004; Hsia et al. 2007; Brohawn et al. 2008; Debler et al. 2008; Brohawn et al. 2009; Leksa et al. 2009; Seo et al. 2009; Whittle and Schwartz 2009), although nearly 2 billion years of evolution have made it difficult at first glance to recognize the common origin of these two groups. However, their common β-propeller and helix-turn-helix repeat structure is still unmistakable (Brohawn et al. 2008; Field and Dacks 2009). In NPCs the “coat” comprises the core scaffold of the NPC, where—analogous to the curved membrane of a vesicle being stabilized by a COP or clathrin coat—it stabilizes the curved pore membrane. These similarities also give a tantalizing glimpse into the deep evolutionary origins of eukaryotes. It seems early proto-eukaryotes distinguished themselves from their prokaryotes by acquiring a membrane-curving protein module, the “proto-coatomer” (likely composed of a simple β-propeller/α-solenoid protein), that allowed them to mold their plasma membranes into internal compartments. Modern eukaryotes diversified this module into many specialized membrane coating complexes, accounting for the evolution of their internal membrane systems (Devos et al. 2004; Devos et al. 2006).
The framework of the NPC serves two key transport purposes: to form a barrier of defined permeability within the pore, and to facilitate transport of selected macromolecules across it. Both processes are dependent on the correct positioning of critical Nups in the NPC architecture (Radu et al. 1995; Strawn et al. 2004; Liu and Stewart 2005). Attached to the inside face of the NPC core scaffold, facing the central tube’s cavity, are groups of nucleoporins termed “linker nucleoporins” (Fig. 1). Together with the inner ring, these seem to form most of the attachment sites for a last set of nucleoporins, termed “FG Nups” (Alber et al. 2007b). These FG Nups, named for their phenylalanine-glycine repeats, are the direct mediators of nucleocytoplasmic transport (Radu et al. 1995; Strawn et al. 2004; Liu and Stewart 2005) (see the following section).
The core NPC scaffold is connected to a set of integral membrane proteins, which form an outer luminal ring in the NE lumen and anchor the NPC into the NE (Nehrbass et al. 1996; Alber et al. 2007a; Alber et al. 2007b) (Fig. 1). Oddly, the membrane nucleoporins seem poorly conserved—if at all—across the eukaryotes. The fact that all the currently known pore membrane proteins in Aspergillus nidulans can seemingly be dispensed with for NPC function and assembly might indicate that there are not strong pressures for their conservation, and that there are other membrane proteins that can serve the role (Liu et al. 2009). This fact also sets up a quandary—if most or all of the NPC’s presumed membrane anchors are dispensible, how then is the NPC reliably anchored to the membrane? Several groups are seeking the answer to this question (Hetzer and Wente 2009).
OPERATION OF THE MACHINE: THE SOLUBLE PHASE
Understanding the transport machine requires resolving both its barrier and binding activities. How the NPC machine balances both of these selective functions has been a challenging mystery. Studies of fluorescently labeled sized dextrans or gold particles microinjected into cells (Feldherr and Akin 1997; Keminer and Peters 1999) have defined the practical permeability limits of the NPC, showing that under physiological time scales, macromolecules greater than 40 kDa in size do not show any measureable redistribution between the nucleus and cytoplasm and thus, no movement through the NPC. Conversely, metal ions, small metabolites, and molecules less than ∼40 kDa in mass or ∼5 nm in diameter can pass relatively freely. NPC permeability is altered in several yeast nup mutants, pinpointing NPC structural elements, including core scaffold components, that are critical to the assembly or maintenance of this barrier (Shulga et al. 2000; Denning et al. 2001; Shulga and Goldfarb 2003; Strawn et al. 2004; Patel et al. 2007). Larger macromolecules overcome this permeability barrier by interacting either directly with the NPC themselves or through soluble transport factors. These macromolecules account for a tremendous variety of cargo including proteins, tRNAs, ribosomal subunits, and viral particles (reviewed in Macara 2001). Overall, the NPC is capable of transporting cargo up to 39 nm in diameter. This is on par with the size of the ribosomal subunits and viral capsids that are known to move as intact complexes (Pante and Kann 2002). Macromolecules larger than this can still be translocated across the NPC, including mRNPs (mRNAs coated with RNA-binding proteins) with masses reaching several hundred thousand daltons. EM images of Balbiani ring mRNP particles associated with the NPC show the posttranscriptional ∼50 nm mRNA–protein particles to rearrange into rodlike structures, decreasing their maximum diameter to ∼25 nm (Mehlin et al. 1992). Thus, cargoes above a limiting diameter must rearrange to pass through the selective barrier of the NPC (Daneholt 2001).
A transport signal and a shuttling receptor for that transport signal are the minimal requirements for any facilitated translocation (reviewed in Mattaj and Englmeier 1998; Pemberton and Paschal 2005). The targeting of proteins into or out of the nucleus requires specific amino acid sequence spans, termed nuclear localization sequences (NLSs) or nuclear export sequences (NESs). All the information required to target a protein to the nucleus is within these short sequences. In fact, fusion of an NLS to a nonnuclear protein is sufficient to mediate its transport and import to the nucleus (Goldfarb et al. 1986). For proteins, there are many distinct types of NLSs and NESs. For example, the classical NLS (cNLS) is the simple five amino acid peptide KKKRK, necessary and sufficient for targeting its attached protein to the nucleus (Goldfarb et al. 1986), whereas many proteins carry a more complex “bipartite” NLS consisting of two clusters of basic amino acids, separated by a spacer of roughly 10 amino acids (Dingwall et al. 1988). However, the full spectrum of sequences recognized by each transport receptor has not yet been carefully and fully defined. The most in depth analysis of NLS structural recognition by a transport receptor and extrapolation to predicting cargoes on a broader genome level has only been reported for one transport receptor (Lee et al. 2006). The key parameters defining an NLS or NES include critical tests for necessity and sufficiency in the endogenous protein. Importantly, some proteins undergo dynamic cycles of nuclear import and export and harbor both NLSs and NESs. This can increase the complexity of identifying the signals. Moreover, the recognition and accessibility of the signals can be controlled by signaling, cell cycle, and developmental events (reviewed in Weis 2003; Terry et al. 2007).
During NPC translocation, soluble transport factors are required to either bring cargo to the NPC or modulate cargo translocation across the NPC. Most of these soluble transport factors come from the family of proteins known as the karyopherins (Table 2). The karyopherins (also called importins, exportins, and tranportins) were the first family of shuttling transport factors discovered. Fourteen karyopherin family members are found in Saccharomyces cerevisiae whereas at least 20 have been found in metazoans (reviewed in Fried and Kutay 2003; Pemberton and Paschal 2005). Most karyopherins bind their cargoes directly. However, in some cases an adaptor protein is needed in addition to the karyopherin to recognize signals. Not only do karyopherins have a cargo-binding domain, they also have an NPC-binding domain(s) as well as a binding domain at the amino-terminus for the small Ras-like GTPase Ran (see the following paragraph) (reviewed in Macara 2001; Harel and Forbes 2004). Overall, karyopherin family members share only modest sequence homology, with the greatest similarity being within their Ran-binding domains (Gorlich et al. 1997). However, a hallmark architecture within the karyopherins, as determined by recent high-resolution structural studies, is the tandem HEAT-repeat fold formed by antiparallel helices connected by a short turn (reviewed in Conti and Izaurralde 2001). The HEAT-repeats arrange to form a superhelical structure, similar to a snail’s shell. This folding is reminiscent of the helix-turn-helix repeats found in the NPC’s core scaffold proteins. This similarity raises the intriguing possibility that karyopherins diverged from a common structure involved in both stationary and soluble phases of transport.
Table 2.
Karyopherin transport factors of yeast and vertebrates*
| S. cerevisiae Karyopherins | Vertebrate Karyopherins | Examples of Cargo(s): (v) –vertebrate, (sc) – S. cerevisiae |
|---|---|---|
| Kap95 | Importin-β1 | Imports via sc-Kap60/v-importin-α adaptor proteins with cNLS; Imports via v-Snurportin the UsnRNPs; with no adaptor, imports v-cargo SREBP-2, HIV Rev, HIV TAT, cyclin B |
| Kap104 | Transportin or Transportin 2 | Imports sc-cargo – Nab2, Hrp1; v-cargo – PY-NLS proteins, mRNA-binding proteins, histones, ribosomal proteins |
| Kap108/Sxm1 | Importin 8 | Imports sc-cargo – Lhp1, ribosomal proteins; v-cargo – SRP19, Smad |
| Kap109/Cse1 | CAS | Imports sc-cargo – Kap60/Srp1; v-cargo – importin αs |
| Kap111/Mtr10 | Transportin SR1 or SR2 | Imports sc-cargo – Npl3, tRNAs; v-cargo – SR proteins, HuR |
| Kap114 | Importin 9 | Imports sc-cargo – TBP, histones, Nap1, Sua7; v-cargo – histones, ribosomal proteins |
| Kap119/Nmd5 | Importin 7 | Imports sc-cargo – Hog1, Crz1, Dst1, ribosomal proteins, histones; v-cargo – Smad, ERK, GR, ribosomal proteins |
| Kap120 | HsRanBP11 | Imports sc-cargo – Rpf1 |
| Kap121/Pse1 | Importin 5/Importin β3/RanBP5 | Imports sc-cargo – Yra1, Spo12, Ste12, Yap1, Pho4, histones, ribosomal proteins; v-cargo – histones, ribosomal proteins |
| Kap122/Pdr6 | - | Imports sc-cargo – Toa1 and Toa2, TFIIA |
| Kap123 | Importin 4 | Imports sc-cargo – SRP proteins, histones, ribosomal proteins; v-cargo - Transition Protein 2, histones, ribosomal protein S3a |
| Kap127/Los1 | Exportin-t | Exports tRNAs |
| Kap142/Msn5 | Exportin 5 | sc-cargo – imports replication protein A; exports Pho4, Crz1, Cdh1; v-cargo - exports pre-miRNA |
| Importin 13 | v-cargo – imports UBC9, Y14; exports eIF1A | |
| Crm1/Xpo1 | CRM1/Exportin 1 | Exports proteins with leucine-rich NES, 60S ribosomal subunits (via NMD3 adaptor), 40S ribosomal subunits |
| — | Exportin 4 | v-cargo – imports SOX2, SRY; exports Smad3, eIF5A |
| — | Exportin 6 | Exports profilin, actin |
| — | Exportin 7/RanBP16 | Exports p50-RhoGAP |
*Based on references cited within and adapted from Tran et al. 2007a and DeGrasse et al. 2009.
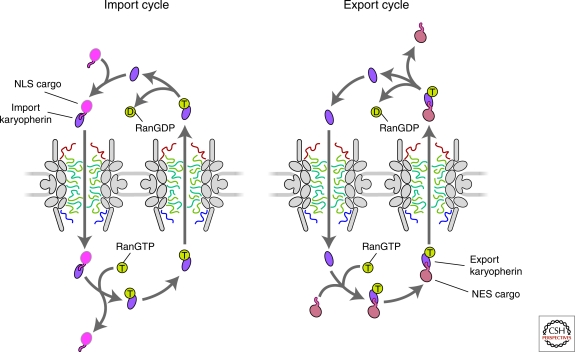
The association and dissociation of a karyopherin-cargo complex is regulated by direct binding of the small GTPase Ran (Fig. 2) (reviewed in Fried and Kutay 2003; Madrid and Weis 2006; Cook et al. 2007). In vitro binding studies show that import complexes are dissociated by RanGTP binding. Conversely, export complexes are formed via RanGTP association (Rexach and Blobel 1995; Floer and Blobel 1996; Chi and Adam 1997; Floer et al. 1997; Kutay et al. 1997a; Kutay et al. 1997b; Nakielny et al. 1999). Based on the localizations of the Ran GTPase activating protein (RanGAP) in the cytoplasm and the Ran guanine nucleotide exchange factor (RanGEF) in the nucleoplasm, cytoplasmic Ran is primarily in the GDP-bound state whereas nucleoplasmic Ran is kept primarily in the GTP-bound state (Fig. 2). The gradient formed from these localizations has been elegantly demonstrated by imaging fluorescence resonance energy transfer-based biosensors (Kalab et al. 2002). The RanGTP gradient across the two faces of the NPC is essential for establishing the directionality of karyopherin-mediated transport.
Figure 2.
The nuclear transport cycle for karyopherins and their cargos. See main text for details.
The pathway for karyopherin-mediated translocation is well described (reviewed in Weis 2003; Terry et al. 2007) (Fig. 2). For import, a specific karyopherin recognizes its cognate cargo in the cytoplasm where RanGTP levels are low. The karyopherin mediates the binding of the import complex to the NPC and facilitates translocation through the NPC. Once the complex moves through the NPC, release and dissociation of the karyopherin-cargo complex are stimulated by RanGTP in the nucleus. The karyopherin bound to RanGTP is then recycled back to the cytoplasm. Finally, GTP hydrolysis of Ran on the cytoplasmic side frees the karyopherin to interact with a second cargo molecule for further cycles of transport. Overall, Ran decreases the affinity of the karyopherin for its cargo (reviewed in Macara 2001; Cook et al. 2007). For export complexes, an analogous process occurs, but in this case, RanGTP binding increases the affinity of the karyopherin β for the export cargo. For example, for the exporting karyopherin Crm1 and an export cargo SPN1 (snurportin 1 adaptor for UsnRNPs), the Crm1 affinities for RanGTP and SPN1 in the ternary RanGTP-Crm1-SPN1 complex are increased ∼1000-fold (Paraskeva et al. 1999; Monecke et al. 2009). Actual movement through the NPC does not require energy input. The Ran affinity switches provide the energy for efficient cargo delivery and release. The only possible exception to this rule involves the import of large cargoes, where the presence of Ran and hydrolyzable GTP may be required for the import of cargoes >500 kDa in vitro (Lyman et al. 2002). It was also originally thought that an individual karyopherin was adapted for either import or export, but not both. However, there are now documented examples of karyopherins functioning in both import and export, although with different cargoes in each direction (Yoshida and Blobel 2001).
In addition to protein import and export, karyopherins can also transport RNAs. For example, the karyopherins Crm1 and exportin-t mediate the export of uridine-rich small nuclear RNAs (U snRNAs) and tRNAs, respectively (Simos et al. 2002; Rodriguez 2004). Transport is accomplished via direct binding of karyopherins to RNA or to signal sequences within the protein components of the RNP complexes. For example, Crm1 does not bind UsnRNAs directly and requires the adaptor PHAX that binds the cap-complex on the RNA (Ohno et al. 2000). However, exportin-t directly interacts with tRNAs (Arts et al. 1998; Hellmuth et al. 1998; Kutay et al. 1998; Lipowsky et al. 1999).
Karyopherins are also involved in the export of some viral RNAs, including their mRNAs (Carmody and Wente 2009). However, the primary mRNP export transport receptor is a nonkaryopherin designated Mex67 in yeast and NXF1 in metazoans, which heterodimerizes with a protein termed respectively Mtr2 or p15/Nxt1 (Erkmann and Kutay 2004). Even though Mex67 is unrelated in sequence and structure to the karyopherin family, it has all the requirements of a transport receptor: cargo binding, nucleocytoplasmic shuttling, and NPC-binding. The stoichiometry of the Mex67-Mtr2 heterodimer per transported mRNP is unknown; Mex67-Mtr2 either is recruited directly to the mRNA or interacts cotranscriptionally with proteins of the mRNP assembly (Erkmann and Kutay 2004; Carmody and Wente 2009). Like karyopherins, Mex67-Mtr2 heterodimers bind directly to FG Nups, although it seems they prefer different subsets of FG Nups to their karyopherin counterparts, which might reflect how the karyopherin mediated transport pathways and mRNP export pathways are kept apart at the NPC (Strawn et al. 2001; Terry et al. 2007; Terry and Wente 2007).
As mRNP is a major source of traffic across the NPC, it is interesting that most of this transit is facilitated by non-karyopherin carriers. Because Ran is not utilized to establish a gradient, directionality in the mRNA export pathway is conferred by proteins that modify the mRNPs as they cross the NPC. Chief among these is the protein Dbp5. Dbp5 is a member of the SF2 helicase superfamily of RNA-dependent ATPases (Snay-Hodge et al. 1998; Tseng et al. 1998) and carries a DEAD/H-box sequence motif. Such DEAD-box proteins are involved in the alteration of RNA structure, such as the facilitation of RNA duplex formation and unwinding or aiding the association and dissociation of RNA-binding proteins (Jankowsky et al. 2001; Fairman et al. 2004; Yang and Jankowsky 2006). Much like the GDP/GTP triggered switches for Ran, DEAD-box proteins potentially use ADP/ATP for nucleotide-dependent conformational switches (Henn et al. 2002; Tran et al. 2007b; Henn et al. 2008; Fan et al. 2009). Dbp5 ATPase activity is activated by Gle1, a protein that exchanges between the nucleus and cytoplasm with a docking site on the cytoplasmic filaments of the NPC (Cole and Scarcelli 2006). Gle1 activation of Dbp5 in turn requires the soluble molecule inositol hexakisphosphate (IP6) to increase the binding affinity of Dbp5 to RNA (Alcazar-Roman et al. 2006; Weirich et al. 2006). This raises the possibility that IP6 is acting as a signaling molecule to somehow regulate the mRNA export machinery (York et al. 1999). The Dbp5 on the cytoplasmic side of the NPC triggers the release of mRNP proteins such as Mex67 and Nab2, effectively remodeling the mRNP protein composition as they exit the NPC as well as imparting directionality to mRNP transport (Tran et al. 2007b). There also seems to be a checkpoint at the NPC basket, to ensure that mRNAs have been correctly spliced and packaged into mRNPs immediately before export. SUMO is a ubiquitin-like molecule that can be attached to proteins as a signal or modulator of their function. The SUMO protease, Ulp1, is retained at the NPC via its interaction with the nuclear basket component Mlp/Tpr, and it has been suggested that sumoylation of mRNPs acts to signal that they are correctly assembled. Conversely, the desumoylation of incorrectly spliced mRNPs at the nuclear basket prevents their export and instead targets them for degradation (Lewis et al. 2007; Xu et al. 2008).
Other nonkaryopherin carriers for NPC translocation exist with the same capacity to dock at various NPC sites via the FG Nups. Ntf2/p10 is a carrier for RanGDP importing Ran to the nucleus. Ntf2/p10 also uses the localization of RanGEF in the nucleus to create a differential affinity for RanGDP versus RanGTP (reviewed in Pemberton and Paschal 2005; Cook et al. 2007). There are also some proteins with karyopherin-like HEAT-repeats that require no receptor and translocate independently of a carrier. The driving force for their accumulation in the nucleus is their affinity for binding targets in the nucleoplasm (Fagotto et al. 1998).
Perhaps the most abundant nuclear transport cargos in many cells are the 60S and 40S ribosomal subunits, on the way from synthesis in the nucleolus to form mature ribosomes in the cytoplasm. Export of ribosomal subunits is still poorly understood, and it seems that unlike the case of a single karyopherin bound to each protein cargo molecule as described earlier, several transport receptors must chaperone each ribosomal subunit across the NPC. The earliest identified ribosomal exporter was Crm1 (Fornerod et al. 1997; Fukuda et al. 1997; Stade et al. 1997). For export of the 60S subunit, the NES is provided by the export adaptor Nmd3 (Johnson et al. 2002). Nmd3 binds reversibly to the 60S subunit while the latter is being assembled in the nucleus. After export into the cytoplasm, a GTPase called Lsg1 appears to mediate the release of Nmd3 from the 60S ribosomal subunit (Zemp and Kutay 2007). In yeast the Mex67-Mtr2 heterodimer has also been suggested to serve as an export receptor for the 60S subunit (Yao et al. 2007). Although more karyopherin-like proteins have recently been identified that play a role in ribosomal export (Oeffinger et al. 2004), other nonkaryopherin receptors like Arx1 have been found, though Arx1 still interacts with the same NPC docking sites as karyopherins (Bradatsch et al. 2007). Why might so many transport factors be required to ferry the ribosomal subunits out of the nucleus? One reason might be explained by the analogy of maneuvering ships out of the harbor. One tugboat is all that is required to move a small barge quickly and efficiently, but on its own does not have the power to move an ocean liner, which instead needs many tugboats together to take it safely out of harbor. Similarly, as the efficiency of passage through the NPC decreases with cargo size (see the preceding section) but increases with binding strength to FG Nups (see the following section), many transport factors are needed to carry large cargos across the NPC. Regardless, the fundamental mechanism for transport in all these cases of different carriers is still dependent on the same features of the NPC machine.
OPERATION OF THE NPC: THE “FG” TRANSLOCATION MACHINERY
Although the cargo binding mechanisms of the transport factors discussed earlier vary, almost all binding sites with the NPC are found in the same class of Nups. These Nups are collectively termed FG Nups because they contain “FG repeat regions.” The NPC core framework provides the correct positioning of these FG Nups so that they flank and fill the central tube. Approximately a third of all Nups contain FG repeat regions, which consist of multiple small hydrophobic clusters containing an FG (Phe-Gly) dipeptide (usually FG, FXFG, or GLFG) separated by ∼20–70 residue hydrophilic linkers (reviewed in Rout and Wente 1994; Allen et al. 2001; Tran and Wente 2006). Binding of these repeats is cooperative, with two to four repeats associating with each transport factor molecule (Bayliss et al. 2002; Grant et al. 2003; Isgro and Schulten 2005; Liu and Stewart 2005). With 5–50 repeats per FG Nup and ∼200 FG Nups per NPC, there is a potential for >1000 transport factor binding sites per NPC. Imaging of the trajectories of a single translocating molecule through the NPC is consistent with movement between multiple binding sites (presumably the many FG repeats) within the NPC (Yang et al. 2004; Kubitscheck et al. 2005; Yang and Musser 2006). FG repeat regions are natively disordered and having no secondary structure they instead form filaments that can diffusively writhe around their attachment sites at the NPC. Although some will be diffuse random coils, others may have enough internal cohesion to form more compact “molten globules” (Denning et al. 2003; Lim et al. 2006; Patel et al. 2007; Krishnan et al. 2008). By forming a dense meshwork in and around the central tube, the FG repeats appear to set up an entropic potential that excludes macromolecules from their vicinity, while permitting the approach of small molecules such as water and nucleotides (Lim et al. 2006). However, karyopherins overcome this barrier as they bind FG Nups and, passing from FG Nup to FG Nup, diffuse readily through the NPC. Indeed, this model is supported by the fact that a simple device with similar properties—a nanotube coated with FG Nups—replicated many of the features of nuclear transport (Jovanovic-Talisman et al. 2009). In terms of the Gibb’s free energy required to cross the NPC, the enthalpy for binding the FG Nups cancels out the entropic barrier of the central tube region, flattening the energy landscape and thus lowering the activation energy of translocation across the NE. This is analogous to how an enzyme or catalyst lowers the activation energy of the transition state permitting a substrate to convert to product. This concept has been termed “virtual gating” (Rout et al. 2000; Rout et al. 2003).
Nevertheless, despite much work over the last few years, how exactly this gating works at the molecular level remains unclear and several models have attempted to explain it. One possibility is that the natively unfolded FG Nups could act as a “polymer brush” where rapid diffusion-driven movements of their unfolded domains would sweep away macromolecules from their vicinity. Although a central tube filled with such brushes would exclude nonbinding macromolecules, transport factors would overcome this exclusion by binding to the FG repeats and so pass through the NPC (Rout et al. 2000; Rout et al. 2003; Lim et al. 2006). In another model, regions of FG repeats may collapse upon binding transport factors, as indicated by atomic force microscopy data (Lim et al. 2007a; Lim et al. 2007b). As they pass through the central tube, transport factors would open up their own passageway through the meshwork of FG repeats. A different view is provided by the “saturated” model (Frey et al. 2006; Frey and Gorlich 2007), originally the “hydrophobic gel” model (Ribbeck and Gorlich 2002). Here it is suggested that the phenylalanines in the FG repeat regions are crosslinked with each other through their Phe residues to form a dense gel of FG repeat filaments. Transport factors pass through this gel by binding the FG repeats, and in doing so they dissolve the crosslinks. Other macromolecules are therefore excluded because they cannot do this. A more recent “reduction in dimensionality”-model posits that the FG repeat regions form a layer coating the inner walls of the central tube (Peters 2005). Although transport factors enter this layer through binding giving them full access to the tube’s volume, nonbinding molecules can only pass through the narrow FG Nup-free middle. Recently, work has emerged that suggests while crossing the NPC, transport factors not only carry cargoes but also enhance the selectivity of the NPC by competing away nonspecific macromolecules, indicating that the transport factors themselves are also important components of the selectivity barrier (Timney et al. 2006; Zilman et al. 2007; Jovanovic-Talisman et al. 2009). Not all of these ideas are mutually exclusive, and “hybrid” models have also been proposed (Strawn et al. 2004; Patel et al. 2007; Krishnan et al. 2008).
FG Nup repeat regions seem to fall into several classes on the basis of their charge density and FG repeat motif type. Moreover, these classes seem to have somewhat different physical behaviors (Krishnan et al. 2008). Each class of FG Nup appears to be preferred by only certain kinds of transport factor. Thus, these different FG Nup classes might serve to mediate multiple, simultaneous, functionally independent transport pathways through the NPC (reviewed in Tran and Wente 2006). Supporting evidence for this has come from in vivo studies using target combinations of different FG region deletions. Certain deletion combinations can affect passage of one transport factor through the NPC without affecting another (Strawn et al. 2004; Terry and Wente 2007). FG Nups organization around the NPC can be categorized as either symmetric or asymmetric. “Symmetric” FG Nups are found on both sides of the NPC closely packed around and within the central tube. “Asymmetric” FG Nups are found primarily on either the nuclear or cytoplasmic side only, and significantly further from the tube (Rout et al. 2000; Alber et al. 2007b). Deletion mutagenesis experiments in yeast have shown that the FG regions of asymmetric Nups are not essential for transport (Strawn et al. 2004; Zeitler and Weis 2004). Instead, they may guide the directionality of transport-factor/cargo complexes by providing a high-affinity binding platform at the far end of a transport factor’s route through the NPC. A transport factor would be drawn to this site and prevented by it from returning through the NPC until Ran terminates the transport reaction, in this way increasing the efficiency of transport (Rout et al. 2003; Strawn et al. 2004; Zilman et al. 2007).
REGULATORS OF TRANSLOCATION: THE NON-FG DETERMINANTS
Unidirectional transport through the NPC can be viewed as being driven by the formation of gradients, in which cargos move from a “source” to a “sink.” Karyopherin-cargo complexes form on one side of the NPC and diffuse down their concentration gradient to the other side, where their concentration is low because they are disassembled there. This “source and sink” process also drives other forms of nucleocytoplasmic transport, such as the attachment of RNA binding proteins on one side of the NPC and their removal on the other side during RNP export (reviewed in Rout et al. 2003; Stewart 2007).
The NPC on its own does not enforce directionality (Nachury and Weis 1999). Single molecule studies show the transport receptor-cargo complexes seem to move randomly within the NPC channel until they exit either face of the NPC (Yang et al. 2004). Thus, in a physiological context, other NPC events and non-FG Nup binding sites are required to provide transport efficiency, regulation, and directionality (Fig. 3). As detailed earlier, there are specific factors for mediating transport directionality and cargo release: The RanGTPase for karyopherins and the Gle1-IP6 activated Dbp5 for mRNA export. These use non-FG NPC docking sites that increase their local concentration at their respective sites of action. Dbp5 and Gle1 bind to distinct but structurally juxtaposed Nups on the cytoplasmic filaments. In budding yeast, Dbp5 interacts with Nup159 (Hodge et al. 1999; Schmitt et al. 1999) and Gle1 with Nup42 (Murphy and Wente 1996; Strahm et al. 1999), with both sets of binding partners fully conserved in metazoans. The NPC can also help to regulate the GTP-bound state of Ran in its vicinity. Thus, mammalian cells have binding sites for sumoylated-RanGAP on the cytoplasmic filament Nup358, likely to help enhance the dissociation of exporting karyopherin complexes as they exit the NPC (Mahajan et al. 1997; Matunis et al. 1998). In organisms like plants that lack Nup358, unique NE associated proteins facilitate the localized anchoring of RanGAP (Xu et al. 2007; Zhao et al. 2008), so it seems that regulating the GTP-bound state in the immediate vicinity of the NPC is important for transport efficiency in many eukaryotes.
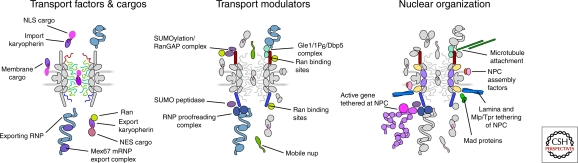
Figure 3.
The main NPC interactors: left, transport factors and their cargos, center, modulators of RNP export, karyopherins or Ran, right, cytoskeletal, nucleoskeletal, and chromosomal elements.
Although the FG domains do not form a gradient of docking sites in the translocation channel, there are non-FG high affinity docking sites located at terminal transport steps that influence efficiency. In particular, two Nups on the opposing cytoplasmic and nuclear faces provide a virtual smorgasbord of binding sites for different factors that regulate cell physiology: Nup358 and Nup153, respectively (Fig. 3). In metazoan cells, the cytoplasmic facing Nup358 provides a high affinity binding site for the mRNA export factor NXF1 on the cytoplasmic face of the NPC (Forler et al. 2004). Similarly, Nup153 provides a high affinity binding site for an importing karyopherin on the nuclear face of the NPC (Moroianu et al. 1997). Although both have FG domains it is the non-FG docking sites that are functionally distinguishing.
Nup358 was originally identified as a RanGTP binding protein (Wu et al. 1995; Yokoyama et al. 1995) and binds sumoylated RanGAP. However, it also has a domain with E3 SUMO1 ligase activity and docks the sumolylation cofactor enzyme Ubc9 to facilitate the addition of SUMO to targets directly at the cytoplasmic filaments (Pichler et al. 2002; Reverter and Lima 2005). Nup358 also harbors a cyclophilin A homologous domain that associates with subunits of the 26S proteasome and might modulate ubiquitin-proteasome function (Wu et al. 1995; Yi et al. 2007). The amino-terminal domain of Nup358 allows interaction with microtubules during interphase as well as with kinetochores in mitosis (Salina et al. 2003; Joseph et al. 2004; Joseph and Dasso 2008).
On the nuclear side of the NPC, Nup153 has a Zn finger Ran binding domain (Nakielny et al. 1999; Higa et al. 2007) attracting Ran to the vicinity of the NPC to regulate karyopherin-based transport events at its nuclear face. In sharp contrast to the Nup358-SUMO1 ligase activity, Nup153 actually binds a SUMO1/sentrin/SMT3 specific peptidase 2, perhaps to help desumolyate misassembled mRNPs (Hang and Dasso 2002; Zhang et al. 2002). Nup153 also possesses a novel single-stranded RNA binding domain (Ball et al. 2004) that is perhaps concomitant with a function in RNP export. Nup153 also plays a role in recruiting the COPI coatomer complex, possibly to mediate NPC mitotic disassembly (Liu et al. 2003). Taken together, Nup358 and Nup153 are but two examples of an emerging paradigm for multifunctional nucleoporins. They demonstrate that the NPC serves as a scaffold for a diversity of processes that are distinct from those strictly associated with simply moving a cargo from one side of the NPC to the other.
Transport can also be modulated at the level of the transport factors and their cargos. A common method is to regulate the binding strength or accessibility of an NLS or NES in a cargo for its cognate karyopherin. This often involves phosphorylation in or near the signal sequence of the cargo. Another method involves modulating the level of transport machinery components, usually particular subtypes of karyopherin. Using this method, transport of particular proteins recognized by that karyopherin is prevented until the karyopherin itself is expressed (reviewed in Poon and Jans 2005).
THE NPC AS A NUCLEAR COMPONENT
Although the most dynamic process associated with the NPC is nucleocytoplasmic transport, the NPC should not be regarded as a static structure. New NPCs are assembled de novo during interphase (D’Angelo et al. 2006) and, in higher eukaryotes at least, the NPC is reversibly disassembled during mitosis (reviewed in Hetzer et al. 2005; Fernandez-Martinez and Rout 2009). There is emerging evidence for multiple non-nucleoporin accessory factors that play roles in NPC biogenesis and maintenance (Fig. 3). This includes proteins that regulate membrane composition and membrane dynamics such as Acc1 and Apq12, respectively (Schneiter et al. 1996; Scarcelli et al. 2007). Changes in the lipid composition or membrane fluidity can impact NPC structure and localization. Chromatin associated proteins, such as ELYS, are required specifically for postmitotic assembly of NPCs (Franz et al. 2007).
Recent studies have found several discrete roles for the reticulon (Rtn) family of membrane-associated proteins in NE disassembly and reassembly during open mitosis. The Rtns are critical for regulating transitions between the endoplasmic reticulum tubular network and the NE sheet (Anderson and Hetzer 2008). However, during interphase, an Rtn1 pool at the NE facilitates de novo NPC assembly (Dawson et al. 2009). This most likely is to provide a transient stabilizer of the highly convex region of the pore membrane during early steps in its formation. It is likely that additional non-NPC accessory factors will continue to be revealed, and this is an active area of research that has been well covered in several recent reviews (Antonin et al. 2008; D’Angelo and Hetzer 2008; Kutay and Hetzer 2008; Lim et al. 2008).
Even after its assembly, many of the interactions holding the bulk of the NPC together are actually surprisingly transient. Upon completion of NPC construction, some Nups remain stably associated, others turn over promptly, and some shuttle rapidly between the nucleoplasm and cytoplasm, spending little time on the NPC itself (Rabut et al. 2004; Dilworth et al. 2001; Dilworth et al. 2005). For example, there are multiple roles for Nups in kinetochore function and mitosis, as well as in the maintenance of the boundaries between silent and active chromatin (Roux and Burke 2006; Kalverda et al. 2008; Lim et al. 2008). Despite these processes requiring Nups, none appear to occur at the NPC itself. Indeed, recent work underscores how a great number of Nup-chromatin interactions occur within the nuclear interior and away from the NPC itself; these interactions have also been shown to be crucial in controlling gene expression during cell differentiation in higher eukaryotes (Capelson et al. 2010; Kalverda et al. 2010). Conversely, the NPC can act as a reservoir for cell cycle proteins when they are inactive. One example, the Mad proteins, are docked to the NPC in interphase but are released when they function in mitosis (Iouk et al. 2002; Scott et al. 2005; Lee et al. 2008).
The NPC also plays an important role in the organization of internal nuclear architecture (Fig. 3) and is used as an attachment site for such nuclear peripheral proteins as lamins and yMlp/vTpr. An exciting recent discovery is that certain inducible genes carry specific sequences that can cause them to associate with the NPC’s vicinity upon transcriptional activation; once there, they retain a “memory” of their active state for some time after they cease transcription, leaving the genes poised for rapid reactivation (Ahmed et al. 2010). This tethering of induced genes to the NPC might also be also aided by factors required for posttranscriptional mRNP processing and export (Dieppois et al. 2006; Chekanova et al. 2008; Rougemaille et al. 2008).
CONCLUSIONS
Our structural and functional understanding of the nuclear transport machinery has blossomed in the past decade, revealing new and unexpected aspects about the NPC and its interacting transport factors. Perhaps most surprising has been the discovery of the evolutionary connections between the NPC and the vesicular trafficking apparatus, the unusual nature of the FG Nup-mediated selective barrier, and the increasing range of connectivity between the NPC and global aspects of cellular physiology and gene expression. Yet as is so often the case, the solving of one question has raised many others. Many of the unanswered questions regarding the mechanisms and regulation of the nuclear transport machinery present exciting challenges for the years ahead.
ACKNOWLEDGMENTS
We acknowledge that many of the primary references for critical studies are included in reviews cited, and appreciate our colleagues’ consideration in this limitation. We thank Renee Dawson and Kelli Mullin for assistance with manuscript preparation. This work was supported by funding from the National Institutes of Health (R01 GM51219 to S. R. W. and R01 GM062427 and R01 GM071329 to M. P. R.).
Footnotes
Editors: Tom Misteli and David L. Spector
Additional Perspectives on The Nucleus available at www.cshperspectives.org
REFERENCES
- Ahmed S, Brickner JH 2007. Regulation and epigenetic control of transcription at the nuclear periphery. Trends Genet 23: 396–402 [DOI] [PubMed] [Google Scholar]
- Ahmed S, Brickner DG, Light WH, Cajigas I, McDonough M, Froyshteter AB, Volpe T, Brickner JH 2010. DNA zip codes control an ancient mechanism for gene targeting to the nuclear periphery. Nat Cell Biol 12: 111–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitchison JD, Rout MP, Marelli M, Blobel G, Wozniak RW 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: Complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p. J Cell Biol 131: 1133–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007a. Determining the architectures of macromolecular assemblies. Nature 450: 683–694 [DOI] [PubMed] [Google Scholar]
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007b. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Alcazar-Roman AR, Tran EJ, Guo S, Wente SR 2006. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nat Cell Biol 8: 711–716 [DOI] [PubMed] [Google Scholar]
- Allen NP, Huang L, Burlingame A, Rexach M 2001. Proteomic analysis of nucleoporin interacting proteins. J Biol Chem 276: 29268–29274 [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Hetzer MW 2008. Reshaping of the endoplasmic reticulum limits the rate for nuclear envelope formation. J Cell Biol 182: 911–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W, Mattaj IW 2005. Nuclear pore complexes: Round the bend? Nat Cell Biol 7: 10–12 [DOI] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E 2008. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett 582: 2004–2016 [DOI] [PubMed] [Google Scholar]
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW 1998. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J 17: 7430–7441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball JR, Dimaano C, Ullman KS 2004. The RNA binding domain within the nucleoporin Nup153 associates preferentially with single-stranded RNA. RNA 10: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R, Leung SW, Baker RP, Quimby BB, Corbett AH, Stewart M 2002. Structural basis for the interaction between NTF2 and nucleoporin FxFG repeats. EMBO J 21: 2843–2853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J Cell Biol 154: 1147–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol 167: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradatsch B, Katahira J, Kowalinski E, Bange G, Yao W, Sekimoto T, Baumgartel V, Boese G, Bassler J, Wild K, et al. 2007. Arx1 Functions as an Unorthodox Nuclear Export Receptor for the 60S Preribosomal Subunit. Mol Cell 27: 767–779 [DOI] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JR, Schwartz TU 2009. The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140: 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody SR, Wente SR 2009. mRNA nuclear export at a glance. J Cell Sci 122: 1933–1937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chekanova JA, Abruzzi KC, Rosbash M, Belostotsky DA 2008. Sus1, Sac3, and Thp1 mediate post-transcriptional tethering of active genes to the nuclear rim as well as to non-nascent mRNP. RNA 14: 66–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi NC, Adam SA 1997. Functional domains in nuclear import factor p97 for binding the nuclear localization sequence receptor and the nuclear pore. Mol Biol Cell 8: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CN, Scarcelli JJ 2006. Transport of messenger RNA from the nucleus to the cytoplasm. Curr Opin Cell Biol 18: 299–306 [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E 2001. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol 13: 310–319 [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E 2007. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem 76: 647–671 [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ 2002. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Hetzer MW 2008. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol 18: 456–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Anderson DJ, Richard E, Hetzer MW 2006. Nuclear pores form de novo from both sides of the nuclear envelope. Science 312: 440–443 [DOI] [PubMed] [Google Scholar]
- Daneholt B 2001. Assembly and transport of a premessenger RNP particle. Proc Natl Acad Sci 98: 7012–7017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson TR, Lazarus MD, Hetzer MW, Wente SR 2009. ER membrane-bending proteins are necessary for de novo nuclear pore formation. J Cell Biol 184: 659–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debler EW, Ma Y, Seo HS, Hsia KC, Noriega TR, Blobel G, Hoelz A 2008. A fence-like coat for the nuclear pore membrane. Mol Cell 32: 815–826 [DOI] [PubMed] [Google Scholar]
- DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning D, Mykytka B, Allen NP, Huang L, Al B, Rexach M 2001. The nucleoporin Nup60p functions as a Gsp1p-GTP-sensitive tether for Nup2p at the nuclear pore complex. J Cell Biol 154: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning DP, Patel SS, Uversky V, Fink AL, Rexach M 2003. Disorder in the nuclear pore complex: The FG repeat regions of nucleoporins are natively unfolded. Proc Natl Acad Sci 100: 2450–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci 103: 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieppois G, Iglesias N, Stutz F 2006. Cotranscriptional recruitment to the mRNA export receptor Mex67p contributes to nuclear pore anchoring of activated genes. Mol Cell Biol 26: 7858–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD 2001. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol 153: 1465–1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD 2005. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol 171: 955–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, Robbins J, Dilworth SM, Roberts B, Richardson WD 1988. The nucleoplasmin nuclear location sequence is larger and more complex than that of SV-40 large T antigen. J Cell Biol 107: 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkmann JA, Kutay U 2004. Nuclear export of mRNA: From the site of transcription to the cytoplasm. Exp Cell Res 296: 12–20 [DOI] [PubMed] [Google Scholar]
- Fagotto F, Gluck U, Gumbiner BM 1998. Nuclear localization signal-independent and importin/karyopherin-independent nuclear import of β-catenin. Curr Biol 8: 181–190 [DOI] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E 2004. Protein displacement by DExH/D "RNA helicases" without duplex unwinding. Science 304: 730–734 [DOI] [PubMed] [Google Scholar]
- Fan JS, Cheng Z, Zhang J, Noble C, Zhou Z, Song H, Yang D 2009. Solution and crystal structures of mRNA exporter Dbp5p and its interaction with nucleotides. J Mol Biol 388: 1–10 [DOI] [PubMed] [Google Scholar]
- Feldherr CM, Akin D 1997. The location of the transport gate in the nuclear pore complex. J Cell Sci 110: 3065–3070 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Rout MP 2009. Nuclear pore complex biogenesis. Curr Opin Cell Biol 21: 603–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field MC, Dacks JB 2009. First and last ancestors: Reconstructing evolution of the endomembrane system with ESCRTs, vesicle coat proteins, and nuclear pore complexes. Curr Opin Cell Biol 21: 4–13 [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G 1996. The nuclear transport factor karyopherin β binds stoichiometrically to Ran-GTP and inhibits the Ran GTPase activating protein. J Biol Chem 271: 5313–5316 [DOI] [PubMed] [Google Scholar]
- Floer M, Blobel G, Rexach M 1997. Disassembly of RanGTP-karyopherin β complex, an intermediate in nuclear protein import. J Biol Chem 272: 19538–19546 [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Matunis MJ 1999. A conserved biogenesis pathway for nucleoporins: Proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol 144: 1097–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E 2004. RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRNA export. Mol Cell Biol 24: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Franz C, Walczak R, Yavuz S, Santarella R, Gentzel M, Askjaer P, Galy V, Hetzer M, Mattaj IW, Antonin W 2007. MEL-28/ELYS is required for the recruitment of nucleoporins to chromatin and postmitotic nuclear pore complex assembly. EMBO Rep 8: 165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Gorlich D 2007. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130: 512–523 [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, Gorlich D 2006. FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314: 815–817 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U 2003. Nucleocytoplasmic transport: Taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390: 308–311 [DOI] [PubMed] [Google Scholar]
- Goldfarb DS, Gariepy J, Schoolnik G, Kornberg RD 1986. Synthetic peptides as nuclear localization signals. Nature 322: 641–644 [DOI] [PubMed] [Google Scholar]
- Gorlich D, Dabrowski M, Bischoff FR, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E 1997. A novel class of RanGTP binding proteins. J Cell Biol 138: 65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Dang T, Pane N, Shevchenko A, Mann M, Forbes D, Hurt E 1997. Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol Biol Cell 8: 2017–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant RP, Neuhaus D, Stewart M 2003. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol 326: 849–858 [DOI] [PubMed] [Google Scholar]
- Hang J, Dasso M 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J Biol Chem 277: 19961–19966 [DOI] [PubMed] [Google Scholar]
- Harel A, Forbes DJ 2004. Importin β: Conducting a much larger cellular symphony. Mol Cell 16: 319–330 [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol 18: 6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Cao W, Hackney DD, De La Cruz EM 2008. The ATPase cycle mechanism of the DEAD-box rRNA helicase, DbpA. J Mol Biol 377: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn A, Shi SP, Zarivach R, Ben-Zeev E, Sagi I 2002. The RNA helicase DbpA exhibits a markedly different conformation in the ADP-bound state when compared with the ATP- or RNA-bound states. J Biol Chem 277: 46559–46565 [DOI] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR 2009. Border control at the nucleus: Biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell 17: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Walther TC, Mattaj IW 2005. Pushing the envelope: Structure, function, and dynamics of the nuclear periphery. Annu Rev Cell Dev Biol 21: 347–380 [DOI] [PubMed] [Google Scholar]
- Higa MM, Alam SL, Sundquist WI, Ullman KS 2007. Molecular characterization of the Ran-binding zinc finger domain of Nup153. J Biol Chem 282: 17090–17100 [DOI] [PubMed] [Google Scholar]
- Hodge CA, Colot HV, Stafford P, Cole CN 1999. Rat8p/Dbp5p is a shuttling transport factor that interacts with Rat7p/Nup159p and Gle1p and suppresses the mRNA export defect of xpo1–1 cells. EMBO J 18: 5778–5788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia KC, Stavropoulos P, Blobel G, Hoelz A 2007. Architecture of a coat for the nuclear pore membrane. Cell 131: 1313–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T, Kerscher O, Scott RJ, Basrai MA, Wozniak RW 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J Cell Biol 159: 807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isgro TA, Schulten K 2005. Binding dynamics of isolated nucleoporin repeat regions to importin-β. Structure 13: 1869–1879 [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM 2001. Active disruption of an RNA-protein interactionby a DExH/D RNA helicase. Science 291: 121–125 [DOI] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J 2002. Nuclear export of ribosomal subunits. Trends Biochem Sci 27: 580–585 [DOI] [PubMed] [Google Scholar]
- Joseph J, Dasso M 2008. The nucleoporin Nup358 associates with and regulates interphase microtubules. FEBS Lett 582: 190–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Liu ST, Jablonski SA, Yen TJ, Dasso M 2004. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr Biol 14: 611–617 [DOI] [PubMed] [Google Scholar]
- Jovanovic-Talisman T, Tetenbaum-Novatt J, McKenney AS, Zilman A, Peters R, Rout MP, Chait BT 2009. Artificial nanopores that mimic the transport selectivity of the nuclear pore complex. Nature 457: 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R 2002. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295: 2452–2456 [DOI] [PubMed] [Google Scholar]
- Kalverda B, Roling MD, Fornerod M 2008. Chromatin organization in relation to the nuclear periphery. FEBS Lett 582: 2017–2022 [DOI] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M 2010. Nucleoporins Directly Stimulate Expression of Developmental and Cell-Cycle Genes Inside the Nucleoplasm. Cell 140: 360–371 [DOI] [PubMed] [Google Scholar]
- Keminer O, Peters R 1999. Permeability of single nuclear pores. Biophys J 77: 217–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan VV, Lau EY, Yamada J, Denning DP, Patel SS, Colvin ME, Rexach MF 2008. Intramolecular cohesion of coils mediated by phenylalanine–glycine motifs in the natively unfolded domain of a nucleoporin. PLoS Comput Biol 4: e1000145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitscheck U, Grunwald D, Hoekstra A, Rohleder D, Kues T, Siebrasse JP, Peters R 2005. Nuclear transport of single molecules: Dwell times at the nuclear pore complex. J Cell Biol 168: 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hetzer MW 2008. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol 20: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Gorlich D 1997a. Export of importin α from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Gorlich D 1997b. Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J 16: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D 1998. Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lee BJ, Cansizoglu AE, Suel KE, Louis TH, Zhang Z, Chook YM 2006. Rules for nuclear localization sequence recognition by karyopherin β 2. Cell 126: 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Sterling H, Burlingame A, McCormick F 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev 22: 2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa NC, Brohawn SG, Schwartz TU 2009. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17: 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis A, Felberbaum R, Hochstrasser M 2007. A nuclear envelope protein linking nuclear pore basket assembly, SUMO protease regulation, and mRNA surveillance. J Cell Biol 178: 813–827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Fahrenkrog B, Koser J, Schwarz-Herion K, Deng J, Aebi U 2007a. Nanomechanical basis of selective gating by the nuclear pore complex. Science 318: 640–643 [DOI] [PubMed] [Google Scholar]
- Lim RY, Huang NP, Koser J, Deng J, Lau KH, Schwarz-Herion K, Fahrenkrog B, Aebi U 2006. Flexible phenylalanine-glycine nucleoporins as entropic barriers to nucleocytoplasmic transport. Proc Natl Acad Sci 103: 9512–9517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim RY, Koser J, Huang NP, Schwarz-Herion K, Aebi U 2007b. Nanomechanical interactions of phenylalanine-glycine nucleoporins studied by single molecule force-volume spectroscopy. J Struct Biol 159: 277–289 [DOI] [PubMed] [Google Scholar]
- Lim RY, Ullman KS, Fahrenkrog B 2008. Biology and biophysics of the nuclear pore complex and its components. Int Rev Cell Mol Biol 267: 299–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D 1999. Coordination of tRNA nuclear export with processing of tRNA. Rna 5: 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SM, Stewart M 2005. Structural basis for the high-affinity binding of nucleoporin Nup1p to the Saccharomyces cerevisiae importin-β homologue, Kap95p. J Mol Biol 349: 515–525 [DOI] [PubMed] [Google Scholar]
- Liu HL, De Souza CP, Osmani AH, Osmani SA 2009. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84–120 complex. Mol Biol Cell 20: 616–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Prunuske AJ, Fager AM, Ullman KS 2003. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell 5: 487–498 [DOI] [PubMed] [Google Scholar]
- Lyman SK, Guan T, Bednenko J, Wodrich H, Gerace L 2002. Influence of cargo size on Ran and energy requirements for nuclear protein import. J Cell Biol 159: 55–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macara IG 2001. Transport into and out of the nucleus. Microbiol Mol Biol Rev 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Weis K 2006. Nuclear transport is becoming crystal clear. Chromosoma 115: 98–109 [DOI] [PubMed] [Google Scholar]
- Mahajan R, Delphin C, Guan T, Gerace L, Melchior F 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell 88: 97–107 [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L 1998. Nucleocytoplasmic transport: The soluble phase. Annu Rev Biochem 67: 265–306 [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Wu J, Blobel G 1998. SUMO-1 modification and its role in targeting the Ran GTPase-activating protein, RanGAP1, to the nuclear pore complex. J Cell Biol 140: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlin H, Daneholt B, Skoglund U 1992. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 69: 605–613 [DOI] [PubMed] [Google Scholar]
- Miller BR, Powers M, Park M, Fischer W, Forbes DJ 2000. Identification of a new vertebrate nucleoporin, Nup188, with the use of a novel organelle trap assay. Mol Biol Cell 11: 3381–3396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R 2009. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science 324: 1087–1091 [DOI] [PubMed] [Google Scholar]
- Moroianu J, Blobel G, Radu A 1997. RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc Natl Acad Sci 94: 9699–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy R, Wente SR 1996. An RNA-export mediator with an essential nuclear export signal. Nature 383: 357–360 [DOI] [PubMed] [Google Scholar]
- Nachury MV, Weis K 1999. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci 96: 9622–9627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J 18: 1982–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol 133: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger M, Dlakic M, Tollervey D 2004. A pre-ribosome-associated HEAT-repeat protein is required for export of both ribosomal subunits. Genes Dev 18: 196–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Pante N, Kann M 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskeva E, Izaurralde E, Bischoff FR, Huber J, Kutay U, Hartmann E, Luhrmann R, Gorlich D 1999. CRM1-mediated recycling of snurportin 1 to the cytoplasm. J Cell Biol 145: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SS, Belmont BJ, Sante JM, Rexach MF 2007. Natively unfolded nucleoporins gate protein diffusion across the nuclear pore complex. Cell 129: 83–96 [DOI] [PubMed] [Google Scholar]
- Pemberton LF, Paschal BM 2005. Mechanisms of receptor-mediated nuclear import and nuclear export. Traffic 6: 187–198 [DOI] [PubMed] [Google Scholar]
- Peters R 2005. Translocation through the nuclear pore complex: Selectivity and speed by reduction-of-dimensionality. Traffic 6: 421–427 [DOI] [PubMed] [Google Scholar]
- Pichler A, Gast A, Seeler JS, Dejean A, Melchior F 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell 108: 109–120 [DOI] [PubMed] [Google Scholar]
- Poon IK, Jans DA 2005. Regulation of nuclear transport: Central role in development and transformation? Traffic 6: 173–186 [DOI] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G 1995. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81: 215–222 [DOI] [PubMed] [Google Scholar]
- Reverter D, Lima CD 2005. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature 435: 687–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G 1995. Protein import into nuclei: Association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83: 683–692 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D 2002. The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J 21: 2664–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez MS, Dargemont C, Stutz F 2004. Nuclear export of RNA. Biol Cell 96: 639–655 [DOI] [PubMed] [Google Scholar]
- Rougemaille M, Dieppois G, Kisseleva-Romanova E, Gudipati RK, Lemoine S, Blugeon C, Boulay J, Jensen TH, Stutz F, Devaux F, et al. 2008. THO/Sub2p functions to coordinate 3′-end processing with gene-nuclear pore association. Cell 135: 308–321 [DOI] [PubMed] [Google Scholar]
- Rout MP, Wente SR 1994. Pores for thought: Nuclear pore complex proteins. Trends Cell Biol 4: 357–365 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD 2001. The nuclear pore complex as a transport machine. J Biol Chem 276: 16593–16596 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Magnasco MO, Chait BT 2003. Virtual gating and nuclear transport: The hole picture. Trends Cell Biol 13: 622–628 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT 2000. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Burke B 2006. From pore to kinetochore and back: Regulating envelope assembly. Dev Cell 11: 276–278 [DOI] [PubMed] [Google Scholar]
- Salina D, Enarson P, Rattner JB, Burke B 2003. Nup358 integrates nuclear envelope breakdown with kinetochore assembly. J Cell Biol 162: 991–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarcelli JJ, Hodge CA, Cole CN 2007. The yeast integral membrane protein Apq12 potentially links membrane dynamics to assembly of nuclear pore complexes. J Cell Biol 178: 799–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C, von Kobbe C, Bachi A, Pante N, Rodrigues JP, Boscheron C, Rigaut G, Wilm M, Seraphin B, Carmo-Fonseca M, et al. 1999. Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p. EMBO J 18: 4332–4347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter R, Hitomi M, Ivessa AS, Fasch EV, Kohlwein SD, Tartakoff AM 1996. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol 16: 7161–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Lusk CP, Dilworth DJ, Aitchison JD, Wozniak RW 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae. Mol Biol Cell 16: 4362–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A 2009. Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex. Proc Natl Acad Sci 106: 14281–14286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Goldfarb DS 2003. Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating. Mol Cell Biol 23: 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulga N, Mosammaparast N, Wozniak R, Goldfarb DS 2000. Yeast nucleoporins involved in passive nuclear envelope permeability. J Cell Biol 149: 1027–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G, Grosshans H, Hurt E 2002. Nuclear export of tRNA. Results Probl Cell Differ 35: 115–131 [DOI] [PubMed] [Google Scholar]
- Siniossoglou S, Lutzmann M, Santos-Rosa H, Leonard K, Mueller S, Aebi U, Hurt E 2000. Structure and assembly of the Nup84p complex. J Cell Biol 149: 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Snay-Hodge CA, Colot HV, Goldstein AL, Cole CN 1998. Dbp5p/Rat8p is a yeast nuclear pore-associated DEAD-box protein essential for RNA export. EMBO J 17: 2663–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stade K, Ford CS, Guthrie C, Weis K 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90: 1041–1050 [DOI] [PubMed] [Google Scholar]
- Stewart M 2007. Ratcheting mRNA out of the nucleus. Mol Cell 25: 327–330 [DOI] [PubMed] [Google Scholar]
- Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F 1999. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J 18: 5761–5777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR 2001. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem 276: 6445–6452 [DOI] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Shulga N, Goldfarb DS, Wente SR 2004. Minimal nuclear pore complexes define FG repeat domains essential for transport. Nat Cell Biol 6: 197–206 [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR 2007. Nuclear mRNA export requires specific FG nucleoporins for translocation through the nuclear pore complex. J Cell Biol 178: 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR 2007. Crossing the nuclear envelope: Hierarchical regulation of nucleocytoplasmic transport. Science 318: 1412–1416 [DOI] [PubMed] [Google Scholar]
- Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP 2006. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J Cell Biol 175: 579–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EJ, Wente SR 2006. Dynamic nuclear pore complexes: Life on the edge. Cell 125: 1041–1053 [DOI] [PubMed] [Google Scholar]
- Tran EJ, Bolger TA, Wente SR 2007a. SnapShot: Nuclear transport. Cell 131: 420. [DOI] [PubMed] [Google Scholar]
- Tran EJ, Zhou Y, Corbett AH, Wente SR 2007b. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:Protein remodeling events. Mol Cell 28: 850–859 [DOI] [PubMed] [Google Scholar]
- Tseng SS, Weaver PL, Liu Y, Hitomi M, Tartakoff AM, Chang TH 1998. Dbp5p, a cytosolic RNA helicase, is required for poly(A)+ RNA export. EMBO J 17: 2651–2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu S, Shah S, Orjalo A, Park M, Fischer WH, Forbes DJ 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J Cell Biol 155: 339–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K 2006. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP6 is required for mRNA export. Nat Cell Biol 8: 668–676 [DOI] [PubMed] [Google Scholar]
- Weis K 2003. Regulating Access to the Genome. Nucleocytoplasmic Transport throughout the Cell Cycle. Cell 112: 441–451 [DOI] [PubMed] [Google Scholar]
- Wente SR 2000. Gatekeepers of the nucleus. Science 288: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Whittle JR, Schwartz TU 2009. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem 270: 14209–14213 [DOI] [PubMed] [Google Scholar]
- Xu XM, Meulia T, Meier I 2007. Anchorage of plant RanGAP to the nuclear envelope involves novel nuclear-pore-associated proteins. Curr Biol 17: 1157–1163 [DOI] [PubMed] [Google Scholar]
- Xu Z, Lam LS, Lam LH, Chau SF, Ng TB, Au SW 2008. Molecular basis of the redox regulation of SUMO proteases: A protective mechanism of intermolecular disulfide linkage against irreversible sulfhydryl oxidation. FASEB J 22: 127–137 [DOI] [PubMed] [Google Scholar]
- Yang W, Musser SM 2006. Nuclear import time and transport efficiency depend on importin β concentration. J Cell Biol 174: 951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Jankowsky E 2006. The DEAD-box protein Ded1 unwinds RNA duplexes by a mode distinct from translocating helicases. Nat Struct Mol Biol 13: 981–986 [DOI] [PubMed] [Google Scholar]
- Yang W, Gelles J, Musser SM 2004. Imaging of single-molecule translocation through nuclear pore complexes. Proc Natl Acad Sci 101: 12887–12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Roser D, Kohler A, Bradatsch B, Bassler J, Hurt E 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol Cell 26: 51–62 [DOI] [PubMed] [Google Scholar]
- Yi H, Friedman JL, Ferreira PA 2007. The cyclophilin-like domain of Ran-binding protein-2 modulates selectively the activity of the ubiquitin-proteasome system and protein biogenesis. J Biol Chem 282: 34770–34778 [DOI] [PubMed] [Google Scholar]
- Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature 376: 184–188 [DOI] [PubMed] [Google Scholar]
- York JD, Odom AR, Murphy R, Ives EB, Wente SR 1999. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285: 96–100 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Blobel G 2001. The karyopherin Kap142p/Msn5p mediates nuclear import and nuclear export of different cargo proteins. J Cell Biol 152: 729–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitler B, Weis K 2004. The FG-repeat asymmetry of the nuclear pore complex is dispensible for bulk nucleocytoplasmic transport in vivo. J Cell Biol 167: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemp I, Kutay U 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett 581: 2783–2793 [DOI] [PubMed] [Google Scholar]
- Zhang H, Saitoh H, Matunis MJ 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol Cell Biol 22: 6498–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Brkljacic J, Meier I 2008. Two distinct interacting classes of nuclear envelope-associated coiled-coil proteins are required for the tissue-specific nuclear envelope targeting of Arabidopsis RanGAP. Plant Cell 20: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilman A, Di Talia S, Chait BT, Rout MP, Magnasco MO 2007. Efficiency, selectivity, and robustness of nucleocytoplasmic transport. PLoS Comput Biol 3: e125. [DOI] [PMC free article] [PubMed] [Google Scholar]