Summary
The hammerhead, hairpin, hepatitis delta virus (HDV), Varkud Satellite (VS), and glmS ribozymes catalyze sequence-specific intramolecular cleavage of RNA. They range between 50 and 150 nucleotides in length, and are known as the “small self-cleaving ribozymes.” Except for the glmS ribozyme that functions as a riboswitch in Gram-positive bacteria, they were originally discovered as domains of satellite RNAs. However, recent studies show that several of them are broadly distributed in genomes of organisms from many phyla. Each of these ribozymes has a unique overall architecture and active site organization. Crystal structures have revealed how RNA active sites can bind preferentially to the transition state of a reaction, whereas mechanistic studies have shown that nucleobases can efficiently perform general acid–base and electrostatic catalysis. This versatility explains the abundance of ribozymes in contemporary organisms and also supports a role for catalytic RNAs early in evolution.
Five classes of self-cleaving ribozyme have been found in modern organisms. Each has a unique architecture, and together they showcase the catalytic versatility of RNA.
1. Introduction
Sequence-specific cleavage and ligation are fundamental nucleic acid reactions. Five ribozymes that catalyze them are known from present-day organisms: the hammerhead (Prody et al. 1986), hairpin (Buzayan et al. 1986a), hepatitis delta virus (HDV) (Sharmeen et al. 1988), Varkud satellite (VS) (Saville and Collins, 1990), and glmS (Winkler et al. 2004) ribozymes. In nature, these small, self-cleaving ribozymes catalyze intramolecular reactions. Although the evolutionary origin of these ribozymes is unknown, they have been central to our understanding of the structural, biochemical, and biological versatility of RNA, and thus to the epistemological construction of the RNA World hypothesis. They provided the first high-resolution glimpses of RNA catalysts (Pley et al. 1994; Scott et al. 1995), and have yielded the most detailed structural descriptions of ribozyme-catalyzed chemical reactions (Chi et al. 2008; Klein et al. 2007a; Rupert et al. 2002). The five ribozymes each possess unique folds and active sites, enabling comparison of structurally distinct RNAs that catalyze the same chemical reaction. These studies led to a paradigm shift from the view that all ribozymes are metalloenzymes (Pyle, 1993) to one in which RNA plays a direct role in chemical catalysis (Bevilacqua and Yajima, 2006; Murray et al. 1998). Discovery (Winkler et al. 2004) and characterization (Klein and Ferré-D'Amaré, 2006) of the glmS ribozyme revealed the first natural ribozyme with a coenzyme. Finally, genomic sequencing and analysis are revealing that ribozymes are far more widespread in nature than originally thought (Martick et al. 2008; Salehi-Ashtiani and Szostak, 2001; Webb et al. 2009; Winkler et al. 2004). This article reviews mechanistic insights derived from the study of the small self-cleaving RNAs, whose importance to ribozyme enzymology is analogous to that of serine proteases, lysozyme, and RNase A to protein enzymology.
2. The internal transesterification reaction and its catalysis
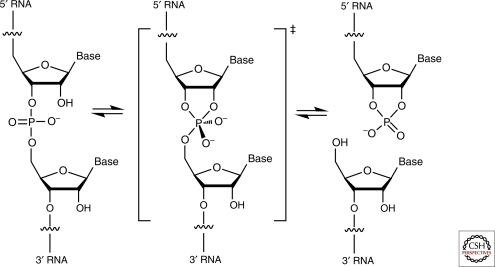
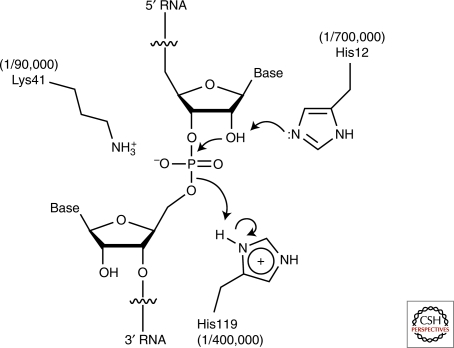
RNA undergoes nonspecific base-catalyzed degradation through an internal transesterification reaction wherein the 2′-O of a ribose attacks the adjacent 3′ phosphate; cleavage products have a 2′,3′-cyclic phosphate and a 5′-OH, respectively (Fig. 1). This reaction is catalyzed by deprotonation of the 2′-OH; hence, its acceleration with increasing pH. The five ribozymes discussed in this article employ the same chemical mechanism, but are highly sequence-specific. RNase A catalyzes RNA cleavage through the same reaction, thus providing a frame of reference. (Unlike the ribozymes, the protein enzyme hydrolyzes the cyclic phosphate in a subsequent step.) Although RNase A shows only modest sequence specificity, it achieves a rate enhancement of ∼1011 over the uncatalyzed reaction. The principal factors responsible are schematized in Figure 2 (Raines 1998). Like all enzymes, RNase A overcomes the entropic and steric penalties of bringing the reactants into the active site in a productive conformation through binding energy. The transesterification reaction proceeds through a concerted SN2-like mechanism wherein the 2′ ribose oxygen, the phosphorus, and the 5′ ribose oxygen are aligned. Richards et al. (1971) determined the structure of the enzyme bound to a dinucleotide mimic inhibitor and discovered that RNase A splays apart the nucleotides flanking the scissile phosphate to achieve alignment. The structure revealed the location of two catalytic histidines and one lysine. His12 functions as a general base catalyst, deprotonating the 2′-OH, and His114 functions as a general acid, protonating the 5′-oxo leaving group. The reaction proceeds through a trigonal bipyramidal oxyphosphorane transition-state whose excess negative charge is stabilized by the ammonium group of Lys41. Mutating these catalytic amino acids greatly impairs RNase A (Fig. 2).
Figure 1.
Internal transesterification reaction catalyzed by the hammerhead, hairpin, HDV, VS, and glmS ribozymes. The concerted cleavage reaction proceeds without intermediates. The hammerhead, hairpin, and VS ribozymes also catalyze the ligation reaction.
Figure 2.
RNase A active site structure and catalytic mechanism. The degree of impairment resulting from site-directed mutations of the catalytic residues (Raines 1998) is indicated in parentheses.
3. Overall structures of self-cleaving ribozymes
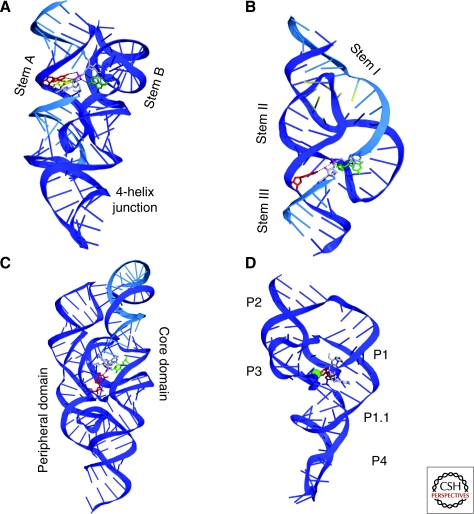
How is an enzyme active site constructed entirely of RNA? Crystal structures of the hairpin, hammerhead, HDV and glmS ribozymes reveal four unique answers (Fig. 3). Each positions the substrate inside an active-site cleft, surrounding it with nucleotides distant in primary sequence. Ribozymes are similar to protein enzymes in that the need to converge multiple functional groups from different parts of the nucleic acid at the active site places a lower bound on their length (for a first-principles discussion about proteins, see de Gennes 1990). RNA is primarily helical; active site formation requires a multi-helical fold, either head-to-tail (coaxial) (Quigley and Rich 1976), or side-by-side. The prevalence of the latter fold is counterintuitive, as it brings the negatively charged backbones of helices into close apposition (Murthy and Rose 2000). Like other structured RNAs, the architecture of self-cleaving ribozymes consists of multihelical junctions, interactions of nonhelical elements (helix-terminal loops and internal bulges), and pseudoknotting.
Figure 3.
Cartoon representations of the overall structures of four self-cleaving ribozymes. (A) The hairpin ribozyme, (B) the hammerhead ribozyme, (C) the glmS ribozyme-riboswitch, and (D) the hepatitis delta virus (HDV) ribozyme. Residues implicated in general acid and base catalysis in the cleavage reaction are green and red, respectively. The scissile phosphate and nucleophilic 2′-O atoms are magenta, flanked by substrate residues shown in light blue.
Multihelical junctions organize the hammerhead, hairpin, and VS ribozymes. The active site of the hairpin ribozyme is formed by apposition of the minor grooves of two irregular helices (stems A and B) (Rupert and Ferré-D'Amaré 2001). One of the strands of stem A carries the scissile phosphate. Although these two helices suffice for activity in vitro (Butcher et al. 1995; Shin et al. 1996), in the natural genomic context of the hairpin ribozyme, they are joined by a four-helix junction that stabilizes the active structure (Murchie et al. 1998) (Fig. 3A). The hammerhead ribozyme is comprised of three helices (stems I, II, and III) arranged into an approximate “γ” shape, with stems II and III stacking coaxially, and stem I packing against stem II (Martick and Scott 2006). In addition to the junction, interactions of nonhelical elements at the tip of stem II and the distal portion of stem I are critical for organizing the RNA (Fig. 3B). The substrate strand is part of stem I, and the active site lies at the three-helix junction (Haseloff and Gerlach 1989; Ruffner et al. 1990; Symons 1997; Uhlenbeck 1987). No crystal structure is available for the VS ribozyme. Modeling based on biochemistry and small-angle-X ray scattering indicates that it is comprised of two three-helix junctions and that it is also stabilized by a long-range interaction between two loops (Lilley 2004). The hammerhead, hairpin and VS ribozymes share the manner in which they bind their substrate. All three have sequence elements that form canonical base pairs with nucleotides on either side of the scissile phosphate on the substrate strand (although not with nucleotides immediately adjacent to the cleavage site). As a result, the substrate is bound as part of an irregular helix that docks with the rest of the ribozyme to form the active molecule. An important consequence of this mode of substrate binding is that after cleavage, both of the product RNA strands may remain bound by the ribozyme (as each forms a short A-form helix with parts of the ribozyme), so that depending on the conditions, these ribozymes can efficiently catalyze the ligation of substrates containing the correct functional groups (a 2′,3′-cyclic phosphate and a 5′-OH) on their termini (Blount and Uhlenbeck 2002; Canny et al. 2007; Fedor 1999; Jones et al. 2001; Saville and Collins 1991).
Pseudoknots organize the structures of the HDV and glmS ribozymes. A pseudoknot is a nucleic acid structure formed by nucleotides in the loop of a stem-loop base pairing with nucleotides outside of the stem-loop (Pleij 1990). The HDV ribozyme has five helices (P1–P4 and P1.1) connected as a nested double pseudoknot (Ferré-D'Amaré et al. 1998; Perrotta and Been 1991). The intricate connectivity of the RNA chain allows this compact (minimal forms are ∼60 nt) and remarkably stable (it remains active in 5M urea) ribozyme to pack two helices stably side-by-side to form the active site (Fig. 3D). The core of the glmS ribozyme is also a nested double pseudoknot (Klein and Ferré-D'Amaré 2006). In addition, this larger ribozyme (∼150 nt) has a peripheral domain that itself contains a pseudoknot. This peripheral domain packs against one of the sides of the core double pseudoknot (Fig. 3C). The HDV and glmS ribozymes position their substrates similarly. Unlike the hammerhead, hairpin, and VS ribozymes, these two ribozymes only base pair with the segment 3′ of the scissile phosphate of their substrates. There are a few non-Watson-Crick interactions between the nucleotide immediately 5′ of the scissile phosphate and the ribozyme core, but no helix is formed between the ribozyme and substrate nucleotides further 5′. As a result, product dissociation occurs readily. Hence, as with RNase A, neither HDV nor glmS ribozymes can relegate cleaved RNA.
4. Active site structures and catalytic mechanisms
Unlike proteins, RNA has no functional groups that are positively charged at neutral pH. The groups with pKa's closest to neutrality are the N1 nitrogens of purines (3.5 and 9.2 for A and G, respectively) and the N3 nitrogens of pyrimidines (4.2 and 9.2 for C and U, respectively). As a polyanion, however, RNA binds cations. In principle, these can function not only as electrostatic catalysts that stabilize negative charges (analogous to Lys41 of RNase A) (Fig. 2), but also as either Lewis acids that perturb the pKa of RNA functional groups bound to them, or as Brønsted acids that exist as hydrates in solution, lowering the pKa of the cation-coordinated waters. Thus, in addition to providing a localized positive charge, tightly bound cations might assist in catalysis by providing hydroxide or hydronium ions to function as reactants or specific base/acid catalysts. Because the Group I intron has been shown to employ tightly bound Mg2+ ions for electrostatic transition-state stabilization and as Lewis acids, it was widely believed that all catalytic RNAs would employ bound metal ions as cofactors (reviewed in Pyle 1993). In all but one of the ribozymes considered here, that assumption proved unnecessary.
5. The hairpin ribozyme: Analogy to RNase A
The hairpin ribozyme was the first self-cleaving ribozyme for which strong evidence was gathered against direct participation of divalent metal ions in catalysis. Like all highly structured RNAs, this ribozyme requires divalent cations for folding under physiological conditions. However, it was found that the hairpin ribozyme remains fully active if magnesium ion is replaced entirely with cobalt (III) hexammine (Hampel and Cowan 1997; Nesbitt et al. 1997; Young et al. 1997). Co(NH3)63+ is isosteric with Mg(H2O)62+, but unlike the water ligands of Mg2+, the amino ligands cannot readily dissociate; therefore, the metal ion cannot chemically participate in catalysis, nor can its ligands be replaced by RNA functional groups to form direct cation–RNA coordinations. Thus, the full activity of the hairpin ribozyme in Co(NH3)63+ in the absence of Mg2+ implies that the RNA, rather than acting as a scaffold for binding divalent cations, must itself instead be directly participating in catalysis. The structure of the ribozyme unambiguously corroborated this prediction.
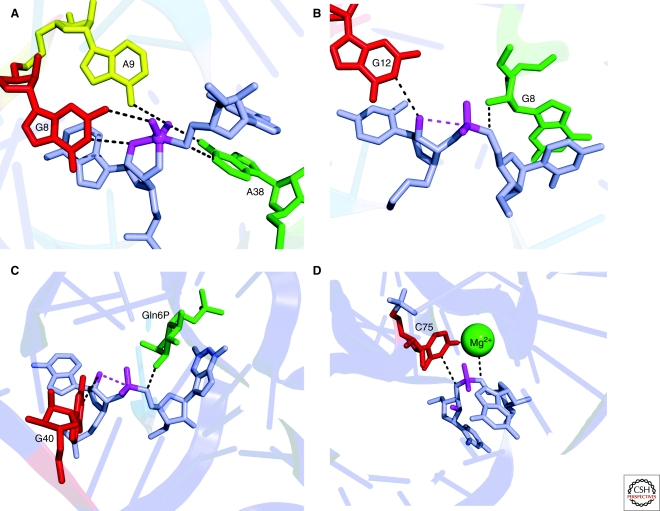
The structure of the hairpin ribozyme was determined in uncleaved, intermediate, and cleaved states. An uncleavable substrate analog was used to obtain the initial-state structure, and an RNA-vanadate complex to mimic the intermediate or transition-state (Rupert and Ferré-D'Amaré 2001; Rupert et al. 2002). The hairpin ribozyme active site possesses some striking similarities to that of RNase A. As in the protein, the substrate nucleotides flanking the scissile phosphate are splayed apart. The nucleotide preceding the scissile phosphate forms a noncanonical pair within stem A, whereas the nucleotide that follows it flips out of stem A and base pairs with an unpaired nucleotide in stem B. This has the effect of bringing the three reactive atoms into a near-in-line conformation (Fig. 4A). Two conserved and functionally important purines, G8 and A38, are positioned on either side of the scissile phosphate, in locations analogous to those of the catalytic histidines of RNase A. Another conserved nucleobase, A9, lies in a position analogous to that of Lys41.
Figure 4.
Active sites of the (A) hairpin, (B) hammerhead, (C) glmS, and (D) HDV ribozymes color-coding as in Figure 3. The moieties thought to contribute to catalysis are labeled.
The hairpin ribozyme transition-state analog structure implicates G8 and A39 as the general base and general acid, respectively, in the cleavage reaction (their roles would be reversed in the ligation reaction). It also indicates that the exocyclic amines of A9 and A38 play a role analogous to the amine of Lys41 in electrostatic transition-state stabilization. Modeling of the pH-dependence of the hairpin ribozyme reaction, and comparison of it to that of the RNase A reaction, not only corroborates this assessment but permits assignment of slightly perturbed pKa values to the general base and general acid of 9.0 and 5.0, respectively (Bevilacqua 2003), consistent with their identification in the crystal structures as a G and an A (but also see Liu et al. 2009). Hence the hairpin ribozyme structure, in a manner consistent with the observed biochemistry, revealed that the RNA itself possesses the necessary structural and chemical capacity to catalyze acid-base phosphodiester chemistry.
6. The hammerhead ribozyme: Variation on a theme
Although the hammerhead has long been the prototype ribozyme and was the first catalytic RNA whose structure was determined, its history has been fraught with experimental discord (Blount and Uhlenbeck 2005). Between its discovery (Prody et al. 1986) and 2003, most attention was focused on fifteen near-invariant nucleotides at the three-helix junction core of a “minimal” hammerhead (Blount and Uhlenbeck 2005; McKay 1996; Scott 1999; Wedekind and McKay 1998). The crystal structures revealed an active site that offered no clear explanation for the invariance of many of the core residues (McKay 1996). The hammerhead ribozyme was originally thought to be a metalloenzyme. Although no obvious mechanism was suggested by metal ions present in the crystal structures of the minimal hammerhead ribozyme, divalent cations were still considered to be the most likely acid/base catalytic components. In 1998, it was shown that the hammerhead, hairpin, and VS ribozymes, unlike the HDV ribozyme, did not require divalent metal ions for catalysis, but instead were catalytically proficient in high concentrations of monovalent cations, including the nonmetallic ammonium ion (Murray et al. 1998). In other words, folding and charge stabilization were sufficient to support catalytic activity of these ribozymes.
In 2003, two groups revealed that “full-length” hammerhead ribozymes derived from natural sequences, which included a previously neglected tertiary contact between helices I and II in a region with little sequence conservation, were up to 1000-fold more active than minimal hammerheads (De la Peña et al. 2003; Khvorova et al. 2003). Subsequent crystal structures of precleavage and postcleavage full-length hammerhead ribozymes that include these tertiary contacts revealed a much more informative active site that is both reminiscent of the hairpin ribozyme active site and intriguingly different (Chi et al. 2008; Martick and Scott 2006).
Crystal structures of full-length hammerhead ribozymes revealed an arrangement of residues in the active site that makes their participation in catalysis clear (Fig. 4B) The invariant G12 hydrogen-bonds to the nucleophilic 2′-O; this conserved guanine is likely the general base in the self-cleavage reaction. A hydrogen bond between a the 2′-OH of a second invariant nucleotide, G8, and the 5′-oxo leaving group was also identified. This implicates the 2′-OH of the ribose of G8 as a general acid catalyst. (The requirement for G at position 8 is due to a Watson-Crick tertiary base pair it forms with the equally invariant C3.) In addition, the crystal structure of a very slowly cleaving G12A hammerhead (whose core structure is essentially identical but whose purine general base has a pKa lowered by ∼5 units) revealed, in the product structure, a potential additional interaction between A9 and the scissile phosphate, reminiscent of the coincidently named A9 of the hairpin ribozyme. However, whether A9 or another entity engages in transition-state stabilization in the hammerhead ribozyme remains speculative.
7. The glmS ribozyme: Co-opting a cofactor
This ribozyme was discovered as an RNA domain that catalyzes site-specific RNA cleavage in the presence of the small-molecule metabolite glucosamine-6-phosphate (GlcN6P) (Winkler et al. 2004). In the absence of GlcN6P, the rate of cleavage of the ribozyme is indistinguishable from that of background hydrolysis. Addition of GlcN6P results in as much as 107-fold increase of reactivity at the specific cleavage site. The glmS ribozyme is not a metalloenzyme, because it is fully active with Co(NH3)63+ (Roth et al. 2006). In principle, GlcN6P could function either as an allosteric activator or as a coenzyme. The structure reveals that the N1 of the conserved nucleotide G40 lies close to the 2′-OH nucleophile, well positioned to serve as a general base. GlcN6P binds on the opposite side of the scissile phosphate, with its amine group in van der Waals contact with the 5′-O leaving group, an arrangement compatible with its function as a general acid (Fig. 4C). If GlcN6P binds to the ribozyme in its protonated (ammonium) form, a positive charge would lie adjacent to the scissile phosphate, where it could stabilize the transition state. Therefore, the structure of the active site of the glmS ribozyme is consistent with a coenzyme function for GlcN6P. Moreover, structures of the glmS ribozyme in different states (precleavage, precleavage bound to GlcN6P, transition-state mimic, postcleavage) do not show any evidence of conformational change of the RNA resulting from GlcN6P binding (Klein and Ferré-D'Amaré 2006; Klein et al. 2007b).
A remarkable feature of the glmS ribozyme is the absolute requirement for both G40 and GlcN6P for catalytic activity. The ribozyme is inactive in the absence of GlcN6P, but is also inactive in the presence of GlcN6P if a G40A mutation is introduced into the ribozyme. Structure determination shows that the mutant adopts the wild-type conformation, and that GlcN6P binds in precisely the same location as to the wild-type (Klein et al. 2007a). Thus, lack of activity of the G40A mutant is not caused by misfolding. The strict requirement for both catalytic functional groups contrasts with the behavior of other enzymes. For instance, site-directed mutation of either catalytic histidine of RNase A to alanine results in an enzyme that is still active, although impaired by 400,000 to 700,000-fold (Raines 1998). Analogously, abasic substitutions of the three catalytic purines in the active site of the hairpin ribozyme results in 9350- and 14,000-fold loss in activity (for A9, G8, and A38, respectively) but each mutant retains activity (Kuzmin et al. 2005; Lebruska et al. 2002). The simultaneous requirement for G40 and GlcN6P by the glmS ribozyme implies that the active-site guanine and the small molecule coenzyme mutually tune each other's chemical properties such that neither is catalytically proficient in the absence of the other.
8. The HDV ribozyme: Nucleobase and metal-ion catalysis
Of the five known self-cleaving ribozymes, the HDV ribozyme is the only one that employs an active-site divalent cation for catalysis. Ironically, this was also the first ribozyme for which compelling evidence for nucleobase participation in catalysis was gathered. Structure determination of the postcleavage form of the HDV ribozyme revealed that the N3 imine of the functionally essential residue C75 (C76 in the antigenomic HDV ribozyme) hydrogen bonds to the 5′-OH leaving group of the reaction. In addition, C75 was found in a region of strongly negative electrostatic potential. These observations suggested that C75 may have an altered pKa, and function as a general acid/base catalyst (Ferré-D'Amaré et al. 1998). This was reinforced by several observations, including the rescue of a C75U mutant by exogenous imidazole, and the shift of the reaction pKa in C76A mutant that matches the difference in pKa between C and A (Perrotta et al. 1999). Kinetic experiments cannot distinguish between general acid and general base functions for C75/76. Strong evidence for the former comes from Das and Piccirilli (2005), who synthesized chemically modified HDV ribozymes in which the leaving group oxygen of the reaction was replaced with a sulfur. These RNAs were expected lose their dependence on C75 if the role of this nucleobase was to protonate the leaving group (stabilizing it), but not if C75 functions as a general base, deprotonating the 2′-OH. Kinetic characterization revealed that the sulfur substitution limits the need for C75. The most definitive evidence for divalent cation participation in HDV ribozyme catalysis comes from kinetic analysis, and from the inactivity of the ribozyme in the presence of Co(NH3)63+ (Nakano et al. 2000). Thus, in the active site of the HDV ribozyme, a magnesium ion-activated water molecule functions as a general base, and the N3 imine of C75 serves as a general acid (Fig. 4D).
9. How widespread are the self-cleaving ribozymes?
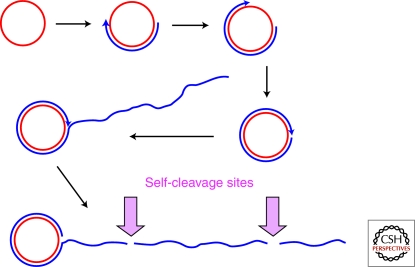
The hammerhead and hairpin ribozymes were discovered embedded within the sense and antisense strands, respectively, of the satellite RNA of tobacco ringspot virus (Buzayan et al. 1986b; Prody et al. 1986), and subsequently in other plant satellite virus RNAs and viroids. These virus-like RNAs are single-stranded, covalently closed circular molecules that are replicated via the rolling-circle mechanism (Fig. 5). The ribozyme domains occur at the interface of two monomeric segments of a concatamer, and catalyze the self-cleavage and self-ligation of the genomic and antigenomic RNA segments. Two variants of the HDV ribozyme were subsequently discovered in the sense and antisense strands of HDV, a single-stranded circular RNA virus that is a satellite of the hepatitis B (DNA) virus and a human pathogen. HDV also replicates by the rolling-circle mechanism, and the ribozymes are responsible for generating unit-length genomes. The VS ribozyme was discovered as part of an abundant mitochondrial RNA of natural isolates of Neurospora. The RNA is encoded by a retroplasmid, and the function of the ribozyme is to process into unit length molecules the RNA concatamers that result from transcription of the plasmid (Collins 2002). Because the biological contexts of the hammerhead, hairpin, HDV, and VS ribozymes are very similar, it was thought that self-cleaving ribozymes would be restricted to this specialized evolutionary niche.
Figure 5.
Rolling-circle replication of satellite RNAs. Single-stranded covalently closed circles of sense (or genomic) RNA (red circle) are replicated by an RNA polymerase of the host, which transcribes processively, generating a linear multimeric concatamer (blue) that is complementary to the monomeric satellite RNA sense-strand. The multimeric complementary copy is then cleaved in to monomeric fragments, and these antigenomic RNAs circularize to produce templates for the second half of the replicative cycle, in which copies of the genomic sense strand are ultimately produced. The cleavage and ligation sites are often comprised of conserved ribozyme sequences that catalyze both phosphodiester bond cleavage and, in the case of circularization of the monomeric fragments, phosphodiester bond ligation. In the case of the satellite RNA of tobacco ringspot virus, the cleavage site in the sense strand is a hammerhead ribozyme, and that in the antisense strand is the hairpin ribozyme. HDV ribozyme sequences are present in both the sense and antisense strands of HDV.
Hints that these ribozymes might be more widespread in nature began to emerge in the 1990s. Circularizing RNA transcripts of repetitive satellite “junk” DNAs from newt (Epstein and Pabon-Pena 1991) and Schistosoma (Ferbeyre et al. 2000) were found to contain hammerhead ribozymes; the latter were found to be exceptionally active. Although genomic scans for known hammerhead sequences aided in discovery of the Schistosoma hammerhead (Bourdeau et al. 1999; Ferbeyre et al. 1998), it turned out that the search criteria were too restrictive. If the implicit requirement that the hammerhead ribozyme sequence be continuous is relaxed, a whole set of discontinuous ribozymes that closely resemble the Schistosoma sequence can be found in the 3′-UTRs of mature mRNAs that code for the clec2 family of proteins in mammals ranging from platypus to mice (Martick et al. 2008). These hammerhead ribozymes are located immediately downstream from the stop codon, are quite active, and capable of deactivating translation in vivo. These findings suggested self-cleaving RNAs that regulate gene expression may be widespread in the genomes of free-living organisms. Moreover, in vitro selection experiments revealed that the minimal hammerhead ribozyme commonly arises, suggesting it might have evolved multiple times independently (Salehi-Ashtiani and Szostak 2001).
The glmS ribozyme was discovered through a bioinformatic search for riboswitches in bacterial genomes (Barrick et al. 2004). Riboswitches, which are not typically catalytic, are gene-regulatory RNAs that bind small molecules and modulate the expression of mRNAs of which they are part (see Breaker 2010 and Garst et al. 2010). The glmS ribozyme-riboswitch is present in the 5′-UTRs of mRNAs that encode the essential enzyme GlcN6P synthase in Gram-positive bacteria. The 5′ termini of mRNAs are capped by a triphosphate in Gram-positive bacteria. GlcN6P-activated self-cleavage of the ribozyme domain exposes a 5′-OH, and this recruits an RNase protein that degrades the ribozyme-cleaved mRNA. Because GlcN6P synthase is an unstable protein, degradation of the mRNA completes a negative feedback loop that modulates intracellular GlcN6P concentration (Collins et al. 2007). The glmS ribozyme is widespread in Gram-positive bacteria, including important human pathogens. It is at present unknown in Gram-negative bacteria, archaea or Eukarya.
The most recent expansion of the known range of self-cleaving ribozymes comes from searches for HDV-like ribozymes. Salehi-Ashtiani et al. (2006) discovered several self-cleaving RNAs in the human genome through in vitro selection. One of these is conserved throughout mammals in an intron of the CPEB3 gene. This ribozyme is closely related to the HDV ribozyme. When Lupták and coworkers performed bioinformatic searches for additional HDV/CPEB3 ribozymes using a structure-based template, they discovered variants of the ribozymes scattered throughout phylogeny, in viruses, bacteria, protists, plants, fungi, and animal phyla including platyhelmynths, nematodes, molluscs, insects, echinoderms, cephalochordates, and fish (Webb et al. 2009). All these RNAs preserve the core structure and catalytic elements of the HDV ribozyme, and many are active in vitro and in vivo. An HDV-like ribozyme from was found to be differentially active in different developmental stages of the African mosquito Anopheles gambiae. HDV-like ribozymes occur in dozens of loci in several organisms. However, the genomic location and numbers of ribozymes vary greatly in sister species. Perhaps related to this is that the cleavage sites of several of them occur at the 5′ termini of transposable elements. Although it is too early to draw conclusions about the origins of these RNAs, it is clear now that self-cleaving ribozymes are common elements of genomes throughout the biosphere.
10. Concluding remarks
The study of the small self-cleaving ribozymes has revealed that remarkably small RNA units can achieve high sequence specificity and catalytic efficiency. The information content required for activity is even lower than indicated by the length of the ribozymes, because the only requirement for some tracts of their sequences is that A-form double helices be able to form. That is, the composition of many of the helical segments of small ribozymes has little effect on catalytic activity. Thus, RNAs capable of biochemical catalysis are likely to be quite common in sequence space, and even a modest repertory of RNAs of random sequence present at the outset of the RNA world may have contained ribozymes. This conclusion is supported by in vitro selection experiments (e.g., Salehi-Ashtiani and Szostak 2001). In addition, the limited sequence requirements of ribozymes would have been important for primordial RNA organisms whose replicative polymerases may have been error-prone (see Robertson and Joyce 2010). In vitro selection experiments also show that small ribozymes can catalyze a broad range of biochemical transformations (reviewed in Wilson and Szostak 1999), as would have been required in the RNA world if ribozymes were responsible for all biochemical catalysis. Thus far, however, all compact ribozymes known from nature catalyze the same overall transformation, RNA cleavage by internal transesterification. At this point in time, it is unclear if this indicates that small ribozymes that catalyze other reactions remain to be discovered in present-day organisms, or that ribozymes with other biochemical functions have become extinct.
If the advent of biochemical catalysis predates the transition from an RNA world to one in which protein enzymes play the predominant role, it is fair to ask what forms of catalysis might be of such a fundamental nature as to be shared by both RNA and protein enzymes. Metal-ion assisted catalysis and general acid–base catalysis both emerge as contenders for this distinction. Group I and Group II self-splicing introns (see Lambowitz and Zimmerly 2010) share with protein enzymes that polymerize nucleic acids the use of catalytic Mg2+ ions (Steitz and Steitz 1993). On the other hand, the similar general acid–base catalytic strategies, employed by the small self-cleaving RNAs and analogous protein enzymes, such as RNase A, are a recurring theme. What is remarkable, in the context of small self-cleaving ribozymes, is the variation within shared catalytic strategies. The hairpin ribozyme is perhaps the most RNase A-like, in that purines play roles analogous to both catalytic histidines of the protein, and both the RNA and the protein employ amine groups to aid in transition-state stabilization. Nonetheless, this is clearly not the only way, and the variability of the acid catalyst in particular indicates a remarkable ability for evolution to fine-tune a ribozyme's catalytic strategy to meet what are presumably different selective pressures. The accelerating pace of discovery of self-cleaving ribozymes in the genomes of contemporary free-living organisms indicates that far from being molecular living fossils, these RNAs are active players in nucleic acid metabolism. Perhaps this is less surprising now, in light of the catalytic versatility of RNA revealed by the study of self-cleaving ribozymes.
Acknowledgments
The authors dedicate this review to the memory of Frederic M. Richards (1925-2009), and gratefully acknowledge the efforts of past and present members of their research groups. Work in the authors' laboratories summarized here was funded in part by the National Institutes of Health (AI043393 to WGS and GM63576 to ARF) the W.M. Keck Foundation (ARF and WGS) and the Howard Hughes Medical Institute (ARF).
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I, et al. 2004. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci 101: 6421–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua PC 2003. Mechanistic considerations for general acid-base catalysis by RNA: revisiting the mechanism of the hairpin ribozyme. Biochemistry 42: 2259–2265 [DOI] [PubMed] [Google Scholar]
- Bevilacqua PC, Yajima R 2006. Nucleobase catalysis in ribozyme mechanism. Current Op Chem Biol 10: 455–464 [DOI] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC 2002. Internal equilibrium of the hammerhead ribozyme is altered by the length of certain covalent cross-links. Biochemistry 41: 6834–6841 [DOI] [PubMed] [Google Scholar]
- Blount KF, Uhlenbeck OC 2005. The structure-function dilemma of the hammerhead ribozyme. Annu Rev Biophys Biomol Struct 34: 410–440 [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Ferbeyre G, Pageau M, Paquin B, Cedergren R 1999. The distribution of RNA motifs in natural sequences. Nucleic Acids Res 27: 4457–4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SE, Heckman JE, Burke JM 1995. Reconstitution of hairpin ribozyme activity following separation of functional domains. J Biol Chem 270: 29648–29651 [DOI] [PubMed] [Google Scholar]
- Buzayan JM, Gerlach WL, Bruening G 1986a. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus statellite RNA. Nature 323: 349–353 [Google Scholar]
- Buzayan JM, Hampel A, Bruening G 1986b. Nucleotide sequence and newly formed phosphodiester bond of spontaneously ligated satellite tobacco ringspot virus RNA. Nucleic Acids Res 14: 9729–9743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker RR 2010. Riboswitches and the RNA world. Cold Spring Harb Perspect Biol doi: 10.1101.cshperspect.a003566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny MD, Jucker FM, Pardi A 2007. Efficient ligation of the Schistosoma hammerhead ribozyme. Biochemistry 46: 3826–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Y-I, Martick M, Lares M, Kim R, Scott WG, Kim S-H, Joyce GF 2008. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. Plos Biol 6: e234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R 2002. The Neurospora varkud satellite ribozyme. Biochem Soc Trans 30: 1122–1126 [DOI] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21: 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Piccirilli JA 2005. General acid catalysis by the hepatitis delta virus ribozyme. Nature Chem Biol 1: 45–52 [DOI] [PubMed] [Google Scholar]
- de Gennes P-G 1990. Minimum numer of aminoacids required to build up a specific receptor with a folded polypeptide chain. In Introduction to polymer dynamics Cambridge, Cambridge University Press, pp. 17–26 [Google Scholar]
- De la Peña M, Gago S, Flores R 2003. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J 22: 5561–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Pabon-Pena LM 1991. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res 19: 1699–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor MJ 1999. Tertiary structure stabilization promotes hairpin ribozyme ligation. Biochemistry 38: 11040–11050 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Smith JM, Cedergren R 1998. Schitosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol 18: 3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré-D'Amaré AR, Zhou K, Doudna JA 1998. Crystal structure of a hepatitis delta virus ribozyme. Nature 395: 567–574 [DOI] [PubMed] [Google Scholar]
- Ferbeyre G, Bourdeau V, Pageau M, Miramontes P, Cedergren R 2000. Distribution of hammerhead and hammerhead-like RNA motifs through the GenBank. Genome Res 10: 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT 2010. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol doi: 10.1101.cshperspect.a003533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampel A, Cowan JA 1997. A unique mechanism for RNA catalysis: The role of metal cofactors in hairpin ribozyme cleavage. Chem Biol 4: 513–517 [DOI] [PubMed] [Google Scholar]
- Haseloff J, Gerlach WL 1989. Sequences required for self-catalyzed cleavage of the satellite RNA of tobacco ringspot virus. Gene 82: 43–52 [DOI] [PubMed] [Google Scholar]
- Jones FD, Ryder SP, Strobel SA 2001. An efficient ligation reaction promoted by a Varkud Satellite ribozyme with extended 5′- and 3′-termini. Nucleic Acids Res 29: 5115–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena S 2003. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol 10: 708–712 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Ferré-D'Amaré AR 2006. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science 313: 1752–1756 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Been MD, Ferré-D'Amaré AR 2007a. Essential role of an active-site guanine in glmS ribozyme catalysis. J Am Chem Soc 129: 14858–14859 [DOI] [PubMed] [Google Scholar]
- Klein DJ, Wilkinson SR, Been MD, Ferré-D'Amaré AR 2007b. Requirement of helix P2.2 and nucleotide G1 for positioning of the cleavage site and cofactor of the glmS ribozyme. J Mol Biol 373: 178–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin YI, Da Costa CP, Cottrell JW, Fedor MJ 2005. Role of an active site adenine in hairpin ribozyme catalysis. J Mol Biol 349: 989–1010 [DOI] [PubMed] [Google Scholar]
- Lambowitz AM, Zimmerly S 2010. Group II introns: mobile ribozymes that invade DNA. Cold Spring Harb Perspect Biol doi: 10.1101.cshperspect.a003616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebruska LL, Kuzmine II, Fedor MJ 2002. Rescue of an abasic hairpin ribozyme by cationic nucleobases. Evidence for a novel mechanism of RNA catalysis. Chem Biol 9: 465–473 [DOI] [PubMed] [Google Scholar]
- Lilley DM 2004. The Varkud satellite ribozyme. RNA 10: 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cottrell JW, Scott LG, Fedor MJ 2009. Direct measurement of the ionization state of an essential guanine in the hairpin ribozyme. Nature Chem Biol 5: 351–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Scott WG 2006. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell 126: 309–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG 2008. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature 454: 899–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay DB 1996. Three-dimensional structure of the hammerhead ribozyme. Nucleic Acids Mol Biol 10: 161–172 [Google Scholar]
- Murchie AIH, Thomson JB, Walter F, Lilley DMJ 1998. Folding of the hairpin ribozyme in its natural conformation achieves close physical proximity of the loops. Mol Cell 1: 873–881 [DOI] [PubMed] [Google Scholar]
- Murray JB, Seyhan AA, Walter NG, Burke JM, Scott WG 1998. The hammerhead, hairpin and VS ribozymes are catalytically proficient in monovalent cations alone. Chem Biol 5: 587–595 [DOI] [PubMed] [Google Scholar]
- Murthy VL, Rose GD 2000. Is counterion delocalization responsible for collapse in RNA folding. Biochemistry 39: 14365–14370 [DOI] [PubMed] [Google Scholar]
- Nakano S-I, Chadalavada DM, Bevilacqua PC 2000. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science 287: 1493–1497 [DOI] [PubMed] [Google Scholar]
- Nesbitt S, Hegg LA, Fedor MJ 1997. An unusual pH-independent and metal-ion independent mechanism for hairpin ribozyme catalysis. Chem Biol 4: 619–630 [DOI] [PubMed] [Google Scholar]
- Perrotta AT, Been MD 1991. A pseudoknot-like structure required for efficient self-cleavage of hepatitis delta virus RNA. Nature 350: 434–436 [DOI] [PubMed] [Google Scholar]
- Perrotta AT, Shih I, Been MD 1999. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science 286: 123–126 [DOI] [PubMed] [Google Scholar]
- Pleij CW 1990. Pseudoknots: A new motif in the RNA game. Trends Biochem Sci 15: 143–147 [DOI] [PubMed] [Google Scholar]
- Pley HW, Flaherty KM, McKay DB 1994. Three-dimensional structure of a hammerhead ribozyme. Nature 372: 68–74 [DOI] [PubMed] [Google Scholar]
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G 1986. Autolytic processing of dimeric plant virus satellite RNA. Science 231: 1577–1580 [DOI] [PubMed] [Google Scholar]
- Pyle AM 1993. Ribozymes: A distinct class of metalloenzymes. Science 261: 709–714 [DOI] [PubMed] [Google Scholar]
- Quigley GJ, Rich A 1976. Structural domains of transfer RNA molecules. Science 194: 796–806 [DOI] [PubMed] [Google Scholar]
- Raines RT 1998. Ribonuclease A. Chem Rev 98: 1045–1065 [DOI] [PubMed] [Google Scholar]
- Richards FM, Wyckoff HW, Carlson WD, Allewell NM, Lee B, Mitsui Y 1971. Protein structure, ribonuclease-S, and nucleotide interactions. Cold Spring Harbor Symp Quant Biol 36: 35–43 [DOI] [PubMed] [Google Scholar]
- Robertson MP, Joyce GF 2010. The origins of the RNA world. Cold Spring Harb Perspect Biol doi: 10.1101.cshperspect.a003608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Nahvi A, Lee M, Jona I, Breaker RR 2006. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA 12: 607–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruffner DE, Stormo GD, Uhlenbeck OC 1990. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry 29: 10695–10702 [DOI] [PubMed] [Google Scholar]
- Rupert PB, Ferré-D'Amaré AR 2001. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature 410: 780–786 [DOI] [PubMed] [Google Scholar]
- Rupert PB, Massey AP, Sigurdsson ST, Ferré-D'Amaré AR 2002. Transition state stabilization by a catalytic RNA. Science 298: 1421–1424 [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Szostak JW 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84 [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW 2006. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313: 1788–1792 [DOI] [PubMed] [Google Scholar]
- Saville BJ, Collins RA 1990. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell 61: 685–696 [DOI] [PubMed] [Google Scholar]
- Saville BJ, Collins RA 1991. RNA-mediated ligation of self-cleavage products from a Neurospora mitochondrial plasmid transcript. Proc Natl Acad Sci USA 88: 8826–8830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WG 1999. Biophysical and biochemical investigations of RNA catalysis in the hammerhead ribozyme. Q Rev Biophys 32: 241–284 [DOI] [PubMed] [Google Scholar]
- Scott WG, Finch JT, Klug A 1995. The crystal structure of an all-RNA hammerhead ribozyme: A proposed mechanism for RNA catalytic cleavage. Cell 81: 991–1002 [DOI] [PubMed] [Google Scholar]
- Sharmeen L, Kuo MY-P, Dinter-Gottlieb G, Taylor J 1988. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol 62: 2674–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Choi JN, Song SI, Song JT, Ahn JH, Lee JS, Choi YD 1996. The loop B domain is physically separable from the loop A domain in the hairpin ribozyme. Nucleic Acids Res 24: 2685–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons RH 1997. Plant pathogenic RNAs and RNA catalysis. Nucl Acids Res 25: 2683–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck OC 1987. A small catalytic oligoribonucleotide. Nature 328: 596–600 [DOI] [PubMed] [Google Scholar]
- Webb C-HT, Riccitelli NJ, Ruminski DJ, Lupták A 2009. Widespread occurrence of self-cleaving ribozymes. Science 326: 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedekind JE, McKay DB 1998. Crystallographic structures of the hammerhead ribozyme: relationship to ribozyme folding and catalysis. Annu Rev Biophys Biomol Struct 27: 475–502 [DOI] [PubMed] [Google Scholar]
- Wilson DS, Szostak JW 1999. In vitro selection of functional nucleic acids. Annu Rev Biochem 68: 611–647 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286 [DOI] [PubMed] [Google Scholar]
- Young KJ, Gill F, Grasby JA 1997. Metal ions play a passive role in the hairpin ribozyme catalyzed reaction. Nucleic Acids Res 25: 3760–3766 [DOI] [PMC free article] [PubMed] [Google Scholar]