Abstract
European funding under framework 7 (FP7) for the virtual physiological human (VPH) project has been in place now for nearly 2 years. The VPH network of excellence (NoE) is helping in the development of common standards, open-source software, freely accessible data and model repositories, and various training and dissemination activities for the project. It is also helping to coordinate the many clinically targeted projects that have been funded under the FP7 calls. An initial vision for the VPH was defined by framework 6 strategy for a European physiome (STEP) project in 2006. It is now time to assess the accomplishments of the last 2 years and update the STEP vision for the VPH. We consider the biomedical science, healthcare and information and communications technology challenges facing the project and we propose the VPH Institute as a means of sustaining the vision of VPH beyond the time frame of the NoE.
Keywords: virtual physiological human, physiome, computational physiology, multi-scale modelling
1. The virtual physiological human vision
The physiome, systems biology, the virtual physiological human (VPH ), personal health systems, biomedical informatics, life science e-infrastructures, systems pharmacology—all these domains share one common issue: the need for integration. To implement biomedical research outputs into clinical practice and healthcare industries, we need to integrate data, information, knowledge and wisdom. We need to integrate data of the same patient stored in different hospitals in different member states or in clinical research databases; we need to integrate the information related to various parts and processes of the human body into a systemic understanding of pathophysiology; we need to integrate the knowledge digitally captured into metadata, ontologies and models in order to respond to the combinatorial explosion of cognitive complexity that integrative research is producing; and we need to integrate the wisdom produced in the research laboratories and in clinical practice, which will be formalized in guidelines, standards and protocols and used to promote translation of basic science and integrative models into healthcare benefits.
This is a huge challenge that, if met, will have a tremendous impact on the life of our citizens, and on the European and international economy. The vision of a ‘digital me’ that contains all my healthcare information, safely managed for access by the various biomedical professionals with my approval, communicated with all my wearable and implanted technology to constantly monitor my health status and informing me, my family and friends, or my healthcare providers of alarming events, supporting the collaboration of various specialists around my complex systemic diseases, and used with all my data to predict the future development of my health in order to facilitate disease prevention and a fully self-aware lifestyle, is a powerful vision. But the challenges are huge.
The VPH framework 7 (FP7) project is addressing this challenge by promoting and facilitating the use of computational models, software tools and web services. The goal is to achieve a more efficient and effective twenty-first century healthcare system and to create new economic opportunities for European healthcare industries. In common with other areas of application of modern scientific methods, medical practice will benefit from technologies in which digital data enables predictable outcomes through quantitative models that integrate physical processes across spatial scales down to the molecular level. We need to promote personalized, predictive, integrative and evidence-based approaches to medicine. These will use computational tools to link individual patient data with virtual population databases via the knowledge of biological processes encoded in mathematical models. The biomedical community also now has the opportunity, thanks to the adoption of new model and data standards and a common set of reference ontologies, to assemble the molecular pieces from 50 years of reductionist science in order to understand genotype–phenotype relationships by linking databases of genetic and proteomic data to anatomy and function at the cell, tissue and organ levels. Biophysically based computational modelling of the human body, applied to human physiology and the diagnosis and treatment of disease, will revolutionize twenty-first century biosciences and medicine. The success of this exciting opportunity is highly dependent both on the development, adoption and integration of information and communications technology (ICT) and e-Health infrastructures throughout Europe, and on the coordination of this effort with other related international initiatives such as the physiome project.
Note that the network of excellence (NoE) is concerned primarily with ICT infrastructure, coordination and training for the VPH, while the VPH initiative (VPH-I) projects themselves are primarily focused on developing and implementing biophysically based computational models into clinical environments via industrial partners. The success of all of these endeavours is of course dependent on the continued progress of (separately funded) biomedical science in revealing the biophysical mechanisms underlying structure and function at all spatial scales.
A roadmap for the VPH project was laid out in 2006 by the strategy for a European physiome (STEP) coordinated action (Fenner et al. 2008; http://www.europhysiome.org/roadmap). The outcome of the first FP7 VPH funding round in 2007 (call 2) was the VPH NoE, three integrated projects (IPs), nine specific targeted research projects (STREPs) and two cooperative actions (CAs), all of which form the core of the European VPH-I. With nearly 3 years of experience behind us (including STEP), it is time to assess our achievements and plans for the short-, medium- and long-term future of the VPH and the NoE.
2. What have we achieved so far?
The importance of establishing a solid foundation for the VPH by creating model and data standards, together with mechanisms for achieving model reproducibility and reuse, was recognized in the STEP roadmap. This, together with the development of plans for dissemination, training and outreach to the communities of researchers, physicians, patients, students, European industry and the public in general, has been the primary focus for the first year of the NoE. Direct engagement with the other VPH projects and clear examples of how standards-based models, software tools and web-based services can be used to facilitate clinical outcomes are the next top priority target. These goals are discussed below, along with proposals for the additional resources and engagements needed to establish digital, personalized and predictive medicine in Europe.
(a). Standards, tools and services
The first stage of the NoE project has been largely concerned with establishing standards for models and data, building model and data repositories for published models, and assembling a toolbox of the existing software programmes (many, but not all, open source) that are relevant to the VPH. A key role for the NoE has been writing the application programming interfaces (APIs) for the markup languages (MLs) that allow the application software packages to read models and data from the repositories. The MLs provide the syntax (the grammar) for encoding models and data. Equally important are the semantic ‘metadata’ that give biological and biophysical meaning to the models and data via biological and biophysical ontologies (structured vocabularies). Mapping a number of existing ontologies onto the modelling framework is therefore another important thread in the immediate future, since it will enable unambiguous links to gene and protein databases. Note that considerable progress has been made over the last 12 months in coordinating the development of VPH standards such as CellML and FieldML with those of the systems biology community, in particular systems biology markup language (SBML).
(b). Dissemination, training and outreach
Many communities will be impacted by the VPH, including, in the short term, biomedical researchers and students; in the medium term, healthcare workers and European industry; and, in the long term, patients and the general public. Dissemination and training is therefore a major responsibility and one that has to be tailored separately for these various audiences. The initial focus for the NoE has been in establishing a website containing full descriptions of the VPH projects and giving access to the VPH modelling and data resources and training programmes, suitable for the first target community. The NoE website is already receiving 13 000 visits per month and the newsletters, which are published at 6 monthly intervals, have been extremely well received. There is clearly a large and growing interest in the VPH, and this needs to be addressed now in the way outreach, lobbying and dissemination are handled. Training is considered a fundamental part of the dissemination strategy, where the VPH NoE will target young and experienced researchers alike. Training has been identified not only as a key development area for the next 12 months, but also as one that has the potential to help sustain the VPH initiative, in the long term. VPH training should be embedded in basic and applied research method courses for higher degree students and as part of informatics education and training for the health workforce.
Other efforts are contributing to the formation of the VPH community: the Biomed Town online community, which hosted the consensus process of the STEP action, has been constantly growing since then and now has nearly 2000 members. Also, the larger VPH projects are contributing to the dissemination of the VPH vision. In addition to their project websites, projects like VPH osteoporosis project (VPHOP) and euHeart are publishing periodic newsletters that reach thousands of stakeholders worldwide.
(c). International linkages
Internationally, the World integrative research initiative agreement (http://www.biomedtown.org/biomed_town/LHDL/Reception/lhpnews/wiri) and the Osaka accord (http://www.biomedtown.org/biomed_town/VPH/wiri/OsakaAccord) have established a worldwide agenda for physiome research under the patronage of the European VPH initiative and the International Union of Physiological Sciences (IUPS) physiome project. Four other important events in the last 12 months have been the participation of a European delegation at the IMAG1 symposium in Montreal in 2008; the CellML/SBML (http://www.cellml.org/community/events/workshop/2009) combined meeting in Auckland, New Zealand, in April 2009; the Virtual tissue conference organized by the USA Environmental Protection Agency in Spring 2009; and the satellite symposium at the IUPS 2009 conference in Kyoto. All these events have been of considerable political relevance, and have strengthened the role of the European VPH community on the international research scene. Note that many of the VPH-I projects have international partners and the NoE itself has ‘international general members’. This formal recognition of international membership is also important for VPH-linked co-funding arrangements in the countries outside Europe.
(d). Virtual physiological human initiative (VPH-I ) projects
The goals of the current 14 VPH projects are summarized briefly below. Although these projects have been running now for only 18 months, it is quite clear that most consortia are on track to deliver what they promised and some preliminary results are already emerging. There are major technological achievements in various areas, including: data collection, management and integration; processing and curation of data into information; reductionist and integrative modelling of pathophysiological processes; and presentation, deployment and end-user applications. It is also notable that there is already an active involvement of companies participating in the VPH consortia, both small to medium enterprises and large corporations, and that the involvement is rapidly moving from their research and development (R&D) departments to their strategic management as the first business scenarios emerge. The clinical partners are providing a vital contribution to many of the VPH projects, participating enthusiastically and with considerable commitment. Note that the NoE clinical advisory board will be playing a more active role from now on and will provide a mechanism for generalizing the lessons learned from the clinical partners of the individual VPH-I projects.
The initial (call 2) VPH projects are targeted as follows:
— euHeart (IP) is a developing open-source code and multi-scale/multi-physics model of heart electromechanics in clinical cardiac diagnostic and device-development applications,
— VPH2 (STREP) is a developing decision-support tool for heart disease,
— preDiCT (STREP) is a developing model of cardiac electrophysiology for drug design and toxicity testing,
— ARTreat (IP) is developing an interventional decision-support system for stenting procedures based on multi-scale patient-specific models of atherosclerotic disease,
— ARCH (STREP) is a developing clinical decision-support tools based on patient-specific predictive modelling of vascular pathologies,
— PASSPORT is developing an open-source multi-scale framework for diagnostics and surgical training in the liver, based on modelling liver cell regeneration,
— IMPPACT (STREP) is developing minimally invasive, patient-specific treatment strategies for liver cancer based on bioengineering multi-scale modelling principles,
— PredictAD (STREP) is developing an evidence-based statistical framework for the diagnosis of Alzheimer’s disease,
— NeoMARK (STREP) is implementing collaborative research networks and tools for the early detection of oral squamous cell carcinoma,
— CONTRACANCRUM (STREP) is using multi-scale modelling techniques to simulate patient-specific cancer treatment outcomes,
— VPHOP (IP) is developing a patient-specific, multi-scale modelling framework for predicting osteoporotic fracture in elderly patients,
— HAMAM (STREP) is establishing a database of curated and annotated imaging data and software tools for breast cancer diagnosis,
— ACTION-Grid (CA) is promoting collaboration in medical/biomedical informatics and grid technologies to promote the interface between ICT and nanotechnology, and
— RADICAL (CA) is investigating security and privacy issues for VPH applications and best practices for medical and genetic data protection in distributed environments.
Nearly all of these projects deal with challenges relating to patient-specific, multi-scale modelling and the implementation of models and software in clinical environments.
(e). Distributed European infrastructure for supercomputing applications virtual physiological human virtual community
The NoE has also taken steps to build a relationship between the NoE and the VPH-I research projects through the development of a distributed European infrastructure for supercomputing applications VPH virtual community. The virtual community, applied for and managed by the NoE on behalf of the VPH-I, provides access to high-performance computing facilities for any VPH-I research project that requires such a facility; currently, over 50 per cent of the projects are being supported in this manner. Moreover, additional European Union (EU)-funded projects working in e-Health-related domains are also being supported.
3. New initiatives
Much of the work currently underway in the NoE will continue and expand in scope over the next 12 months, but here we describe a number of new initiatives that are currently underway.
(a). Workflows and driving clinical problems
Over the next 12 months, the NoE will work closely with the other VPH-I projects to define how the models, data and tools are used in biomedical and clinical workflows—the sequence of operations that start, for example, with clinical data (complying with standards relevant for privacy, security and ethics) and, by using the tools and models, end with a clinically useful diagnostic index or treatment strategy. In some cases, this will require resources to support the real-time computational needs of medical diagnoses, and the NoE is therefore currently trialling high-performance grid-based compute services.
(b). Training and dissemination
Another essential and urgent step is the implementation of workshops and summer schools to train people in the use of VPH models and software. Such activities will form an important part of wider, pan-European process directed towards the introduction of systematic educational activities with the aim of ensuring that academia, medicine and industry throughout Europe have a workforce that is appropriately equipped to meet the possibilities offered by this new and important discipline. Key to this initiative will be the VPH virtual academy. This web-based facility, accessed by a portal via the VPH NoE website, will provide a focus for interaction between training providers, course developers, young researchers wishing to develop a career in physiological modelling, established researchers seeking training as part of a commitment to life-long learning, and representatives from the major employment sectors. VPH training will need to be responsive to the changing needs of employers, and the Virtual Academy will provide an environment to engage with and obtain feedback from industry, healthcare and professional bodies.
The aim for the next 12 month period is to foster a training community by involving a critical mass of students from institutions across Europe. In this initial phase, the Academy will be directed towards gathering information on training needs, promoting VPH-related training activities and courses, and providing a point of contact between potential employers, training providers and newly qualified researchers. The Biomed Town VPH portal, using Web 2.0 technologies and approaches could be used to engage and build communities.
(c). RICORDO project
A recently funded VPH project called RICORDO will research and prototype a communal ontology-based annotation and repository communication strategy that supports the interoperability of VPH data and models across different biological scales. A new community standard for the representation of multi-scale biological entities will be developed and integrated into the VPH ToolKit development strategy. The medical relevance of such an advance resides in: (i) the use of a coherent multi-scale anatomy standard2 to refer to any body structure or location regardless of scale (an aspect that is also of relevance to electronic health record (EHR)-keeping and clinical document architecture standards) and (ii) the demonstration of an interoperability plan to connect patient-specific radiological images to mathematical models of physiology, as well as to disease-related genomic and molecular data.
(d). Multi-scale visualization project
Another recently funded VPH project called multi-scale visualization (MSV) will provide a formal conceptual taxonomy for multi-scale spatio-temporal visualization for both spatial multi-scale and temporal multi-scale problems, sometimes involving multiple datasets. It will also develop better visualization algorithms for handling large multi-scale datasets and implement these in an open-source software library.
4. Sustaining the virtual physiological human
Fourteen new projects related to the VPH were launched in call 2; five more were funded in call 4, and many others will be funded in call 6. Currently, the only coordination of these related endeavours is via the VPH NoE, which is also pursuing its own specific goals such as the VPH ToolKit. In order to transform the VPH vision into a reality for European industries and healthcare services, we need a long-term coordination action in order to
— coherently strategize and periodically revise the concrete research and technological development goals that should make the vision come true,
— sustain the standardization and interoperability efforts,
— sustain the further development, maintenance and provision of tools, services, databases and other infrastructure for common use,
— monitor the development, adoption and impact of VPH technologies,
— sustain the global adoption of VPH-based protocols that have proved effective, and
— provide training and re-training in the use of VPH technologies.
All these activities cannot be properly scoped by the NoE or by any other initiative that is limited in time; they require the attention of a permanent organization, capable of ensuring continuity over actions that may last for decades. This requires that the issue of sustainability of the VPH initiative be addressed. In our view, the best way to achieve this will be to establish a non-profit European ‘VPH Institute’ with a mandate to support the maintenance of VPH databases and the continued development of standards and business-friendly open-source software.
(a). The virtual physiological human Institute
There is a growing consensus among VPH stakeholders that in order to ensure long-term sustainability of the VPH initiative, it is necessary to establish a collective identity in the form of an international, non-governmental, non-profit organization, hereafter referred to as the VPH Institute.
The scope of this institute will be to sustain the coordination of all academic, governmental, industrial and societal stakeholders’ efforts towards the common goal of developing an integrative biomedical science and technology that makes the VPH vision practically possible, effective, sustainable and ethical. This general scope will be pursued according to a detailed strategic plan periodically revised by the Institute’s governing body, which will include actions in specific contexts to
— provide a strategic think-tank capable of conducting road-mapping exercises and of developing strategic recommendations for the European Research System around the concept of integrative biomedicine,
— support standardization processes to ensure interoperability of integrative technologies and the integrability of data, information, knowledge and wisdom captured in digital format,
— foster the collaborative development, assessment, deployment and maintenance of tools, services and methods for biomedical integrative research and clinical practice,
— manage, directly or through its member organizations, shared infrastructures to support the integrative approach in research and clinical practice; this includes repositories of data and models, benchmark collections, infrastructures for the collaborative development, etc.,
— conduct, under the mandate of governmental agencies and research-funding charities, studies to assess the status, the level of adoption and the impact of integrative methods and technologies,
— organize actions for positive lobbying and public information around the concept of integrative medicine,
— provide for the heterogeneous collection of stakeholders that are involved with research, technological development and clinical practice, a single voice for the citizens of Europe, their elected representatives, and any other institutional body that can sustain the development, assessment and deployment of the integrative biomedicine vision, and
— offer ‘VPH-certified’ courses for using the VPH tools and services.
The VPH Institute will be an organization whose members are other legally established organizations. Membership will be open to any organization whose legal status and statutes are compatible with the membership rules defined by the VPH Institute statute. To start the statutory process, a pro tempore board will be formed by inviting a small number of VPH researchers that have been involved with the VPH initiative from the very beginning, are recognized authorities by the community and are willing to allocate significant time to the statutory process. The only duty of this pro tempore board will be the drafting of the statute on which the VPH Institute will be incorporated. As soon as the incorporation is completed, a membership campaign will be launched, the pro tempore board will be disbanded and a new board of directors will be appointed according to the statutory procedures.
We expect to have the pro tempore board formed by the end of 2010, the draft statute by the end of 2011 and the VPH Institute fully operational in 2012, with at least 1 year of overlapping with some VPH projects funded in call 2, including the VPH NoE, which will ensure a smooth handover.
(b). Virtual physiological human conference series
The ICT-Bio meeting (http://ec.europa.eu/information_society/newsroom/cf/itemdetail.cfm?item_id=3956) held in Brussels in October 2008 was the kick-off meeting for the VPH. The International Conference on Computational Biology (ICCB) 2009 meeting held in Bologna in September 2009 (http://www.iccb2009.org) was a second integrative biosciences VPH meeting. The NoE steering group are now planning a VPH conference series that will provide an annual event for the VPH community and also help to coordinate international physiome activities.
5. What are the biomedical science challenges?
The VPH project will achieve important outcomes within the lifetime of the current NoE by introducing computational modelling into the diagnosis and treatment of some diseases (particularly cardiovascular and orthopaedic diseases), but the real impact in the long term will be to transform European healthcare into a more personalized, predictive and preventive process (see §6). The resources needed to achieve this long-term goal must be realistically assessed and, in particular, we must now instigate projects to fill identified gaps in the necessary know-how and infrastructure.
(a). Physiological databases
One current gap is the lack of comprehensive web-accessible databases of physiological data, encoded with well-established data and metadata standards. Such data provide numerical parameters for use in computational models. This need was expressed in §3.2.3 of the VPH STEP roadmap. One standard, digital imaging and communications in medicine (DICOM), does exist for medical image data. Others such as C3D (http://www.c3d.org) are well-established binary data formats for specialist communities (biomechanics, animation and gait analysis in the case of C3D). A more general purpose metadata standard (BioSignalML) is being developed for annotating physiological time-dependent signals encoded in a wide variety of existing specialist standards. But even this represents a small fraction of what is needed. A major effort is now needed by the physiology community to identify the types of physiological data that are available and to begin the development of a broad range of data standards and data repositories; as a first step, example datasets are being collected from the VPH-I and VPH NoE-exemplar projects. The tools for interpreting these data are being developed by the VPH and Physiome Projects.
(b). Genetics
The challenge of connecting modern genetics theory and methodology with multi-scale physiological models, such that one can address and understand the genetics of complex diseases in a population context, defines a large and ambitious research topic that needs to be given specific attention in the coming years if we are to hope for tailored clinical treatments based on simulation studies of the genetic profile of the individual requiring treatment. Many challenges in personalized medicine reflect a lack of understanding of what is called the genotype–phenotype map, i.e. the aggregated phenotypic effects across different length and time scales of different constellations of genetic variation (genotypes). The biomedical genetics community is now facing serious challenges concerning the overall applicability of the genome-wide association study approach when it comes to drug development and personalized medicine. The VPH initiative may be of substantial help to bring in a remedy for this situation by offering a new R&D paradigm that has the potential to provide mechanistic model descriptions of the phenotypic effects of genetic variation. Such causally cohesive genotype–phenotype models are very advanced multi-scale physiological models with an explicit link to genetic information, and with the capacity to describe how genetic variation manifests in phenotypic variation at various systemic levels up to the tissue, organ and whole-organism level.
(c). Clinical data
The re-use of clinical data remains a key challenge. As pointed out in the STEP roadmap, there is a need for a ‘global VPH security that makes possible the federation of clinical databases located behind hospital firewalls into the VPH framework’ (VPH research roadmap §12.1.5; www.europhysiome.org/roadmap). In a recent report, Deloitte writes: ‘Most life sciences (LS) Research & Development functions are under increasing pressure to improve innovation, reduce development inefficiencies and advance product safety. Patient-level data, collected through EHR systems, offer one promising avenue for redefining Research & Development and revolutionizing the LS value chain. Globally aggregated, patient-level data could support the identification of disease mechanisms and new discovery areas, accelerate the termination of unsuccessful compounds, decrease patient recruitment cycle times for clinical trials and improve drug safety surveillance through continuous monitoring (Deloitte Development LLC 2009).
Interoperability is the key to the effective re-use of clinical data. Currently, data exchange tends to be ad hoc, and no facility exists to support organized data exchange between multiple independent repositories (clinical, industrial and research). Candidate technologies capable of providing a data infrastructure that can facilitate VPH-wide data exploration, exchange and interoperability need to be explored and evaluated. A viable data infrastructure must support many activities (curation, etc.) giving data prospectors the freedom to revolutionize clinical procedures from the data they obtain, and yet issuing data providers with necessary assurances that their data will not be abused. Facilities for secure download/upload must be supported and data providers require further assurances that data users are appropriately authenticated and authorized to use the data. The VPH Institute is one mechanism by which formal authorization procedures could be managed and policed while also providing guidance on recommended procedures (e.g. anonymization) that are important to the clinic. The Institute is also well placed to be at the vanguard of standardization efforts, since it will be astutely aware of the needs of the community and could recommend appropriate strategies that maximize opportunities for data interoperability. For instance, the systemized nomenclature of medicine clinical terms (SNOMED CT) is a comprehensive clinical terminology that is starting to become a standard in many clinical communities, including adoption as the language of the Care records service of the National Health Service in the UK, and is thus a target for adoption by the VPH initiative. This is but one aspect of a wider initiative, which includes such fundamental efforts as the harmonization of nomenclature (modelling and clinical) through adoption of formalized ontologies, recommendations for data formats and data interchange (e.g. DICOM) and suitable representation on international standards committees (e.g. health level 7; HL7). The standards activity is necessarily multi-faceted, and must recognize the competing demands of research, industry and the clinic. This is an opportunity for VPH advancement that would be sensitive to the dangers of over-regulation.
Finally, there are important legal issues to be tackled. By its very nature, the VPH crosses the scientific and national boundaries. Differing interpretations of data protection law (e.g. EU directive 95/46/EC) between member states discourage collaborative sharing of data for patient benefit. This is compounded by jurisdictional uncertainty owing to a lack of legal precedents in this area. The ethical considerations relating to sharing of patient data are also formidable and could easily become a stumbling block to the progress of the VPH. The Institute would be a proponent for change, guiding and focusing effort in both areas, ensuring that these issues are given the priority they deserve in order to secure the vision of personalized and integrative medicine that is the goal of the VPH.
6. What are the healthcare challenges?
Major diseases, such as cancer and neurological and cardiovascular diseases, are complex in nature involving environmental, life style, ageing and genetic components. A major challenge for the future is how to integrate the knowledge of all these different components into robust and fully reliable computer models and ‘in silico’ environments that will help in the development and testing of new therapies and better disease prediction and prevention tools in healthcare. The progressive advance in computing power and associated information technology offers the potential to deliver tailored clinical treatments based on simulation studies of the genetic profile of the individual requiring treatment.
(a). The needs
The European healthcare system, including its biomedical research and technological development component is a huge, complex and highly articulated system. Because of the peculiar political history of the EU, it is not a surprise that such a system is highly fragmented, not only between member states, but also between regions, districts and even single hospitals. However, in spite of this extreme heterogeneity, there are some common requirements that are emerging in a number of analysis documents produced by very different sources (Deloitte Development LLC 2007, 2009; US Department of Health & Human Services 2007; Price Waterhouse Cooper 2008; Brynjolfsson 2008; Apoteket and Stockholm County Council, Sweden 2006; Johansen 2006; European Commission, Information Society 2006; Barry & Gartner 2008). Such requirements can be summarized in three keywords: personalized, predictive and integrative healthcare. A fourth keyword, affordable, is implicit, as the sustainability of healthcare systems is becoming the main issue in a number of member states dealing with a constantly ageing population. More specifically, these common needs are: to maximize the yield of biomedical research expenditure; to achieve personalized healthcare for individuals and groups (women, children, etc.); to improve the reliability, repeatability and the timeliness of medical decisions; to integrate digital health information on a global scale; and to resolve the individual society conflict around the privacy of health data. It should be noted that, at this stage, these needs are very hard to quantify because the information is fragmented over dozens of reports produced by dozens of different medical specialities, and much effort is required to elaborate into a single coherent framework a detailed and quantifiable description of needs. To address these issues, it might be appropriate for the European Commission to consider funding a specific support action to collect, organize and compose all these evidences into a fully justified and quantified needs analysis.
(b). The ‘digital me’ vision
The vision we have is of a ‘digital me’, a coherent digital representation of each patient that is used as an integrative framework for the consolidation within the European research system of fundamental and translational integrative biomedical research and the provision to European citizens of an affordable personalized, predictive and integrative medicine.
(c). Personalized, predictive and integrative healthcare
A new generation of medical technologies are needed to integrate the data available on a patient to support a more personalized diagnosis, prognosis, treatment planning and monitoring, and to develop new drugs, therapies, medical devices, assistive and diagnostic technologies that are optimized for specific groups of patients (age, gender, co-morbidity, etc.). Diagnostic workflows are required, not on predefined general protocols, but on the prediction of risk obtained by models that combine both population and patient-specific information.
(d). Ageing
Age is still the best predictor for most complex diseases. This implies that if we are to create individualized models describing the development and maintenance of complex disease situations on given genetic backgrounds in a way that is of broad practical utility for medicine, we have to incorporate the effects of ageing in multi-scale and multi-physics models. By offering in silico representations of ageing tissues and organs upon which one can systematically study the effects of various chemical treatments, such individualized models would also define the foundation for a new and sorely needed drug-targeting paradigm. The making of multi-scale physiological models capturing the ageing process defines a daunting long-term theoretical–experimental research programme. But activity in this direction should be invoked in the coming years such that one can get a clearer view of what the major challenges are.
7. What are the information and communications technology challenges?
The VPH physiome project aims to provide a systematic framework for understanding physiological processes in the human body in terms of anatomical structure and biophysical mechanisms at multiple length and time scales. The importance of establishing a solid foundation for the VPH by creating model and data standards, together with mechanisms for achieving model reproducibility and reuse, was recognized in the STEP roadmap. The framework includes modelling languages for encoding systems of differential-algebraic equations (CellML, http://www.cellml.org; SBML, http://www.sbml.org) and the spatially varying fields used with systems of partial differential equations (FieldML, http://www.fieldml.org). In both cases, the parameters and variables in the mathematical models are annotated with the metadata that provides the biological meaning. The languages encourage modularization and have import mechanisms for creating complex models from modular components. Model repositories have been established, together with freely available open-source software tools to create, visualize and execute the models. The CellML repository (http://www.cellml.org/models) now includes models for a wide variety of subcellular processes.
(a). Model and data-encoding standards
The development of a biophysically based mathematical model is a creative endeavour, often requiring a great deal of insight into the physical processes being modelled and personal judgment about the approximations needed to satisfy Einstein’s maxim that ‘a model should be as simple as possible but no simpler’. Once created, however, a model should stand independent of its creator and be reproducible and testable by others. The model and data files that together can demonstrate reproducibility of a model on an automated basis, are called the reference description of the model. The issue of robustness and reproducibility is particularly important when a model representing some small component of physiological function is incorporated into a more comprehensive model—and especially one that is to be used in a clinical setting. To be worthy of reuse in this fashion, each independently developed component should be demonstrably ‘correct’ for the function it claims to represent, in the sense both of biological validity—it matches some aspect of biological reality—and mathematical validity—for example, it has consistent units and does not violate physical principles such as conservation of mass or charge.
The general strategy for developing the modelling standards is as follows:
— develop MLs for encoding models, including metadata and data,
— develop APIs based on the MLs,
— develop libraries of open-source tools that can read and write the ML-encoded files,
— develop data and model repositories based on MLs,
— develop reference descriptions to demonstrate model reproducibility, and
— implement web services for a variety of tasks including access to automated scripts to run the models and compare results against experimental data, optimize parameter values for new experimental data and provide sensitivity analyses for changes in model parameters. A useful way of viewing the development of standards is shown in table 1, where progress in developing a specification of the minimal requirements for data, models and the simulation experiments are shown, along with the standards for the syntax of the data, models or simulation experiments and the ontologies for annotating the semantic meaning of terms in the data, models or simulation experiments.
Table 1.
The minimum information standards, syntax and semantics being developed for data, models and simulation experiments. Note that the best example of an e-Health technology that is already in widespread use is the picture archiving communications systems, usually based on the use of DICOM image encoding standard. Others close to maturity are electronic transfer of prescriptions (ETP), computer-based patient records (CPR), electronic medical records (EMR) and electronic health records.
| data | models | simulation | |
|---|---|---|---|
| minimal requirements | not available | MIRIAM (http://www.ebi.ac.uk/miriam/) | MIASE (http://www.ebi.ac.uk/compneur-srv/miase) |
| standard formats | PDB (http://www.rcsb.org/pdb/home/home.do)DICOM (http://medical.nema.org)BioSignalML (http://www.embs.org/techcomm/tc-cbap/biosignal.html) | SBML, CellML FieldML | SED-ML (http://www.ebi.ac.uk/compneur-srv/sed-ml) |
| ontologies | GO (http://www.geneontology.org), BioPAX (http://www.biopax.org), FMASBO (http://www.ebi.ac.uk/sbo)OPB | GO, BioPAX, FMA SBO, OPB | KiSAO (http://www.ebi.ac.uk/compneur-srv/kisao) |
(b). Model curation
There must be a concerted effort towards reproducibility, interoperability and the re-use of VPH models, including both future models and legacy, published models. This requires adoption of a consensus set of standards for the metadata that describe the models and of MLs for their mathematical description. An integral aspect of interoperability will be the tagging of model variables and parameters with identifiers from reference ontologies such as the foundational model of anatomy (FMA), gene ontology (GO) and appropriate ontologies for units, physics-based quantities, physiological processes, etc.). These must be adopted not only by the VPH community but also by the curators of the massive existing gene- and protein-databases in order to enable vertical multi-scale linking of models at the physiological scale (organs, tissues, cells) to the wealth of medically relevant molecular data. Model curation is a long-term task that spans a wide spectrum of disciplines and will require a major effort, but it is crucial for the success of the VPH vision. Sustainability of model repositories and software (including version control, archiving, technical upgrades and provision for updating and expansion) will also be a major expense and limiting factor in the community acceptance of VPH models.
(c). Top-down or bottom-up?
The VPH of course needs to encompass both ‘top-down’ and ‘bottom-up’ approaches. A good example of top-down is the Pharmaco-Kinetic-Pharmaco-Dynamic (PKPD) modelling community. PK deals with the advection, distribution, metabolism and excretion of drugs and PD deals with the dynamics of how the drugs affect receptors. PKPD models accommodate human variability in an empirical fashion and treat body compartments with highly lumped approximations. The ‘bottom-up’ approach of modelling biophysical mechanisms at the subcellular level is the realm of the systems biology community. The anatomically and biophysically based approaches of the VPH project are being designed to link these approaches. The model repositories based on the CellML and SBML standards already contain many models of both types.
(d). The challenges of integrative multi-scale modelling
Biological systems are characterized by multiple space and time scales, and there is a substantial need for new multi-scale modelling algorithms to help bridge between the large range of spatial and temporal scales involved in the VPH. For instance, very often the macroscopic properties of a tissue, such as the bone stress–strain curve, or the diffusion coefficient of a drug or a chemical in the interstitial space of skeletal muscle, are related to microscopic effects. In this case, one needs to complement the macroscopic phenomenological description with a microscopic view of the problem. The basic idea is to model the theoretical input to a coarse-grained model from a more detailed microscopic model, bypassing the necessity of empirical description. As an example, homogenization and volume-averaging methods can be used to obtain the strain energy function for bone tissue starting from the basic information about a ‘reference’ microscopic cell. Analogously, mass transfer in biomaterials or within a polymeric scaffold for tissue growth, can be effectively described at the macroscale by using macroscopic parameters that are obtained on the basis of a microscopic ‘cell problem’ (i.e. on the reference elementary volume describing the scaffold). Finally, biological systems are often characterized by processes lasting a few seconds, such as metabolic biochemical reactions, and processes that fully develop only after days, months or years, such as atherosclerosis, angiogenesis or the onset of an aneurysm. Biological systems are also characterized by the interaction of many different physical processes at each spatial scale. For example, at the organ/tissue level, analysis of the beating heart couples large deformation mechanics of the myocardium with the reaction–diffusion equations governing the spread of electrical excitation, and with the equations of fluid mechanics for blood flow both within the ventricles and within the coronary vasculature. At the subcellular level, the analysis of cardiac myocyte function requires the coupling of ion channel electrophysiology, calcium transport, myofilament mechanics, pH regulation and complex networks for signal transduction, metabolism and gene regulation.
(e). The challenge of model reduction
With minimal information and model-encoding standards now in place, it is time to address the extremely important question of ‘model reduction’—how to automatically reduce the number of variables and parameters in a model when it is used under prescribed conditions. Some processes, for example, may be sufficiently fast in comparison with the time scale of events being modelled that they can be assumed to be instantaneous. Others may be approximated as being at steady state. Some groups of reactions could be considered as a single module. In some cases, there may be substantial computational gains to be made by automated reduction in the number of parameters or variables associated with describing a spatial field. Thus, a complex model that captures the biophysical and anatomical detail of some aspect of the physiology and anatomy of the body could be reduced to a simpler and therefore computationally more tractable problem under certain conditions.
(f). The challenge of dealing with stochastic processes
Another modelling challenge, which has so far received relatively little attention, is that of incorporating stochastic behaviour into the multi-scale VPH models. At a molecular level, stochastic behaviour can be a reflection of Brownian (thermal) motion, but at higher spatial scales it can be a reflection of unknown mechanisms—i.e. ignorance. It is very important that the consequences of this uncertainty in, for example, parameter values, be quantified.
(g). The challenge of multi-scale simulation and visualization software
Visualization of the output of complex systems models and the human–computer interface issues inherent in user interaction with such models is a new and significant area of research. Complex systems models are likely to have many input and output variables and may produce non-intuitive data representing, for example, emergent behaviours that are not easily represented by classical graphical or text-based methods. This is a rapidly moving area of cross-disciplinary research that may need specific funding under future calls for VPH project proposals. The above challenges provide worthwhile and challenging problems for the mathematics and computational science communities and some VPH funding should be directly targeted to attract their expertise.
8. A strategy for the virtual physiological human
(a). Timelines
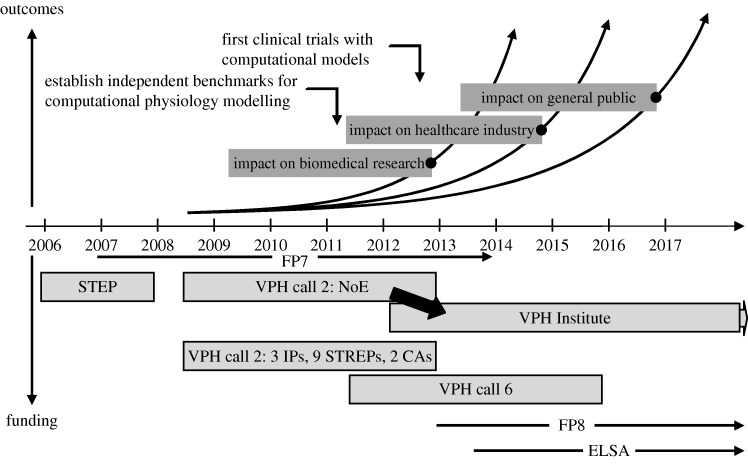
A timeline for the STEP project (a framework 6 initiative), the NoE and other VPH projects funded under the first call 2 of FP7 and the recently funded international call 4 projects, is shown in figure 1, together with anticipated future calls. The establishment of a VPH Institute is also indicated. The anticipated impact that the VPH activities will have is also illustrated, first on biomedical research, then on industry and finally on the general public.
Figure 1.
Timelines for VPH funding calls, the VPH projects and their impact on various communities. Sustainability of the NoE, and hence ongoing support for the VPH and the healthcare industries that depend on it, will be achieved through the VPH Institute. FP8 is framework 8 and ELSA is the European large scale action.
(b). The next steps
The VPH is a grand challenge. We propose the following specific actions.
(i). 2009–2010: establish a collective identity
It is important that the multitude of players involved can speak with a single voice in a few strategic situations. This requires the creation of a collective identity around the VPH brand name. The VPH community is already working in this direction and will have a VPH Institute fully operation in early 2011.
(ii). 2010–2012: definition and quantification of needs
The STEP experience showed that when properly managed by a motivated consortium, and when embraced by a lively and motivated community, a road-mapping exercise can be of great value to capture and quantify needs and to develop a vision around them. In particular, we recommend three road-mapping actions:
— road-mapping coordinated support action (CSA) on VPH (FET; ‘future and emerging technologies’ call 5),
— road-mapping CSA in integrative health research, and
— road-mapping CSA in health e-infrastructures.
These actions should be sustained by other units of the European Commission (namely the Information society and media directorate-general (DGINFSO) FET proactive, Directorate general for research in the European Commission (DGRTD) Systems Biology and DGINFSO e-infrastructures as part of the Capacities programme) and should involve significant portions of the traditional constituencies of these units, as it is necessary to include in the action a number of complementary expertises that are well represented in these constituencies.
Similar actions should be undertaken to push the VPH agenda as high as possible in those European institutions that fund fundamental research such as the European Science Foundation or the European Research Council. The general strategy to adopt towards these units and their constituencies should be inclusive, not invasive. We need to make clear that we do not want as VPH researchers to start designing e-infrastructures, running web-bench biology experiments or develop fundamental research in computer science, mathematics or physics. We recognize that there are neighbourhood communities that can do this much better than we can. What we offer is a common goal towards which all these skills and those we represent as a VPH community can join forces. In the continuum of skills and interests, we need to find among VPH researchers those who are working closer to the fence with each of these communities, and support them as ambassadors, towards the formation of mixed consortia that can run these road-mapping exercises in a qualified and representative way. It is equally clear that, in each of these neighbourhood communities, we need to find the experts who are fascinated by the challenge we pose, and who are not afraid of the change that this would necessarily require to their research practice.
(iii). 2011–2014: disseminate and structure
As the results from these road-mapping exercises emerge, it will be necessary to coordinate and organize them into an operational plan to tackle this European large scale action. This action will have to find substantial dedicated funding at the European level, but at the same time will be sustained and nurtured by a number of funding actions in the various neighbourhood domains that will take place as part of framework 8. Another important dimension will be played by member-state funding agencies that case by case will support horizontal or vertical initiatives. Preparing such composite and structured action requires, in our opinion, a specific coordination action, which will play a preparatory role in this direction.
(iv). 2014–2019: European large scale action on personalized, predictive and integrative healthcare
These 5 years will make it possible to face this grand challenge at all levels (research, technological development, implementation, assessment, deployment) only if the necessary critical mass is reached in terms of skills, resources and commitment of all stakeholders. We hope this document will serve as the first step towards the creation of such a critical mass.
Acknowledgements
Many people have contributed to this document, which formed the core of a report to the European Commission. The main task of drafting the document and seeking feedback was undertaken by Peter Hunter and Marco Viceconti. All the other authors have made substantive contributions in the form of corrections, suggested improvements or additional text. We are also grateful to members of the NoE Steering Committee and Scientific Advisory Board for their suggestions. We sought feedback on earlier drafts of the document from the VPH-I community generally, including the project leaders for all the currently funded VPH projects. We hope that we have addressed most of their concerns but we do acknowledge two deficiencies in the current review, which we hope to address in the next update of the document in 2010. These are the requests to have more detail on the connection with the systems biology community and more specific recommendations for funding calls. We felt that both of these issues required more thought and community feedback than that was possible in the timeframe allowed for the present document.
The Interagency Modelling and Analysis Group (IMAG) coordinates multi-scale modelling initiatives from various United States agencies including the National Institutes of Health, National Science Foundation, National Aeronautics and Space Administration, Department of Energy, Department of Defense, United States Department of Agriculture and United States Department of Veteran Affairs.
Discussed extensively by the VPH NoE (http://www.vph-noe.eu/vph-repository/doc_download/48-vph-anatomy-framework).
One contribution of 13 to a Theme Issue ‘The virtual physiological human: computer simulation for integrative biomedicine I’.
References
- Apoteket and Stockholm County Council, Sweden. eRecept, an ePrescribing application. 2006. See http://ec.europa.eu/information_society/activities/health/docs/events/opendays2006/ehealthimpact-7-2.pdf .
- Barry R., Gartner H. Stop the bleeding: use IT to achieve sustained value in healthcare. 2008. See http://www.gartner.com/DisplayDocument?doc_cd=144534 .
- Brynjolfsson E. Investing in the IT that makes a competitive difference. 2008. See http://hbr.harvardbusiness.org/2008/07/investing-in-the-it-that-makes-a-competitivedifference/ar/1 .
- Deloitte Development LLC. The benefits from translating biomedical research into the health care system. Report to Bio21 Australia. 2007. See http://www.bio21.com.au/admin/file/content14/c4/Insight%20Economics%20Report_March%202007.pdf .
- Deloitte Development LLC. Secondary uses of electronic health record (EHR) data in life sciences. 2009. See http://www.deloitte.com/assets/Dcom-UnitedStates/Local%20Assets/Documents/us_lshc_Secondary%20Uses%20of%20EHR_0409.pdf .
- European Commission, Information Society. eHealth is worth it—the economic benefits of implemented eHealth solutions at ten European sites. 2006. See http://ec.europa.eu/information_society/activities/health/docs/publications/ehealthimpactsept2006.pdf .
- Fenner J. W., et al. The EuroPhysiome, STEP and a roadmap for the virtual physiological human. Phil. Trans. R. Soc. A. 2008;366:2979–2999. doi: 10.1098/rsta.2008.0089. ( ) [DOI] [PubMed] [Google Scholar]
- Johansen I. E-Health and implementation of EHR. 2006. See http://www.ehealthbenchmarking.org/2006/images/stories/06_johansen_denmark.pdf .
- Price Waterhouse Cooper. Pharma 2020: marketing the future: which path will you take? 2008. See http://www.pwc.com/gx/en/forms/gxengallspharma2020.jhtml?opendocument&doc=virtual .
- US Department of Health & Human Services. Personalized health care: opportunities, pathways, resources. 2007. See http://www.hhs.gov/myhealthcare/news/presonalized-healthcare-9-2007.html .