Abstract
The current study investigates the direct effects of in utero vinclozolin exposure on the developing F1 generation rat testis transcriptome. Previous studies have demonstrated that exposure to vinclozolin during embryonic gonadal sex determination induces epigenetic modifications of the germ line and transgenerational adult onset disease states. Microarray analyses were performed to compare control and vinclozolin treated testis transcriptomes at embryonic day 13, 14 and 16. A total of 576 differentially expressed genes were identified and the major cellular functions and pathways associated with these altered transcripts were examined. The sets of regulated genes at the different development periods were found to be transiently altered and distinct. Categorization by major known functions of altered genes was performed. Specific cellular process and pathway analyses suggest the involvement of Wnt and calcium signaling, vascular development and epigenetic mechanisms as potential mediators of the direct F1 generation actions of vinclozolin.
Keywords: Sex Determination, Vinclozolin, Endocrine Disruptor, Epigenetic, Transgenerational
1. INTRODUCTION
Previously, embryonic vinclozolin exposure at the time of sex determination was shown to promote heritable adult onset diseases such as increased spermatogenic cell apoptosis, decreased sperm motility, prostate disease, kidney disease and tumor development [1–3]. To elucidate the mechanism of vinclozolin action the current study examines the direct (i.e. immediate) actions of vinclozolin on the F1 generation developing embryonic gonad. Vinclozolin induced transcriptional alterations during gonadal sex determination and early testis development was examined. Sex determination and testis development are complex processes required for the perpetuation of mammalian species. Germ cells originate in the yolk sac and begin to migrate to the genital ridge at embryonic day (E) 10 - E11 in the rat [4, 5]. Gonadogenesis occurs at E12 and gonadal sex determination is initiated at E12.5. Subsequently, in chromosomal males carrying the Sex determining Region of the Y chromosome (SRY), Sertoli cells differentiate and aggregate with primordial germ cells [5, 6]. Testis cords begin to form at E14 and are surrounded by peritubular cells that migrate from the mesonephros. Extracellular matrix that forms the basal lamina of the testis cords is secreted, and vasculature and Leydig cells form in the subsequent few days of development [7–10]. There is also a dramatic cellular proliferative event in the developing testis [11, 12]. At puberty, the testis cords become seminiferous tubules and spermatogenesis is initiated. Proper development of the testis is critical for full reproductive capacity of the male [4, 5]. Mutation of genes involved in the key events of embryonic testis development can cause sub-fertility, intersex disorders, or sex reversal [reviewed in [13–15]]. The embryonic environment and exposures to chemicals such as endocrine disrupting compounds can also alter development and decrease reproductive capacity [16].
Endocrine disrupting compounds can act as hormone agonists or antagonists and have been implicated in a host of reproductive abnormalities in wildlife and human populations [17–21]. Several endocrine disrupting compounds have been shown to inhibit male sexual development [22–24]. Vinclozolin is one such compound that is used as a fungicide for turfgrass, ornamental plants, grapes and other fruits and vegetables [24]. Vinclozolin and its major metabolites, enanilide and butenoic acid, act as anti-androgens by inhibiting androgen receptor (AR) activity [24, 25]. AR activity is required for male reproductive development and expression of AR first appears in testis and reproductive tract tissues of the rat between E14 and E15 [26–28]. Vinclozolin exposure of neonates has been shown to affect both embryonic development and adult onset disease states. Maternal oral vinclozolin treatment of rats between E12 and postnatal day 3 revealed malformations in the reproductive tracts and renal systems of male offspring with highest sensitivity between E14 and E19 [18, 29]. Embryonic vinclozolin exposure during gonadogenesis and sex determination (from E8-E14 through maternal intraperitoneal injection) caused increased spermatogenic cell apoptosis in pubertal and adult rat testis and decreased sperm numbers and motility in males for three generations [1–3, 30]. As the animals age numerous transgenerational adult onset disease states develop including prostate disease, kidney disease, immune abnormalities, male infertility and tumor development [2]. These adult onset transgenerational disease phenotypes appear to involve an altered epigenetic programming of the male germ line and are initiated by vinclozolin exposure of the first F1 generation embryonic testis [1]. Another anti-androgenic compound flutamide was found to cause a similar F1 generation phenotype as vinclozolin, but not the transgenerational phenotype [31]. Therefore, alternative mechanisms of action for vinclozolin must be considered during the initial exposure of the F1 generation embryonic testis.
Vinclozolin exposure from E8-E14 of rat testis embryonic development has the potential to interfere with several concurrent events critical to testis function including gonadogenesis, sex determination, testis differentiation, and epigenetic programming of the male germ line. Transcriptional regulation during rat sex determination and testis differentiation between E13 and E16 has been investigated [32]. This previous study produced a list of 109 candidate transcripts involved in normal testis morphogenesis processes such as cellular differentiation, proliferation, focal contact, RNA localization, and development. Specific signaling pathways were not found to be highly affected in that study, but specific genes identified suggest regulation of several cellular processes. Specific genes identified include known players in testis development such as Sry, Vanin, Fgf9, and Amh, and many novel gene candidates including Tgfb3, Sfrp4, Wnt5a, Deltex4, and Jag1. These genes appear to play a role in sexual differentiation and may be potential targets for the action of vinclozolin. Epigenetic mechanisms should also be considered as they have previously been implicated as causative in some adult onset diseases [33–36], and in the action of vinclozolin on the testis transcriptome at E16 leading to adult onset and transgenerational disease [1, 2, 37]. The current study investigates the testis transcriptome following the E8-E14 vinclozolin treatment at the testis differentiation stages of E13, E14, and E16. Changes in gene expression were identified that provide insight into the direct actions of vinclozolin on testis development. The objective of the current study was to identify the direct pharmacologic effects of vinclozolin on the F1 generation developing testis transcriptome, and not to do risk assessment or toxicology studies for vinclozolin.
2. MATERIALS AND METHODS
2.1 Animals
Sprague–Dawley rats were kept in a temperature controlled environment and given food and water ad libitum. Estrous cycles of female rats were monitored by cellular morphology from vaginal smears [38]. Rats in early estrus were bred overnight and mating confirmed by sperm positive smears, denoted as day 0 of pregnancy. Pregnant rats were injected with 100mg/kg/day of vinclozolin, or DMSO vehicle only, daily on days E8-E14 of pregnancy to expose pups in utero during this time. Animals were euthanized at E13, E14, and E16 of pregnancy, and embryonic gonads were collected for RNA isolation and histology. Sex was determined by PCR using primers specific for Sry on genomic DNA isolated from embryo tails as previously described [39]. Therefore, in addition to the histological presence of testis cords in E14 and E16 the sex was confirmed with SRY PCR, in particular for the E13 testis. All procedures were approved by the Washington State University Animal Care and Use Committee.
2.2. Organ Cultures
Embryonic cultures undergo similar testis morphogenesis after 3 days of culture as that seen in vivo [39, 40]. Rat gonads from E13 embryos were dissected with mesonephros intact and cultured 3 days as previously described [40, 41]. Briefly, gonads were placed in drops of medium on Millicell CM filters (Millipore, Bedford, MA, USA) floating on 0.4 ml of CMRL 1066 medium (Gibco BRL) supplemented with penicillin–streptomycin, insulin (10 µg/ml), L-glutamine (350 µM), transferrin (10 µg/ml), and BSA (0.01%). Gonadal pairs were split with one gonad treated with 50µM vinclozolin in DMSO and the contra-lateral gonad with DMSO only as a vehicle control. Alternatively, one gonad was treated with 5µM hydroxy flutamide with ethanol and the contra-lateral in ethanol only. Media and treatments were changed daily and given fresh vinclozolin or hydroxy-flutamide. Gonads were maintained in culture for 3 days at which time testis cords formed and testes were used for histological analysis or separated from mesonephros and used in RNA collections.
2.3 Histology
Tissue specimens were fixed in Bouin’s solution for 1 h and embedded in paraffin using standard procedures. Serial sections of 4 µm were stained with hematoxylin and eosin (H&E) using standard procedures by the Histology Core Laboratory of the Center for Reproductive Biology, Washington State University. Sections were visualized by light microscopy.
2.4 RNA Preparation
Sprague–Dawley rat embryonic gonads without mesonephros from E13, E14, and E16 control and treated testes were collected (a total of 6 conditions). Stage of development was confirmed by counting tail somites of each embryo. E13 gonads cultured for three days with 50µM vinclozolin, 5µM flutamide, or vehicle controls of DMSO or ethanol respectively were also collected (total of 4 additional conditions). Two separate gonadal pools of 10–30 gonads were collected for each treatment group for replicate analysis and stored in TRIZOL (Invitrogen) at −80°C until RNA extraction following the manufacturer’s protocol. Each separate RNA pool was used for a single microarray chip, for a total of 12 chips for in vivo analysis and 8 for in vitro analysis. High quality RNA samples were assessed with gel electrophoresis and required a minimum OD260/280 ratio of 1.8.
2.5 Microarray and Statistical Analysis
At least 3 µg RNA per sample was delivered to the Center for Reproductive Biology, Genomics Core Laboratory, Washington State University for processing. Briefly, RNA was transcribed into cDNA, and cDNA transcribed into biotin-labeled RNA. Biotin-labeled RNA was then hybridized to the rat RAE230 2.0 arrays containing 31,099 transcripts (Affymetrix, Santa Clara, CA, USA) and labeled with phycoerythrin-coupled avidin. Hybridized chips were visualized on an Affymetrix Scanner 3000 (Affymetrix). CEL files containing raw data were then processed and analyzed using R software and Bioconductor packages [42, 43]. Theses CEL files have also been deposited with the NCBI gene expression and hybridization array data repository (GEO, # pending, http://www.ncbi.nlm.nih.gov/geo) and can be accessed at www.skinner.wsu.edu.
Microarray hybridization data were examined for physical anomalies on the chip by pseudochip and residual error visualizations. Analysis continued when no anomalies were identified. For all further analyses, microarrays were split into three experiments in vivo; E13, E14, and E16 vinclozolin treated and control (12 chips), in vitro vinclozolin treated and control (4 chips), and in vitro flutamide treated and control (4 chips). Each separate experiment used a different gonadal collection from different animals and separate chips. Quality Assurance of microarray data was completed using Bioconductor packages AffyQCReport. Hybridization and housekeeping controls, RNA degradation, sample clustering, NUSE plots, LPE plots, and RLE plots all showed high quality data for each analysis (not shown) and no chips were removed.
Array signal values were calculated using RMA, GCRMA, PLIER, and MAS 5. MAS 5 pre-processing was chosen for further analysis for its ability to best separate background from signal and standardize samples in this experiment. Gene expression per chip for in vivo conditions was scaled to a signal value of 210, calculated as the median of average 2% trimmed mean chip values. The calculated target value for conditions in the in vitro vinclozolin treated testis experiment was 218, and 107 for conditions of in vitro flutamide treated testis experiment. Signal values correlate to the amount of transcript in the sample.
To determine the effects of vinclozolin treatment on gene expression during sexual differentiation, a linear model was applied to each gene. First, unexpressed or unchanged genes were removed by filtering in an unbiased manner. Present-marginal-absent (PMA) calls were determined using a P value cut off for absent of greater then 0.06, and less then 0.04 for presence. Unexpressed genes were then defined as less than a signal cut off value at which 99.5% of genes absent across all samples and were excluded. This value of log2 6.68 for in vivo treated conditions (6.972 for in vitro vinclozolin treated conditions, and 6.638 for in vitro flutamide conditions) was used to filter the data on expression value and removed 15,930 genes presumed absent leaving 15,169 genes for further analysis. A filter on range was also applied to remove genes that did not change. Genes with a range of less then 1 across all chips in the experiment were excluded leaving 8,259 genes for further analysis. Differential expression analysis was performed on the filtered gene list using a linear model on log2 signal values which considered time, treatment, and the interaction between time and treatment to identify significantly changing genes. Comparisons of interest were extracted through contrasts (mediated t-statistic) where a raw P value of 0.05 was considered significant.
Post filtering and visualization of data was achieved using GeneChip Operating Software (GCOS) version 1.4 (Affymetrix) and GeneSpring version GX 7.3.1 (Silicon Genetics, Redwood City, CA, USA). Sample biased post filters were applied to obtain more conservative gene lists. Specifically, a gene was kept for consideration if it was expressed above 104 in at least one treatment group in a comparison. Also, for each pair of control and treated groups (i.e. E13, E14, E16, and Cultured E13) genes changing by a factor of less then 1.5 were removed.
The microarray quantitation of gene expression is superior to any quantitative PCR procedure due to the use of over 10 different oligonucleotide probes for each gene and required comparable results with all probes prior to quantitation to eliminate probe bias and allow optimal statistics, in contrast to QPCR that uses two probes and not able to account for this variable. As previously reported [32], similar observations were made between microarray and quantitative PCR. For this reason confirmation with quantitative PCR was not performed or required.
For genes annotation, Affymetrix annotation file Rat230_2.na27.annot.scv was used unless otherwise specified. The microarray data is available at GEO, NCBI and at www.skinner.wsu.edu.
3. RESULTS
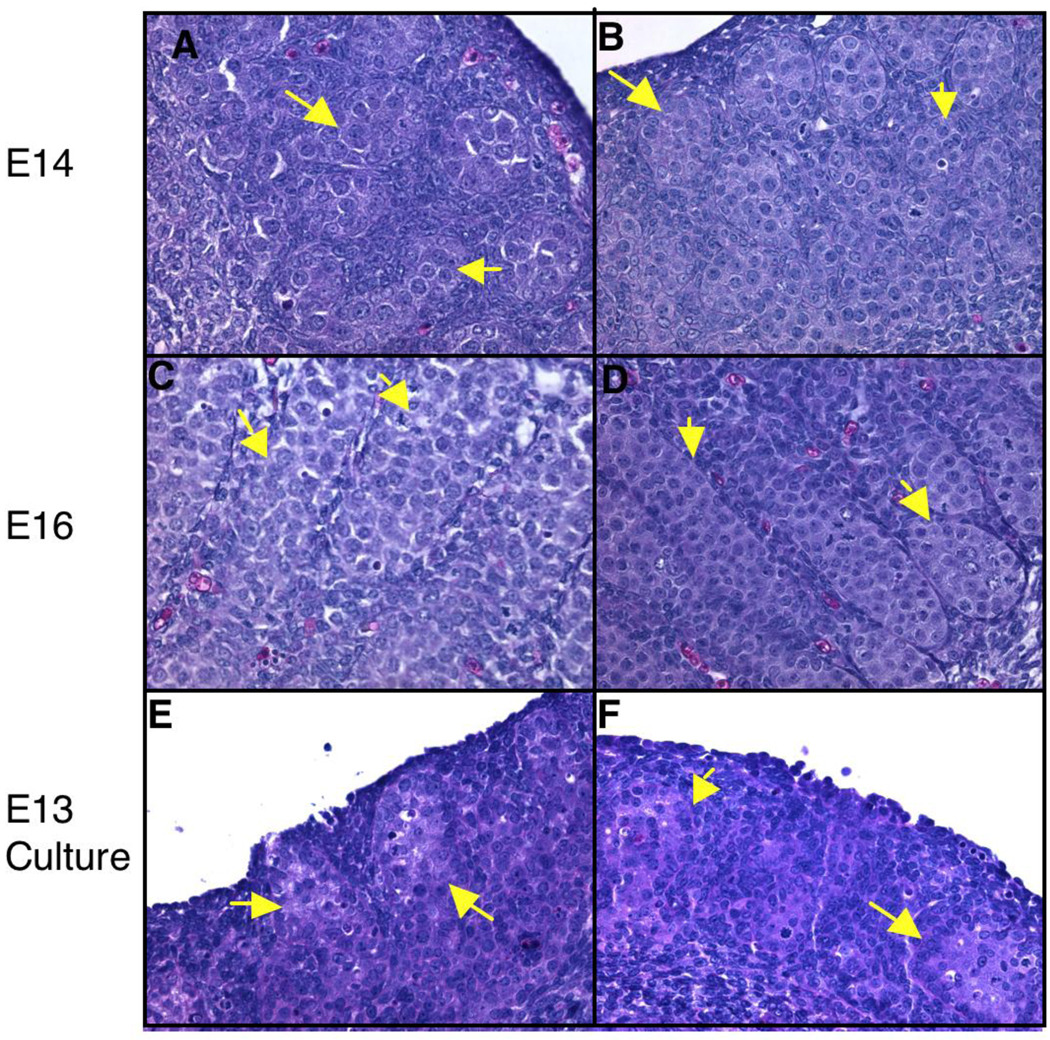
In the present study, embryos were exposed in utero to vinclozolin (daily 100 mg/kg/day maternal intraperitoneal injections from E8-E14) and vehicle (DMSO) control. Embryonic testes were dissected from E13, E14, and E16 rats and RNA isolated to evaluate transcriptional changes during gonadal morphogenesis. Histological analysis of E13, E14, and E16 revealed no significant difference in morphology between control and vinclozolin treated animals (Fig. 1A–D). At E13 the morphological changes associated with gonadal sex determination have not begun, so gender was identified by PCR for Sry. Therefore, control and vinclozolin treated E13 testis both lack visible structures and were indistinguishable (data not shown). At E14 pre-Sertoli cell/germ cell aggregates were visible and testis cords were seen forming in both control and vinclozolin treated animals (Fig. 1A and B). Testis cords by E16 have formed with Sertoli and germ cells surrounded by peritubular cells in the testis of both control and vinclozolin treated animals (Fig 1C and D). The lack of morphological changes seen in the testis cross-sections between control and vinclozolin treated animals suggests vinclozolin does not alter morphological processes of embryonic testis differentiation.
Figure 1.
Histological sections of vinclozolin treated and control testis (H&E stained). A. E14 control. B. E14 treated. C. E16 control. D. E16 treated. E. E13 cultured control. F. E13 cultured vinclozolin treated. Arrows indicate forming cords which contain large round darker stained germ cells surrounded by lighter stained Sertoli cells.
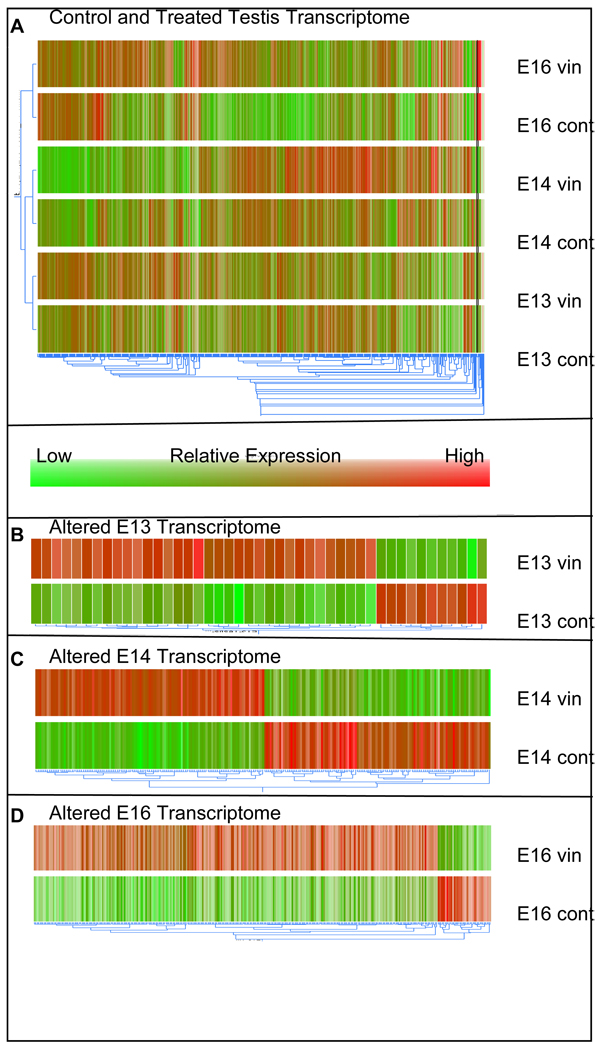
Although the previously reported transgenerational adult phenotypes occur after embryonic vinclozolin exposure [1–3], the current study focuses on the direct actions (i.e. immediate effects during exposure) of vinclozolin on the F1 generation. Microarray analyses were used to determine the transcriptome changes in the embryonic testis after vinclozolin exposure. Samples of E13, E14, and E16 control and treated testis RNA were hybridized to the RAE230 2.0 Affymetrix microarray chips for analysis. The microarray chips contain probe sets for 31,099 transcripts and allow for the majority of the rat transcriptome to be evaluated. Quality assurance analysis (data not shown) of raw array data revealed high quality data and all chips were used in further analysis using the Bioconductor packages Affymetrix Quality Control Report. A dendrogram with a condition tree showing relatedness of treatment groups was generated in GeneSpring from transcripts expressed in at least one condition of the in vivo treated or control samples (Fig. 2A). The condition tree reveals that control and vinclozolin treated samples for each individual time period cluster closest together indicating that their transcription profiles are similar. The E13 and E14 conditions also cluster closer to each other than to E16 conditions. Together this shows that transcriptional changes with treatment for each time period are more subtle then transcriptional changes between the time periods E13, E14 and E16 during testis differentiation. Furthermore, the comparison of E13, E14, and E16 samples agrees with the findings of a previous study of transcriptional changes during gonadal development with E16 clustered distally to E13 and E14 samples [32]. This supports the validity of the approach in the current study and demonstrates the large number of transcriptional changes involved in testis morphogenesis.
Figure 2.
Dendrogram of in vivo vinclozolin treated and control testis. A. All genes expressed in any of the six conditions (E13, E14, and E16 control and treated) for in vivo exposed analysis. Condition tree to the left shows relatedness of the 6 conditions. B. Genes altered by vinclozolin treatment at E13 (45 total genes). C. Genes altered by vinclozolin treatment at E14 (241 total genes). D. Genes altered by vinclozolin treatment at E16 (316 total genes). Gene trees below dendrograms indicate relative relatedness of transcript expression patterns between conditions.
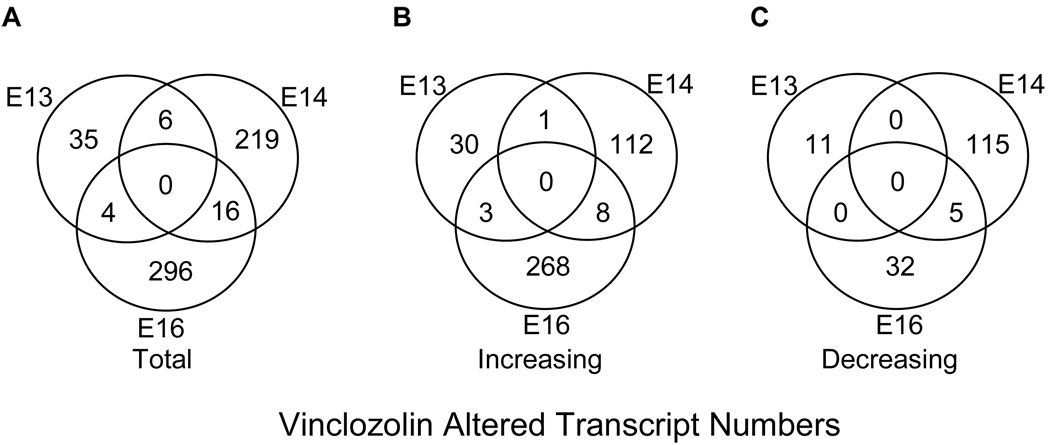
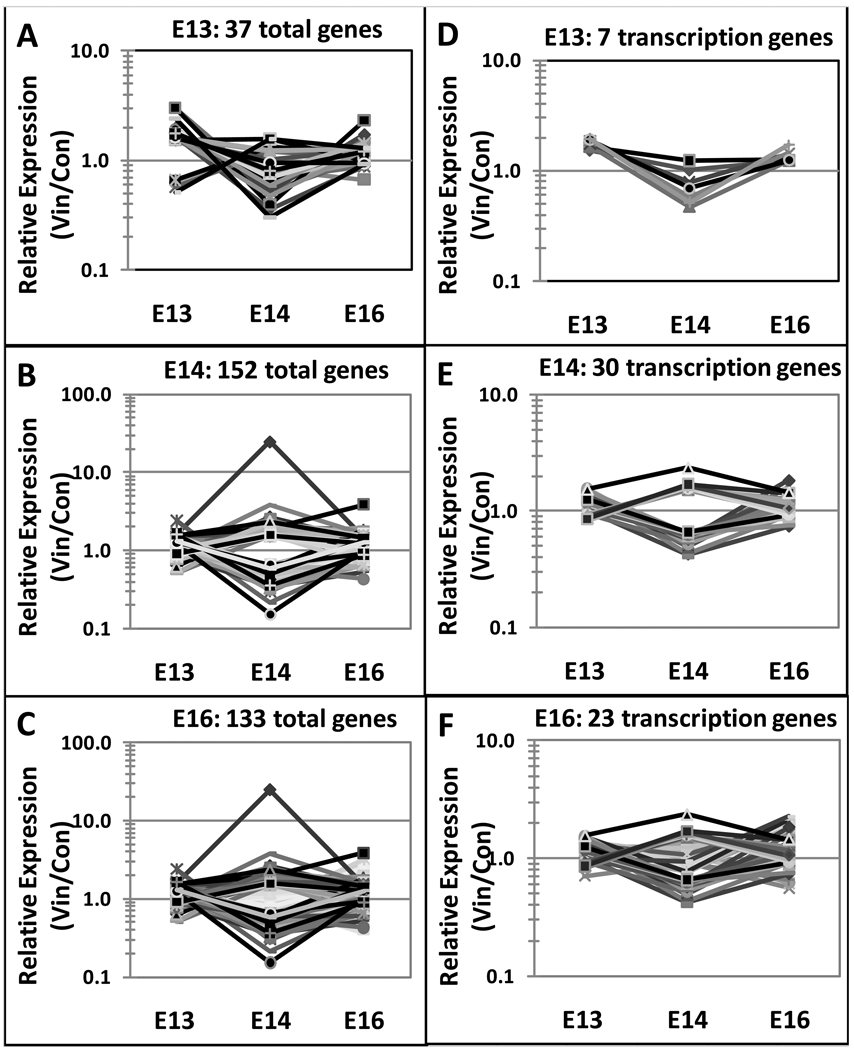
Genes altered in the differentiating testis due to vinclozolin treatment were identified by extracting genes significantly changed between vinclozolin treated and control samples with a 1.5 or higher fold change for E13, E14, and E16. This revealed a total of 576 altered genes (Fig. 3A) representing approximately two percent of transcripts on the RAE230 2.0 microarray chip. Interestingly, nearly all genes identified are altered at only one time point with only 26 transcripts altered in any two time points and none altered in all three (Fig. 3A). These transiently altered genes, excluding ESTs, for each developmental time are shown in Figure 4A–C. The developmental pattern of gene expression is generally distinct for each time point. The complete gene lists and genes appearing in multiple lists are indicated in the comparative cross list column of Supplemental Tables S1 – S3. Forty-five genes had altered expression at E13. These genes are listed in Supplemental Table S1 categorized by major gene function. At E14 a total of 241 genes had altered expression (Supplemental Table S2). At E16 there were 316 altered genes (Supplemental Table S3). A dendrogram clustering altered transcripts for E13, E14, and E16 separately shows that an equal number of genes are increasing and decreasing with treatment at E14 (Fig. 2C), while at E13 and E16 there are more genes that increase after vinclozolin treatment than decrease (Fig. 2B and D). A total of 422 transcripts increased after vinclozolin treatment with 34 increased at E13, 121 at E14, and 279 at E16 (Fig. 3B). Only 163 total transcripts decreased after vinclozolin treatment with 11 decreased at E13, 120 at E14, and 37 at E16 (Fig. 3C). Together these observations suggest a general increase in transcriptional abundance following vinclozolin treatment.
Figure 3.
Venn diagrams of significantly altered transcripts with at least a 1.5 fold change in E13, E14, and/or E16 after in vivo vinclozolin exposure. A. All 576 vinclozolin altered transcripts from in vitro treated E13, E14, and E16. B. 422 increased transcripts. C. 163 decreased transcripts.
Figure 4.
Developmental expression of vinclozolin altered genes at E13, E14 and E16 time points for each individual period altered gene sets (A) E13, (B) E14, (C) E16. Similar analysis of the transcription factor category of genes are presented in (D) E13, (E) E14, (F) E16. Observations indicate the altered gene sets are generally unique to the time period.
Categorization by major known function of all 576 genes altered by in utero vinclozolin exposure reveals that transcription, signaling, cytoskeletal and extra cellular matrix associated transcripts are highly represented (Fig. 5). Development, translation and protein modification, protein binding, epigenetic, proteolysis, growth factor, immune response, cell cycle, and electron transport were also represented by multiple transcripts. Interestingly, transcripts that play a role in protein translation and modification are over-represented at E13 suggesting greater sensitivity of this cellular process to vinclozolin treatment prior to gonadal differentiation. The specific genes within the various functional categories are presented in Supplemental Tables S1 – S3. The gene category with the largest number of genes is the transcription factor category and the developmental pattern of expression of this gene set is shown in Figure 4D–F.
Figure 5.
Vinclozolin altered genes for E13, E14, and E16 categorized by major known function. For each time period a gene is only represented once, for each category a gene is represented for each time point in which it appears, with minimal overlap. The specific categories and genes are presented in Supplemental Tables S1–S3.
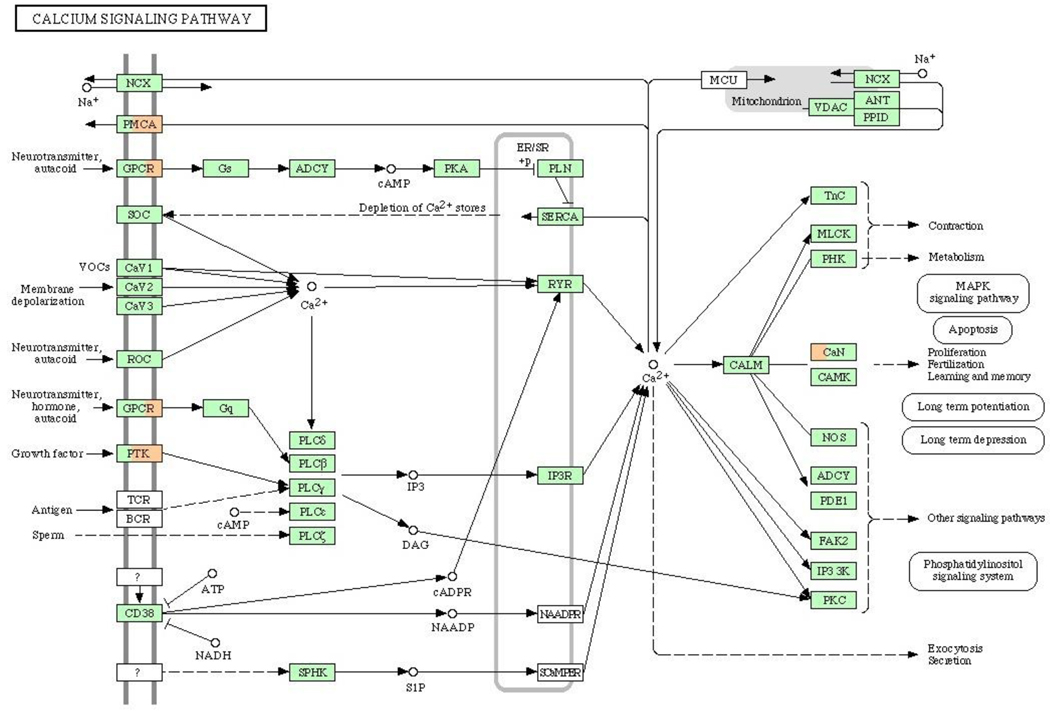
To identify cellular signaling pathways affected by vinclozolin, KEGG Pathway Analysis [44] was performed on the 576 genes found to be altered by in vivo vinclozolin exposure. Ten pathways were affected by three or more genes of the 576 genes altered by in vivo vinclozolin treatment at least at one time point. These pathways are listed in Table 1 with the number of genes and the associated impact factors (as calculated in KEGG Pathway Analysis) indicated for each time point. Impact factor calculation by KEGG considers the total number of genes in the pathway, the number of genes from the submitted list appearing in the pathway, and the roles those genes play in the pathway. The majority of pathways affected by vinclozolin have altered gene expression from the later time points of E14 and E16. Observations suggest that vinclozolin may initiate a cascade of events affecting a larger number of signaling pathways as development progressed. Calcium signaling is affected at E13, E14, and E16 by vinclozolin treatment, Figure 6. MAPK, Gap junction, and cell adhesion pathways are also among those affected by vinclozolin altered genes at E14 and E16, Supplemental Figures S1 and S2. The majority of these pathways are affected by the same small subset of genes of the 576 transcripts appearing in a represented pathway. This suggests that a few genes may be responsible for a large number of signaling mediated effects including altered transcript levels of other identified genes. Therefore specific known pathways may not be as critical in mediation of vinclozolin altered expression as various cellular processes or gene-gene interactions in general.
Table 1.
Pathways Affected by Vinclozolin Altered Genes
| E13 | E14 | E16 | ||||
|---|---|---|---|---|---|---|
| Pathway Name | Impact Factor |
#Genes Altered |
Impact Factor |
#Genes Altered |
Impact Factor |
#Genes Altered |
| Calcium signaling pathway | 4.39 | 1 | 4.80 | 3 | 6.04 | 4 |
| Leukocyte transendothelial migration | 2.75 | 1 | 4.67 | 3 | 5.57 | 3 |
| Neuroactive ligand-receptor interaction | 2.61 | 1 | 0.73 | 2 | 2.33 | 3 |
| Regulation of actin cytoskeleton | 3.03 | 1 | 4.62 | 4 | 3.66 | 1 |
| Focal adhesion | 2.91 | 1 | 5.43 | 3 | 9.86 | 1 |
| MAPK signaling pathway | 1.33 | 2 | 3.67 | 4 | ||
| Gap junction | 10.14 | 4 | 8.32 | 3 | ||
| Cell adhesion molecules (CAMs) | 20.15 | 2 | 264.81 | 3 | ||
| Tight junction | 7.97 | 1 | 6.78 | 3 | ||
| Cytokine-cytokine receptor interaction | 0.76 | 1 | 3.54 | 3 | ||
Impact Factor indicates relative significance of pathways. Impact Factor is generated by KEGG pathway analysis software. Higher values indicate pathways of greater relative significance.
Figure 6.
The Calcium Signaling Pathway (KEGG, Kanehisa Laboratory in Kyoto University, Japan) with vinclozolin altered genes indicated. Green boxes indicate genes which are present on the Affymetrix Rat 230-2.0 chip, white – not present on the chip, orange – up-regulated by vinclozolin, blue – down-regulated by vinclozolin. Each affected gene box divided on 3 parts: left part represent E13 gene list, central part – E14, and right part – E16 gene list.
In the current study, organ cultures were used to compare the actions of vinclozolin in vivo and in vitro. Gonads isolated at E13 and cultured for three days develop testis cords, and this system has been used as an in vitro research prototype for testis development [30, 45, 46]. Previously, 50 µM vinclozolin did not induce morphological changes in cultured testis [30] and so this concentration was used for the current in vitro studies (Fig 1 E and F). The lack of change in testis histology in treated cultures compared to controls is desirable so that transcriptional changes could be compared to in vivo treatments that also lacked morphological change compared to controls. This comparison was made to determine if vinclozolin altered transcription was similar between the treated organ cultures and in vivo samples. The anti-androgen flutamide was also used to treat cultured testes at a concentration of 5 µM, that did not cause morphological change (data not shown), to help determine if vinclozolin was acting by an anti-androgenic mechanism. Organ cultures treated with vinclozolin versus vehicle control revealed 19 altered transcripts (Supplemental Table S4). Three of these transcripts were also among the 576 altered by in vivo vinclozolin treatment (marked with a “V” in the “cross listed” column of supplemental tables) including Sap18 and two ESTs. Organ cultures treated with flutamide versus vehicle control revealed 43 altered transcripts (Supplemental Table S5). Four of these transcripts were also among the 576 altered by in vivo vinclozolin treatment (marked with an “F” in “Cross listed” column of supplemental tables) and included Pdgfb, Ncor1, Mthfd21, and an EST. The transcripts altered by in vitro vinclozolin or flutamide treatments did not share any common transcripts. Therefore vinclozolin may not be acting through an anti-androgenic mechanism, as previously reported [31].
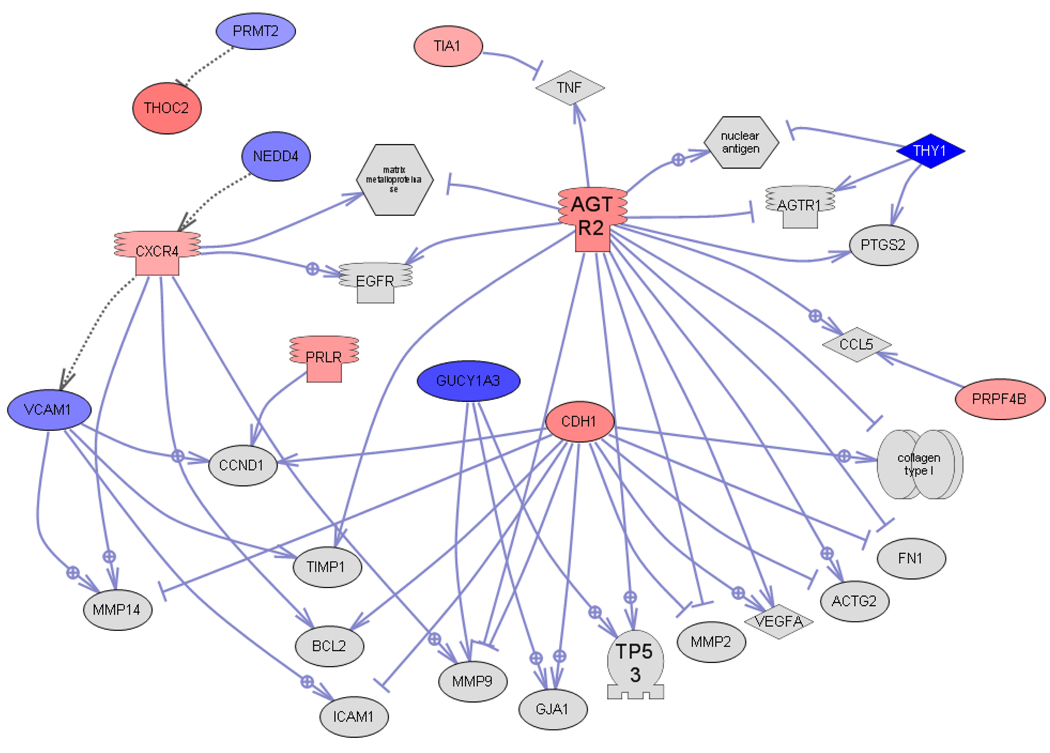
Based on the findings of the current study a list of 36 candidate genes for future investigation of vinclozolin action on differentiating testis was compiled (Table 2). These genes were chosen for their higher fold changes, interesting cellular functions, or appearance in multiple lists compared in this study. The candidate list includes genes that affect epigenetics, extracellular matrix, signaling, and development. The interactions between these genes were determined with global literature review analysis software (Pathway Studio, Ariadne Genomics Inc., Rockville, MD) and the gene network is shown in Figure 7. From the 36 genes 12 were found to interact in the network. Observations suggest the integration of pathways and cellular processes with this network of vinclozolin altered genes. The present study presents this list of candidate genes for consideration in future studies of the mechanism of direct action of vinclozolin on testis development of the F1 generation male.
TABLE 2.
Candidate Genes for Vinclozolin Action on Differentiating Testis
| Category | Gene | Expression Ratio V/C |
Cross Listed* |
GeneBank |
|---|---|---|---|---|
| Transcription | THO complex 2 (Thoc2, predicted) | 3 | E13 | AI639237 |
| Signaling | ||||
| Protein phosphatase 1, regulatory subunit 3C (Ppp1r3c) | 0.5 | A, E14 | BM390827 | |
| Guanylate cyclase 1, soluble, alpha 3 (Gucy1a3) | 0.4 | A, E14 | M57405 | |
| SH3 domain binding protein CR16 (Cr16)**/// WAS/WASL interacting protein family, member 3 (Wipf3) |
2.3 | A, E14 | U25281 | |
| Calcium/calmodulin-dependent protein kinase II, delta (Camk2d) | 1.7 | E13 | X77194 | |
| Rho guanine nucleotide exchange factor17 (Arhgef17, pred) | 0.6 | E13 | BI274338 | |
| Sushi domain containing 3 (Susd3, predicted) | 2.2 | E14 | AI407337 | |
| Receptors | Angiotensin II receptor, type 2 (Agtr2) | 2.4/0.4 | E13, E14 | BF552873 |
| Platelet derived growth factor receptor beta (Pdgfrb) | 1.9/3.9 | F, E14, E16 | BF283421 | |
| Protein Modification |
Protease, serine, 35 (Prss35) | 0.3 | A, E14 | AA866443 |
| PRP4 pre-mRNA processing factor 4 homolog B (Prpf4b, yeast) | 1.9 | E13 | BE105398 | |
| EF-hand calcium binding domain 2 (Efcab2, predicted) | 1.6 | E16 | BI299709 | |
| Immune | ||||
| Response | Thymus cell antigen 1, theta (Thy1) | 0.2 | E14 | AI145313 |
| RT1 class II, locus Bb (RT1-Bb) | 24.6 | E14 | AI715202 | |
| Growth Factors, Chemokines |
Insulin-like 3 (Insl3) | 1.9 | E16 | AF139918 |
| Prolactin receptor (Prlr) | 2 | E16 | M57668 | |
| Spondin 1 (Spon1) | 1.8 | E16 | M88469 | |
| Chemokine (C-X-C motif) receptor 4 (Cxcr4) | 1.7 | E16 | U54791 | |
| Epigenetics | Non-coding RNA expressed in the brain, repeat sequence, clone3** /// EST |
0.3 | A, E14 | BG670822 |
| SNF2 histone linker PHD RING helicase (Shprh, predicted) | 0.5 | A, E14 | BE104039 | |
| Chromogranin B (Chgb) | 0.2 | E14 | NM_012526 | |
| Non-coding RNA expressed in the brain, repeat sequence, clone 12**/// EST |
0.4 | E16 | BE099838 | |
| Enhancer of polycomb homolog 2 (Epc2, Drosophila, predicted) | 0.6 | E14 | AW918173 | |
| Similar to GTL2, imprinted maternally expressed untranslated (RGD1566401) |
3 | E13 | NM_019161 | |
| Hypermethylated in cancer 2 (Hic2, predicted) | 1.6 | E14 | BI284294 | |
| Cytotoxic granule-associated RNA binding protein 1 (Tia1) | 1.7 | E16 | BE104820 | |
| Chromodomain helicase DNA binding protein 6 (Chd6) | 2.1 | E16 | BF413591 | |
| HMT1 hnRNP methyltransferase-like 1 (S. cerevisiae)(Hrmt1l1)** /// | 0.7 | E16 | BG670963 | |
| Protein N-arginine methyltransferase 2 (Prmt2) | ||||
| Development | ||||
| Neural expressed developmentally down-regulated 4 (Nedd4) | 0.6 | E14 | BI277849 | |
| Adenomatosis polyposis coli 2 (Apc2, predicted) | 0.3 | E14 | BI294552 | |
| SRY-box containing gene 7 (Sox7, predicted) | 1.6 | E14 | AI058292 | |
| Synaptic vesicle glycoprotein 2b (Sv2b) | 2.4 | E16 | BG672437 | |
| Cytoskeleton- ECM |
Vascular cell adhesion molecule 1 (Vcam1) | 0.6 | E13,E14,C,A | NM_012889 |
| Osteoglycin (Ogn) | 0.3 | E14 | BG664221 | |
| Myosin-binding protein H (Mybphl) | 2.6 | E16 | AW252385 | |
| Cadherin 1 (Cdh1) | 2.5 | E16 | NM_031334 |
For any gene identified in multiple lists, the other lists in which it appears are indicated in the Cross Listed column. E13= altered by vinclozolin at E13, E14= altered by vinclozolin at E14, E16= altered by vinclozolin at E16, V= altered by vinclozolin in organ cultures, F= altered by flutamide in organ cultures, A= appeared in a list of F1 E16 vinclozolin altered transcripts published by [37] and C=appeared in a list of testis differentiation candidates published by [32]
gene names from Affymetrix annotation Rat230_2.na23.annot as they were referenced in List A; the rest from annotation Rat230_2.na27.annot
Figure 7.
Gene interaction sub-network of shortest connections to common targets for the 36 candidate regulatory genes obtained by global literature analysis (Pathway Studio 7.0, Ariadne Genomics Inc., Rockville MD). Only 12 connected genes from the list of 36 are shown, the rest are not connected and not shown. Node shapes code: oval and circle – protein; crescent – protein kinase and kinase; diamond – ligand; irregular polygon – phosphatase; circle/oval on tripod platform – transcription factor; ice cream cone – receptor. Red color represents up-regulated genes, blue color – down-regulated genes, grey – common targets which are not in the input list; arrows with plus sign show positive regulation/activation, arrows with minus sign – negative regulation/inhibition.
4. DISCUSION
The current study investigates vinclozolin induced alterations in gene expression during embryonic testis morphogenesis. Previous studies have shown that intraperitoneal injection of 100mg/kg/day vinclozolin from E8-E14 of pregnancy resulted in increased apoptosis in spermatogenic cells at P20 and P60, decreased sperm number, and decreased sperm motility in male offspring [30]. These phenotypes were also shown to persist for at least three subsequent generations [1–3]. The transgenerational adult disease phenotypes are presumed to result from changes induced during the time of direct embryonic exposure which are coincident with testis development. The current study was designed to investigate the direct mechanisms of vinclozolin action on F1 generation embryonic testis. Vinclozolin is used as a pharmacologic agent to promote alterations in testis development and study the mechanisms of action. Therefore this study used IP injection to control dose, versus gavage, and used a higher dose than expected in the environment. This study was not designed to perform risk assessment information on vinclozolin toxicology. Future toxicology studies will need to consider mode of administration and dose in the context of risk assessment, but the current study focuses on potential mechanism of action.
Histological analysis of testes from E13 (not shown), E14, and E16 embryos (Fig. 1A–D), showed no apparent morphological changes between control and treated testis. Both control and treated testes appeared to develop normally as previously described [32]. This suggests few or no vinclozolin induced alterations are due to gross morphological processes being disrupted. Results of this study also indicate that the transcriptomes of control and treated testes are more similar than the developmental changes between E13, E14, or E16 testes. This was expected due to the lack of morphological changes with treatment.
To assess vinclozolin induced transcriptome alterations, the transcripts with statistically significant changes in expression between control and treated samples were identified for E13, E14, and E16 testis. The lack of overlap in the 576 altered transcripts between the developmental time points may be due to the dramatic differences in the transcriptomes throughout testicular differentiation and development. Alternatively, a cascade of events initiated by vinclozolin treatment leading to new alterations in transcription over time may explain the lack of overlapping alterations. The ratios of increasing to decreasing transcripts suggest a general increase in transcriptional activity following vinclozolin treatment (Fig. 2). This general trend would be consistent with vinclozolin causing a cascade of events mediated by transcriptional regulators or post transcriptional modifiers leading to increased transcript abundance.
Cellular processes shown to be affected by vinclozolin are also consistent with a potential role of vinclozolin in transcriptional alteration. Interestingly, genes known to affect cellular processes such as oxidative stress that are associated with toxicology were not identified following vinclozolin treatment in this study. This suggests that vinclozolin may not cause adult onset disease through classic toxicological mechanisms. The cellular functions most likely affected by vinclozolin altered genes appear to be transcription followed by signaling, extra cellular matrix, and metabolism. At E13, the earliest time point studied, RNA processing and translation is the highest represented function. In a previous microarray study of normal sex determination and gonadal development extra cellular matrix, signaling, and metabolism were highly represented, but not transcription and translation [32]. Combined observations suggest vinclozolin action is mediated at least in part through altering transcriptional regulation and possibly altered translation.
To determine if genes regulated during gonadal sex determination and testis development were altered by vinclozolin treatment, the 576 transcripts found to be altered by in vivo vinclozolin treatment in the current study were compared to candidate genes previously implicated in sex determination and testis development [32]. In this previous study 109 genes that were regulated between E13, E14, and E16 testis were identified as candidates for regulation of male gonadal sex determination and testis differentiation. Surprisingly, of these 109 genes only three (Spon1, Spock2, and Ogn) appear to be altered by vinclozolin during testis differentiation (indicated with a “C” in the “Cross listed” column of Supplemental Tables S1–S3). This suggests limited involvement of those genes regulated during gonadal sex determination and testis differentiation in the mechanism of vinclozolin action on embryonic gonads.
In addition to the in vivo vinclozolin treatments, the effect of in vitro treatments on E13 cultured gonads was investigated using microarray analysis. This was done to compare in vivo and in vitro vinclozolin altered transcripts and determine if the treated organ cultures could be used as a model system to study the effects of vinclozolin on testis development. In addition, the anti-androgenic compound flutamide was used to treat organ cultures. Microarray analysis results were compared to vinclozolin treated organ cultures to determine if flutamide and vinclozolin had similar effects. Different transcripts were altered by in vitro vinclozolin treatment then by flutamide treatment. Observations suggest vinclozolin and flutamide are not acting in the same manner. A previous study demonstrated flutamide did not promote a transgenerational phenotype in comparison to vinclozolin [31]. Caution must be taken in drawing conclusions about organ culture experiments due to the low doses chosen for analysis. Doses were selected that did not affect histology between control and treated samples. These in vitro doses may be too low and not significantly alter testis development. Another possible reason for the small number of altered transcripts in the organ cultures may be that testes were not exposed from E8-E14, but rather starting at E13 for three days in culture. Thus the treatment did not overlap with the formation of the indifferent gonad or germ cell reprogramming, and may not induce the same alterations as seen in vivo. Observations suggest that treated cultures are not an overly useful model as applied in this study. However, the results suggest flutamide and vinclozolin treatments alter different transcripts and may be acting through different mechanisms.
From the in vivo analyses a number of genes with interesting and potentially significant functions were chosen to highlight as candidates altered by vinclozolin in the differentiating testis. This list contains 36 vinclozolin altered genes chosen for interesting expression patterns, larger changes in expression, or intriguing functional or pathway associations. Many of the candidates identified as altered by vinclozolin in the embryonic testis have known functions identified in previous studies. Knowledge of the previously identified functions for these candidate genes provides insight into potential functions in the embryonic testis. A gene interaction network of these genes identified a subset of genes potentially involved in regulating multiple pathways and cellular processes. The functional roles of these genes in vinclozolin actions needs to be investigated.
An epigenetic mechanism of action for vinclozolin has been proposed to explain the adult onset and transgenerational phenotypes associated with exposure [1, 3, 37]. The list of 576 vinclozolin altered genes from this study was compared to a previous study of E16 control and vinclozolin treated testis for F1, F2, and F3 generation animals which identified 1597 altered transcripts between control and treated F1 testes [37]. A total of 124 transcripts appeared in both studies (indicated by an A in the “cross listed” column of supplemental tables), many of which are associated with epigenetic functions consistent with the previous study. A total of twelve transcripts with epigenetic related functions were altered by vinclozolin in the current study.
These epigenetic genes include non-coding RNAs, Prp4b which is known to play a role in RNA processing, and Thoc2 which has been shown to associate with ribonucleoprotein complexes and be important for mRNA quality control [47, 48]. Another transcript, Camk2d, is known to phosphorylate histone deacetylases that affect histone localization on DNA [49, 50]. Camk2d histone phosphorylation is a calcium-dependant process and is known to affect vascular smooth muscle cell proliferation and migration [51, 52]. Gtl2 is known to be a maternally imprinted region involved in micro RNA stabilization [53, 54]. Gtl2 is methylated in the male germ line during germ cell differentiation correlating with the time of treatment in this study [55]. The DNA methylation patterns that arise have been shown to persist to spermatogonia and spermatocytes [56]. The RNA stabilization role and epigenetic imprinting of Gtl2 persists in the adult in the germ line that makes the Gtl2 gene particularly interesting for future studies of vinclozolin action on differentiating testis. Four transcripts altered at E14 are known to be sensitive to hypermethylation suppression. These include Ppp1r3c, a serine/threonine protein phosphatase implicated in diverse cellular processes [57], the tumor suppressor Hic2 [58, 59] and Sox7 that is known to bind to beta-catenin and inhibit transcriptional activity [60, 61]. The fourth gene, Apc2, can also affect beta-catenin function in cadherin signaling and is important for cellular proliferation and differentiation [62, 63]. At E16, several known epigenetic regulators were altered by vinclozolin. Chd6 is an ATP dependant chromatin remodeling enzyme with DNA dependent ATPase activity [64]. Prmt1 has been identified in embryonic germ cells [65], is a coactivator of the androgen receptor (AR), and binds and methylates hnRNPQ for internalization of insulin receptors [66, 67]. A role for these genes in mediation of vinclozolin induced disease or altered testis development is currently unknown. Further investigation may help in understanding how potential epigenetic regulators or epigenetically regulated genes can affect testis development ultimately leading to adult onset disease.
Increased spermatogenic cell apoptosis is one adult onset testis disease phenotype induced by vinclozolin. Although vinclozolin treatment between E8-E14 was previously shown to increase apoptosis in pubertal and adult rat testis, the molecular mechanisms that lead to this apoptosis phenotype has not been investigated. Perhaps a reduction in cellular apoptotic protection in the embryonic testis persists until puberty. Nedd4 is a ubiquitin ligase important for protein trafficking and signal transduction [68–72]. Because a loss of Nedd4 translocation leads to a loss of apoptotic protection, Nedd4 is another interesting candidate for mediation of vinclozolin action in the embryo that may lead to increased spermatogenic cell apoptosis in pubertal and adult rats. Other transcripts affecting apoptosis were also altered by vinclozolin. Hrmt1l1 is a protein arganine methyltransferase which can interact with ribonucleoprotein complexes and promote apoptosis through NF-KappaB transcription [73, 74]. Tia1 is an alternative splicing factor that can also promote apoptosis by affecting Fas receptor splice variants [75, 76]. The role of these transcripts during testis differentiation and there potential role in mediating vinclozolin induced increases in spermatogenic cell apoptosis later in life would also be interesting to further investigate.
Although the current study was not designed to investigate transgenerational phenomena or mechanisms, the direct actions of vinclozolin studied do indirectly lead to transgenerational phenotypes [1, 2]. The observations and genes identified in the current study provide insights into the potential mechanism of action of vinclozolin that promote the epigenetic reprogramming of the germ line required for the transgenerational phenotype. Recently two chemical companies that produce the compounds of interest have published reports regarding vinclozolin actions. The company that produces vinclozolin (BASF, Germany) found that oral administration (gavage) of the same dose used IP [1] did not have transgenerational effects nor major effects in the F1 generation animals [77]. Compounds administered through oral gavage treatment generally have an order of magnitude lower circulating dose than IP injection, such that the lack of effect [77] was likely due to insufficient dose [1]. Therefore, in regards to toxicology, this study suggests vinclozolin at the dose used may not be a significant risk factor [77]. However, our studies use vinclozolin as a pharmacologic agent to promote the transgenerational phenotype and study mechanism [1], and do not perform risk assessment or classic toxicology experiments. A second study repeated the vinclozolin experiment [1] using a more inbred CD-Sprague Dawley (Charles River) rat line, versus the outbred Harlan Sprague Dawley line [1]. This study did not obtain a dramatic first generation or transgenerational phenotype [78]. Previously we reported that the inbred Fisher rat line did not respond as well as the outbred Harlan Sprague Dawley line [1, 3], and have recently found the CD-Sprague Dawley response is also not as robust (unpublished observations). The inbred status of the line may be a factor in the efficiency of promoting the phenotype. Recently we have repeated the original observation [1] with the outbred Harlan Sprague Dawley line [31]. In addition, several recent studies confirm the ability of environmental agents to promote transgenerational phenotypes [79, 80], as well as an independent confirmation of the epigenetic transgenerational actions of vinclozolin [81].
Vinclozolin treatment affects expression of multiple transcripts implicating epigenetics, vascular development, cellular apoptosis, transcription and signaling by calcium, insulin, Wnt, and AR. The present study has provided a list totaling 576 candidate genes for further investigation, of which 36 of the more interesting altered transcripts have been highlighted. In addition, the processes and pathways most affected by vinclozolin treatment have been identified. This study has provided a better understanding of the direct effects of vinclozolin treatment on the F1 generation developing testis.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the assistance and helpful discussions of Drs. Ingrid Sadler-Riggleman, Eric Nilsson and Ramji Bhandari. We thank Ms. Heather Johnson for assistance in preparation of the manuscript. We acknowledge the assistance of the Genomics and Bioinformatics Core Laboratories at the Center for Reproductive Biology at WSU and UI. The current address for Dr. Matthew Anway is the Department of Biological Sciences, University of Idaho, Moscow, ID 83844-3051. The current address for Dr. Tracy Clement is National Institute of Environmental Sciences, Research Triangle Park, NC 27709. This research was supported by NIH grants to MKS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jost A, Magre S, Agelopoulou R. Early stages of testicular differentiation in the rat. Hum Genet. 1981;58:59–63. doi: 10.1007/BF00284150. [DOI] [PubMed] [Google Scholar]
- 5.Ginsburg M, Snow MH, McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110:521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 6.Magre S, Agelopoulou R, Jost A. [Sertoli cells and organogenesis of the fetal testis (author's transl)] Ann Endocrinol (Paris) 1980;41:531–537. [PubMed] [Google Scholar]
- 7.Tung PS, Skinner MK, Fritz IB. Cooperativity between Sertoli cells and peritubular myoid cells in the formation of the basal lamina in the seminiferous tubule. Ann N Y Acad Sci. 1984;438:435–446. doi: 10.1111/j.1749-6632.1984.tb38304.x. [DOI] [PubMed] [Google Scholar]
- 8.Kanai Y, Kawakami H, Takata K, Kurohmaru M, Hirano H, Hayashi Y. Involvement of actin filaments in mouse testicular cord organization in vivo and in vitro. Biol Reprod. 1992;46:233–245. doi: 10.1095/biolreprod46.2.233. [DOI] [PubMed] [Google Scholar]
- 9.Yao HH, Aardema J, Holthusen K. Sexually dimorphic regulation of inhibin beta B in establishing gonadal vasculature in mice. Biol Reprod. 2006;74:978–983. doi: 10.1095/biolreprod.105.050286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Merchant-Larios H, Moreno-Mendoza N. Mesonephric stromal cells differentiate into Leydig cells in the mouse fetal testis. Exp Cell Res. 1998;244:230–238. doi: 10.1006/excr.1998.4215. [DOI] [PubMed] [Google Scholar]
- 11.Mittwoch U, Delhanty JD, Beck F. Growth of differentiating testes and ovaries. Nature. 1969;224:1323–1325. doi: 10.1038/2241323a0. [DOI] [PubMed] [Google Scholar]
- 12.Chubb C. Genes regulating testis size. Biol Reprod. 1992;47:29–36. doi: 10.1095/biolreprod47.1.29. [DOI] [PubMed] [Google Scholar]
- 13.Wilhelm D, Koopman P. The makings of maleness: towards an integrated view of male sexual development. Nat Rev Genet. 2006;7:620–631. doi: 10.1038/nrg1903. [DOI] [PubMed] [Google Scholar]
- 14.Wilhelm D, Palmer S, Koopman P. Sex determination and gonadal development in mammals. Physiol Rev. 2007;87:1–28. doi: 10.1152/physrev.00009.2006. [DOI] [PubMed] [Google Scholar]
- 15.Vaiman D, Pailhoux E. Mammalian sex reversal and intersexuality: deciphering the sex-determination cascade. Trends Genet. 2000;16:488–494. doi: 10.1016/s0168-9525(00)02126-0. [DOI] [PubMed] [Google Scholar]
- 16.Sharpe RM. Hormones and testis development and the possible adverse effects of environmental chemicals. Toxicol Lett. 2001;120:221–232. doi: 10.1016/s0378-4274(01)00298-3. [DOI] [PubMed] [Google Scholar]
- 17.Andersson AM, Jorgensen N, Main KM, Toppari J, Rajpert-De E, Leffers Meyts H, Juul A, Jensen TK, Skakkebaek NE. Adverse trends in male reproductive health: we may have reached a crucial 'tipping point'. Int J Androl. 2008;31:74–80. doi: 10.1111/j.1365-2605.2007.00853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolf CJ, LeBlanc GA, Ostby JS, Gray LE., Jr Characterization of the period of sensitivity of fetal male sexual development to vinclozolin. Toxicol Sci. 2000;55:152–161. doi: 10.1093/toxsci/55.1.152. [DOI] [PubMed] [Google Scholar]
- 19.Rempel MA, Schlenk D. Effects of environmental estrogens and antiandrogens on endocrine function, gene regulation, and health in fish. Int Rev Cell Mol Biol. 2008;267:207–252. doi: 10.1016/S1937-6448(08)00605-9. [DOI] [PubMed] [Google Scholar]
- 20.Ketata I, Denier X, Hamza-Chaffai A, Minier C. Endocrine-related reproductive effects in molluscs. Comp Biochem Physiol C Toxicol Pharmacol. 2008;147:261–270. doi: 10.1016/j.cbpc.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 21.Schoeters G, Den Hond E, Dhooge W, van Larebeke N, Leijs M. Endocrine disruptors and abnormalities of pubertal development. Basic Clin Pharmacol Toxicol. 2008;102:168–175. doi: 10.1111/j.1742-7843.2007.00180.x. [DOI] [PubMed] [Google Scholar]
- 22.Peets EA, Henson MF, Neri R. On the mechanism of the anti-androgenic action of flutamide (alpha-alpha-alpha-trifluoro-2-methyl-4'-nitro-m-propionotoluidide) in the rat. Endocrinology. 1974;94:532–540. doi: 10.1210/endo-94-2-532. [DOI] [PubMed] [Google Scholar]
- 23.Ostby J, Kelce WR, Lambright C, Wolf CJ, Mann P, Gray LE., Jr The fungicide procymidone alters sexual differentiation in the male rat by acting as an androgen-receptor antagonist in vivo and in vitro. Toxicol Ind Health. 1999;15:80–93. doi: 10.1177/074823379901500108. [DOI] [PubMed] [Google Scholar]
- 24.Kelce WR, Monosson E, Gamcsik MP, Laws SC, Gray LE., Jr Environmental hormone disruptors: evidence that vinclozolin developmental toxicity is mediated by antiandrogenic metabolites. Toxicol Appl Pharmacol. 1994;126:276–285. doi: 10.1006/taap.1994.1117. [DOI] [PubMed] [Google Scholar]
- 25.Wong C, Kelce WR, Sar M, Wilson EM. Androgen receptor antagonist versus agonist activities of the fungicide vinclozolin relative to hydroxyflutamide. J Biol Chem. 1995;270:19998–20003. doi: 10.1074/jbc.270.34.19998. [DOI] [PubMed] [Google Scholar]
- 26.You L, Casanova M, Archibeque-Engle S, Sar M, Fan LQ, Heck HA. Impaired male sexual development in perinatal Sprague-Dawley and Long-Evans hooded rats exposed in utero and lactationally to p,p'-DDE. Toxicol Sci. 1998;45:162–173. doi: 10.1093/toxsci/45.2.162. [DOI] [PubMed] [Google Scholar]
- 27.Bentvelsen FM, Brinkmann AO, van der Schoot P, van der Linden JE, van der Kwast TH, Boersma WJ, Schroder FH, Nijman JM. Developmental pattern and regulation by androgens of androgen receptor expression in the urogenital tract of the rat. Mol Cell Endocrinol. 1995;113:245–253. doi: 10.1016/0303-7207(95)03593-v. [DOI] [PubMed] [Google Scholar]
- 28.Quigley CA, De Bellis A, Marschke KB, el-Awady MK, Wilson EM, French FS. Androgen receptor defects: historical, clinical, and molecular perspectives. Endocr Rev. 1995;16:271–321. doi: 10.1210/edrv-16-3-271. [DOI] [PubMed] [Google Scholar]
- 29.Gray LE, Jr, Ostby JS, Kelce WR. Developmental effects of an environmental antiandrogen: the fungicide vinclozolin alters sex differentiation of the male rat. Toxicol Appl Pharmacol. 1994;129:46–52. doi: 10.1006/taap.1994.1227. [DOI] [PubMed] [Google Scholar]
- 30.Uzumcu M, Suzuki H, Skinner MK. Effect of the anti-androgenic endocrine disruptor vinclozolin on embryonic testis cord formation and postnatal testis development and function. Reprod Toxicol. 2004;18:765–774. doi: 10.1016/j.reprotox.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Anway MD, Rekow SS, Skinner MK. Comparative anti-androgenic actions of vinclozolin and flutamide on transgenerational adult onset disease and spermatogenesis. Reprod Toxicol. 2008;26:100–106. doi: 10.1016/j.reprotox.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clement TM, Anway MD, Uzumcu M, Skinner MK. Regulation of the gonadal transcriptome during sex determination and testis morphogenesis: comparative candidate genes. Reproduction. 2007;134:455–472. doi: 10.1530/REP-06-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007;23:297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 35.Ozanne SE, Constancia M. Mechanisms of disease: the developmental origins of disease and the role of the epigenotype. Nat Clin Pract Endocrinol Metab. 2007;3:539–546. doi: 10.1038/ncpendmet0531. [DOI] [PubMed] [Google Scholar]
- 36.Fleming TP, Kwong WY, Porter R, Ursell E, Fesenko I, Wilkins A, Miller DJ, Watkins AJ, Eckert JJ. The embryo and its future. Biol Reprod. 2004;71:1046–1054. doi: 10.1095/biolreprod.104.030957. [DOI] [PubMed] [Google Scholar]
- 37.Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics. 2008;91:30–40. doi: 10.1016/j.ygeno.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Uzumcu M, Dirks KA, Skinner MK. Inhibition of platelet-derived growth factor actions in the embryonic testis influences normal cord development and morphology. Biol Reprod. 2002;66:745–753. doi: 10.1095/biolreprod66.3.745. [DOI] [PubMed] [Google Scholar]
- 39.Levine E, Cupp AS, Skinner MK. Role of neurotropins in rat embryonic testis morphogenesis (cord formation) Biol Reprod. 2000;62:132–142. doi: 10.1095/biolreprod62.1.132. [DOI] [PubMed] [Google Scholar]
- 40.Cupp AS, Uzumcu M, Skinner MK. Chemotactic role of neurotropin 3 in the embryonic testis that facilitates male sex determination. Biol Reprod. 2003;68:2033–2037. doi: 10.1095/biolreprod.102.012617. [DOI] [PubMed] [Google Scholar]
- 41.Cupp AS, Kim GH, Skinner MK. Expression and action of neurotropin-3 and nerve growth factor in embryonic and early postnatal rat testis development. Biol Reprod. 2000;63:1617–1628. doi: 10.1095/biolreprod63.6.1617. [DOI] [PubMed] [Google Scholar]
- 42.Team RDC. R: A language and environmentfor Statistical computing. Australia: Vienna; 2008. [Google Scholar]
- 43.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero R. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H, Kim KH. Retinoic acid inhibits rat XY gonad development by blocking mesonephric cell migration and decreasing the number of gonocytes. Biol Reprod. 2004;70:687–693. doi: 10.1095/biolreprod.103.023135. [DOI] [PubMed] [Google Scholar]
- 46.Bott RC, McFee RM, Clopton DT, Toombs C, Cupp AS. Vascular endothelial growth factor and kinase domain region receptor are involved in both seminiferous cord formation and vascular development during testis morphogenesis in the rat. Biol Reprod. 2006;75:56–67. doi: 10.1095/biolreprod.105.047225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Assenholt J, Mouaikel J, Andersen KR, Brodersen DE, Libri D, Jensen TH. Exonucleolysis is required for nuclear mRNA quality control in yeast THO mutants. RNA. 2008;14:2305–2313. doi: 10.1261/rna.1108008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Rubio M, Chavez S, Huertas P, Tous C, Jimeno S, Luna R, Aguilera A. Different physiological relevance of yeast THO/TREX subunits in gene expression and genome integrity. Mol Genet Genomics. 2008;279:123–132. doi: 10.1007/s00438-007-0301-6. [DOI] [PubMed] [Google Scholar]
- 49.Bossuyt J, Helmstadter K, Wu X, Clements-Jewery H, Haworth RS, Avkiran M, Martin JL, Pogwizd SM, Bers DM. Ca2+/calmodulin-dependent protein kinase IIdelta and protein kinase D overexpression reinforce the histone deacetylase 5 redistribution in heart failure. Circ Res. 2008;102:695–702. doi: 10.1161/CIRCRESAHA.107.169755. [DOI] [PubMed] [Google Scholar]
- 50.Little GH, Bai Y, Williams T, Poizat C. Nuclear calcium/calmodulin-dependent protein kinase IIdelta preferentially transmits signals to histone deacetylase 4 in cardiac cells. J Biol Chem. 2007;282:7219–7231. doi: 10.1074/jbc.M604281200. [DOI] [PubMed] [Google Scholar]
- 51.House SJ, Ginnan RG, Armstrong SE, Singer HA. Calcium/calmodulin-dependent protein kinase II-delta isoform regulation of vascular smooth muscle cell proliferation. Am J Physiol Cell Physiol. 2007;292:C2276–C2287. doi: 10.1152/ajpcell.00606.2006. [DOI] [PubMed] [Google Scholar]
- 52.Mercure MZ, Ginnan R, Singer HA. CaM kinase II delta2-dependent regulation of vascular smooth muscle cell polarization and migration. Am J Physiol Cell Physiol. 2008;294:C1465–C1475. doi: 10.1152/ajpcell.90638.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Braem C, Recolin B, Rancourt RC, Angiolini C, Barthes P, Branchu P, Court F, Cathala G, Ferguson-Smith AC, Forne T. Genomic matrix attachment region and chromosome conformation capture quantitative real time PCR assays identify novel putative regulatory elements at the imprinted Dlk1/Gtl2 locus. J Biol Chem. 2008;283:18612–18620. doi: 10.1074/jbc.M801883200. [DOI] [PubMed] [Google Scholar]
- 54.Kircher M, Bock C, Paulsen M. Structural conservation versus functional divergence of maternally expressed microRNAs in the Dlk1/Gtl2 imprinting region. BMC Genomics. 2008;9:346. doi: 10.1186/1471-2164-9-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiura H, Komiyama J, Shirai M, Obata Y, Ogawa H, Kono T. DNA methylation imprints on the IG-DMR of the Dlk1-Gtl2 domain in mouse male germline. FEBS Lett. 2007;581:1255–1260. doi: 10.1016/j.febslet.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 56.Li JY, Lees-Murdock DJ, Xu GL, Walsh CP. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84:952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 57.Bonazzi VF, Irwin D, Hayward NK. Identification of candidate tumor suppressor genes inactivated by promoter methylation in melanoma. Genes Chromosomes Cancer. 2009;48:10–21. doi: 10.1002/gcc.20615. [DOI] [PubMed] [Google Scholar]
- 58.Bertrand S, Pinte S, Stankovic-Valentin N, Deltour-Balerdi S, Guerardel C, Begue A, Laudet V, Leprince D. Identification and developmental expression of the zebrafish orthologue of the tumor suppressor gene HIC1. Biochim Biophys Acta. 2004;1678:57–66. doi: 10.1016/j.bbaexp.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Deltour S, Pinte S, Guerardel C, Leprince D. Characterization of HRG22, a human homologue of the putative tumor suppressor gene HIC1. Biochem Biophys Res Commun. 2001;287:427–434. doi: 10.1006/bbrc.2001.5624. [DOI] [PubMed] [Google Scholar]
- 60.Guo L, Zhong D, Lau S, Liu X, Dong XY, Sun X, Yang VW, Vertino PM, Moreno CS, Varma V, Dong JT, Zhou W. Sox7 Is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol Cancer Res. 2008;6:1421–1430. doi: 10.1158/1541-7786.MCR-07-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takash W, Canizares J, Bonneaud N, Poulat F, Mattei MG, Jay P, Berta P. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Res. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dhir M, Montgomery EA, Glockner SC, Schuebel KE, Hooker CM, Herman JG, Baylin SB, Gearhart SL, Ahuja N. Epigenetic regulation of WNT signaling pathway genes in inflammatory bowel disease (IBD) associated neoplasia. J Gastrointest Surg. 2008;12:1745–1753. doi: 10.1007/s11605-008-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitfield JF. Calcium, calcium-sensing receptor and colon cancer. Cancer Lett. 2009;275:9–16. doi: 10.1016/j.canlet.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 64.Lutz T, Stoger R, Nieto A. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. FEBS Lett. 2006;580:5851–5857. doi: 10.1016/j.febslet.2006.09.049. [DOI] [PubMed] [Google Scholar]
- 65.Buhr N, Carapito C, Schaeffer C, Kieffer E, Van Dorsselaer A, Viville S. Nuclear proteome analysis of undifferentiated mouse embryonic stem and germ cells. Electrophoresis. 2008;29:2381–2390. doi: 10.1002/elps.200700738. [DOI] [PubMed] [Google Scholar]
- 66.Iwasaki H. Involvement of PRMT1 in hnRNPQ activation and internalization of insulin receptor. Biochem Biophys Res Commun. 2008;372:314–319. doi: 10.1016/j.bbrc.2008.05.051. [DOI] [PubMed] [Google Scholar]
- 67.Meyer R, Wolf SS, Obendorf M. PRMT2, a member of the protein arginine methyltransferase family, is a coactivator of the androgen receptor. J Steroid Biochem Mol Biol. 2007;107:1–14. doi: 10.1016/j.jsbmb.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 68.Li H, Xu LL, Masuda K, Raymundo E, McLeod DG, Dobi A, Srivastava S. A feedback loop between the androgen receptor and a NEDD4-binding protein, PMEPA1, in prostate cancer cells. J Biol Chem. 2008;283:28988–28995. doi: 10.1074/jbc.M710528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cao XR, Lill NL, Boase N, Shi PP, Croucher DR, Shan H, Qu J, Sweezer EM, Place T, Kirby PA, Daly RJ, Kumar S, Yang B. Nedd4 controls animal growth by regulating IGF-1 signaling. Sci Signal. 2008;1 doi: 10.1126/scisignal.1160940. ra5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Monami G, Emiliozzi V, Morrione A. Grb10/Nedd4-mediated multiubiquitination of the insulin-like growth factor receptor regulates receptor internalization. J Cell Physiol. 2008;216:426–437. doi: 10.1002/jcp.21405. [DOI] [PubMed] [Google Scholar]
- 71.Vecchione A, Marchese A, Henry P, Rotin D, Morrione A. The Grb10/Nedd4 complex regulates ligand-induced ubiquitination and stability of the insulin-like growth factor I receptor. Mol Cell Biol. 2003;23:3363–3372. doi: 10.1128/MCB.23.9.3363-3372.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peruzzi F, Prisco M, Morrione A, Valentinis B, Baserga R. Anti-apoptotic signaling of the insulin-like growth factor-I receptor through mitochondrial translocation of c-Raf and Nedd4. J Biol Chem. 2001;276:25990–25996. doi: 10.1074/jbc.M103188200. [DOI] [PubMed] [Google Scholar]
- 73.Ganesh L, Yoshimoto T, Moorthy NC, Akahata W, Boehm M, Nabel EG, Nabel GJ. Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis. Mol Cell Biol. 2006;26:3864–3874. doi: 10.1128/MCB.26.10.3864-3874.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kzhyshkowska J, Schutt H, Liss M, Kremmer E, Stauber R, Wolf H, Dobner T. Heterogeneous nuclear ribonucleoprotein E1B–AP5 is methylated in its Arg-Gly-Gly (RGG) box and interacts with human arginine methyltransferase HRMT1L1. Biochem J. 2001;358:305–314. doi: 10.1042/0264-6021:3580305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kuwasako K, Takahashi M, Tochio N, Abe C, Tsuda K, Inoue M, Terada T, Shirouzu M, Kobayashi N, Kigawa T, Taguchi S, Tanaka A, Hayashizaki Y, Guntert P, Muto Y, Yokoyama S. Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode. Biochemistry. 2008;47:6437–6450. doi: 10.1021/bi7024723. [DOI] [PubMed] [Google Scholar]
- 76.Izquierdo JM, Valcarcel J. Fas-activated serine/threonine kinase (FAST K) synergizes with TIA-1/TIAR proteins to regulate Fas alternative splicing. J Biol Chem. 2007;282:1539–1543. doi: 10.1074/jbc.C600198200. [DOI] [PubMed] [Google Scholar]
- 77.Schneider S, Kaufmann W, Buesen R, van Ravenzwaay B. Vinclozolin--the lack of a transgenerational effect after oral maternal exposure during organogenesis. Reprod Toxicol. 2008;25:352–360. doi: 10.1016/j.reprotox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 78.Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol Appl Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 79.Waterland RA, Travisano M, Tahiliani KG, Rached MT, Mirza S. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond) 2008;32:1373–1379. doi: 10.1038/ijo.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Salian S, Doshi T, Vanage G. Impairment in protein expression profile of testicular steroid receptor coregulators in male rat offspring perinatally exposed to Bisphenol A. Life Sci. 2009 doi: 10.1016/j.lfs.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 81.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2010;139:373–379. doi: 10.1530/REP-09-0340. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.