Abstract
N-Acylethanolamines (NAE) are fatty acid derivatives, some of which function as endocannabinoids in mammals. NAE metabolism involves common (phosphatidylethanolamines, PEs) and uncommon (N-acylphosphatidylethanolamines, NAPEs) membrane phospholipids. Here we have identified and quantified more than a hundred metabolites in the NAE/endocannabinoid pathway in mouse brain and heart tissues, including many previously unreported molecular species of NAPE. We found that brain tissue of mice lacking fatty acid amide hydrolase (FAAH −/−) had elevated PE and NAPE molecular species in addition to elevated NAEs suggesting that FAAH activity participates in the overall regulation of this pathway. This perturbation of the NAE pathway in brain was not observed in heart tissue of FAAH −/− mice indicating that metabolic regulation of the NAE pathway differs in these two organs and the metabolic enzymes that catabolize NAEs are most likely differentially distributed and / or regulated. Targeted lipidomics analysis, like that presented here, will continue to provide important insights into cellular lipid signaling networks.
Keywords: FAAH, N-Acylethanolamines, Lipid profiling, Lipid signaling, Lipid mediators, Mass spectrometry
Introduction
Endocannabinoids are endogenous lipid mediators that bind to and activate cannabinoid receptors in mammals to regulate a wide range of physiological and behavioral processes [De Petrocellis and Di Marzo, 2009; Di Marzo et al., 2000]. The first endocannabinoid discovered was anandamide [Devane et al., 1992], or N-arachidonylethanolamine (a type of N-acylethanolamine, 20:4 NAE). It was promptly revealed that anandamide and other members of its class are generated as part of a metabolic pathway involving the common membrane lipid phosphatidylethanolamine (PE) and an unusual N-acylated derivative of PE, N-acylphosphatidylethanolamine (NAPE; reviewed in [Schmid et al., 1996]). Although earlier research had established the metabolic relationship between these lipid classes in mammals and characterized the enzyme activities involved [Schmid et al., 1990], molecular identification of fatty acid amide hydrolase (FAAH) was a critical step, leading to the ability to perturb the NAE metabolic pathway in order to further elucidate the pathway's physiological functions [Ahn et al., 2008; Cravatt et al., 1996; Giang and Cravatt, 1997].
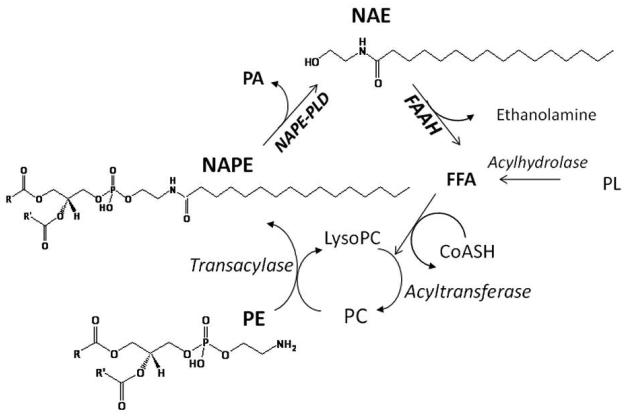
The overall pathway, termed the N-acylation-phosphodiesterase pathway, has more recently been designated the “endocannabinoid signaling pathway” to reflect its physiological function [Piomelli, 2003; Wilson and Nicoll, 2002]. However, this designation does not indicate the role of the pathway during response to ischemia; in fact, most of the early metabolic studies of this pathway were related to observations that both NAE and NAPE content were elevated markedly in ischemic brain and heart tissues [Epps et al., 1980; Epps et al., 1979; Natarajan et al., 1981; Natarajan et al., 1986]. The overall endocannabinoid pathway, the structures of representative principal metabolite classes, and key enzymatic steps are summarized in Fig 1.
Figure 1.
N-acylethanolamine (NAE) metabolism in mammals. NAEs are derived from the hydrolysis of a minor membrane lipid, N-acylphosphatidylethanolamine (NAPE), by a phospholipase D (PLD) and are hydrolyzed by a fatty acid amide hydrolase (FAAH) into ethanolamine and free fatty acid (FFA). NAPEs are synthesized from free fatty acid (FFA) via coordinated acyltransferase-transacylase reaction in which the sn-1-O-acyl moiety of phosphatidylcholine (PC) generated is transferred to the N-atom of phosphatidylethanolamine (PE).
Although the function of the endocannabinoid pathway relies mostly on the production and turnover of bioactive NAEs, the precursor pools of NAPEs can determine the prevailing mix of endogenous NAEs [Schmid et al., 1996]. Indeed, a concept called the “entourage effect” has been proposed to explain how abundant NAE molecular species that do not activate cannabinoid receptors can influence the activity of minor components, such as 20:4 NAE, that do activate receptors. To date, the many ethanolamine lipid components of the NAE-metabolic pathway have rarely been profiled comprehensively and concomitantly [Astarita et al., 2009; Astarita and Piomelli, 2009; Walker et al., 2005]. Here, we employ a targeted lipidomics approach to exhaustively identify and quantify the major and minor PE, NAPE, and NAE species in mouse heart and brain tissues and examine the impact of the loss of FAAH in FAAH −/− mice [Cravatt et al., 2001] on the metabolites of the endocannabinoid pathway.
In the current work, O-diacyl and O-alk(en)yl,O-acyl PE and NAPE molecular species were identified directly in lipid extracts by electrospray ionization mass spectrometry (ESI-MS) using neutral loss scans and quantifying in relation to internal standards. NAEs were identified by GC-MS after chromatographic clean-up and quantified by isotope dilution MS, using procedures similar to those developed elsewhere [Fontana et al., 1995]. Our results detail the metabolites of this important lipid signaling pathway. Moreover, the results suggest that ablation of FAAH in brain tissue results in dysregulation throughout the pathway, not just in anandamide content as previously reported, while similar effects were not apparent in heart tissue [Clement et al., 2003; Cravatt et al., 2001]. Comprehensive profiling approaches like those utilized here enable understanding of the metabolic regulation of important signaling pathways.
Experimental Procedures
Animals and Tissue Extracts
FAAH −/− mice were kindly provided by Dr. Benjamin F. Cravatt, The Scripps Research Institute, La Jolla, CA, and were genotyped as described previously [Cravatt et al., 2001]. Wild type littermate control mice were used for comparisons. Animals were kept in a temperature-controlled room with 12-h light/dark cycles and free access to food and water. Animal use and care was reviewed and approved by the Institutional Animal Care and Use Committee. For each treatment, after euthanasia, brains and heart tissue from five animals were removed and flash frozen in liquid nitrogen.
Lipid Extractions
Tissues were homogenized with hot 2-propanol (70°C) with glass beads by beating for five 30 s bursts. Tissue homogenate in 2-propanol was combined with chloroform (0.45 g FW : 2 mL 2-propanol : 1 mL chloroform) and incubated on ice for 30 min before extracting overnight at 4°C. Monophasic lipid extracts were partitioned with 1 part chloroform and 2 parts 1 M KCl in water. The lower organic phase was collected and washed three additional times with 2 parts 1 M KCl in water. Extracts were stored under nitrogen at −80°C until further analysis.
NAE Quantification
One-third of the total extract was fractionated by normal-phase HPLC (HP1100 Series; Agilent, Wilmington DE) using an Alltech (Deerfield, IL) semi-preparative silica column. NAEs were derivatized to TMS-ethers and analyzed by GCMS (Model 6890 GC, coupled with a 5973 Mass Selective Detector; Agilent, Wilmington DE), as described previously [Venables et al., 2005], with identification via selective ion monitoring and quantification in relation to deuterated internal standards (16:0 NAE and 20:4 NAE). NAE content of each sample was normalized to the sample fresh weight (FW). Statistical significance (P-value < 0.05) was determined using an unpaired Student's t-test.
Phospholipid analysis and quantification
An automated electrospray ionization (ESI)-tandem mass spectrometry approach was used, and data acquisition was carried out as described previously [Devaiah et al., 2006] with modifications. Dried mouse brain and heart lipid extracts were dissolved in 1 ml chloroform/methanol (9:1). An aliquot of 2 to 30 μl of extract, equivalent to 0.5-2 mg tissue FW, was used. Precise amounts of internal standards, obtained and quantified as previously described [Welti et al., 2002], were added in the following quantities (with some small variation in amounts in different batches of internal standards): 0.66 nmoles of di14:0 PC, di24:1 PC, 13:0 lysoPC, and19:0 lysoPC, 0.36 nmoles of di14:0 PE, di24:1 PE, 14:0 lysoPE, 18:0 lysoPE, di14:0 PA, and di20:0 (phytanoyl) PA, 0.24 nmol di14:0 PS and di20:0 (phytanoyl) PS, 0.20 nmoles of 16:0-18:0 PI, and 0.16 nmoles of di18:0 PI. Solvent was added to the sample/standard mixture so that the final ratio of chloroform/methanol/300 mM ammonium acetate in water was approximately 300:665:35, and the final volume was 1.3 ml.
Mass spectra were acquired on a triple quadrupole MS/MS system (API 4000, Applied Biosystems, Foster City, CA). Unfractionated samples were introduced by continuous infusion into the Turbo V ESI source at 30 μl/min using an autosampler (LC Mini PAL, CTC Analytics AG, Zwingen, Switzerland) fitted with the required injection loop for the acquisition time. Sequential precursor and neutral loss scans of the extracts produce a series of spectra with each spectrum revealing a set of lipid species containing a common head group fragment. Lipid species were detected in positive mode with the following scans: PC and lysoPC, [M + H]+ ions with Precursors of 184.1 (Pre 184.1); PE and lysoPE, [M + H]+ ions with Neutral Loss of 141.0 (NL 141.0); PI, [M + NH4]+ ions with NL 277.0; PS, [M + H]+ ions with NL 185.0; and PA, [M + NH4]+ ions with NL 115.0. The ion spray voltage was set at +5.5 kV, the source temperature at 100ºC, the curtain gas at 20 (arbitrary units), and the ion source gases at 45 (arbitrary units); the interface heater was on. Declustering potentials were +100 V. Entrance potentials were +15 V for PE and +14 V for PC, PI, PA, and PS. Exit potentials were +11 V for PE and +14 V for PC, PI, PA, and PS. The collision gas, nitrogen, was set at 2 (arbitrary units). The collision energies were +28 V for PE, +40 V for PC, and +25 V for PI, PS and PA. The mass analyzers were adjusted to a resolution of 0.7 u full width at half height. For each spectrum, 8 to 80 continuum scans were collected in multiple channel analyzer (MCA) mode at a scan speed of 50 or 100 u per s.
The data were smoothed, the background of each spectrum was subtracted, and the peaks were centroided and integrated using a custom script and Applied Biosystems Analyst software. Peaks corresponding to the target lipids in these spectra were identified, the data were corrected for isotopic overlap, and molar amounts were calculated in comparison to the internal standards in the same lipid class. A sample containing internal standard mixture only was also run through the same series of scans to correct for chemical or instrumental noise. The molar amounts of each lipid metabolite detected in the “internal standards-only” spectra were subtracted from the molar amounts of each metabolite calculated from the experimental mouse sample spectra. The “internal standards-only” spectra were used to correct the data from the following 9 samples run on the instrument. Finally, the data were adjusted to account for the fraction of sample analyzed and normalized to the sample fresh weight. Data were reported as nmol or μmol of each detected lipid metabolite/g tissue FW. Measured metabolite species include both diacyl and alk(en)yl,acyl glycerophospholipids. Although these two groups may exhibit some differences in mass spectral response, values for the alk(en)yl,acyl (as well as diacyl) species were quantified in relation to the diacyl/monoacyl phospholipid internal standards, and no response correction factors were employed for the alk(en)yl,acyl species.
NAPE standard acquisition and synthesis
C17:0 fatty acid chloride was purchased from Nu-Check Prep (Elysian, MN); 1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine (di16:0 PE) and 1,2-dioleoyl-sn-glycero-3-phospho(N-arachidonoyl)ethanolamine (N-20:4 di18:1 PE) were from Avanti Polar Lipids (Alabaster, AL); 1-2-dipalmitoyl-sn-glycero-3-phospho(N-palmitoyl)ethanolamine (N-16:0 di16:0 PE) was from Sigma-Aldrich (St. Louis, MO); and TLC plates (10×20 cm, scored, HPTLC-GHL silica gel, 150 μm) were from Analtech (Newark, DE). Other organic solvents and reagents were of the highest purity commercially available.
The NAPE molecular species N-17:0 di16:0 PE was prepared as described previously [Astarita et al., 2008], with modifications. Briefly, C17:0 fatty acid chloride was reacted with a two-fold molar excess of di16:0 PE. The reaction was conducted in dichloromethane at 25°C for 2 h, using triethylamine as a catalyst. The products were washed with water and fractionated by silica gel TLC (chloroform/methanol/ammonia, 65:35:4) to separate the desired product NAPE species from unreacted PE and fatty acid chloride. The calculated retention factors for NAPE and PE were 0.60 and 0.40, respectively. Both the purchased and synthesized NAPE species were quantified by phosphate assay [Ames, 1966].
NAPE analysis and quantification
As noted above in the phospholipid analysis section, dried mouse brain and heart lipid extracts were dissolved in chloroform/methanol (9:1). The same sample solutions were used for both phospholipid and NAPE analysis. The synthesized N-17:0 di16:0 PE was employed as an internal standard to quantify the NAPE species in the sample extracts. N-17:0 di16:0 PE (1.1 nmole) was added to an aliquot of extract equivalent to 6-25 mg tissue FW. Solvent was added to the sample/standard mixture so that the final ratio of chloroform/methanol/300 mM ammonium acetate in water was 300:665:35, and the final volume was 1.3 ml.
Mass spectra were acquired on a triple quadrupole MS/MS system (API 4000 QTrap, Applied Biosystems, Foster City, CA). Unfractionated samples were introduced by continuous infusion into the Turbo V ESI source at 30 μl/min using an autosampler (LC Mini PAL, CTC Analytics AG, Zwingen, Switzerland) fitted with the required injection loop for the acquisition time. Sequential neutral loss scans produced a series of spectra, with each spectrum revealing a set of lipid species containing a common ammoniated N-fatty amide head group fragment corresponding to each common fatty acid. Lipids were detected in positive ion mode as [M + NH4]+ ions with the following scans: N-16:0 species with Neutral Loss of 396.3 (NL 396.3), N-18:2 species with NL 420.3, N-18:1 species with NL 422.3, N-18:0 species with NL 424.3, N-20:4 species with NL 444.3, N-22:6 species with NL 468.3, and N-22:5 species with NL 470.3. A scan for N-17:0 species (NL 410.3) was included in order to detect the internal standard. The declustering potential was set at +60 V, the entrance potential at +8 V, and the exit potential at +15 V. The ion spray voltage was set at +5.5 kV. The collision gas, nitrogen, was set at 2 (arbitrary units), and the collision energy was +45 V. The source temperature, curtain gas, ion source gases, interface heater, and mass analyzers were adjusted as for phospholipid analysis. For each spectrum, 100 cumulative scans were collected in multiple channel analyzer (MCA) mode at a scan speed of 50 u per s.
The data were smoothed, the background of each spectrum was subtracted, and the peaks were centroided and integrated using a custom script and Applied Biosystems Analyst software. Peaks corresponding to the target lipids in each N-acyl class (each spectrum) were identified, the data were corrected for isotopic overlap due to the diacylglycerol portion of NAPE, and molar amounts were quantified relative to the N-17:0 di16:0 PE internal standard. A sample containing only the N-17:0 di16:0 PE standard was subjected to the same series of scans. The molar amounts of each lipid metabolite detected in the “standard-only” spectra were subtracted from the molar amounts of each metabolite calculated from the experimental mouse sample spectra to correct for chemical or instrumental noise. The “standard-only” spectra were used to correct the data from the following 9 samples run on the instrument. Finally, the data were corrected for isotopic overlap between head groups (NL fragments), adjusted to account for the fraction of sample analyzed, and normalized to the sample fresh weight. Due to the presence in the NL 444.3 scan of peaks with m/z inconsistent with NAPEs in the m/z range of N-20:4-e40:7, N-20:4-e40:6, N-20:4-40:8, and N-20:4-40:7 NAPEs, these compounds were not measured. The interfering compounds were not present in the spectra of mouse heart tissue. Data were reported as mass spectral signal normalized to N-17:0 di16:0 PE/g tissue FW; the amount of signal produced by 1 pmol of N-17:0 di16:0 PE is 1. Data were evaluated for possible outliers using the Q-test (Shoemaker et al., 1974) on NAPE lipid class totals; one FAAH (−/−) mouse brain replicate (out of 5) was determined to be an outlier and was removed from calculations. Measured species include both N-acylated diacyl and N-acylated alk(en)yl,acyl glycerophospholipids. As described for the phospholipid species, no response correction factors were employed for the alk(en)yl,acyl species.
Validation of NAPE quantitation method
Three experiments were performed to validate the above sequential NL scanning method for NAPE quantitation,. To assess the linearity of the response, varied amounts (10-1000 pmol) of the purchased N-16:0 di16:0 PE and N-20:4 di18:1 PE were analyzed in the presence of 200 pmol of the synthesized N-17:0 di16:0 PE internal standard. An abbreviated mass spectral analysis with scans for N-16:0, N-20:4, and N-17:0 NAPEs was performed on the API 4000 mass spectrometer. Responses of the N-16:0 di16:0 PE and N-20:4 di18:1 PE signals relative to the N-17:0 standard were measured.
The ability of ESI-MS/MS NAPE profiling to detect small changes in NAPE molecular species levels was also evaluated. Again, purchased N-16:0 di16:0 PE and N-20:4 di18:1 PE were utilized. Stepwise-increasing known amounts of these two species (4, 8, 20, and 60 pmol for the N-16:0 compound; 5, 10, 25, and 75 pmol for the N-20:4 compound) were spiked into samples containing roughly a 10 mg FW equivalent of a mixture of WT and FAAH −/− mouse brain extract. Each sample also contained 100 pmol of the synthesized N-17:0 di16:0 PE internal standard. An abbreviated mass spectral analysis with scans for N-16:0, N-20:4, and N-17:0 NAPEs was performed on the API 4000 QTrap mass spectrometer. Responses of the targeted N-16:0 and N-20:4 compounds relative to the N-17:0 standard were calculated.
The variation in mass spectral response due to variation in the amount of biological sample analyzed was also examined. Samples were prepared with varying amounts of a mixture of WT and FAAH −/− mouse brain extract, approximately equivalent to 3.5, 7, 14, and 28 g FW, and 200 pmol of the synthesized N-17:0 di16:0 PE internal standard. The full mass spectral analysis, with scans for N-16:0, N-18:2, N-18:1, N-18:0, N-20:4, N-22:6, N-22:5, and N-17:0, was performed on the API 4000 QTrap mass spectrometer. Data were processed as described above and reported as mass spectral signal of each targeted NAPE species relative to the N-17:0 di16:0 PE internal standard.
Results
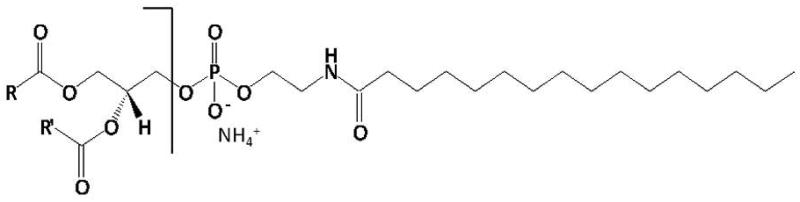
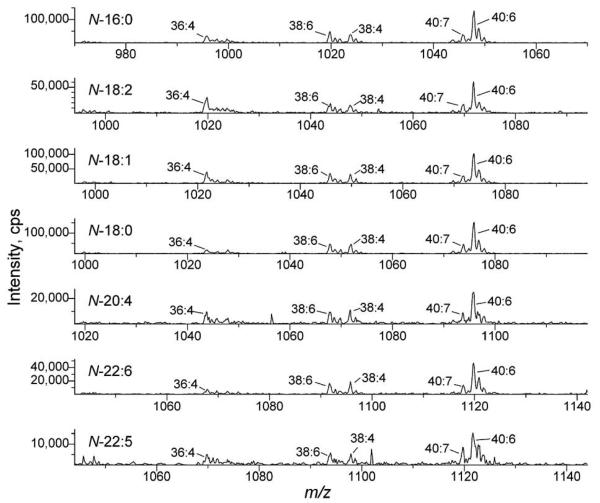
In previous work, methods for analysis of NAEs by GC-MS [Venables et al., 2005] and phospholipids by direct infusion electrospray ionization mass spectrometry [Welti et al., 2007] have been developed. Here, in order to comprehensively analyze lipids in the NAE biosynthetic pathway, we developed a method for direct infusion electrospray ionization mass spectrometry analysis of NAPEs. In solvent containing ammonium acetate, NAPEs form positively charged ammonium adducts [M + NH4]+ by electrospray ionization. By collision induced dissociation, the ammonium adducts of NAPEs can be fragmented into a neutral head group fragment and a charged diradylglycerol (i.e. diacylglycerol or alk(en)yl,acylglycerol) fragment (Fig. 2). Scanning for neutral loss of an ammoniated N-fatty amide head group in a triple quadrupole mass spectrometer produces a spectrum revealing the NAPE molecular species that contain the fragment, in other words, a spectrum of the NAPE class (Fig. 3). Sequential scanning for neutral loss of ammoniated N-fatty amide head groups corresponding to each common fatty acid provides a method to detect the molecular species of NAPE in all N-acyl classes (Fig. 3).
Figure 2.
Collision induced fragmentation of the ammonium adduct of N-16:0 PE.
Figure 3.
Neutral loss scans targeting analysis of NAPE species with varying N-acyl groups. From top to bottom panels, the scans target N-16:0 PE (NL 396.3), N-18:2 PE (NL 420.3), N-18:1 PE (NL 422.3), N-18:0 PE (NL 424.3), N-20:4 PE (NL 444.3), N-22:6 PE (NL 468.3), and N-22:5 PE (NL 470.3) in mouse heart extract corresponding to 6-25 mg FW. The labels indicate the diacyl component (total acyl carbons: total carbon-carbon double bonds) of each detected NAPE.
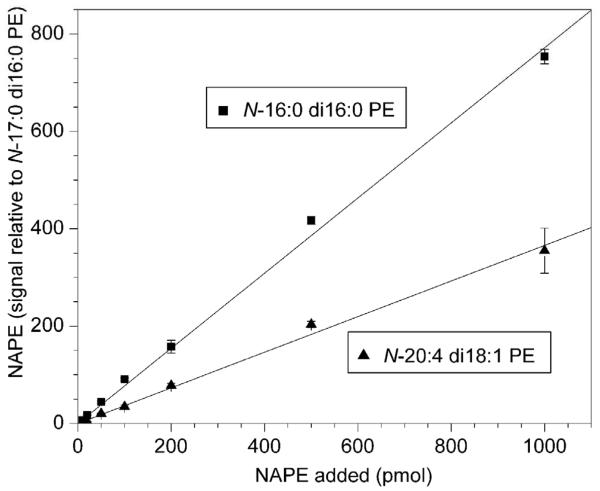
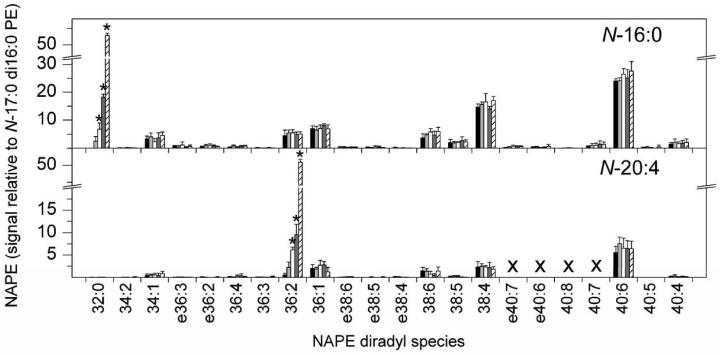
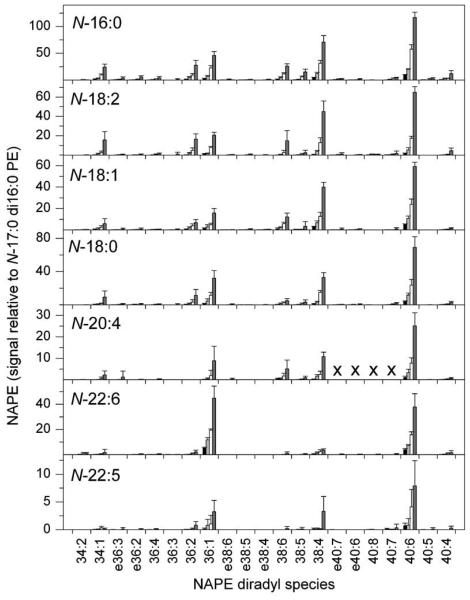
To determine whether neutral loss scanning provides an appropriate method for quantification, the linearity of the mass spectral response of the neutral loss scans was investigated (Fig. 4). The signals for two pure NAPEs were determined in relation to the signal from a known quantity of an internal standard, N-17:0 di16:0 PE. The response was determined to be linear, but both N-16:0 di16:0 PE and N-20:4 di18:1PE produced somewhat less signal per mole than N-17:0 di16:0 PE. This suggests that different NAPE molecular species vary somewhat in their ability to ionize and/or to undergo fragmentation under particular fixed mass spectral conditions. On the other hand, a spike-in experiment in which pure NAPE species were added to a biological mixture of lipids showed that their mass spectral signals, normalized to the N-17:0 di16:0 PE internal standard, were approximately proportional to the amount of each NAPE species added (Fig. 5). Finally, varying amounts of the biological sample while holding the level of internal standard constant resulted in normalized mass spectral responses proportional to the amount of sample added (Fig. 6). Taken together, the data in Fig. 3 through 6 show that, although response factors for individual NAPE molecular species vary (Fig. 3), the NAPE analysis provides a reliable means to compare levels of NAPE species among samples.
Figure 4.
Linearity of the mass spectral response. Varying amounts of N-16:0 di16:0 PE and N-20:4 di18:1 PE were combined with 200 pmol of internal standard N-17:0 di16:0 PE. Neutral loss signals for each NAPE molecular species are presented in relation to the signal from the internal standard; the amount of signal produced by 1 pmol of N-17:0 di16:0 PE is 1. N = 5 ± SD.
Figure 5.
The ability of ESI-MS/MS lipid profiling to detect small changes in NAPE molecular species. The lipid species in roughly 10 mg FW of mouse brain were analyzed and, to demonstrate the ability of the lipid profiling methodology to detect changes, small, known amounts of specific lipid species were added to the extract. One hundred pmol of internal standard N-17:0 di16:0 PE was added to each sample to provide quantitation. Neutral loss scans were performed as described in Experimental Procedures. Signals for each NAPE molecular species are presented in relation to the signal of internal standard N-17:0 di16:0 PE; the amount of signal produced by 1 pmol of N-17:0 di16:0 PE is 1. The additions of the lipid species indicated by the asterisks varied over a 15-fold range (with bars for each species representing no addition, 1x addition, 2x addition, 5x addition, and 15x addition). Specifically, the black bars (first series) represent the composition of the extract. The light gray bars (second series) represent extract with 4 pmol N-16:0 di16:0(32:0) PE and 5 pmol N-20:4 di18:1(36:2) PE added. The white bars (third series) represent extract with 8 pmol N-16:0 di16:0(32:0) PE and 10 pmol N-20:4 di18:1(36:2) PE added. The dark gray bars (fourth series) represent extract with 20 pmol N-16:0 di16:0(32:0) PE and 25 pmol N-20:4 di18:1(36:2) PE added. The hatched bars (fifth series) represent extract with 60 pmol N-16:0 di16:0(32:0) PE and 75 pmol N-20:4 di18:1(36:2) PE added. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. “X” indicates molecular species that were not determined (see Experimental Procedures). N = 5 ± SD.
Figure 6.
Variation in mass spectral response with variation in amount of biological sample. Varying amounts of mouse brain extract were combined with 200 pmol of internal standard N-17:0 di16:0 PE. The amounts of mouse brain extract corresponded to approximately 3.5 mg FW (black bars, first series), 7 mg FW (light gray bars, second series), 14 mg FW (white bars, third series), and 28 mg FW (dark gray bars, fourth series). Neutral loss signals for each NAPE molecular species are presented in relation to the signal from the internal standard; the amount of signal produced by 1 pmol of N-17:0 di16:0 PE is 1. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. “X” indicates molecular species that were not determined (see Experimental Procedures). N = 5 ± SD.
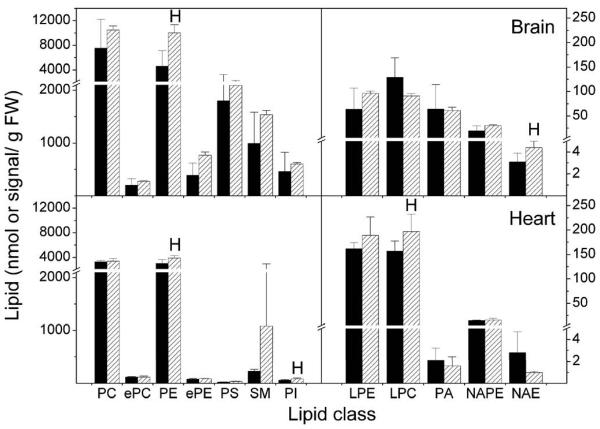
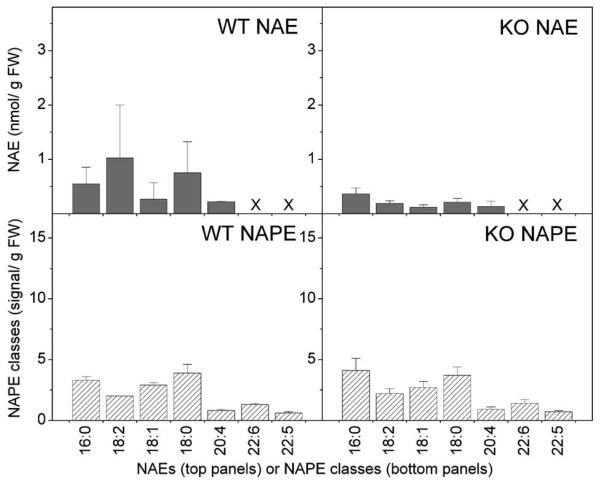
Endocannabinoid metabolism is influenced by both phospholipase D (PLD)-mediated hydrolysis of NAPE and FAAH-mediated breakdown of NAEs (Fig. 1). We examined the effect of FAAH disruption on the content and composition of NAE and various phospholipids, including NAPE and PE, in brain and heart tissue of wild type and FAAH −/− mice. Total levels are summed from the individual molecular species. The data show that lipid content was generally higher in brain than in the heart tissue (Fig. 7). Compared to brain, heart phospholipids had little PS or PA. Both total NAE and PE content were significantly elevated in brain tissue of FAAH −/− mice compared to the wild type controls. On the other hand, in heart tissue there was a significant increase only in PE, PI, and lysoPC content, while NAE content was the same between FAAH −/− and control mice.
Figure 7.
Endogenous phospholipids and ethanolamine-containing NAE metabolites. Total lipid extracts of brain and heart tissue of wild type (WT, black bars) and FAAH −/− (KO, hatched bars) mice were analyzed for phospholipid, including PE and NAPE and NAE content as described in Experimental Procedures. Right panels use right-hand scale and left panels use left-hand scale. NAPEs are expressed as normalized signal/g FW; all others are expressed as nmol/g FW. N = 4 or 5 ± SD. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT. “H” indicates that the KO level is significantly higher than the wild type value.
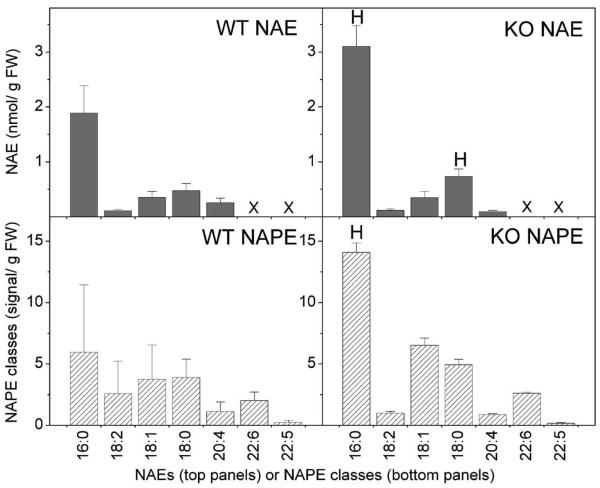
To examine the relationship between NAE composition and the NAPE precursor pool, NAPEs were quantified according to N-acyl head group and these NAPE classes were compared to the principal NAE types. Absolute quantities of NAPE and NAE and their composition were quite different between brain and heart tissue of mice (Fig. 8 and 9). The significantly higher level of NAE in the brain tissue of FAAH −/− mice, as compared to wild type mice, was attributable mostly to 16:0 NAE (Fig. 8); the level of N-16:0 PE was also significantly higher in the brain tissue of FAAH −/− mice, as compared to wild type mice. The concentration of 18:0 NAE species was higher in brain tissue of FAAH −/− mice as well. In contrast to predominant 16:0 NAEs and N-16:0 PEs in brain, heart tissue did not reveal a prevalent NAPE class or NAE type. Murine hearts showed no differences between FAAH −/− and wild type animals in NAE types or NAPE classes. This suggests that FAAH disruption has a considerably greater effect on steady-state levels of endocannabinoid pathway metabolites in brain tissue than in heart tissue. Major differences in steady-state anandamide levels in brain extracts between FAAH −/− mice and wildtype littermates were not evident in our studies, which was inconsistent with previous reports quantifying 15-fold higher anandamide levels in brain tissues of FAAH−/− mice compared to FAAH +/+ mice (Clement et al., 2003 Cravatt and Lichtman, 2004). Differences may be due to organ preparation (euthanasia rather than decapitation) and/or tissue extraction procedures since anandamide levels seem to be particularly sensitive to preparation methods (Muccioloi and Stella, 2008). Nonetheless, the principal saturated NAE types (NAE18:0 and NAE16:0) quantified in our samples showed a significant elevation in brain tissues of knockout mice as expected (Fig. 8).
Figure 8.
Brain NAE species (dark gray bars) and NAPE classes (hatched bars) in wild type (WT) and FAAH −/− (KO) mice. N = 4 or 5 ± SD. “X” indicates species that were not determined. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT. “H” indicates that the KO level is significantly higher than the wild type value.
Figure 9.
Heart NAE species (dark gray bars) and NAPE classes (hatched bars) in wild type (WT) and FAAH −/− (KO) mice. N = 4 or 5 ± SD. “X” indicates species that were not determined. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT.
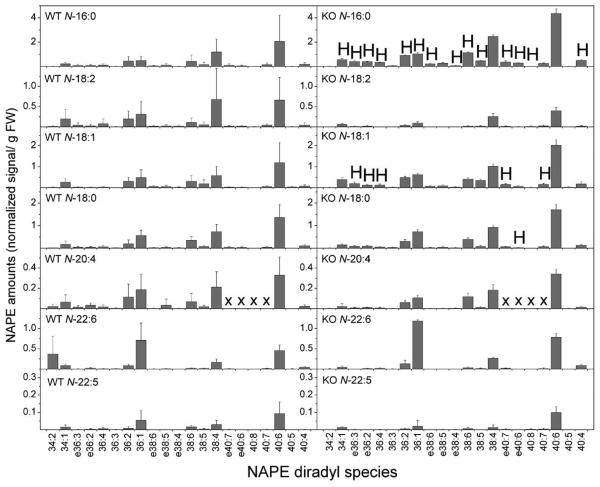
Although useful for visualizing potential NAE precursors, grouping of NAPE species by common N-acyl chains (Fig. 8 and 9) does not reveal the remarkable complexity of NAPEs. Therefore, a detailed molecular species profile of NAPE was generated for brain and heart tissue of wild type and FAAH −/− mice (Figs. 10 and 11). In brain tissue of mice, the most abundant molecular species with 16:0 at the N-position contained PE 36:2, 36:1, 38:6, 38:4, and 40:6, where the species are indicated by total acyl carbons:total carbon-carbon double bonds in the combined acyl chains in the 1 and 2 positions on the glycerol (Fig. 10). Many N-16:0-containing species were significantly elevated in knockout mice compared to wild type, suggesting that the elevation of the N-16:0 PE molecular species in FAAH −/− mice affected those molecular species already prevalent in wild type mice. N-18:2-containing NAPE molecular species were much less abundant overall; only 5 molecular species were quantified at more than 0.1 nmol/g FW and none of these were elevated in FAAH −/− brain tissue (Fig. 10). N-18:1 and N-18:0 molecular species were somewhat more abundant and distributed among diradyl species in a manner similar to N-16:0 PEs; several molecular species were significantly higher in the FAAH −/− knockout tissues (Fig. 10). The anandamide-containing (N-20:4) NAPE pool was relatively minor in terms of overall abundance, and this subgroup showed no differences between wild type and FAAH −/− in terms of quantity or composition. Similarly the N-22:6 PE molecular species were not very abundant, and the N-22:5 PE class was very minor in brain tissue, with only a few molecular species identified in this subgroup. There were no significant differences in N-22:6 and N-22:5 PE species between wild type and FAAH −/− mice.
Figure 10.
Detailed profile of NAPE molecular species from brain tissue of wild type (WT) and FAAH −/− (KO) mice. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. “X” indicates molecular species that were not determined (see Experimental Procedures). N = 4 or 5 ± SD. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT. “H” indicates that the KO level is significantly higher than the wild type value.
Figure 11.
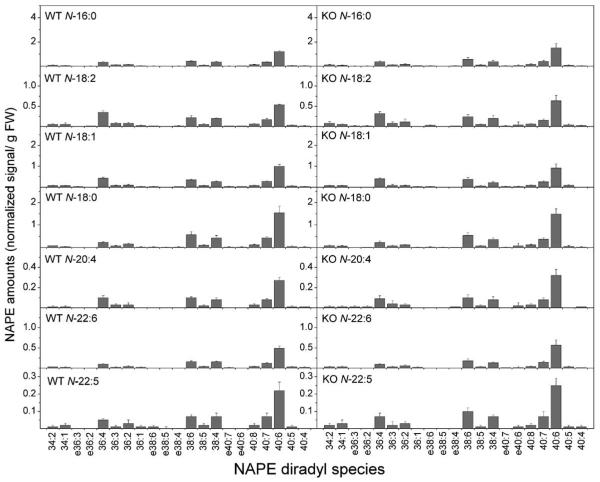
Detailed profile of NAPE molecular species from heart tissue of wild type (WT) and FAAH −/− (KO) mice. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. N = 4 or 5 ± SD. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT.
Similar to brain tissue, murine heart tissue showed complexity in the molecular species composition of NAPE (Fig. 11). Some differences were immediately evident between heart and brain NAPE molecular species. First, the content of all NAPE molecular species in heart tissue was two- to four-fold less than in the brain tissue, and, as suggested from the NAPE class data in Fig. 9, there was no difference in molecular species content of any NAPE type in heart tissue of FAAH −/− mice compared to wild type (Fig. 11).
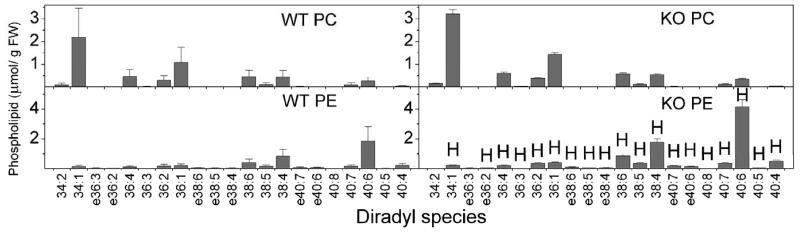
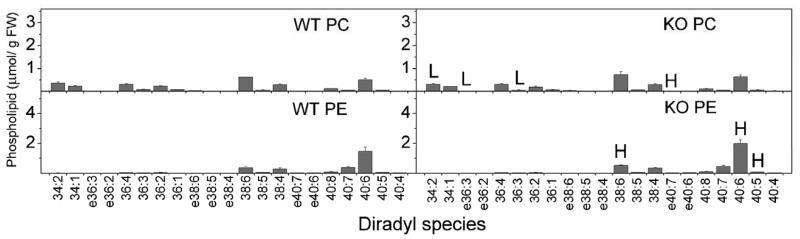
Comparisons of PC and PE compositions of brain and heart (Fig. 12 and 13) show that the diradyl compositions of these two major phospholipid classes are quite different from each other. In brain, 40:6 PE was the most abundant diradyl species for N-acylation with 36:1, 38:6, and 38:4 species also being prominent (Fig. 10). In heart, 40:6 and 38:6 were most prominent with 38:4 and 36:4 also abundant (Fig. 11). These NAPE compositions resemble the PE compositions (Fig. 12 and 13), supporting the notion that NAPE is derived from PE (Fig. 1).
Figure 12.
PC and PE species from brain tissue of wild type (WT) and FAAH −/− (KO) mice. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. N = 4 or 5 ± SD. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT. “H” indicates that the KO level is significantly higher than the wild type value.
Figure 13.
PC and PE species from heart tissue of wild type (WT) and FAAH −/− (KO) mice. Designations for diradyl molecular species are: numbers only represent diacyl species as total acyl carbons: total carbon-carbon double bonds; e before a number indicates an ether-linked (alk(en)yl,acyl) species as total alk(en)yl + acyl carbons: total carbon-carbon double bonds, including any vinyl ether bonds present. N = 4 or 5 ± SD. Significance (P < 0.05) was determined by unpaired Student's t-test for KO vs WT. “H” indicates that the KO level is significantly higher and “L” indicates that the KO level is significantly lower than the wild type value.
Discussion
Considerable prior evidence has established the metabolic relationship of PE, its N-acylation to form NAPE, and the hydrolysis of NAPE to form the bioactive NAEs (Fig. 1). Several NAE types have been identified as endogenous ligands for the cannabinoid receptors while other types act on other cellular targets [Hansen et al., 2000; Schmid and Berdyshev, 2002; Schmid et al., 2002]. The hydrolysis of the NAEs (endocannabinoids and others) by FAAH terminates their lipid mediator functions [Ahn et al., 2008; McKinney and Cravatt, 2005]. Given the broad range of activities of the NAEs, the enzymes of this pathway represent important therapeutic targets. Often less appreciated are the large numbers of metabolites that are generated in this overall N-acylation-phosphodiesterase-FAAH pathway, many of which are cellular lipids with other functions and only some of which are identified and quantified for their lipid mediator functions.
Targeted lipidomics approaches permit comprehensive analysis of metabolites involved in a specific lipid metabolic pathway. Recently, sensitive and high-throughput analytical tools have been established to unravel the biological significance of individual molecules of the endocannabinoid system in the context of the interconnected network of their precursors and derivatives [Astarita and Piomelli, 2009; Astarita et al., 2009; Bisogno et al., 2009]. However, these studies were limited mostly to endocannabinoids that occur in minor quantities relative to the other non-cannabimimetic members of the large N-acylethanolamide family. Since we do not fully understand the nature of selective biosynthesis or degradation of a specific NAE type or its precursors (Fig. 1), especially under the influence of genetic perturbations of this regulatory pathway, we used a targeted lipidomics approach to elucidate changes in the profiles of PE, NAPE and NAE in brain and heart tissue of wild type and FAAH −/− mice.
Comparison of NAE metabolites and major and minor phospholipids between brain and heart tissue of mice revealed some similarities, but also several tissue-specific differences. The most abundant metabolite of the NAE regulatory pathway in brain and heart tissue was PE (Fig. 1 and 7). It appears that the relative abundance of specific PE molecular species in the PE pool determines the molecular composition of NAPE rather than a preferential substrate selectivity of N-acyltransferase (Fig. 10 vs. 12 and 11 vs. 13). Furthermore, brain tissue of FAAH −/− mice showed accumulation of already abundant PE and NAPE molecular species rather than synthesis and accumulation of new or less abundant molecular species or remodeling of the amide-linked fatty acids of NAPE. These data suggest that lack of FAAH in brain tissue results in accumulation of NAE species, which may in turn affect the content but not the composition of the NAE precursor pool. Although total PE was significantly higher in the heart tissue of FAAH −/− mice when compared with wild type (Fig. 7), fewer PE molecular species contributed significantly to the overall increase in PE content in heart than in brain (Fig. 12 and 13). The lesser FAAH-specific effect on NAE and its metabolites in heart tissue (Figs. 7-13) was not surprising because FAAH activity was reported to be negligible in heart tissue of mice [Ueda et al., 2000].
Although the total NAE content was similar between the two tissue types, it is interesting to note that total PE and NAPE levels are lower in heart tissue (Fig. 7). Furthermore, N-16:0 PE and 16:0 NAE were the most predominant species in brain tissue while heart tissue did not show preference for any specific type (Fig. 8 and 9). It is possible that N-acylethanolamine-hydrolyzing acid amidase, a second NAE-degrading amidase expressed in the heart and brain [Tsuboi et al. 2005], may exhibit differential expression and activity in these tissues, potentially leading to some of the tissue-specific differences seen in levels of NAE species in FAAH −/− mice. The higher levels and distinctive composition of NAPE may suggest additional brain NAPE roles, such as maintenance of physical properties of membrane domains [Brites et al., 2004] and influence on signaling processes in the brain [Terova et al., 2005], in addition to serving as the precursor for NAE synthesis.
Comparative analysis of major and minor phospholipids revealed higher levels of most phospholipids in brain tissue compared to heart, as expected, perhaps due to overall higher lipid content in the brain (Fig. 7). However, in FAAH −/− mice, brain tissue showed accumulation only in the PE and NAE contents, as discussed earlier. On the other hand, FAAH −/− heart tissue showed an increase not only in PE but also in phosphatidylinositol (PI) and lysoPC. In fact lysoPC content was higher in wild type heart tissue than in brain (Fig. 7). LysoPC is a byproduct of the trans-acylase reaction and might serve, together with other 1-lyso-phospholipids, as a precursor for the formation of 2-arachidonoylglycerol (2-AG; Di Marzo et al., 1996). Additionally, hydrolysis of PI by a PI-specific phospholipase A1 can also generate 2-AG (Suguira et al., 1995). Even though elimination of FAAH activity did not directly affect the NAE metabolites in heart tissue of mice, it may have influenced alternate endocannabinoid metabolic pathways by affecting the phospholipid composition.
Collectively, our targeted lipidomics results imply a metabolic N-acylation-phosphodiesterase pathway for acylethanolamides that is dominated by major cellular lipid constituents, but in which minor metabolites also play roles. Perturbation of FAAH may influence metabolite pools in this entire pathway, beyond simply the direct substrates for this enzyme; however, this influence by FAAH on NAPE and PE molecular species content is tissue-specific. Metabolic profiling tools like these developed here, applied to different physiological or pathological situations that have been attributed to endocannabinoid function, may help to reveal important new tissue-specific metabolic targets for therapeutic intervention at points of control not yet discovered.
Acknowledgements
We would like to thank Mary R. Roth for expert technical assistance. This work was supported by a seed grant from the University of North Texas and by a grant from the US Dept of Energy, Office of Basic Energy Sciences (DE-FG02-05ER15647). This study was supported in part by NIH grants MD001633 from NCMHD (R.S.D.), EY014227, AG010485, AG022550 and AG027956 (P.K.) as well as by The Garvey Texas Foundation and the Felix and Carmen Sabates Missouri Endowed Chair in Vision Research (P.K.). Instrument acquisition and method development at the Kansas Lipidomics Research Center was supported by NSF grants MCB 0455318 and DBI 0521587, K-INBRE (NIH Grant P20 RR16475 from the INBRE program of the National Center for Research Resources), and NSF EPSCoR grant EPS-0236913 with matching support from the State of Kansas through Kansas Technology Enterprise Corporation and Kansas State University.
Abbreviations
- AG
arachidonoylglycerol
- ePC
alk(en)yl, acyl glycerophosphocholine
- ePE
alk(en)yl, acyl glycerophosphoethanolamine
- ESI
electrospray ionization
- FAAH
fatty acid amide hydrolase
- FFA
free fatty acid
- FW
fresh weight
- KO
knockout
- LPC
lysophosphatidylcholine
- LPE
lysophosphatidylethanolamine
- NAE
N-acylethanolamine
- NAPE
N-acylphosphatidylethanolamine
- NL
neutral loss
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PI
phosphatidylinositol
- PLD
phospholipase D
- Pre
precursor
- PS
phosphatidylserine
- SM
sphingomyelin
- WT
wild type
- X:Y
designates carbon chain length: total number of carbon-carbon double bonds
References
- Ahn K, McKinney MK, Cravatt BF. Enzymatic pathways that regulate endocannabinoid signaling in the nervous system. Chem Rev. 2008;108:1687–707. doi: 10.1021/cr0782067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN. Assay of Inorganic Phosphate, Total Phosphate and Phosphatases. In: Neufeld E, Ginsburg V, editors. Methods in Enzymology, Vol. VIII: Complex Carbohydrates. Academic Press; New York: 1966. pp. 115–118. [Google Scholar]
- Astarita G, Ahmed F, Piomelli D. Identification of biosynthetic precursors for the endocannabinoid anandamide in the rat brain. J Lipid Res. 2008;49:48–57. doi: 10.1194/jlr.M700354-JLR200. [DOI] [PubMed] [Google Scholar]
- Astarita G, Geaga J, Ahmed F, Piomelli D. Targeted lipidomics as a tool to investigate endocannabinoid function. Int Rev Neurobiol. 2009;85:35–55. doi: 10.1016/S0074-7742(09)85004-6. [DOI] [PubMed] [Google Scholar]
- Astarita G, Piomelli D. Lipidomic analysis of endocannabinoid metabolism in biological samples. J Chromatogr B Analyt Technol Biomed Life Sci. 2009 doi: 10.1016/j.jchromb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisogno T, De Petrocellis L, Di Marzo V. Methods for measuring endocannabinoid production and expression and activity of enzymes involved in the endocannabinoid system. In: Murphy EJ, Rosenberger TA, editors. In Lipid-Mediated Signaling. CRC Press; Boca Raton: 2009. pp. 109–150. [Google Scholar]
- Brites P, Waterham HR, Wanders RJA. Functions and biosynthesis of plasmalogens in health and disease. Biochim Biophys Acta. 2004;1636:219–231. doi: 10.1016/j.bbalip.2003.12.010. [DOI] [PubMed] [Google Scholar]
- Clement AB, Hawkins EG, Lichtman AH, Cravatt BF. Increased seizure susceptibility and proconvulsant activity of anandamide in mice lacking fatty acid amide hydrolase. J Neurosci. 2003;23:3916–23. doi: 10.1523/JNEUROSCI.23-09-03916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Demarest K, Patricelli MP, Bracey MH, Giang DK, Martin BR, Lichtman AH. Supersensitivity to anandamide and enhanced endogenous cannabinoid signaling in mice lacking fatty acid amide hydrolase. Proc Natl Acad Sci U S A. 2001;98:9371–6. doi: 10.1073/pnas.161191698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–7. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- Cravatt BF, Lichtman AH. The endogenous cannabinoid system and its role in nociceptive behavior. J. Neurobiol. 2004;61(1):149–60. doi: 10.1002/neu.20080. [DOI] [PubMed] [Google Scholar]
- De Petrocellis L, Di Marzo V. An introduction to the endocannabinoid system: from the early to the latest concepts. Best Pract Res Clin Endocrinol Metab. 2009;23:1–15. doi: 10.1016/j.beem.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Devaiah SP, Roth MR, Baughman E, Li M, Tamura P, Jeannotte R, Welti R, Wang X. Quantitative profiling of polar glycerolipid species and the role of phospholipase Dα1 in defining the lipid species in Arabidopsis tissues. Phytochemistry. 2006;67:1907–24. doi: 10.1016/j.phytochem.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, Griffin G, Gibson D, Mandelbaum A, Etinger A, Mechoulam R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–9. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, Bisogno T, De Petrocellis L. Endocannabinoids: new targets for drug development. Curr Pharm Des. 2000;6:1361–80. doi: 10.2174/1381612003399365. [DOI] [PubMed] [Google Scholar]
- Di Marzo V, De Petrocellis L, Sigiura T, Waku K. Potential biosynthetic connections between two eicosanoids, anandamide and 2- arachidonylglycerol, in mouse neuroblastoma cells. Biohcem Biophys Res Commun. 1996;227(1):281–8. doi: 10.1006/bbrc.1996.1501. [DOI] [PubMed] [Google Scholar]
- Epps DE, Natarajan V, Schmid PC, Schmid HO. Accumulation of N-acylethanolamine glycerophospholipids in infarcted myocardium. Biochim Biophys Acta. 1980;618:420–30. doi: 10.1016/0005-2760(80)90260-x. [DOI] [PubMed] [Google Scholar]
- Epps DE, Schmid PC, Natarajan V, Schmid HH. N-Acylethanolamine accumulation in infarcted myocardium. Biochem Biophys Res Commun. 1979;90:628–33. doi: 10.1016/0006-291x(79)91281-6. [DOI] [PubMed] [Google Scholar]
- Fontana A, Di Marzo V, Cadas H, Piomelli D. Analysis of anandamide, an endogenous cannabinoid substance, and of other natural N-acylethanolamines. Prostaglandins Leukot Essent Fatty Acids. 1995;53:301–8. doi: 10.1016/0952-3278(95)90130-2. [DOI] [PubMed] [Google Scholar]
- Giang DK, Cravatt BF. Molecular characterization of human and mouse fatty acid amide hydrolases. Proc Natl Acad Sci U S A. 1997;94:2238–42. doi: 10.1073/pnas.94.6.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen HS, Moesgaard B, Hansen HH, Petersen G. N-Acylethanolamines and precursor phospholipids - relation to cell injury. Chem Phys Lipids. 2000;108:135–50. doi: 10.1016/s0009-3084(00)00192-4. [DOI] [PubMed] [Google Scholar]
- McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–32. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- Muccioli GG, Stella N. An optimized GC-MS method detects nanomolar amounts of anandamide in mouse brain. Anal Biochem. 2008;373:220–228. doi: 10.1016/j.ab.2007.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan V, Reddy PV, Schmid PC, Schmid HH. On the biosynthesis and metabolism of N-acylethanolamine phospholipids in infarcted dog heart. Biochim Biophys Acta. 1981;664:445–8. doi: 10.1016/0005-2760(81)90067-9. [DOI] [PubMed] [Google Scholar]
- Natarajan V, Schmid PC, Schmid HH. N-acylethanolamine phospholipid metabolism in normal and ischemic rat brain. Biochim Biophys Acta. 1986;878:32–41. doi: 10.1016/0005-2760(86)90341-3. [DOI] [PubMed] [Google Scholar]
- Piomelli D. The molecular logic of endocannabinoid signalling. Nat Rev Neurosci. 2003;4:873–84. doi: 10.1038/nrn1247. [DOI] [PubMed] [Google Scholar]
- Schmid HH, Berdyshev EV. Cannabinoid receptor-inactive N-acylethanolamines and other fatty acid amides: metabolism and function. Prostaglandins Leukot Essent Fatty Acids. 2002;66:363–76. doi: 10.1054/plef.2001.0348. [DOI] [PubMed] [Google Scholar]
- Schmid HH, Schmid PC, Berdyshev EV. Cell signaling by endocannabinoids and their congeners: questions of selectivity and other challenges. Chem Phys Lipids. 2002;121:111–34. doi: 10.1016/s0009-3084(02)00157-3. [DOI] [PubMed] [Google Scholar]
- Schmid HH, Schmid PC, Natarajan V. N-acylated glycerophospholipids and their derivatives. Prog Lipid Res. 1990;29:1–43. doi: 10.1016/0163-7827(90)90004-5. [DOI] [PubMed] [Google Scholar]
- Schmid HH, Schmid PC, Natarajan V. The N-acylation-phosphodiesterase pathway and cell signalling. Chem Phys Lipids. 1996;80:133–42. doi: 10.1016/0009-3084(96)02554-6. [DOI] [PubMed] [Google Scholar]
- Sigiura T, Kondo S, Sukagawa A, Nakane S, Shinoda A, Itoh K, Yamashita A, Waku K. 2-Arachidonylglycerol: a possible endogenous cannabinoid receptor ligand. Biochem Biophys Res Commun. 1995;215(1):89–97. doi: 10.1006/bbrc.1995.2437. [DOI] [PubMed] [Google Scholar]
- Shoemaker JP, Garland CW, Steinfeld JI. Experiments in Physical Chemistry. McGraw-Hill; New York: 1974. pp. 34–39. [Google Scholar]
- Tan B, Bradshaw HB, Rimmerman N, Srinivasan H, Yu YW, Krey JF, Monn MF, Chen JS, Hu SS, Pickens SR, Walker JM. Targeted lipidomics: discovery of new fatty acyl amides. Aaps J. 2006;8:E461–5. doi: 10.1208/aapsj080354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terova B, Heczko R, Slotte JP. On the importance of the phosphocholine methyl groupsgroups for sphingomyelin/cholesterol interactions in membranes: a study with ceramide phosphoethanolamine. Biophys J. 2005;88(4):2661–9. doi: 10.1529/biophysj.104.058149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi K, Sun YX, Okamoto Y, Araki N, Tonai T, Ueda N. Molecular characterization of N-acylethanolamine-hydrolyzing acid amidase, a novel member of the choloylglycine hydrolase family with structural and functional similarity to acid ceramidase. J Biol Chem. 2005;280(12):11082–92. doi: 10.1074/jbc.M413473200. [DOI] [PubMed] [Google Scholar]
- Ueda N, Puffenbarger RA, Yamamoto S, Deutsch DG. The fatty acid amide hydrolase (FAAH) Chem Phys Lipids. 2000;108(1-2):107–121. doi: 10.1016/s0009-3084(00)00190-0. [DOI] [PubMed] [Google Scholar]
- Vellani V, Petrosino S, De Petrocellis L, Valenti M, Prandini M, Magherini PC, McNaughton PA, Di Marzo V. Functional lipidomics. Calcium-independent activation of endocannabinoid/endovanilloid lipid signalling in sensory neurons by protein kinases C and A and thrombin. Neuropharmacology. 2008;55:1274–9. doi: 10.1016/j.neuropharm.2008.01.010. [DOI] [PubMed] [Google Scholar]
- Venables BJ, Waggoner CA, Chapman KD. N-Acylethanolamines in selected legumes. Phytochemistry. 2005;66:1913–1918. doi: 10.1016/j.phytochem.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Walker JM, Krey JF, Chen JS, Vefring E, Jahnsen JA, Bradshaw H, Huang SM. Targeted lipidomics: fatty acid amides and pain modulation. Prostaglandins Other Lipid Mediat. 2005;77:35–45. doi: 10.1016/j.prostaglandins.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Welti R, Li W, Li M, Sang Y, Biesiada H, Zhou H, Rajashekar CB, Williams TD, Wang X. Profiling membrane lipids in plant stress responses: Role of phospholipase Dα in freezing-induced lipid changes in Arabidopsis. J Biol Chem. 2002;277:31994–2002. doi: 10.1074/jbc.M205375200. [DOI] [PubMed] [Google Scholar]
- Welti R, Shah J, Li W, Li M, Chen J, Burke JJ, Fauconnier ML, Chapman K, Chye ML, Wang X. Plant lipidomics: discerning biological function by profiling plant complex lipids using mass spectrometry. Front Biosci. 2007;12:2494–506. doi: 10.2741/2250. [DOI] [PubMed] [Google Scholar]
- Wilson RI, Nicoll RA. Endocannabinoid signaling in the brain. Science. 2002;296:678–82. doi: 10.1126/science.1063545. [DOI] [PubMed] [Google Scholar]