Abstract
The licensed anthrax vaccine and many of the new anthrax vaccines being developed are based on protective antigen (PA), a nontoxic component of anthrax toxin. For this reason, an understanding of the immune response to PA vaccination is important. In this study, we examined the antibody response elicited by PA-based vaccines and identified the domains of PA that contribute to that response in humans as well as nonhuman primates (NHPs) and rabbits, animal species that will be used to generate efficacy data to support approval of new anthrax vaccines. To this end, we developed a competitive enzyme-linked immunosorbent assay (ELISA), using purified recombinant forms of intact PA and its individual domains. We found that PA-based vaccines elicited IgG antibodies to each of the four PA domains in all three species. We also developed a competitive toxin neutralization assay, which showed that rabbits, NHPs, and humans all have functional antibody populations that bind to domains 1, 3, and 4. While the domain specificities of the antibody responses elicited by PA-based vaccines were similar in humans, NHPs, and rabbits, competitive assays suggested that humans may have a more significant secondary population of IgG antibodies that bind to partially unfolded or incorrectly folded PA. These findings provide information that will be useful when linking animal protection data to humans via an antibody bridge to establish efficacy of new anthrax vaccines.
The encapsulated, toxigenic species Bacillus anthracis causes cutaneous, gastrointestinal, and inhalational anthrax disease in humans (4). In 2001, B. anthracis was deliberately released through the United States postal system, leading to 22 confirmed cases of anthrax, of which 5 were fatal (14). The manifestations of anthrax are believed to be caused primarily by the effects of B. anthracis tripartite toxin composed of the binding component known as protective antigen (PA) as well as catalytic components, lethal factor (LF) and/or edema factor (EF). After PA binds to cellular receptors, it is cleaved by furin (11). The remaining PA (PA63) heptamerizes, at which point LF and/or EF can bind to form lethal toxin (LT) or edema toxin (ET), respectively (3). The toxin is then internalized, and LF/EF is released into the cytosol after endosomal acidification. LF is a zinc metalloprotease that has downstream effects on mitogen-activated protein kinase kinase signaling, whereas EF is an adenylate cyclase that increases cellular concentrations of cyclic AMP.
PA contains four domains (20), each with a unique role in toxin function. Domain 1 (residues 1 to 258) contains the furin recognition site RKKR, which is cleaved to release the N-terminal PA20(1-167) fragment. After removal of PA20, the remaining portion of domain 1 (domain “1b”) forms the LF/EF binding site. Domains 2 (residues 259 to 487) and 3 (residues 488 to 595) are involved in heptamerization, and domain 3 also seems to have a role in effector binding (16, 20). Domain 2 is responsible for formation of the pore through which the effector molecules traverse to enter the cytosol. Domains 2 and 4 (residues 596 to 735) participate in the binding of PA to the cellular receptors (15, 16).
Vaccination against B. anthracis is thought to be one of the most effective prophylactic measures for anthrax. Currently, one FDA-licensed vaccine for anthrax is available called “anthrax vaccine adsorbed” (AVA), or BioThrax. This vaccine is derived from the cell-free supernatant of a nonencapsulated strain of B. anthracis adsorbed to aluminum adjuvant. PA is the primary immunogen in AVA (2, 12). Immunization with purified PA has been shown to be protective in animal models (8, 13, 18), and therefore new-generation anthrax vaccines are being developed which are composed of purified preparations of recombinant PA (rPA) (2, 10, 12, 23, 29).
Because it is not feasible or ethical to test the efficacy of new anthrax vaccines in humans, the FDA will rely on the “Animal Rule” for licensure of these vaccines (26). This rule allows for the bridging of protection in relevant animal models to humans through comparison of the vaccine-induced immune response (26). The two animal models that have been determined to be appropriate for use in demonstrating efficacy of anthrax vaccines are nonhuman primates (NHPs) and rabbits (5-7, 21, 22). In order to make a strong link between protection in these animal models and human efficacy using an antibody bridge, it is important to know not only that the antibody response in relevant animal models is quantitatively similar to that of humans but also that it is qualitatively similar. While it is known that PA holoprotein (PA83) is protective in animals and elicits an immune response in humans, it is currently unknown whether the individual domains of PA are each immunogenic in both humans and relevant animal models. Because the domains have different roles in toxin function, differences in the immunogenic profiles of these domains could imply various mechanisms of protection in humans versus animals.
Previous work has evaluated the role of certain of the domains in immunogenicity (1) and protection from challenge in mice (9). Other studies have examined the ability of monoclonal antibodies generated from AVA-vaccinated humans to bind to certain regions of PA (PA20, PA63, or domain 4) (25). However, the contribution of each of the individual domains to the antibody response induced by PA-based vaccines has not been evaluated in humans. In addition, little is known about the domain specificity of antibodies elicited by vaccination with PA-based vaccines in NHPs and rabbits, the two animal models being proposed for Animal Rule approval of rPA vaccines, compared to that of humans.
In this study, we sought to determine whether humans, rabbits, and NHPs have qualitatively similar or distinctly different responses to PA with respect to domain specificity of the antibodies generated by vaccination with PA-based vaccines. To this end, we cloned, expressed, and purified each of the four PA domains individually. Using competitive assay formats, we assessed the ability of total IgG antibodies present in sera from animals and humans immunized with PA-based vaccines to bind to PA and its individual domains. We also examined the domain specificity of neutralizing antibodies generated by immunization with PA-based vaccines. The data presented in this paper highlight the similarities and differences in immunogenicity of anthrax vaccines in animals and humans and may help in evaluation of these vaccines under the Animal Rule.
MATERIALS AND METHODS
Strains, plasmids, and reagents.
Chemi-competent TOP10 Escherichia coli cells and the pTRChis2c expression plasmid were obtained from Invitrogen (Carlsbad, CA). Chemi-competent E. coli BL21 cells were generated in our laboratory as described by Mandel and Higa (17). Recombinant PA (NR-164), anti-rPA rabbit reference polyclonal serum (NR-3839), anti-rPA nonhuman primate reference polyclonal serum (NR-9329), anti-AVA human reference serum (AVR801), anti-PA monoclonal antibody DD-508, and murine macrophage-like cell line J774A.1 cells (NR-28) were obtained from the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH (Bethesda, MD). The rabbit anti-AVA serum pool used in this study was obtained by combining equal volumes of sera from five animals vaccinated with AVA. Briefly, each animal received 0.125 ml of AVA subcutaneously (s.c.) at zero, 14, and 28 days. Serum was harvested at 42 days and pooled at equal volumes. The human anti-rPA serum pool used in this study was obtained by combining equal volumes of sera from nine individuals immunized with rPA vaccine that were obtained through the NIH/NIAID, Division of Microbiology and Infectious Diseases (DMID) Study 01-003, under contract no. N01-AI-25461. Each subject received at least two doses of an experimental rPA vaccine. Primers were generated by the CBER Facility for Biotechnology Resources (Bethesda, MD). All chemical reagents were obtained from Fisher Scientific (Pittsburgh, PA), unless otherwise noted. This study complied with all relevant guidelines and FDA's policies regarding the use of human subjects in research.
Cell culture.
The murine macrophage-like cell line J774A.1 was grown in Dulbecco's modified Eagle medium (DMEM; containing a high glucose concentration and sodium pyruvate) supplemented with 5% heat-inactivated fetal bovine serum, 2 mM glutamine, penicillin (25 U/ml), streptomycin sulfate (25 μg/ml), and 10 mM HEPES. Cell culture reagents were obtained from GIBCO (Carlsbad, CA).
Cloning of PA domains.
The individual protein domains of PA, which are composed of amino acids 1 to 258 (domain 1), 259 to 487 (domain 2), 488 to 595 (domain 3), and 596 to 735 (domain 4) (20), were amplified from the B. anthracis Sterne strain using PCR (refer to Table 1 for primers), with a C-terminal six-histidine tag and stop codon added in frame with the coding sequence. The amplified DNA fragments were then cloned into the appropriate restriction sites of the expression vector pTRChis2c downstream of the lac operon. Ligated vectors were transformed into E. coli TOP10 cells, and selection for positive colonies was performed using ampicillin. The presence of inserts was confirmed by restriction digestion, and clones were verified by sequencing. Plasmids were then subcloned into the E. coli strain BL21 for expression.
TABLE 1.
Primers used in this study
| Primer name | Sequence (5′ to 3′)a |
|---|---|
| Domain 1 for | CGCGGATCCAGAAGTTAAACAGGAG |
| Domain 1 rev | AAAACTGCAGTCAATGATGATGATGATGATGAGCTGCCACAAG |
| Domain 2 for | CGGGATCCAGCAGCTTATCCGATTGTACATGTAGAT |
| Domain 2 rev | CCCAAGCTTTCAATGATGATGATGATGATGTGTTTCTTGAATTTG |
| Domain 3 for | GGATCCACTGCACGTATCATTTTTAATGGTAA |
| Domain 3 rev | CCCAAGCTTTCAATGATGATGATGATGATGACGTTTATCGCGAATCAGAATATT |
| Domain 4 for | CGCGGATCCATTTCATTATGATAGAAATAACATAGCAGTTGGG |
| Domain 4 rev | CCCAAGCTTTCAATGATGATGATGATGATGTCCTATCTCATAGCC |
Restriction sites are underlined.
Expression of recombinant PA domain proteins.
For each recombinant protein, 250-ml cultures were inoculated 1:50 from an overnight culture and grown to an optical density at 600 nm (OD600) of 0.5 to 0.6 prior to induction. Noninduced samples were removed from each culture prior to the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). The growth conditions for each recombinant domain were optimized for maximal expression and yield. The growth conditions were as follows. E. coli cells producing domain 1 were grown in Super broth at 37°C and induced with 1 mM IPTG, and the cells were transferred to room temperature for an additional 4 h of growth. E. coli cells producing domain 2 were grown in Luria broth (LB) at 37°C and induced overnight, at the same temperature, with the addition of 0.75 mM IPTG. E coli cells producing either domain 3 or domain 4 were grown in LB at 30°C and then induced with 1 mM IPTG for 4 h.
Purification of recombinant PA domain proteins.
Purification of domains 1 and 4 was as follows. After growth as described above, cultures were divided in half and centrifuged for 10 min at 5,000 × g, and pellets were retained. Each cell pellet (comprised of the cells from 125 ml of culture) was resuspended in 2 ml of lysis buffer (1 mg/ml lysozyme, 1% Triton X-100, 25% sucrose, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride [PMSF], 5 mM β-mercaptoethanol, 4 μM leupeptin, 4 μM pepstatin) and incubated on ice for 30 min. Resuspended pellets were then sonicated, and cellular debris was removed by centrifugation at 13,000 × g for 10 min. One milliliter of cell lysate was then mixed with 100 μl of HisPur cobalt beads (Pierce, Rockford, IL) and incubated at 4°C overnight on a rotator. The following day, the beads were pelleted by centrifugation at 700 × g and then washed three times according to the manufacturer's instructions for batch method purification. His-tagged recombinants were eluted with five 100-μl aliquots of elution buffer (50 mM sodium phosphate, 300 mM sodium chloride, 150 mM imidazole, pH 7.4).
Purification of domains 2 and 3 was carried out using the hybrid protocol for the ProBond purification system (Invitrogen) according to the manufacturer's instructions. This protocol was designed for insoluble proteins and provides conditions that encourage the refolding of the protein on the column. Briefly, pelleted cells were lysed under denaturing conditions as per the manufacturer's instructions. Lysates were then applied to Ni-nitrilotriacetic acid (NTA) columns and washed twice with denaturing binding buffer, twice with denaturing wash buffer, and four times with native wash buffer. Proteins were eluted using native elution buffer, as per the manufacturer's instructions.
For all recombinant proteins, purity was confirmed by SDS-PAGE and the proteins were quantified using bicinchoninic acid (BCA; Pierce) according to the manufacturer's instructions. Proteins were then stored at −70°C until use.
Competitive ELISA.
Ninety-six-well plates (Maxisorp; Nalge Nunc International, Rochester, NY) were coated with 1 μg/ml PA (NR-164) in carbonate binding buffer (100 μl, pH 9.6) overnight at room temperature (RT). The plates were then washed three times with wash buffer (0.85% sodium chloride, 0.05% Tween 20) and blocked with 2% nonfat dry milk in phosphate-buffered saline (PBS) for 2 h at 37°C. Competitor-serum mixtures were prepared in a separate 96-well plate. To prepare these mixtures, test sera were diluted in dilution buffer (PBS, 0.05% Tween 20, 2% fetal bovine serum) to the concentration that resulted in approximately 1/2 maximal binding (OD405 of ∼1.0) in a standard (noncompetitive) enzyme-linked immunosorbent assay (ELISA), and various amounts of competitor proteins (PA83 or individual recombinant domains) were then mixed with the diluted serum to give 5-fold dilutions of the competitor across the plate, beginning with a maximal concentration of 600 nM. These plates containing the competitor-serum mixtures were incubated at RT for 1 h, and the contents were then transferred to the blocked and washed ELISA plates. The plates were incubated at RT for 2 h, washed three times, and the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-rabbit IgG at 1:2,000 [Millipore, Temecula, CA] or monoclonal anti-human IgG at 1:1,000, used for both NHP and human assays, clone HP6043 [Hybridoma Reagent Laboratory, Baltimore, MD]) was then added, and the mixture was incubated for another 2 h at RT. Plates were washed a final time, and ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (KPL, Gaithersburg, MD)] was added for color development. After 10 min, the reaction was stopped through the addition of an equal volume of ABTS stop solution (KPL). Optical densities were then read at 405 nm (OD405) on a VersaMax microplate reader (Molecular Devices, Sunnyvale, CA).
Sandwich competitive ELISA.
The sandwich competitive ELISA was performed as described above with the following modifications. Competitor-serum mixtures were prepared in 96-well plates as described above for the competitive ELISA and incubated at 4°C overnight. Separate 96-well plates (MaxiSorp) were coated overnight with monoclonal antibody DD-508 at a concentration of 2.5 μg/ml in PBS. This concentration of DD-508 was chosen because when used to capture PA83 (1 μg/ml), it produced binding curves in a sandwich ELISA similar to those observed in a standard ELISA using PA83 (1 μg/ml) adsorbed to the plate (data not shown). The monoclonal antibody-coated plates were then blocked and washed as described above for the competitive ELISA. PA at 1 μg/ml was then added to the DD-508-coated wells, and the plates were incubated at RT for 1 h. After washing, the competitor-serum samples were transferred to the ELISA plates. From this point, the assay continued as for the competitive ELISA, including the incubation times, wash steps, dilution of secondary antibodies, and development. OD values for the sandwich ELISA were corrected by subtracting the OD405 for DD-508 incubated with serum and secondary antibody but no PA (to eliminate nonspecific reactivity), and then corrected values were analyzed as described in the “Data analysis” section of Materials and Methods.
Macrophage lysis assay and competitive TNA.
Before using the recombinant PA domains in competitive toxin neutralization assays (TNAs) to analyze the contribution of each domain to anthrax vaccine-induced immunity, each was tested in a macrophage lysis assay to evaluate if there was any interference with the action of LT (PA plus LF) in J774A.1 cells. Briefly, each domain was 2-fold diluted in a 96-well round-bottom plate in DMEM, starting with a concentration of 600 nM. Different dilutions of the domains and the LT solution (50 ng/ml PA plus 40 ng/ml LF, the concentration of LT that kills approximately 95% of cells) were added simultaneously to a 96-well cell plate (40,000 cells/well), and plates were incubated for 4 h at 37°C. Following incubation, MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] was added to plates, and after 2 h of incubation, cells were lysed by addition of solubilization buffer (90% isopropanol, 0.5% [wt/vol] SDS, 38 mM HCl). The plates were read for OD using a microplate reader at OD570, and the percentage of killing was calculated by comparing the ODs of the treated cells with those of nontreated, healthy cells. Interference of PA domains in J774A.1 killing was calculated by comparing the percentages of LT-killed cells in the presence and absence of each domain.
For competitive TNAs, different concentrations of purified recombinant domains 1, 3, and 4 were incubated with diluted serum samples (NR-3839, rabbit; NR-9329, NHP; and AVR801, human [each diluted in DMEM]) overnight at 4°C. The next day, the domain-serum combinations were preincubated with a constant amount of LT (PA, 50 ng/ml PA plus 40 ng/ml LF) for 30 min. Mixtures were then transferred to a 96-well cell plate (40,000 cells/well), and plates were incubated for 4 h at 37°C. Following incubation, TNAs were performed essentially as described previously (28). The plates were read using a microplate reader at OD570, and the percentage of competition for each domain was calculated as described below.
Data analysis.
All data were plotted using GraphPad Prism software (version 5; GraphPad Software, La Jolla, CA). For competitive ELISA and TNA, data were plotted with a four-parameter logistic (4PL) or two-site competition curve fit of OD (y axis) versus the logarithm of the competitor concentration (x axis), as indicated. The upper asymptote for each data set was then used to normalize its respective data set, so that the upper asymptote became 100% and 0% was equivalent to an OD of 0. Normalized data were then plotted as percent IgG binding to plate (left y axis) versus log competitor concentration (x axis). Percent competition was calculated by subtracting percentage of IgG binding to the plate from 100% (right y axis). For competitive TNA, the data were plotted as bar graphs with “percent competition” as the y axis. Where quantification of percent competition by domains was performed, this value at 600 nM competitor was determined (mean of duplicates).
Statistical analysis.
Statistical analyses were performed using GraphPad Prism. One-way analysis of variance (ANOVA) on the percent competition at 600 nM competitor protein was carried out to evaluate species responses to soluble PA83. A P value of <0.05 was considered significant. For competitive ELISAs and TNAs using PA domains, the 95% confidence interval (CI) of the mean percent competition at 600 nM competitor was determined. Competition was deemed to be significant if the 95% CI of the percent competition did not include zero.
RESULTS
Assessing domain specificity of anthrax vaccine-induced antisera using a competitive ELISA format.
In order to determine which domain(s) contributes to the immunogenicity of PA upon vaccination with PA-based vaccines, we developed a competitive ELISA in which the ability of the individual domains to compete with PA for binding to the IgG antibodies found in sera from humans, NHPs, or rabbits immunized with PA-based vaccines (either AVA or rPA) was assessed. In this assay, increasing concentrations of competitor (either PA or individual domains) were mixed with antisera generated by immunization with a PA-based vaccine. The ability of the serum, in the presence of the competitor, to bind to PA coating a 96-well plate was then determined.
Results obtained using PA83 as competitor.
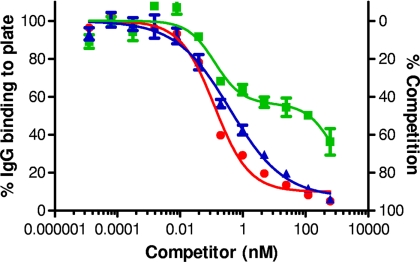
We first examined the ability of PA83 to act as a competitor. We would expect that at high concentrations of PA83 competitor, approximately 100% competition should be achieved. As seen in Fig. 1, when PA83 was used as the competitor, nearly complete competition was obtained with both rabbit and NHP antisera, with a lower asymptote that approached zero on the left y axis (percent IgG binding) and 100% on the right y axis (percent competition). Surprisingly however, human antiserum mixed with PA83 competitor gave what appeared to be biphasic curves, and approximately 35% of maximal binding remained, even at the highest concentration of competitor used (600 nM) (Fig. 1). The level of inhibition achieved with 600 nM PA83 seen with either rabbit or NHP serum was significantly higher than that observed with the human serum (P = 0.0038; ANOVA). This disparity was not due to differences in secondary antibody, as the same anti-human IgG-HRP conjugate was used for both human and NHP samples. Also, because nonimmune human serum did not give a detectable OD405 reading over buffer controls, the lower level of competition by PA83 observed with human serum was not attributable to nonspecific binding (data not shown).
FIG. 1.
Analysis of immune sera in a competitive ELISA using PA83 as the competitor. Diluted rabbit anti-rPA serum (NR-3839) (▴), NHP anti-rPA serum (NR-9329) (•), and human anti-AVA serum (AVR801) (▪) were mixed with soluble PA83 at the indicated concentrations prior to addition to a PA83-coated ELISA plate and analyzed as described in Materials and Methods. Nonlinear-fit curves were generated for OD405 readings, and readings were normalized to the OD upper asymptote as 100% as described in Materials and Methods. Rabbit and NHP curves were fit to a 4PL model, while the human serum curve was fit to a two-site competition model. Shown are graphs generated from duplicate plates with the mean ± standard error of the mean (SEM) shown for each point. The figure is representative of three independent experiments.
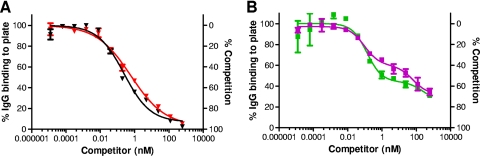
We further examined this phenomenon. Because the rabbit and NHP sera examined were generated by immunization with rPA vaccine and the human serum was obtained from individuals immunized with AVA, we first evaluated whether the vaccine type (AVA versus rPA) was the cause of this observation. As shown in Fig. 2A, similar competition curves were obtained when sera from rabbits immunized with either rPA or AVA were examined. In this case, essentially complete competition was observed. Competition curves obtained when sera from humans immunized with either rPA or AVA were examined were also similar to each other in that the level of competition seen at 600 nM PA83 was incomplete (Fig. 2B) and the biphasic shape of the curves remained with both vaccine types. We were unable to evaluate this comparison in NHPs due to unavailability of anti-AVA NHP sera. These results suggest that the lack of complete competition seen in the competitive ELISA when human antiserum is examined does not appear to be vaccine specific.
FIG. 2.
Analysis of anti-AVA and anti-rPA sera in a competitive ELISA using PA83 as the competitor. (A) Rabbit anti-rPA serum (NR-3839) (red ▾) or a rabbit anti-AVA serum pool (black ▾) or (B) a human anti-rPA serum pool (purple ▪) or human anti-AVA serum (AVR801) (green ▪) was mixed with the indicated concentrations of PA83 competitor and used in the competitive ELISA as described in Materials and Methods. Nonlinear-fit curves were generated for OD405 readings, and readings were normalized to the OD upper asymptote as 100%. In panel A, rabbit competition curves were fit to a 4PL model, while in panel B, human competition curves were fit to a two-site competition model. Graphs were generated from duplicate plates, with each point showing the mean ± SEM, and are representative of three independent experiments.
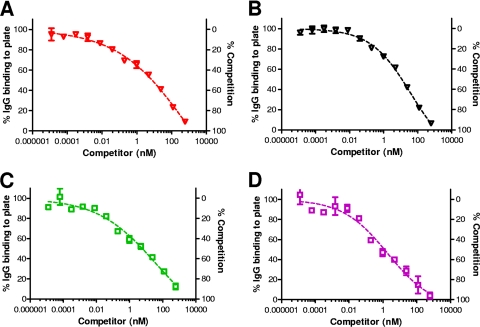
Research has shown that a small population of antigen may become fully or partially denatured during the process of adsorption to plastic surfaces such as 96-well plates (19, 27). We therefore investigated whether unfolding or denaturation of some of the PA coating the 96-well plate might expose denatured epitopes to which human IgG antibodies bind. These IgG antibodies would be expected to bind poorly, or not at all, to the native PA83 used as competitor but would bind to any denatured PA bound to the plate. If the PA were bound to the plate in such a way that all molecules retained their native structure, then soluble PA83 should be able to efficiently compete for all IgG antibodies that recognize the captured PA. To this end, we developed a “sandwich” competitive ELISA in which PA, instead of being bound directly to the plate, is captured by a monoclonal antibody (DD-508) coating the plate that selectively binds the native form of PA, as determined by reactivity to PA on dot blots but lack of reactivity to PA on Western blots (data not shown); thus, PA retains its native conformation. Results from these sandwich competitive ELISAs are shown in Fig. 3. The curves generated for the rabbit sera (both anti-rPA serum and anti-AVA serum) show essentially complete competition at 600 nM PA83 (Fig. 3A and B). When human sera were examined, essentially complete inhibition was also observed in the sandwich competitive ELISA (Fig. 3C and D). Moreover, when human sera were examined in the sandwich competitive ELISA, the curves lost their biphasic appearance (compare Fig. 2B to Fig. 3C and D). These results are consistent with the idea that the human antisera may contain a population of IgG antibodies that react to unfolded or incorrectly folded PA but poorly or not at all to the native form of PA. This population of IgG antibodies—manifested in the biphasic appearance of the curves—only becomes apparent in the competitive ELISA when high concentrations of soluble PA competitor are used, possibly because only a small amount of denatured PA is present in the PA preparation and competitive levels of this form of the protein are only achieved at high concentrations of the preparation. In the sandwich competitive ELISA, this effect is negated by evaluating only those anti-PA IgG antibodies that bind to the protein in its native form.
FIG. 3.
Analysis of rabbit and human antisera in a sandwich competitive ELISA using PA83 as competitor. Assays used either a rabbit anti-rPA serum (NR-3839) (A), a rabbit anti-AVA serum pool (B), a human anti-rPA serum pool (C), or human anti-AVA serum (AVR801) (D). Nonlinear-fit curves were generated and data were normalized to the OD upper asymptote as 100% as described in Materials and Methods. Graphs were generated from duplicate plates, with each point indicating the mean ± SEM, and are representative of three independent experiments.
Results obtained using individual domains as competitor.
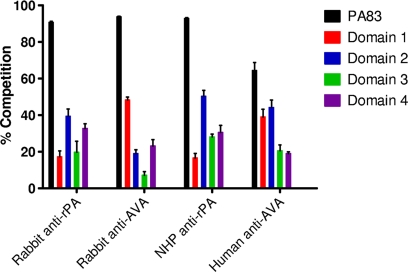
Competitive ELISAs were next carried out using individual PA domains as competitors premixed with antisera generated by PA-based vaccines prior to use in a PA ELISA. In this study, we used rabbit anti-AVA and anti-rPA, NHP anti-rPA, and human anti-AVA sera. Figure 4 shows the extent of competition obtained when 600 nM competitor was used. For all sera examined, the competition observed for each of the individual domains is significantly different from zero (P < 0.05) since the 95% confidence interval does not include zero. These results indicate that IgG antibodies to each of the individual domains are present in all sera examined.
FIG. 4.
Competitive ELISA using purified recombinant domains as competitors. Competitive ELISAs were carried out as described in Materials and Methods. Rabbit anti-rPA serum (NR-3839), rabbit anti-AVA serum pool, NHP anti-rPA (NR-9329), and human anti-AVA serum (AVR801) were used in each assay and mixed with either PA83 or purified recombinant domain as indicated. For each serum-competitor combination, the percent competition at 600 nM competitor protein was plotted as the mean ± SEM. The data shown were generated from three independent experiments.
Assessing domain specificity of anthrax vaccine-induced antisera, using a competitive TNA format.
In order to determine the domain specificity of functional (toxin-neutralizing) antibodies found in sera from animals or humans immunized with PA-based vaccines, we developed a competitive TNA. In the typical format, the TNA examines the ability of antibodies to neutralize the cytocidal activity of LT (PA plus LF) on J774A.1 cells. The competitive TNA is designed to assess the ability of individual PA domains to compete with PA for binding to the neutralizing antibodies. Any competition by an individual domain would indicate that functional antibodies specific to that domain are present.
Before we used an individual domain as an antibody competitor in the competitive TNA, we needed to ascertain that the domain, in the absence of any antibodies, did not interfere with LT action since such interference could not be differentiated from antibody neutralization of the toxin in the assay. Such a dominant-negative effect might occur if an individual domain were able to interact with native PA monomers and interfere with subsequent toxin action in some way: e.g., by interfering with heptamer formation, by interfering with pore formation, etc. In order to test for dominant-negative behavior, macrophage lysis assays were performed in which individual domains were combined with PA83 in the presence of LF and then applied to J774A.1 cells. The concentration of PA plus LF that we used in this assay has been shown previously to kill 95% of the cell population (28); therefore, any decrease in cytocidal activity in the presence of the domain compared to in its absence is due to a dominant-negative activity of the domain. We found that domain 2 interfered with LT activity, while domains 1, 3, and 4 did not (data not shown). Therefore, only domains 1, 3, and 4 could be used in the competitive TNA.
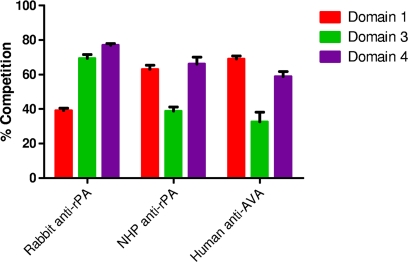
We examined the ability of the individual domains 1, 3, and 4 to compete with PA83 for binding to neutralizing antibodies present in sera from rabbits immunized with rPA, NHPs immunized with rPA, and humans immunized with AVA. We found that all sera examined contained functional antibodies to each of the domains (Fig. 5). In each case, the level of competition observed was significant (P < 0.05) since the lower limit of the 95% CI was greater than zero. We note that for each serum sample examined, if the competition levels observed for each of the domains are added together, the sum is greater than 100%. We are not sure why this may be, but perhaps a number of epitopes cross domain borders; for example, an epitope could span the junction between domains 3 and 4, and thus competition with either domain would negate the neutralizing activity of the antibodies that react to that epitope. Alternatively, antibodies might bind in a synergistic manner to neutralize toxin action in this assay (M. Ngundi and D. Burns, unpublished results); thus, loss of one epitope might decrease the action of antibodies that bind to other epitopes on the protein, resulting in an overall greater-than-expected decrease in neutralization.
FIG. 5.
Competitive TNA using domains 1, 3, and 4 as competitors. Rabbit anti-rPA serum (NR-3839), NHP anti-rPA serum (NR-9329), and human anti-AVA serum (AVR801) were tested in a competitive TNA using the indicated purified recombinant domains. Each domain was analyzed for its ability to prevent antibody neutralization of LT in J774.A1 cells. Plotted is the mean ± SEM of percent competition at 600 nM competitor for three independent experiments.
DISCUSSION
In this study, we examined the PA domain-specific response after anthrax vaccination using PA-based vaccines in humans as well as rabbits and NHPs—the two animal species proposed for Animal Rule-based anthrax vaccine approval. Information regarding the domain specificity of the antibody response to PA-based vaccines has been limited. Flick-Smith et al. suggested that the epitopes most important for protection within PA are found in domain 4 (9). Abboud and Casadevall, on the other hand, demonstrated that, upon vaccination with various recombinant domain constructs, mice respond strongly to domain 1 (1). However, both of these studies immunized mice with the purified, recombinant domains themselves (or combinations thereof), not PA83, which could lead to immune responses that are different from those achieved by vaccination with PA. Previous work indentified antibodies specific for domain 1 or domain 4 in humans immunized with AVA by examining the specificity of isolated monoclonal antibodies or through the use of competitive ELISAs (24, 25, 30). However, those studies were limited since only recombinant domains 1 and 4 or proteolytic PA fragments (PA20 and PA63) were tested, which does not address antibody specificity for individual domains 2 and 3. Our work expands on previous findings in the following ways: (i) by including domains 2 and 3; (ii) by examining sera generated by immunization with both rPA vaccine and AVA; (iii) by examining the domain specificity of functional (i.e., neutralizing) antibodies generated by vaccination; and (iv) by producing data relevant for Animal Rule-based evaluations through the assessment of the domain-specific antibody levels generated by PA-based vaccines in rabbits and NHP.
In order to determine the role of each PA domain individually in the vaccine-induced immune response, we cloned, expressed, and purified each of the four PA domains separately. We then used the purified recombinant domains as competitors in a competitive ELISA format and in a competitive TNA format, in order to identify the domains recognized by antibodies generated after vaccination with PA-based vaccines. Using a competitive ELISA, we found that each domain plays a role in immunogenicity of PA-based vaccines in that IgG antibodies are generated against all four domains, regardless of species. Using a competitive TNA format, we were able to demonstrate that neutralizing antibodies to domains 1, 3, and 4 are generated by vaccination with PA-based vaccines, regardless of species. We were not able to determine whether neutralizing antibodies specific for domain 2 were present in sera from individuals immunized with PA-based vaccines because of the dominant-negative effect of domain 2. This similarity in response suggests that mechanisms of antibody neutralization of toxin action likely do not differ dramatically between species. This information provides important support for the use of an antibody bridge to link protection in animals to human efficacy under the Animal Rule.
Ideally, we would have liked to compare the domain specificities of total IgG antibodies induced by vaccination in the different species in a more quantitative manner. However, using increasing concentrations of competitor up to 600 nM, the largest amount that was technically feasible to use because of volume and production constraints, we found that percent competition continued to increase; thus, we were not able to ascertain that the maximum level of competition had been achieved. We therefore limited our interpretation to qualitative measures: i.e., whether any competition was generated by the addition of soluble PA domains.
The crystal structure of PA monomers shows that each domain is exposed and is potentially accessible to the host immune system. Therefore, an antibody response to each of the domains upon vaccination with PA-based vaccines is not surprising. After heptamerization of PA, a significant amount of surface becomes buried—mostly parts of domains 1 and 2 (20); however, PA-based vaccines are composed of PA that would not be expected to have heptamerized since the PA is either in the form that is present in the bacterial culture supernatant (for AVA, see references 2 and 12) or a purified form of PA83 (for rPA, see references 2, 10, 12, 23, and 29). Our finding that a subset of the antibodies present in the immune sera specific for either domain 1, 3, or 4 were able to neutralize toxin action is consistent with the fact that each domain plays a critical role in toxin action, whether it be effector binding (domain 1) (20), participation in oligomer formation (domain 3) (16, 20), or receptor binding (domain 4) (15, 16). The fact that neutralizing antibodies to at least three individual PA domains are present in immune sera suggests that each of these domains might participate in generating a protective antibody response in both animals and humans after vaccination.
In developing the competitive ELISA, we observed that complete competition of human IgG antibodies could not be obtained using soluble PA83 as the competitor, even at high concentrations, whereas when combined with either rabbit or NHP sera, PA83 at the same concentration was able to nearly completely abrogate IgG antibody binding to the PA83-coated ELISA plate. The incomplete competition in human serum was not due to vaccine differences, as sera from AVA- and rPA-vaccinated humans had the same pattern. However, the difference in competition levels could be lessened through the use of a sandwich competitive ELISA that selectively detected IgG antibodies specific for native PA83. Also, the biphasic nature of the curve was lost in the sandwich competitive ELISA. These findings suggest that humans have at least two significant populations of anti-PA IgG antibodies: one that reacts with native PA83 and one that reacts only with unfolded or incorrectly folded PA83. It is plausible that rabbits and NHPs also have IgG antibodies specific for unfolded or incorrectly folded forms of the protein; however, in these animals, the population is less apparent than in humans, as evidenced by more complete abrogation of binding to plate-bound antigen by soluble PA83 with rabbit and NHP sera and a lack of a biphasic appearance to the competition curves. It is unclear why humans may have a more significant population of IgG antibodies specific for unfolded or denatured PA than the other species. It is possible that humans respond to a slightly different set of B-cell epitopes than the other species.
The data presented herein highlight the similarities and some differences in the immune responses of animals and humans to PA-based vaccines. The overall immunogenicities of the PA domains were comparable in rabbits, NHPs, and humans in that all domains were recognized by IgG antibodies present in immune sera from each of the species. Moreover, neutralizing antibodies to domains 1, 3, and 4 were generated by immunization with PA-based vaccines in each of the species. We noted one difference between the species in that we were able to detect a more apparent population of IgG antibodies specific for unfolded or incorrectly folded PA in immune sera of humans compared to that from rabbits and NHPs. The presence of similar domain specificities of the IgG antibodies induced by vaccination with PA-based vaccines in the different species strengthens the use of an antibody bridge to link animal protection data to human efficacy. However, our finding that a more noticeable proportion of human IgG antibodies may be specific for unfolded or incorrectly folded PA compared to that found in sera of vaccinated rabbits or NHPs cannot be ignored and adds a cautionary note to making a simple link between animal protection data and human efficacy using total antibody levels. Use of neutralizing antibodies as a bridge might better circumvent this difference between species, since neutralizing antibodies would be expected to bind specifically to the native, active form of the protein. We believe that this information may help in evaluation of new PA-based vaccines using the Animal Rule.
Acknowledgments
This work was supported in part by CBER/FDA-NIAID/NIH Interagency Agreement YI-AI-6153-01. R.B. was partially funded by a program at the Center for Biologics Evaluation and Research administered by the Oak Ridge Institute for Science and Education. The following reagents were obtained from the NIH Biodefense and Emerging Infections Research Resources Repository, NIAID, NIH: anthrax LF, recombinant from Bacillus anthracis, NR-142; anthrax PA, recombinant from Bacillus anthracis, NR-164 and NR-140; J774A.1 monocyte/macrophage (mouse) working cell bank, NR-28; rabbit anti-PA reference serum pool, NR-3839; NHP anti-PA reference serum pool, NR-9329; human anti-AVA reference serum pool, AVR801; and mouse anti-PA monoclonal antibody DD-508.
We thank Leslie Wagner for expert assistance in generating the rabbit serum samples.
Footnotes
Published ahead of print on 14 July 2010.
REFERENCES
- 1.Abboud, N., and A. Casadevall. 2008. Immunogenicity of Bacillus anthracis protective antigen domains and efficacy of elicited antibody responses depend on host genetic background. Clin. Vaccine Immunol. 15:1115-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baillie, L. W. J. 2006. Past, imminent and future human medical countermeasures for anthrax. J. Appl. Microbiol. 101:594-606. [DOI] [PubMed] [Google Scholar]
- 3.Collier, R. J., and J. A. T. Young. 2003. Anthrax toxin. Annu. Rev. Cell Dev. Biol. 19:45-70. [DOI] [PubMed] [Google Scholar]
- 4.Dixon, T. C., M. Meselson, J. Guillemin, and P. C. Hanna. 1999. Anthrax. N. Engl. J. Med. 341:815-826. [DOI] [PubMed] [Google Scholar]
- 5.FDA. 2002. Transcripts of Workshop, Anthrax Vaccines: Efficacy Testing and Surrogate Markers of Immunity. http://www.fda.gov/cber/minutes/workshop-min.htm.
- 6.FDA. 2007. Transcripts of Workshop, Anthrax: Bridging Correlates of Protection in Animals to Immunogenicity in Humans. http://www.fda.gov/cber/minutes/workshop-min.htm.
- 7.Fellows, P. F., M. K. Linscott, B. E. Ivins, M. L. M. Pitt, C. A. Rossi, P. H. Gibbs, and A. M. Friedlander. 2001. Efficacy of a human anthrax vaccine in guinea pigs, rabbits, and rhesus macaques against challenge by Bacillus anthracis isolates of diverse geographical origin. Vaccine 19:3241-3247. [DOI] [PubMed] [Google Scholar]
- 8.Flick-Smith, H. C., J. E. Eyles, R. Hebdon, E. L. Waters, R. J. Beedham, T. J. Stagg, J. Miller, H. O. Alpar, L. W. J. Baillie, and E. D. Williamson. 2002. Mucosal or parenteral administration of microsphere-associated Bacillus anthracis protective antigen protects against anthrax infection in mice. Infect. Immun. 70:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flick-Smith, H. C., N. J. Walker, P. Gibson, H. Bullifent, S. Hayward, J. Miller, R. W. Titball, and E. D. Williamson. 2002. A recombinant carboxy-terminal domain of the protective antigen of Bacillus anthracis protects mice against anthrax infection. Infect. Immun. 70:1653-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedlander, A. M., S. L. Welkos, and B. E. Ivins. 2002. Anthrax vaccines. Curr. Top. Microbiol. Immunol. 271:33-60. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, V. M., K. R. Klimpel, N. Arora, M. A. Henderson, and S. H. Leppla. 1995. Proteolytic activation of bacterial toxins by eukaryotic cells is performed by furin and by additional cellular proteases. Infect. Immun. 63:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabenstein, J. D. 2008. Vaccines: countering anthrax: vaccines and immunoglobulins. Clin. Infect. Dis. 46:129-136. [DOI] [PubMed] [Google Scholar]
- 13.Ivins, B., P. Fellows, L. Pitt, J. Estep, J. Farchaus, A. Friedlander, and P. Gibbs. 1995. Experimental anthrax vaccines: efficacy of adjuvants combined with protective antigen against an aerosol Bacillus anthracis spore challenge in guinea pigs. Vaccine 13:1779-1784. [DOI] [PubMed] [Google Scholar]
- 14.Jernigan, D. B., P. L. Raghunathan, B. P. Bell, R. Brechner, E. A. Bresnitz, J. C. Butler, M. Cetron, M. Cohen, T. Doyle, M. Fischer, C. Greene, K. S. Griffith, J. Guarner, J. L. Hadler, J. A. Hayslett, R. Meyer, L. R. Petersen, M. Phillips, R. Pinner, T. Popovic, C. P. Quinn, J. Reefhuis, D. Reissman, N. Rosenstein, A. Schuchat, W. J. Shieh, L. Siegal, D. L. Swerdlow, F. C. Tenover, M. Traeger, J. W. Ward, I. Weisfuse, S. Wiersma, K. Yeskey, S. Zaki, D. A. Ashford, B. A. Perkins, S. Ostroff, J. Hughes, D. Fleming, J. P. Koplan, and J. L. Gerberding. 2002. Investigation of bioterrorism-related anthrax, United States, 2001: epidemiologic findings. Emerg. Infect. Dis. 8:1019-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacy, D. B., D. J. Wigelsworth, R. A. Melnyk, S. C. Harrison, and R. J. Collier. 2004. Structure of heptameric protective antigen bound to an anthrax toxin receptor: a role for receptor in pH-dependent pore formation. Proc. Nat. Acad. Sci. U. S. A. 101:13147-13151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Little, S. F., J. M. Novak, J. R. Lowe, S. H. Leppla, Y. Singh, K. R. Klimpel, B. C. Lidgerding, and A. M. Friedlander. 1996. Characterization of lethal factor binding and cell receptor binding domains of protective antigen of Bacillus anthracis using monoclonal antibodies. Microbiology 142:707-715. [DOI] [PubMed] [Google Scholar]
- 17.Mandel, M., and A. Higa. 1970. Calcium-dependent bacteriophage DNA infection. J. Mol. Biol. 53:159-162. [DOI] [PubMed] [Google Scholar]
- 18.Miller, J., B. W. McBride, R. J. Manchee, P. Moore, and L. W. Baillie. 1998. Production and purification of recombinant protective antigen and protective efficacy against Bacillus anthracis. Lett. Appl. Microbiol. 26:56-60. [DOI] [PubMed] [Google Scholar]
- 19.Ngai, P. K., F. Ackermann, H. Wendt, R. Savoca, and H. R. Bosshard. 1993. Protein A antibody-capture ELISA (PACE): an ELISA format to avoid denaturation of surface-adsorbed antigens. J. Immunol. Methods 158:267-276. [DOI] [PubMed] [Google Scholar]
- 20.Petosa, C., R. J. Collier, K. R. Klimpel, S. H. Leppla, and R. C. Liddington. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833-838. [DOI] [PubMed] [Google Scholar]
- 21.Phipps, A. J., C. Premanandan, R. E. Barnewall, and M. D. Lairmore. 2004. Rabbit and nonhuman primate models of toxin-targeting human anthrax vaccines. Microbiol. Mol. Biol. Rev. 68:617-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pitt, M. L. M., S. Little, B. E. Ivins, P. Fellows, J. Boles, J. Barth, J. Hewetson, and A. M. Friedlander. 1999. In vitro correlate of immunity in an animal model of inhalational anthrax. J. Appl. Microbiol. 87:304. [DOI] [PubMed] [Google Scholar]
- 23.Pittman, P. R., P. H. Gibbs, T. L. Cannon, and A. M. Friedlander. 2001. Anthrax vaccine: short-term safety experience in humans. Vaccine 20:972-978. [DOI] [PubMed] [Google Scholar]
- 24.Reason, D., J. Liberato, J. Y. Sun, W. Keitel, and J. H. Zhou. 2009. Frequency and domain specificity of toxin-neutralizing paratopes in the human antibody response to anthrax vaccine adsorbed. Infect. Immun. 77:2030-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reason, D. C., A. Ullal, J. Liberato, J. Sun, W. Keitel, and J. Zhou. 2008. Domain specificity of the human antibody response to Bacillus anthracis protective antigen. Vaccine 26:4041-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenthal, S. R., and J. C. Clifford. 2002. Development of vaccines for bio-warfare agents. Dev. Biol. (Basel) 110:99-105. [PubMed] [Google Scholar]
- 27.Schwab, C., and H. R. Bosshard. 1992. Caveats for the use of surface-adsorbed protein antigen to test the specificity of antibodies. J. Immunol. Methods 147:125-134. [DOI] [PubMed] [Google Scholar]
- 28.Verma, A., M. M. Ngundi, B. D. Meade, R. De Pascalis, K. L. Elkins, and D. L. Burns. 2009. Analysis of the Fc gamma receptor-dependent component of neutralization measured by anthrax toxin neutralization assays. Clin. Vaccine Immunol. 16:1405-1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Williamson, E. D., I. Hodgson, N. J. Walker, A. W. Topping, M. G. Duchars, J. M. Mott, J. Estep, C. LeButt, H. C. Flick-Smith, H. E. Jones, H. Li, and C. P. Quinn. 2005. Immunogenicity of recombinant protective antigen and efficacy against aerosol challenge with anthrax. Infect. Immun. 73:5978-5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou, J., A. Ullal, J. Liberato, J. Sun, W. Keitel, and D. C. Reason. 2008. Paratope diversity in the human antibody response to Bacillus anthracis protective antigen. Mol. Immunol. 45:338-347. [DOI] [PMC free article] [PubMed] [Google Scholar]