Abstract
During the initial pandemic influenza H1N1 virus outbreak, assays such as hemagglutination inhibition and microneutralization provided important information on the relative protection afforded by the population's cross-reactivity from prior infections and immunizations with seasonal vaccines. However, these assays continue to be limited in that they are difficult to automate for high throughput, such as in pandemic situations, as well as to standardize between labs. Thus, new technologies are being sought to improve standardization, reliability, and throughput by using chemically defined reagents rather than whole cells and virions. We now report the use of a cell-free and label-free flu antibody biosensor assay (f-AbBA) for influenza research and diagnostics that utilizes recombinant hemagglutinin (HA) in conjunction with label-free biolayer interferometry technology to measure biomolecular interactions between the HA and specific anti-HA antibodies or sialylated ligands. We evaluated f-AbBA to determine anti-HA antibody binding activity in serum or plasma to assess vaccine-induced humoral responses. This assay can reveal the impact of antigenic difference on antibody binding to HA and also measure binding to different subtypes of HA. We also show that the biosensor assay can measure the ability of HA to bind a model sialylated receptor-like ligand. f-AbBA could be used in global surveillance laboratories since preliminary tests on desiccated HA probes showed no loss of activity after >2 months in storage at room temperature, indicating that the same reagent lots could be used in different laboratories to minimize interlaboratory assay fluctuation. Future development of such reagents and similar technologies may offer a robust platform for future influenza surveillance activities.
Vaccination, the cornerstone of public health intervention, helps prevent influenza morbidity and mortality. Effective vaccines induce protective immunity which is correlated with the presence of virus-specific antibodies (Abs) in serum that are directed against the external coat proteins of the virion, hemagglutinin (HA) and, to a lesser extent, neuraminidase (NA). HA is the principal antigen on the viral surface, and neutralizing antibodies are usually directed to hypervariable epitopes located in or near the HA receptor-binding site (RBS) and act to prevent host infection by blocking virus binding to the host cell (17). For this reason, induction of HA-specific antibodies that interfere with virus entry is used as a correlate of vaccine protective efficacy.
Influenza viruses are characterized by their rapid antigenic change as a result of their high mutation frequency. Therefore, the composition of influenza virus vaccines requires frequent updates (every 2 years on average for H3N2) to match their antigenicity as closely as possible to that of the variant viruses most prevalent in the population (25). Analysis of antibody responses that correlate with protective immunity to influenza virus vaccination is an important element in the assessment of the potential impact of viral antigenic drift. The hemagglutination inhibition (HI) test is the most widely used serological test for the detection of anti-influenza virus antibodies (13, 36) and is used routinely to determine the serological outcome of vaccinations. The assay itself is technically simple but difficult to automate and standardize. In addition, interpretation of results can be affected by virus passage, antibody source (species), and variability between red blood cells from different species (19, 36).
The continuing but sporadic human infections with H5N1 avian influenza viruses reported since 1997 have revealed that HI assays are less sensitive in detecting antibodies against avian influenza viruses (2, 16, 23, 24, 28) than are alternative assays such as the microneutralization (MN) test (24). The MN test, however, is technically very demanding and is currently performed only as a reference test on a small number of serum specimens. As virus-neutralizing activity of antiserum is mediated in part by blocking virus-receptor interactions, the results of the HI test often correlate well with those of the MN test. Recent international laboratory network studies showed large intra- and interlaboratory assay variations for both the HI and MN assays (26), although for H5N1, such variability can be reduced through the availability of an antibody standard (27). Despite limited reproducibility between labs, the HI and MN tests still provide the best available data to inform global surveillance and aid in decisions to update the seasonal influenza virus vaccine composition. Indeed, their use in recent months has been critical in assessing immunity to the pandemic H1N1 virus in the human population (4, 20). It is clear that new technologies and assays are urgently needed to improve sensitivity, accuracy, and sample throughput, as well as reproducibility between labs. In addition, assays need to be flexible and adaptable to be able to analyze antibody responses to emerging viruses from sources other than avian populations (12).
Here, we report on a flexible label-free and cell-free assay to determine the relative functional avidity of polyclonal serum antibodies binding to the major virion coat protein, HA. The flu antibody biosensor assay (f-AbBA) presented here utilizes an established recombinant baculovirus expression system for producing HA (32) in conjunction with label-free biolayer interferometry (BLI) technology from Fortebio Inc., an optical technique that analyzes the interference pattern of white light reflected from a layer of immobilized protein on the tip of a fiber optic biosensor (1, 7). Macromolecules binding to the biosensor tip produce an increase in optical thickness at the biosensor tip, which results in a wavelength shift (measured in nanometers) that can be followed in real time, allowing one to determine binding kinetics. BLI technology minimizes interference from extraneous materials present in solution. Only molecules that bind to or dissociate from the biosensor surface produce a signal change. Sample preparation time is reduced because crude mixtures such as cell lysates, patient serum, or hybridoma supernatants can be assayed. The platform is designed to simultaneously analyze eight samples as a dip-and-read format using commercially available 96-well plates. We have used it to measure the biomolecular interactions of recombinant HA with reactive antibodies from patient sera in real time, without the need for secondary reagents for detection. Results have good correlation with existing serological methods but with improved sensitivity and reproducibility. By comparing binding to different HAs, we can determine the polyclonal specificity, while limited studies with human sera from prior vaccination studies reveal a linear correlation between binding observed with the biosensor and corresponding HI titers as well as some cross-reactivity with the 2009 pandemic H1N1 virus HA. Initial stability tests suggest that reagents may be prepared and desiccated on probes and stored at ambient temperature for extended periods of time without loss of function, offering different laboratories the chance to utilize the same reagents and thus improve interlaboratory assay variation.
MATERIALS AND METHODS
Interferometry equipment and binding assays.
An Octet Red instrument (Fortebio, Inc., Menlo Park, CA) was used for all assay development and subsequent binding studies. Binding to the biosensor tip is measured as a wavelength shift (in nanometers). Data were analyzed using the system software or exported as a Microsoft Excel file for analysis and presentation in other software packages. Unless stated otherwise, data analysis is reported only for association data collected for 5 min.
Ferret sera.
For ferret sera against the H3 viruses A/Sydney/5/97 and Brisbane/10/07, 6- to 8-month-old male castrated ferrets (Triple F Farms, Sayre, PA) seronegative in the HI test for currently circulating human influenza A (H1N1 and H3N2) and influenza B viruses were used to generate sera. Ferrets were infected by intranasal inoculation, and blood was collected 15 days later. Sera from individual animals were tested by HI and used without pooling. All other sera were obtained from the Viral Surveillance and Diagnosis Branch at the CDC using viruses provided through the WHO Global Influenza Surveillance Network.
As with the HI assay, f-AbBA is also subject to interference from sialylated components in the sera that may also bind to the HA and affect results. Thus, all the sera used in this study were pretreated with Vibrio cholerae neuraminidase, also known in the influenza field as receptor-destroying enzyme (RDE) (Denka Seiken Co. Ltd., Tokyo, Japan), according to a previously described procedure (35) and were diluted to working dilutions of 1:10 with phosphate-buffered saline (PBS), pH 7.2, containing 100 μg/ml bovine serum albumin (Sigma, St. Louis, MO). Table 1 shows a complete list of sera used in these studies.
TABLE 1.
Recombinant HA and ferret sera used in this study
| Subtypea | Virus(es) used for recombinant HA | Accession no. | Abbreviation used in this study | Homologous ferret serumb | Heterologous ferret serum or sera |
|---|---|---|---|---|---|
| H1N1 | New Caledonia/20/1999 | AJ344014 | H1-NC99 | Anti-H1-NC99 | |
| Solomon Islands/3/2006 | EU124177 | H1-SI06 | Anti-H1-SI06 | ||
| Brisbane/59/2007 | CY030230 | H1-BR07 | Anti-H1-BR07 | ||
| H3N2 | California/7/2004 | EU103820 | H3-CA04 | Anti-H3-CA04 | Anti-H3-Syd97 (Sydney/5/1997) |
| Hiroshima/52/2005 (Wisconsin/67/2005-like virus) | EU283414 | H3-WI05-L | Anti-H3-WI05-L | ||
| Brisbane/10/2007 | EU199250 | H3-BR07 | Anti-H3-BR07 | ||
| Pandemic H1N1 | California/4/2009 | FJ966082 | H1pdm | Anti-H1pdm | |
| H5N1 | Vietnam/1203/2004 | EF541403 | H5-VN04 clade 1 | Anti-H5-VN04 | Anti-H5-INDO05 clade 2.1 (Indonesia/5/2005), anti-H5-BHG05 clade 2.2 (BHG/Qinghai/1A/2005), anti-H5-ANH05 clade 2.3.4 (Anhui/1/2005), anti-H5-EG06 clade 2.2 (Egypt/1162/2006) |
All HA genes were cloned and expressed in a baculovirus expression system.
Homologous sera are listed as such if the equivalent recombinant HA was expressed for this study.
Human serum specimens.
Human serum samples were obtained between October and December 2007 from 12 individuals who were vaccinated with the 2007-2008 influenza virus vaccine for the northern hemisphere (18). Six patients (TIV1 to TIV6) were vaccinated with the trivalent inactivated virus (TIV) vaccine Fluzone (Sanofi Pasteur), while the other six (LAIV1 to LAIV6) received the live attenuated influenza virus (LAIV) vaccine, Flumist (Medimmune). Paired pre- and postvaccination sera were collected at day 0 and between days 21 and 33 postvaccination. Anonymous human serum samples from the general population (GP1 to GP10) were also obtained from Connie M. Westhoff (American Red Cross, Philadelphia, PA). Sera were collected in the months of November and December 2008. All sera were pretreated with RDE as described previously (35) and diluted to working stocks with PBS. For isotyping experiments, murine anti-human isotype antibodies against IgG1, IgG2, IgG3, IgG4, IgA, IgE, and IgM were all from Invitrogen (Carlsbad, CA).
Recombinant HA expression and purification.
By using the cloning strategy from previous studies (29-32), the cDNAs encoding the HA ectodomains of various seasonal and pandemic-potential influenza viruses were cloned into the baculovirus transfer vector pAcGP67-A (BD Biosciences, San Jose, CA). Transfection and virus amplification were carried out as described previously (29-32). For protein expression from Trichoplusia ni (Hi5) cells (Invitrogen, Carlsbad, CA), 10-stack CellSTACK culture chambers (Corning Inc., Corning, NY) were used and soluble HA was recovered from the culture supernatant and purified by metal affinity chromatography and gel filtration chromatography as described previously (29-32).
Biosensor preparation.
Recombinant HAs were diluted to 40 to 100 μg/ml in 1× kinetics buffer (PBS containing 0.02% Tween 20, 0.005% sodium azide, and 100 μg/ml bovine serum albumin, purchased from Fortebio as 10× stock) and were coupled either online (during the assay) or offline (prior to running the assay) to streptavidin-coated biosensors. To achieve this, biosensors were precoupled to penta-His biotin conjugate (Qiagen, Valencia, CA) or biotin nitrilotriacetic acid (BNTA) (Biotium, Hayward, CA), according to instructions provided by Fortebio.
HI assays.
HI assays were performed in Nunc V-bottomed 96-well microtiter plates (Thermo Fisher Scientific, Rochester, NY) with 0.5% turkey erythrocytes, as previously described (35). Assays were performed without knowledge of results from biosensor experiments. For the neutralizing antibody assay, experiments were carried out as for serum analysis except that an additional step with a mixture containing fetuin (Sigma-Aldrich), at a concentration of 0.5 μg/ml, was included.
Stability of HA on biosensor probes.
H5-VN04 HA was coupled to biosensors via a penta-His biotin conjugate (Qiagen, Valencia, CA). After a brief washing step in 1× kinetics buffer, H5-VN04 HA-loaded sensors were incubated in different storage solutions for 2 min and then air dried for 5 to 10 min. Biosensors were stored at room temperature in the dark in a desiccated storage cabinet. For performance studies with time, multiple biosensors were prepared and stored for more than 2 months. At times indicated, they were analyzed for interaction with the anti-H5N1 monoclonal antibody B519M (Abcam, Cambridge, MA) and/or fetuin (Sigma, St. Louis, MO).
RESULTS
Recombinant HA expression in a baculovirus system.
Recombinant HA was cloned and expressed according to methods described previously (29-32). For assay development, HAs selected to be components of recent seasonal influenza virus vaccines (Table 1), as well as pandemic (H1N1pdm) and pandemic-potential (H5N1) subtypes, were expressed. Standard procedures for purification were employed for all targets, and only the ion-exchange step resulted in slight variability in elution times between subtypes. For each preparation using 10 CellSTACK culture chambers, yields were variable but in the range of 3 to 8 mg (H1), 6 to 12 mg (H3), 10 to 30 mg (H5), and 3 to 7 mg (H1pdm). All proteins were expressed as trimers with a “foldon” sequence (32) and a His tag at the extreme C terminus of the construct to enable protein purification. Protein preparations were analyzed for purity and homogeneity by SDS-PAGE, size exclusion chromatography, dynamic light scattering using a Dynapro plate reader (Wyatt Technologies), and glycan microarray analysis. Rather than coupling the HA to the biosensor in a random fashion, the His tag was used for subsequent assay development. The His tag, positioned at the membrane-proximal end of the HA, orientates the molecule for optimal antibody accessibility to its antigenic sites.
Instrument performance.
The instrument was assessed for variability in binding recombinant H1pdm HA to the biosensors as well as variability in anti-H1pdm ferret serum binding to the bound anti-His-tag/HA complexes. Results showed a good correlation in both intra- and interchannel variation with an overall coefficient of variation (CV) of 9.2% for loading recombinant HA at 100 μg/ml (Table 2) and 9.1% for subsequent binding of homologous RDE-treated ferret serum at a 1-in-80 dilution (Table 3).
TABLE 2.
Instrument variability for recombinant H1pdm HA binding to biosensors
| Channel | Binding signal (nm) |
CV (%) | |||||
|---|---|---|---|---|---|---|---|
| After 600 s, by expt: |
Mean | SD | |||||
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | ||||
| 1 | 0.480 | 0.537 | 0.534 | 0.561 | 0.528 | 0.035 | 6.5 |
| 2 | 0.484 | 0.541 | 0.558 | 0.573 | 0.539 | 0.039 | 7.2 |
| 3 | 0.486 | 0.563 | 0.598 | 0.590 | 0.559 | 0.051 | 9.2 |
| 4 | 0.508 | 0.581 | 0.615 | 0.626 | 0.583 | 0.053 | 9.1 |
| 5 | 0.420 | 0.480 | 0.505 | 0.522 | 0.482 | 0.045 | 9.3 |
| 6 | 0.504 | 0.551 | 0.601 | 0.605 | 0.565 | 0.047 | 8.4 |
| 7 | 0.482 | 0.527 | 0.566 | 0.534 | 0.527 | 0.035 | 6.6 |
| Overalla | 0.540 | 0.050 | 9.2 | ||||
The overall number of probes was 28.
TABLE 3.
Instrument variability for binding of RDE-treated anti-H1pdm ferret serum to immobilized recombinant HA
| Channel | Binding signal (nm) |
CV (%) | |||||
|---|---|---|---|---|---|---|---|
| After 300 s, by expt: |
Mean | SD | |||||
| Expt 1 | Expt 2 | Expt 3 | Expt 4 | ||||
| 1 | 0.667 | 0.701 | 0.736 | 0.723 | 0.707 | 0.030 | 4.3 |
| 2 | 0.654 | 0.737 | 0.795 | 0.754 | 0.735 | 0.059 | 8.1 |
| 3 | 0.689 | 0.772 | 0.816 | 0.827 | 0.776 | 0.063 | 8.1 |
| 4 | 0.565 | 0.613 | 0.676 | 0.676 | 0.632 | 0.054 | 8.5 |
| 5 | 0.669 | 0.687 | 0.762 | 0.740 | 0.714 | 0.044 | 6.1 |
| 6 | 0.657 | 0.662 | 0.709 | 0.631 | 0.664 | 0.033 | 4.9 |
| Overalla | 0.705 | 0.064 | 9.1 | ||||
The overall number of probes was 24.
Assay development with ferret sera to seasonal H3N2 viruses.
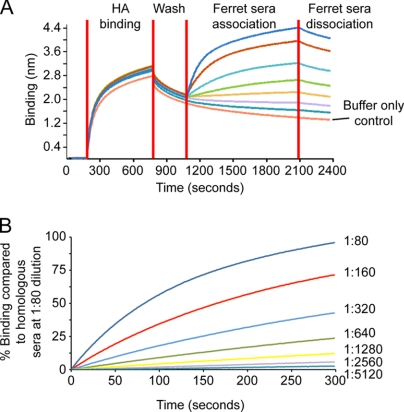
To assess the sensitivity and specificity of detecting antibody binding to HA using the Octet Red system, we initially chose to work on the H3 subtypes using the HA from the virus selected for use in the current seasonal vaccine. To look at sensitivity, H3-BR07 HA was loaded during each run to assess biosensor performance on each of the eight channels on the instrument. On the Octet Red system, eight samples were analyzed in parallel and results revealed comparable binding of the HA to each of the 8 biosensors (Fig. 1 A). For antibody association, anti-H3-BR07 ferret serum was used in seven 2-fold dilutions from 1:80 down to 1:5,120. The eighth channel, a control with no antibody, allows for background subtraction during data processing. Results for the 1:80 dilution were used as a reference to calculate the percentage of observed binding, and for endpoint analysis, we have initially used an empirical 10% binding cutoff value to distinguish significant binding from background. (This cutoff will change as more samples are analyzed and may well differ between different subtypes.) Thus, simplified plots can be generated from which association of the ferret sera with the bound recombinant HA can be detected down to a dilution limit of 1:1,280 (Fig. 1B). The same serum revealed a titer of 1,280 in the hemagglutination inhibition (HI) assay with homologous H3-BR07 virus, suggesting that this platform/assay format is as sensitive as the HI test for detecting HA-specific antibodies in convalescent ferret serum.
FIG. 1.
Detection of antibody binding to recombinant HA by f-AbBA. (A) HA from Brisbane/10/2007 (H3-BR07) was bound and analyzed, in parallel, to eight Octet Red biosensors (HA binding), and after washing to remove excess HA (wash), the biosensors were incubated with 2-fold dilutions of anti-BR07 ferret sera (association from 1:80 to 1:5,120). Each curve is a separate dilution of serum. (B) After data processing, results clearly show that serum binding can be detected down to 1:1,280 (based on the 10% cutoff criterion described in the text). The instrument detects binding to the biosensor tip, which results in a wavelength shift (measured in nanometers).
To address specificity, including comparison of results with the traditional HI assay, different seasonal H3N2 influenza virus strains were selected for expression. In particular, strains used in vaccines from previous seasons (H3-WI05-L and H3-CA04) (Table 1) were chosen due to the availability of ferret sera. Homologous ferret sera (anti-H3-BR07, anti-H3-WI05-L, and anti-H3-CA04), including antiserum to an older H3 virus, Sydney/5/1997 (anti-H3-Syd97), were used. For all assays, binding of ferret antisera (2-fold dilutions from 1:80 down to 1:5,120) to the recombinant HA was assessed for a period of 5 min and results for homologous sera were used as a reference to calculate the percentage of observed binding.
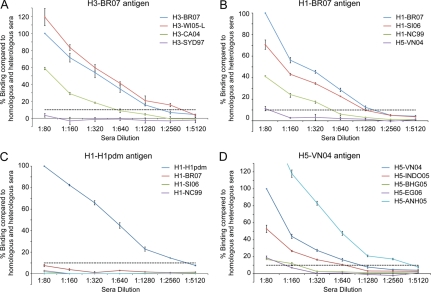
Analysis of ferret serum binding to these H3 HAs revealed detectable binding of ferret sera to all HAs tested, and dilution detection limits were comparable to that detected with an independent HI assay (Fig. 2 A and Table 4 ). Overall, binding using the recombinant HA/Octet Red system was detected at concentrations ∼1- to 2-fold more dilute than those with equivalent HI assay results, with for example the anti-H3-WI05-L sera performing well against all HAs/viruses tested (Table 4). For all three of the H3 HAs tested, there was no observable binding to anti-H3-Syd97 serum and no titer was detected in the HI assay. Recombinant H3-CA04 tested against anti-H3 ferret sera was the only unusual result that was detected on the Octet Red system, at a 2- to 4-fold-lower dilution than that for the HI assay. Thus, results with H3 HAs show a good correlation between HI titers and sensitivity of detection of binding on this platform (Table 4).
FIG. 2.
HA antibody binding detection specificity and sensitivity by f-AbBA. Recombinant HAs from H3-BR07 (A), H1-BR07 (B), H1pdm (C), and H5-VN04 (D) were probed with ferret sera raised against homologous and heterologous viruses. Data are presented as percentages of binding compared to those of homologous sera at 1:80 dilution (100%). The highest dilution with binding above 10% (indicated by a horizontal dashed line above the x axis) was the empirical endpoint, and values are presented in tabular form for direct comparison with titers from an HI assay using equivalent viruses (Table 4). Results correspond to the mean and standard error from 3 independent replicate assays.
TABLE 4.
Comparison of f-AbBA detection limits in H3, H1, and H5 HAs with corresponding HI titers using equivalent viruses
| Antigen | Ferret antiserum to influenza virus | f-AbBA dilutiona | Virus/HI titerb |
|---|---|---|---|
| H3 | |||
| H3-BR07 | H3-BR07 | 1,280 | 1,280 |
| H3-WI05-L | 2,560 | 2,560 | |
| H3-CA04 | 320 | 320 | |
| H3-Syd97 | <80 | <10 | |
| H3-WI05-L | H3-BR07 | 320 | 320 |
| H3-WI05-L | 2,560 | 2,560 | |
| H3-CA04 | 640 | 320 | |
| H3-Syd97 | <80 | <10 | |
| H3-CA04 | H3-BR07 | 160 | 640 |
| H3-WI05-L | 1,280 | 2,560 | |
| H3-CA04 | 2,560 | 5,120 | |
| H3-Syd97 | <80 | <10 | |
| H1 | |||
| H1-BR07 | H1-BR07 | 1,280 | 640 |
| H1-SI06 | 640 | 160 | |
| H1-NC99 | 320 | <10 | |
| H5-VN04 | 80 | NDc | |
| H1-SI06 | H1-BR07 | 1,280 | 320 |
| H1-SI06 | 640 | 640 | |
| H1-NC99 | 640 | <10 | |
| H5-VN04 | <80 | ND | |
| H1-NC99 | H1-BR07 | 320 | 160 |
| H1-SI06 | 320 | 160-320 | |
| H1-NC99 | 1,280 | 2,560 | |
| H5-VN04 | <80 | ND | |
| H5 | |||
| H5-VN04 | VN04 clade 1 | 640 | 160 |
| INDO05 clade 2.1 | 640 | 80 | |
| BHG05 clade 2.2 | 160 | <10 | |
| EG06 clade 2.2 | 80 | 80 | |
| ANH05 clade 2.3.4 | 2,560 | 160-320 |
Maximum dilution that gave >10% binding signal compared to homologous serum binding at 1:80 dilution.
Virus titer determined by HI assay.
ND, not determined.
Assay performance with ferret sera to seasonal H1N1 viruses.
To determine the applicability of this assay for other seasonal viruses, we next looked at H1 HAs and found that profiles similar to those of H3 HAs could be produced (Fig. 2B) with overall ∼2- to 4-fold more binding than that in equivalent HI assays (Table 4), with the H1-NC99 serum being an outlier. We also analyzed cross-reactivity of a pandemic-potential H5N1 HA (H5-VN04), a different subtype but phylogenetically distantly related to H1 (9). As expected with this HA, results revealed little cross-reactivity to H1 ferret sera (results not shown). Conversely, there was no binding of any H1 HA to the anti-H5-VN04 sera (Table 4).
Assay performance with ferret sera to H1N1pdm and other pandemic-potential viruses.
With the ongoing H1N1pdm as well as the threat from other influenza virus subtypes such as H5N1, the flexibility of this assay to adapt to new strains was assessed. We thus expanded studies to use a pandemic H1 HA (H1pdm) (Table 1). In preliminary experiments with the current pandemic H1N1 virus, expression of CA409 HA produced trimeric protein, and as expected, it reacted well with homologous ferret sera but not with seasonal H1N1 sera (Fig. 2C). Compared to H1 HAs, similar results were produced with ∼2- to 4-fold more binding (f-AbBA dilution, 2,560) compared to an equivalent HI assay (titer, 640 to 1,280). Thus, as the H1N1pdm viruses evolve, we fully expect the HAs from these viruses to perform, in this assay, similarly to seasonal H1 HAs.
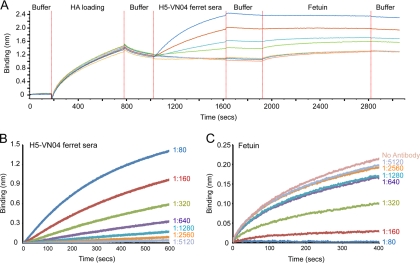
For most of the H5 sera tested in this system, the clade 1 H5-VN04 HA revealed detectable binding at higher dilutions (∼2- to 8-fold) than what was seen with HI assays (Fig. 2D and Table 4). This 2- to 8-fold difference can be reproduced using this platform by prebinding different dilutions of ferret sera to the HA probes and then using fetuin, a 68-kDa sialic acid-containing receptor surrogate that is commonly used in competition assays to study influenza virus receptor binding (10, 11), to assess the serum's ability to inhibit fetuin binding to the receptor-binding site. Results with this receptor binding inhibition assay correlate well with the HI assay in that although binding is detected down to dilutions of 640, fetuin binding is inhibited effectively only at dilutions of 1:160 or less (Fig. 3). Thus, a high proportion of antibodies against H5 HA appear to be nonneutralizing. Anti-H5-ANH05 serum, raised against the clade 2.3.4 A/Anhui/1/2005 virus, was an obvious outlier in that it bound ∼8-fold better than did H5-VN04 homologous sera, even though it gave only slightly better HI titers than did the homologous sera (Fig. 2D and Table 4).
FIG. 3.
Sensitivity in detection of binding anti-H5-VN04 serum and its ability to inhibit fetuin binding. (A) H5-VN04 HA was bound to the Octet Red biosensors (HA binding), and after washing to remove excess HA (wash), the biosensors were incubated with 2-fold dilutions of anti-H5-VN04 ferret sera (association; 1:80 to 1:5,120), followed by incubation with fetuin (0.5 μg/ml). (B and C) After subtraction of the buffer-only control, results clearly show that serum binding (B) can be detected down to 1:1,280, while fetuin binding (C) can be detected only when the ferret serum is at dilutions of 1:160 or more. This serum has a titer of 160 in HI assays (Table 4).
Reactivity of HA to human patient sera.
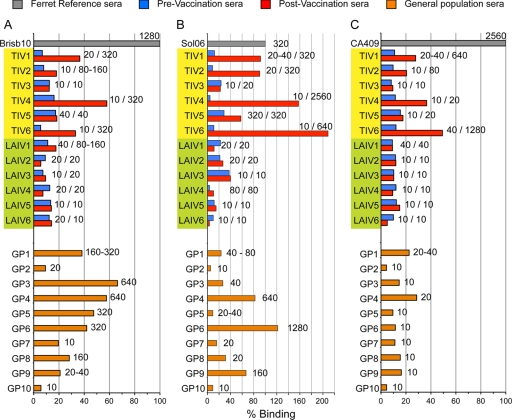
Seasonal H1 and H3 (H1-SI06 and H3-BR07), as well as a pandemic H1 (H1pdm) HA, were used to probe human sera from vaccination studies (from 2007-2008 seasonal vaccine) as well as banked general population sera (collected during the last week of November and the first week of December 2008). To increase throughput, only one dilution, 1:80, of each serum sample was used for analysis. Results are presented as percentage of antibody bound after 5 min, standardized with ferret sera against the homologous virus (100%). In agreement with other antibody assays such as the HI assay, postvaccination sera revealed significant increases (>2-fold over prevaccination sera) in HA binding among the TIV vaccinees and little to no increase in binding with sera derived from LAIV vaccinations (Fig. 4 A and B) (6, 8, 15, 34). Indeed, vaccination with TIV produced similar response profiles for binding to H3 and H1 HAs with the best responses to the H1-SI06 HA (TIV6, TIV4, and TIV1) being mirrored in the H3 binding experiment. However, general population sera produced much more variable binding responses between the H3-BR07 and H1-SI06 experiments, with a number of H3-positive sera having much-reduced binding to H1-SI06 (Fig. 4A and B). The same sera were also analyzed for reactivity to the pandemic H1 HA (H1pdm), and reduced binding was observed among both the LAIV vaccinees and general population sera, with only 2 samples binding >20% (GP1 and GP4), while 4 out of 6 people vaccinated with the TIV (TIV1, TIV2, TIV4, and TIV6) had reactivity to the pandemic H1 HA, postvaccination, with binding of >20% (Fig. 4C). These sera were also best responders to the seasonal vaccine. In order to correlate these results with currently used assays, the human sera were analyzed in HI assays with viruses from which the recombinant HAs were derived. In general, for all three HAs tested, HI titers were comparable with what was seen in the f-AbBA although there were some discrepancies (Fig. 4A to C; HI titers are presented in the graphs). Due to the number of specimens tested in this pilot study, it is not possible to establish any reliable correlation between binding and HI titers. Many more sera will be needed to establish a correlation between HI titers and percent binding.
FIG. 4.
Sensitive and specific detection of antibody to HA in human serum. Binding of human sera to a seasonal H3 virus (BR07) (A), a seasonal H1 virus (Sol06) (B), and a pandemic H1 virus (CA409) (C). General population sera randomly collected by the American Red Cross (GP1 to GP10) as well as 12 pairs of pre-and postvaccination sera were RDE treated and analyzed at a 1:80 dilution. TIV1 to TIV6 (highlighted in yellow) received the TIV vaccine, while LAIV1 to LAIV6 (highlighted in green) received the LAIV vaccine. Results, presented as percentage of antibody bound, were calculated for the binding level after 5 min, standardized with ferret sera against the homologous virus (100%). The ferret sera from the H1-SI06 response bound at a reduced level compared to three of the patient sera tested. Values next to the bars represent corresponding pre- and postvaccination titers (single titers for GP1 to GP10 sera) determined independently by HI assay using turkey red blood cells. Results shown are representative of 2 or 3 experiments.
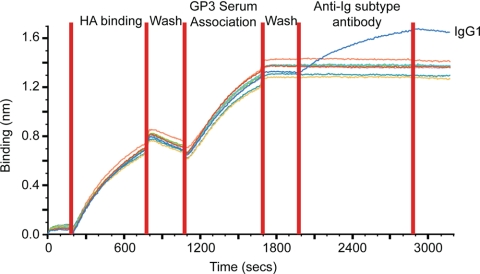
Further information can be gleaned from this assay by including a step in the assay to determine the Ig class of the antibodies binding to the recombinant HA. For example, serum GP3 bound well to H3-BR07, and when probed with anti-human Ig subclass antibodies, only IgG1 could be detected, in line with the previously published findings (8, 15) (Fig. 5).
FIG. 5.
Isotyping the immune response to vaccination. Recombinant H3-BR07 HA (100 μg/ml) was bound to the Octet Red biosensors (HA binding), and after washing to remove excess HA (wash), the biosensors were incubated with human sera (GP3) at a 1:80 dilution, followed by incubation with various anti-human immunoglobulin antibodies (IgG1, IgG2, IgG3, IgG4, IgA, IgE, and IgM), all at 1 μg/ml. Similar results were also obtained with GP3 and TIV1 (results not shown).
HA stability experiments.
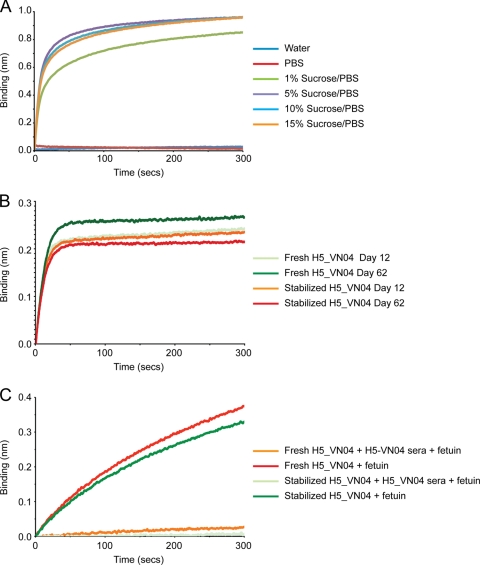
Use of a single lot of ready-to-use biosensors containing the same HA preparation and manufactured under uniform conditions would help to improve reproducibility and throughput analysis. This approach would be facilitated by storage of biosensor probes with desiccated HA. As a proof of concept, studies were performed to assess HA stability on the desiccated biosensors. H5-VN04 HA was coupled to streptavidin-coated biosensors via a penta-His biotin conjugate. H5-VN04 was selected since a number of monoclonal antibodies are commercially available. Biosensors were prepared under a number of storage conditions and stored for 3 days prior to analysis with H5-VN04 sera. Suitable storage conditions were identified using a sucrose-PBS storage buffer (Fig. 6 A). PBS alone or water revealed no binding of the sera to the biosensors. However, more than 5% (wt/vol) sucrose solution appeared to maintain the ability of the HA to bind to sera. A solution of 10% sucrose in PBS was chosen for further study with monoclonal antibodies. HA-bound biosensors were stored for up to ∼80 days, during which time they were screened for binding to neutralizing monoclonal antibodies or fetuin. Results showed that after 62 days probes with stabilized HA bound to monoclonal antibodies similarly to probes loaded with HA on the day of the analysis (Fig. 6B), indicating the presence of these functional epitopes. At day 76, probes were also assessed for their ability to bind to glycans. Experiments were performed where H5-VN04-loaded probes were treated with fetuin with or without pretreatment with H5-VN04 ferret sera (1:80 dilution). Results clearly showed that both freshly loaded and stabilized H5-VN04 HA probes could bind to ferret serum, which blocks subsequent interaction with fetuin (Fig. 6C). Results thus indicate that H5-VN04-loaded probes can be successfully stored for at least 3 months without significant loss of performance, since antigenic sites and receptor binding properties appear to be maintained at comparable levels. Future work will clarify whether this strategy for biosensor preparation and storage is also applicable to other subtypes.
FIG. 6.
Assessment of H5-VN04 stability upon drying/storage during a 2- to 3-month period. H5-VN04-coupled biosensors were dried in different storage buffers, stored for 3 days, and assessed for binding to the homologous polyclonal ferret serum identified formulations containing sucrose as a potential condition to store HA-coupled biosensors in a dry format (A). Ten percent sucrose was selected for experiments to assess longer-term storage of biosensors. At days 12 and 62, biosensors were used to detect binding to a monoclonal antibody (B519M) that can neutralize by HI and thus detects a neutralizing antigenic epitope in the HA (B). Results show only a minimal loss of binding compared to that for biosensors loaded with freshly prepared recombinant HA. The viability of the dried HA after 76 days was also assessed by the biosensor's ability to bind the glycoprotein fetuin with and without preincubation with anti-H5-VN04 ferret serum (which detects a functional receptor binding site) (C).
DISCUSSION
In humans, both humoral and cellular immune effector mechanisms play important roles in host protection against influenza. However, the humoral immunity response is the most reliable correlate of protection against influenza in humans and includes mucosal and serum antibody responses. Response to the HA protein is most important since HA-specific antibodies can neutralize the virus by preventing it from initiating an infection. Neutralization involves blocking binding of the virus to host cells but may also work at other steps involved in the entry and uncoating of the virus (21, 33).
These assays in which the biosensor probe is loaded with HA analyze the polyclonal antibody response to HA but are different from the HI and MN assays in that they measure the total antibody response and cannot distinguish between antibodies binding near the RBS (as in the case of the HI test) and other nonneutralizing antigenic sites. Although nonspecific effects such as binding serum glycoproteins and glycolipids containing sialic acid may affect this assay, we minimize these effects by both RDE treatment and dilution (at 1:80) of the sera prior to analysis. In addition the assay could be focused further to highlight only bound antibodies by utilizing an additional step using an anti-human Ig polyclonal reagent, similar to the isotyping experiment shown in Fig. 5. However, comparison of the reactivity of ferret sera in this assay to that in a corresponding HI assay revealed a good correlation, although there was, in general, increased detection in the biosensor assay (from 2- to 8-fold depending on the HA being analyzed). Comparing results from H1, H3, and H5 revealed the best correlation to be that with the H3 HAs analyzed (Table 4).
The increased disparity between the detection level observed and HI results for the H1 and H5 HAs may be due to the presence of non-HI antibodies. Results with the H5-VN04 HA suggest this to be the case, as although binding of the sera was detected down to dilutions of 1:640 to 1,280, fetuin binding to the receptor-binding site could be inhibited effectively only at dilutions of 1:160 (Fig. 3). Thus, our H5 (and H1) antisera appear to contain a higher proportion of non-HI antibodies than do antisera to H3 HA. The ability of an antibody to block access to the receptor-binding site, as described for Fig. 3, is the basis of an HI assay. However, this assay requires an available HA, and currently, it takes 4 to 5 weeks to produce recombinant HA in quantities sufficient for analysis. Thus, such a biosensor assay cannot replace the HI in its current format, particularly in a pandemic situation. However, its future development to define reagents that can bind to human-adapted seasonal and pandemic virus HAs could be useful in situations where it can complement current assays such as analyzing a patient's immune response to infection and/or vaccination.
The assay platform was further characterized by a limited analysis of human sera from vaccine efficacy trials with both TIV and LAIV, as well as anonymous human serum samples from the Red Cross (collected during November and December 2008). Although the two vaccines stimulate different compartments of the immune system (3), both prevent laboratory-confirmed influenza (3) and reduce viral shedding and the severity of respiratory symptoms (5). TIV vaccines produce a significantly higher level of neutralizing serum IgG (particularly IgG1) and IgA antibody responses than do LAIV vaccines (6, 8, 15, 34). Our results are consistent with all these observations in that a greater difference between pre- and postvaccination sera was seen with the sera collected from people receiving the TIV vaccine (Fig. 4). Interestingly, a number of the sera also from the general population had appreciable binding to H3-BR07 and H1-BR07 (e.g., GP4 and GP6), suggesting that these people may have been vaccinated, while one sample (GP5) bound to H3-BR07 only, suggesting possible exposure to a circulating H3 virus.
The serum antibody response to influenza virus infection or vaccination is commonly measured using the HI test and can be detected in approximately 80% of subjects after natural influenza virus infection with an HI titer of ≥40 (14, 22). Indeed, an HI titer of 40 is correlated with protective immunity to antigenically homologous viruses (14). Based on this pilot study comprising only a limited number of positive samples, it is not possible to establish a reliable correlation between biosensor binding data and HI titers. However, as more reagents and human sera are screened, assay performance can be subjected to a more rigorous examination and accuracy/significance will be determined.
With respect to influenza, the next generation of assays should be able to detect antibodies in serum and plasma, with high sensitivity and minimal sample processing, while minimizing cross-reactivity and nonspecific binding. Biolayer interferometry technology was selected for study/development because this platform satisfies a number of these criteria. In addition, there are no long incubation steps in the analysis, which improves sample throughput. Indeed, assuming a 5-min analysis time to assess antibody binding to the recombinant HA, up to 96 samples (including controls) can be analyzed in approximately 1 h using the current system. By using such a short analysis time, multiple HAs on separate probes can be used to analyze a single sample in the same assay to look at cross-reactivity to other HAs from similar and different subtypes. Finally, biosensors are disposable, thus reducing the logistics for biosensor regeneration, which might lead to cross-contamination and/or loss of performance.
In order to compare results with different sera and HAs, we thus chose to present binding during each run calculated as a percentage of the maximum binding observed with homologous ferret sera at a 1:80 dilution. In this way, variation between experiments can be normalized using sera as a positive control. Interestingly, not all of the homologous ferret sera resulted in a maximum binding response for all HAs tested (for example, in Fig. 2A, anti-H3-WI05-L binds consistently better to the H3-BR07 HA than does the homologous anti-H3-BR07 serum). Similarly, human sera from vaccinees (TIV4 and TIV6) and the general population (GP6) all bound to H1-SI06 at a level higher than did the ferret sera normalized at the 100% level (Fig. 4). Thus, other antibody sources such as those in hyperimmunized sheep sera with maximal-avidity binding could be considered possible alternatives to ferret sera.
Ultimately, the usefulness of any assay for medium-/high-throughput analysis in the public health arena requires reagents to be in a ready-to-use format and preferably have a long shelf life. Preliminary tests on H5 HA stability proved that recombinant HA can be prepared on probes and stored at room temperature for more than 2 months without loss of performance, as measured by neutralizing monoclonal antibody binding as well as binding of fetuin, a sialic acid-containing receptor surrogate. Further development of this methodology for other HAs is required; this technology could help to standardize methodology by minimizing inter- and intralaboratory variability, improving reagents, and introducing assay automation. Future development of such reagents and alternative technologies to include multiplexing might also offer a robust platform for future influenza surveillance activities.
Acknowledgments
We thank Jan Mabry, Amanda Balish, Xiyan Xu, and Alexander Klimov for providing ferret antisera and Connie M. Westhoff (American Red Cross, Philadelphia, PA) for providing general population human sera. We thank Li-Mei Chen for providing viral stocks and the WHO Global Influenza Surveillance Network for providing viruses.
This work was funded in part by a grant to J.S. from the Department of Health and Human Services National Vaccine Program Office.
Several HA proteins used in this study will be made available through the Influenza Reagent Resource, established by the Centers for Disease Control and Prevention (http://www.influenzareagentresurce.org).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention or the Agency for Toxic Substances and Disease Registry.
Footnotes
Published ahead of print on 21 July 2010.
REFERENCES
- 1.Abdiche, Y., D. Malashock, A. Pinkerton, and J. Pons. 2008. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 377:209-217. [DOI] [PubMed] [Google Scholar]
- 2.Beare, A. S., and R. G. Webster. 1991. Replication of avian influenza viruses in humans. Arch. Virol. 119:37-42. [DOI] [PubMed] [Google Scholar]
- 3.Beyer, W. E., A. M. Palache, J. C. de Jong, and A. D. Osterhaus. 2002. Cold-adapted live influenza vaccine versus inactivated vaccine: systemic vaccine reactions, local and systemic antibody response, and vaccine efficacy. A meta-analysis. Vaccine 20:1340-1353. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2009. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb. Mortal. Wkly. Rep. 58:521-524. [PubMed] [Google Scholar]
- 5.Clements, M. L., R. F. Betts, E. L. Tierney, and B. R. Murphy. 1986. Serum and nasal wash antibodies associated with resistance to experimental challenge with influenza A wild-type virus. J. Clin. Microbiol. 24:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clements, M. L., and B. R. Murphy. 1986. Development and persistence of local and systemic antibody responses in adults given live attenuated or inactivated influenza A virus vaccine. J. Clin. Microbiol. 23:66-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Concepcion, J., K. Witte, C. Wartchow, S. Choo, D. Yao, H. Persson, J. Wei, P. Li, B. Heidecker, W. Ma, R. Varma, L. S. Zhao, D. Perillat, G. Carricato, M. Recknor, K. Du, H. Ho, T. Ellis, J. Gamez, M. Howes, J. Phi-Wilson, S. Lockard, R. Zuk, and H. Tan. 2009. Label-free detection of biomolecular interactions using BioLayer interferometry for kinetic characterization. Comb. Chem. High Throughput Screen. 12:791-800. [DOI] [PubMed] [Google Scholar]
- 8.El-Madhun, A. S., R. J. Cox, and L. R. Haaheim. 1999. The effect of age and natural priming on the IgG and IgA subclass responses after parenteral influenza vaccination. J. Infect. Dis. 180:1356-1360. [DOI] [PubMed] [Google Scholar]
- 9.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gambaryan, A., R. Webster, and M. Matrosovich. 2002. Differences between influenza virus receptors on target cells of duck and chicken. Arch. Virol. 147:1197-1208. [DOI] [PubMed] [Google Scholar]
- 11.Gambaryan, A. S., and M. N. Matrosovich. 1992. A solid-phase enzyme-linked assay for influenza virus receptor-binding activity. J. Virol. Methods 39:111-123. [DOI] [PubMed] [Google Scholar]
- 12.Garten, R. J., C. T. Davis, C. A. Russell, B. Shu, S. Lindstrom, A. Balish, W. M. Sessions, X. Xu, E. Skepner, V. Deyde, M. Okomo-Adhiambo, L. Gubareva, J. Barnes, C. B. Smith, S. L. Emery, M. J. Hillman, P. Rivailler, J. Smagala, M. de Graaf, D. F. Burke, R. A. Fouchier, C. Pappas, C. M. Alpuche-Aranda, H. Lopez-Gatell, H. Olivera, I. Lopez, C. A. Myers, D. Faix, P. J. Blair, C. Yu, K. M. Keene, P. D. Dotson, Jr., D. Boxrud, A. R. Sambol, S. H. Abid, K. St. George, T. Bannerman, A. L. Moore, D. J. Stringer, P. Blevins, G. J. Demmler-Harrison, M. Ginsberg, P. Kriner, S. Waterman, S. Smole, H. F. Guevara, E. A. Belongia, P. A. Clark, S. T. Beatrice, R. Donis, J. Katz, L. Finelli, C. B. Bridges, M. Shaw, D. B. Jernigan, T. M. Uyeki, D. J. Smith, A. I. Klimov, and N. J. Cox. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirst, G. K. 1942. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J. Exp. Med. 75:47-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hobson, D., R. L. Curry, A. S. Beare, and A. Ward-Gardner. 1972. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J. Hyg. (Lond.) 70:767-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hocart, M. J., J. S. Mackenzie, and G. A. Stewart. 1990. Serum IgG subclass responses of humans to inactivated and live influenza A vaccines compared to natural infections with influenza A. J. Med. Virol. 30:92-96. [DOI] [PubMed] [Google Scholar]
- 16.Kayali, G., S. F. Setterquist, A. W. Capuano, K. P. Myers, J. S. Gill, and G. C. Gray. 2008. Testing human sera for antibodies against avian influenza viruses: horse RBC hemagglutination inhibition vs. microneutralization assays. J. Clin. Virol. 43:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Knossow, M., and J. J. Skehel. 2006. Variation and infectivity neutralization in influenza. Immunology 119:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monto, A. S., S. E. Ohmit, J. G. Petrie, E. Johnson, R. Truscon, E. Teich, J. Rotthoff, M. Boulton, and J. C. Victor. 2009. Comparative efficacy of inactivated and live attenuated influenza vaccines. N. Engl. J. Med. 361:1260-1267. [DOI] [PubMed] [Google Scholar]
- 19.Ndifon, W., J. Dushoff, and S. A. Levin. 2009. On the use of hemagglutination-inhibition for influenza surveillance: surveillance data are predictive of influenza vaccine effectiveness. Vaccine 27:2447-2452. [DOI] [PubMed] [Google Scholar]
- 20.Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team et al. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360:2605-2615. [DOI] [PubMed] [Google Scholar]
- 21.Okuno, Y., Y. Isegawa, F. Sasao, and S. Ueda. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J. Virol. 67:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Potter, C. W., and J. S. Oxford. 1979. Determinants of immunity to influenza infection in man. Br. Med. Bull. 35:69-75. [DOI] [PubMed] [Google Scholar]
- 23.Profeta, M. L., and G. Palladino. 1986. Serological evidence of human infections with avian influenza viruses. Brief report. Arch. Virol. 90:355-360. [DOI] [PubMed] [Google Scholar]
- 24.Rowe, T., R. A. Abernathy, J. Hu-Primmer, W. W. Thompson, X. Lu, W. Lim, K. Fukuda, N. J. Cox, and J. M. Katz. 1999. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J. Clin. Microbiol. 37:937-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell, C. A., T. C. Jones, I. G. Barr, N. J. Cox, R. J. Garten, V. Gregory, I. D. Gust, A. W. Hampson, A. J. Hay, A. C. Hurt, J. C. de Jong, A. Kielso, A. I. Klimov, T. Kageyama, N. Komadina, A. S. Lapedes, Y. P. Lin, A. Mosterin, M. Obuchi, T. Odagiri, A. D. Osterhaus, G. F. Rimmelzwaan, M. W. Shaw, E. Skepner, K. Stohr, M. Tashiro, R. A. Fouchier, and D. J. Smith. 2008. Influenza vaccine strain selection and recent studies on the global migration of seasonal influenza viruses. Vaccine 26(Suppl. 4):D31-D34. [DOI] [PubMed] [Google Scholar]
- 26.Stephenson, I., R. G. Das, J. M. Wood, and J. M. Katz. 2007. Comparison of neutralising antibody assays for detection of antibody to influenza A/H3N2 viruses: an international collaborative study. Vaccine 25:4056-4063. [DOI] [PubMed] [Google Scholar]
- 27.Stephenson, I., A. Heath, D. Major, R. W. Newman, K. Hoschler, W. Junzi, J. M. Katz, J. P. Weir, M. C. Zambon, and J. M. Wood. 2009. Reproducibility of serologic assays for influenza virus A (H5N1). Emerg. Infect. Dis. 15:1250-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stephenson, I., J. M. Wood, K. G. Nicholson, and M. C. Zambon. 2003. Sialic acid receptor specificity on erythrocytes affects detection of antibody to avian influenza haemagglutinin. J. Med. Virol. 70:391-398. [DOI] [PubMed] [Google Scholar]
- 29.Stevens, J., O. Blixt, L. M. Chen, R. O. Donis, J. C. Paulson, and I. A. Wilson. 2008. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J. Mol. Biol. 381:1382-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stevens, J., O. Blixt, L. Glaser, J. K. Taubenberger, P. Palese, J. C. Paulson, and I. A. Wilson. 2006. Glycoarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J. Mol. Biol. 355:1143-1155. [DOI] [PubMed] [Google Scholar]
- 31.Stevens, J., O. Blixt, T. M. Tumpey, J. K. Taubenberger, J. C. Paulson, and I. A. Wilson. 2006. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 312:404-410. [DOI] [PubMed] [Google Scholar]
- 32.Stevens, J., A. L. Corper, C. F. Basler, J. K. Taubenberger, P. Palese, and I. A. Wilson. 2004. Structure of the uncleaved human H1 hemagglutinin from the extinct 1918 influenza virus. Science 303:1866-1870. [DOI] [PubMed] [Google Scholar]
- 33.Sui, J., W. C. Hwang, S. Perez, G. Wei, D. Aird, L. M. Chen, E. Santelli, B. Stec, G. Cadwell, M. Ali, H. Wan, A. Murakami, A. Yammanuru, T. Han, N. J. Cox, L. A. Bankston, R. O. Donis, R. C. Liddington, and W. A. Marasco. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 16:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Treanor, J. J., K. Kotloff, R. F. Betts, R. Belshe, F. Newman, D. Iacuzio, J. Wittes, and M. Bryant. 1999. Evaluation of trivalent, live, cold-adapted (CAIV-T) and inactivated (TIV) influenza vaccines in prevention of virus infection and illness following challenge of adults with wild-type influenza A (H1N1), A (H3N2), and B viruses. Vaccine 18:899-906. [DOI] [PubMed] [Google Scholar]
- 35.World Health Organization. 2002. WHO manual on animal influenza diagnosis and surveillance. World Health Organization, Geneva, Switzerland.
- 36.Zambon, M. 1998. Laboratory diagnosis of influenza, p. 291-313. In K. G. Nicholson, R. G. Webster, and A. J. Hay (ed.), Textbook of influenza. Blackwell, Oxford, United Kingdom.