Abstract
The human oral cavity contains a number of different habitats, including the teeth, gingival sulcus, tongue, cheeks, hard and soft palates, and tonsils, which are colonized by bacteria. The oral microbiome is comprised of over 600 prevalent taxa at the species level, with distinct subsets predominating at different habitats. The oral microbiome has been extensively characterized by cultivation and culture-independent molecular methods such as 16S rRNA cloning. Unfortunately, the vast majority of unnamed oral taxa are referenced by clone numbers or 16S rRNA GenBank accession numbers, often without taxonomic anchors. The first aim of this research was to collect 16S rRNA gene sequences into a curated phylogeny-based database, the Human Oral Microbiome Database (HOMD), and make it web accessible (www.homd.org). The HOMD includes 619 taxa in 13 phyla, as follows: Actinobacteria, Bacteroidetes, Chlamydiae, Chloroflexi, Euryarchaeota, Firmicutes, Fusobacteria, Proteobacteria, Spirochaetes, SR1, Synergistetes, Tenericutes, and TM7. The second aim was to analyze 36,043 16S rRNA gene clones isolated from studies of the oral microbiota to determine the relative abundance of taxa and identify novel candidate taxa. The analysis identified 1,179 taxa, of which 24% were named, 8% were cultivated but unnamed, and 68% were uncultivated phylotypes. Upon validation, 434 novel, nonsingleton taxa will be added to the HOMD. The number of taxa needed to account for 90%, 95%, or 99% of the clones examined is 259, 413, and 875, respectively. The HOMD is the first curated description of a human-associated microbiome and provides tools for use in understanding the role of the microbiome in health and disease.
The microorganisms found in the human oral cavity have been referred to as the oral microflora, oral microbiota, or more recently as the oral microbiome. The term microbiome was coined by Joshua Lederberg “to signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space and have been all but ignored as determinants of health and disease” (37). The term microbiome has been embraced by the Human Microbiome Project and investigators who believe an understanding of human health and disease is impossible without fully understanding the collective microbiome/human “superorganism.” This work describes the identification and phylogeny of the most prevalent oral taxa.
The oral cavity, or mouth, includes several distinct microbial habitats, such as teeth, gingival sulcus, attached gingiva, tongue, cheek, lip, hard palate, and soft palate. Contiguous with the oral cavity are the tonsils, pharynx, esophagus, Eustachian tube, middle ear, trachea, lungs, nasal passages, and sinuses. We define the human oral microbiome as all the microorganisms that are found on or in the human oral cavity and its contiguous extensions (stopping at the distal esophagus), though most of our studies and samples have been obtained from within the oral cavity. Studies have shown that different oral structures and tissues are colonized by distinct microbial communities (2, 39). Approximately 280 bacterial species from the oral cavity have been isolated in culture and formally named. It has been estimated that less than half of the bacterial species present in the oral cavity can be cultivated using anaerobic microbiological methods and that there are likely 500 to 700 common oral species (47). Cultivation-independent molecular methods, primarily using 16S rRNA gene-based cloning studies, have validated these estimates by identifying approximately 600 species or phylotypes (47; http://www.homd.org).
The oral cavity is a major gateway to the human body. Food enters the mouth and is chewed and mixed with saliva on its way to the stomach and intestinal tract. Air passes through the nose and mouth on the way to the trachea and lungs. Microorganisms colonizing one area of the oral cavity have a significant probability of spreading on contiguous epithelial surfaces to neighboring sites. Microorganisms from the oral cavity have been shown to cause a number of oral infectious diseases, including caries (tooth decay), periodontitis (gum disease), endodontic (root canal) infections, alveolar osteitis (dry socket), and tonsillitis. Evidence is accumulating which links oral bacteria to a number of systemic diseases (58), including cardiovascular disease (6, 32), stroke (31), preterm birth (46), diabetes (25), and pneumonia (4).
For most of the history of infectious diseases, medical practitioners focused on individual organisms in pure culture through the perspective of Koch's postulates. With the realization that essentially all surfaces of humans, animals, plants, and inanimate objects, which have air or water interfaces, are covered by complex microbial biofilms (26), microbiologists have refocused on microbial communities (30). Caries, periodontitis, otitis media, and other infections are now recognized to be caused by consortia of organisms in a biofilm rather than a single pathogen (30). It is the premise of the NIH-supported Human Microbiome Project (51, 64) that we need to know the identity of all of the major organisms comprising the human microbiome to fully understand human health and disease and that we must have tools to rapidly identify members of the microbiome to carry out meaningful clinical research. Culture-independent approaches, like the 16S rRNA gene-based molecular cloning methods, have largely replaced cultivation studies for this task, as the molecular methods can reveal the identities of currently uncultivated microorganisms.
While 16S rRNA gene clone studies have revealed the hidden diversity of the microbial world, clone sequences with no taxonomic anchors currently fill GenBank, and articles refer to novel phylotypes by reference to cryptic clone numbers (69). Taxa known only as 16S rRNA phylotypes cannot be formally named, as naming requires growth and full phenotypic characterization. Candidatus status can be used to name uncultivated or mixed-culture organisms but still requires characterization significantly beyond a 16S rRNA sequence. Thus, there is a critical need for a provisional naming system for species/phylotypes of the human microbiome, so that investigators and the literature can point to provisionally named taxa rather than clone sequences.
The first goal of this research was to develop a provisional taxonomic scheme for the unnamed human oral bacterial isolates and phylotypes and provide this information in an online publicly available database, namely, the Human Oral Microbiome Database (HOMD) (www.homd.org). The second goal was to analyze the 36,043 16S rRNA gene oral clone sequences available from our laboratories to determine the number of clones observed for each human oral taxon and to identify additional taxa not included in the initial setup of the HOMD.
MATERIALS AND METHODS
This report is based on analysis of 16S rRNA gene sequences from 36,043 clones and over 1,000 isolates. Both clone-based and culture-based studies were performed under appropriate Institutional Review Board approval. The 16S rRNA gene sequences, from both clone- and culture-based studies, were obtained in our laboratories over the past 20 years. The protocols listed below represent our current methods.
Bacterial culture studies.
The bacterial isolates came from studies targeting a wide range of oral health and disease statuses, including periodontitis, caries, endodontic infections, and noma. The strains were provided mostly by the laboratories of Tanner, Socransky, and Wade and from the culture collection of Lillian V. Holdeman Moore and the late W. E. C. (Ed) Moore.
Fresh isolates or strains were cultured on BHI+HK (brain heart infusion agar [Becton, Dickinson and Co., Sparks, MD] at 26 g, yeast extract at 5 g, and hemin at 2.5 mg, and menadione at 250 μg in 500 ml H2O plus 25 ml sheep's blood [Northeast Laboratory Services, Winslow, ME]), FAA (fastidious anaerobe agar [Acumedia Manufacturers, Inc. Lansing, MI] at 26 g, yeast extract at 5 g, and hemin at 2.5 mg in 500 ml H2O plus 25 ml sheep's blood), and BUA (Biolog universal agar [Biolog Inc., Hayward, CA] at 26 g in 500 ml H2O supplemented with 25 ml sheep's blood). Subsequent passages were performed using the medium on which the strain grew best. Strain identification was performed by the “Touch PCR” method, where a wire probe or pipette tip was just touched to the colony and a minute sample was transferred directly to a tube for amplification of the 16S rRNA gene operons (see PCR details below). If this quick method did not work, a loopful of cells was collected and placed in 50 μl of 50 mM Tris buffer, pH 7.6, with 1 mM EDTA and 0.5% Tween 20. Proteinase K (200 μg/ml) was added and incubated at 55°C for 2 h. Proteinase K was inactivated by being heated at 95°C for 5 min. A total of 1 μl of this preparation was used for PCR.
16S rRNA gene clone library-based studies.
Table S1 in the supplemental material describes the study source for all clones. Specific details on patient populations, sampling protocols, and sequencing methods used in published studies are given in the references listed in this table. In brief, 16S rRNA gene clone libraries were created from and analyzed in unpublished studies and the following published studies: treponemes in a subject with severe destructive periodontitis (10); treponemes from several subjects with periodontitis and acute necrotizing ulcerative gingivitis (ANUG) (18); subgingival plaque from healthy subjects and subjects with periodontitis, HIV periodontitis, and acute necrotizing ulcerative gingivitis (ANUG) (47); dental plaque from children with caries (7); endodontic lesions (45); subjects with advanced noma lesions (49); subjects with necrotizing ulcerative periodontitis in HIV-positive subjects (1, 50); dorsum tongue microbiota in subjects with halitosis (36); dental caries in adults (44); normal biota of healthy subjects at subgingival, supragingival, dorsal tongue, ventral tongue, hard palate, vestibule, and tonsil sites (2); periodontitis in adults (15); aggressive periodontitis (22); caries-active and caries-free twins (14); root caries in elderly subjects (52); ventilator-associated pneumonia (5).
DNA purification from clinical samples.
Dental plaque from teeth or subgingival periodontal pockets was collected using sterile Gracey curettes. Plaque from the curette was transferred into 100 μl of TE buffer (50 mM Tris-HCl, pH 7.6; 1 mM EDTA). Bacteria on soft tissues were sampled using nylon swabs. The material from the swab was dispersed into 150 μl of TE buffer. DNA extraction was performed using the UltraClean microbial DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA) by following the manufacturer's instructions for the isolation of genomic DNA from Gram-positive bacteria.
16S rRNA gene amplification.
Purified DNA samples were generally amplified with universal primers F24/Y36 to construct broad-coverage libraries. The sequences of primers are given in Table S2 in the supplemental material. Additional libraries seeking expanded coverage of Bacteroidetes/TM7/SR1 groups or Spirochaetes/Synergistetes groups were amplified with F24/F01 or F24/M98 selective primers, respectively. PCR was performed in thin-walled tubes using a PerkinElmer 9700 Thermo Cycler. The reaction mixture (50 μl, final volume) contained 1 μl of the purified DNA template, 20 pmol of each primer, 40 nmol of deoxynucleoside triphosphates (dNTPs), 2.5 unit of Platinum Taq polymerase (Invitrogen, Carlsbad, CA), and 5 μl 10× PCR buffer (200 mM Tris-HCl, pH 8.4; 500 mM KCl). A hot-start protocol was used in which samples were preheated at 94°C for 4 min, followed by amplification using the following conditions: denaturation at 94°C for 45 s, annealing at 60°C for 45 s, and elongation at 72°C for 2 min, with an additional 1 s for each cycle. Thirty cycles were performed, followed by a final elongation step at 72°C for 15 min. Amplicon size and amount were examined by electrophoresis in a 1% agarose gel stained with SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA) and visualized under UV light. After verification that a strong amplicon of the correct size was produced, a preparative gel was run, and the full-length amplicon band was cut out and DNA purified using a Qiagen gel extraction kit (Qiagen, Valencia, CA).
Cloning procedures.
Size-purified 16S rRNA gene amplicons were cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) by following the manufacturer's instructions. Transformation was performed using competent Escherichia coli TOP10 cells provided by the manufacturer. Transformed cells were plated onto Luria-Bertani agar plates supplemented with kanamycin (50 μg/ml) and incubated overnight at 37°C.
Library screening.
Approximately 90 colonies were picked for each library and were placed into tubes containing 40 μl of 10 mM Tris-HCl, pH 8.0. A total of 1 μl of the cell suspension was used directly as the template for PCR with Invitrogen vector M13 (−21) forward and M13 reverse primers. Electrophoresis on a 1% agarose gel was used to verify the correct amplicon size. PCR product for preliminary sequencing with primer Y31 (positions 519 to 533, reverse) was treated with exonuclease and shrimp alkaline phosphatase to remove primers and dNTPs. Five microliters of PCR product was combined with 0.4 μl exonuclease I (10 U/μl; USB Corporation, Cleveland, OH) and 0.4 μl shrimp alkaline phosphatase (1 U/μl; USB Corporation, Cleveland, OH). The reaction mixture was incubated at 37°C for 15 min and then deactivated at 85°C for 15 min. The PCR products from clones chosen for full sequencing with eight additional primers were further concentrated and purified using QIAquick PCR purification kits (Qiagen, Valencia, CA).
16S rRNA gene sequencing.
Purified DNA was sequenced using an ABI Prism cycle sequencing kit (BigDye Terminator cycle sequencing kit) on an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, CA). The sequencing primers (see Table S2 in the supplemental material) were used in quarter-dye reactions by following the manufacturer's instructions.
16S rRNA data analysis.
Sequence information determined using primer Y31 (positions 519 to 533, reverse) allows preliminary identification of clones. Clones or strains whose sequences appeared novel (differing by more than 7 bases from previously identified oral reference sequences in the first 500 bp) were fully sequenced on both strands (approximately 1,540 bases) using 6 to 8 additional sequencing primers (see Table S2 in the supplemental material). Sequences were assembled from the ABI electropherogram files using Sequencher (Gene Codes Corporation, Ann Arbor, MI).
Aligned 16S rRNA database.
All full-length human oral 16S rRNA gene sequences which we believed represented novel taxa and those of named human oral species available in GenBank were entered into a new Aligned Reference Sequence Database. More than 100 nonoral sequences were also entered to link oral phylogenetic clusters to named taxa. The basic program set for data entry, editing and sequence alignment, secondary structure comparison, similarity matrix generation, and phylogenetic tree construction was written by F. E. Dewhirst in Microsoft QuickBasic and has been previously described (48) (the program is available from F. E. Dewhirst). Trees for this work were made by exporting aligned sequences from our database into MEGA version 4 (60). Similarity matrices were corrected for multiple base changes at single positions by the method of Jukes and Cantor (33). Similarity matrices were constructed from the aligned sequences by using only those sequence positions for which 95% of the strains had data. Phylogenetic trees were constructed using the neighbor-joining method of Saitou and Nei (56). Bootstrapping was performed using 1,000 resamplings.
Creation of the human oral microbiome 16S rRNA gene reference set.
The sequences for named species, isolates, and clones were obtained primarily by sequencing efforts in our laboratories or from GenBank. The list of named oral organisms was compiled from the literature and relied heavily on literature reports from investigators at the Forsyth Institute (20, 21, 59, 61, 62) and from Lillian Holdeman Moore and W. E. C. Moore (41, 42, 43), formerly at the Anaerobe Laboratory at the Virginia Polytechnic Institute. To the initial list of oral microorganisms, we added exogenous pathogens, such as Corynebacterium diphtheriae, Bordetella pertussis, Treponema pallidum, Neisseria gonorrhoeae, and several other species which are causative agents of oral lesions and diseases. For the 16S rRNA gene sequences of strains or clones that did not match the named species, we created novel 16S rRNA gene-based phylotypes. We define a phylotype as a cluster of full-length 16S rRNA gene sequences that have greater than 98.5% similarity to one another (≤23 base mismatches per 1,540 bases) and have less than 98.5% similarity to neighboring taxa (species or phylotypes). Each species and phylotype was assigned a human oral taxon (HOT) number, starting at 001. Prior to assigning the HOT numbers, all provisional sequences were compared, and those with sequences having greater than 98.5% similarity were merged into single taxa, except for validly named species, which retained individual HOT numbers regardless of rRNA gene sequence similarity. Sequences were checked for the possibility of being chimeric using multiple methods. Neighbor-joining trees were generated using the first 600 bases and compared with trees using the last 900 bases. Taxa that changed position in the two trees were further examined with the Chimera Check program at the Ribosomal Database Project (11) and with Mallard (3). The first and last 100 bases of all clone sequences described below were analyzed by BLASTN analysis against the HOMD Reference Set. The distance between end matches was captured from a distance matrix file for all full-length HOMD references sequences. All sequences with ends being more than 10% different were rejected as chimeric. The script for this program is available from us. The HOMD reference sequences and clone sequences were rescreened using Chimera Slayer (courtesy of Brian Haas, the Broad Institute [http://sourceforge.net]).
Analysis of 16S rRNA gene sequences obtained from clone studies.
Clone sequences were subject to BLASTN analysis against the HOMD Reference Sequence Set (version 10). Because the first 500 bases of the 16S rRNA molecule generally contain almost half the variability of the full sequence, a match cutoff of 98% similarity with 95% coverage was used as the identification criteria. Those sequences that did not match a human oral taxon sequence were subject to BLASTN analysis against all sequences at the Ribosomal Database Project (RDP; release 10, update 3) (11) and Greengenes (16). Because many clone queries can match the same RDP or Greengenes subject match, a set of unique reference matches was generated. Because this set could contain multiple entries representing a single phylotype, the external database match sequences were clustered as described below. Unique human oral taxon numbers were assigned to each unique phylotype. The clones that did not meet the match criteria to the HOMD, RDP, or Greengenes were clustered into novel taxa defined by the 98% identity with 95% coverage criteria. Clustering was performed by first sorting the clones by length. The first clone was considered a novel taxon and placed in a first taxon folder, and its sequence was declared a reference sequence. Each succeeding clone sequence was compared by BLASTN to the reference sequence(s) and, if matched, added to that taxon folder. If the BLASTN match failed, the clone sequence was used to establish a new taxon folder and added to the reference sequence list. The scripts for these analyses can be obtained from T. Chen. Extended human oral taxon numbers (A01 to H70) were assigned for each novel cluster/folder.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences for the 34,753 clones analyzed are available for download at the Human Oral Microbiome Database website (http://www.homd.org) and from GenBank under accession numbers GU397556 to GU432434. GenBank accession numbers for each taxon in the seven phylogenetic tree figures are included with each taxon label. Additional full-length 16S rRNA gene sequences deposited for this work include GenBank accession numbers FJ577249 to FJ577261, FJ717335, FJ717336, FJ7173350, GQ131410 to GQ131418, and GU470887 to GU470911.
RESULTS AND DISCUSSION
Human Oral Microbiome Database.
The backbone of the HOMD is its set of reference 16S rRNA gene sequences, which are used to define individual human oral taxa and to create the phylogenetic and taxonomic structures of the database. The initial reference set of 16S rRNA gene sequences for the HOMD consisted of over 800 full-length sequences (each of which is greater than 1,500 bases) of named oral bacteria from the oral microbiological literature and strain and clone phylotypes generated from our sequencing and cloning studies. After entering and aligning these sequences in our RNA database, neighbor-joining trees were generated. A total of 78 chimeras were identified and removed from the database. Using a 98.5% sequence similarity cutoff for defining a phylotype, the approximately 800 sequences were placed in 619 taxa. Each taxon was given a human oral taxon (HOT) number (arbitrary number starting at 001). All of the human oral taxa were placed in a full taxonomic classification, including domain, phylum, class, order, family, genus, and species, which can be viewed in Table S4 in the supplemental material. Placement of species in higher taxa was based solely on tree position. For example, Eubacterium saburreum was placed in the family Lachnospiraceae and not Eubacteriaceae. Eubacterium saburreum is not closely related to the type strain of the genus Eubacterium, Eubacterium limosum, and will need to be placed in a new genus and subsequently renamed. The HOMD taxonomy is one of several selectable taxonomies available at the Greengenes website (16) (http://greengenes.lbl.gov). When the HOMD is selected as the taxonomy, the operational taxonomic unit (OTU) numbers equal the human oral taxon numbers.
A summary of the phylogenetic distribution of these 619 taxa is presented in Table 1. It is notable that 65.6% of the taxa have been cultured. This percentage greatly exceeds that in many natural environments where less than 1% has been cultivated. The six major phyla, Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria, Spirochaetes, and Fusobacteria, contain 96% of the taxa. The remaining phyla, Euryarchaeota, Chlamydia, Chloroflexi, SR1, Synergistetes, Tenericutes, and TM7, contain the remaining 4% of the taxa.
TABLE 1.
Phylogenetic distribution of 619 taxa in HOMD version 10
| Phylum | No. (%) of: |
|||
|---|---|---|---|---|
| Taxaa | Named speciesb | Unnamed cultivated taxac | Unnamed uncultivated taxad | |
| Bacteria | ||||
| Firmicutes | 227 (36.7) | 120 (52.9) | 45 (19.8) | 62 (27.3) |
| Bacteroidetes | 107 (17.3) | 39 (36.4) | 27 (25.2) | 41 (38.3) |
| Proteobacteria | 106 (17.1) | 70 (66.0) | 9 (8.5) | 27 (25.5) |
| Actinobacteria | 72 (11.6) | 37 (51.4) | 25 (34.7) | 10 (13.9) |
| Spirochaetes | 49 (7.9) | 11 (22.4) | 3 (6.1) | 35 (71.4) |
| Fusobacteria | 32 (5.2) | 12 (37.5) | 4 (12.5) | 16 (50.0) |
| TM7 | 12 (1.9) | 0 (0.0) | 0 (0.0) | 12 (100.0) |
| Synergistetes | 10 (1.6) | 2 (20.0) | 0 (0.0) | 8 (80.0) |
| Chlamydiae | 1 (0.2) | 1 (100.0) | 0 (0.0) | 0 (0.0) |
| Chloroflexi | 1 (0.2) | 0 (0.0) | 0 (0.0) | 1 (100.0) |
| SR1 | 1 (0.2) | 0 (0.0) | 0 (0.0) | 1 (100.0) |
| Archaea | ||||
| Euryarchaeota | 1 (0.2) | 1 (100.0) | 0 (0.0) | 0 (0.0) |
| Total | 619 (100) | 293 (47.3) | 113 (18.3) | 213 (34.4) |
Taxa refer to named species and to phylotypes with or without cultivable members. Phylotypes are defined as clusters whose members have 98.5% full 16S rRNA sequence similarity. The data in this table are based on those in HOMD version 10.
Named species are those with validly published names.
Unnamed cultivable taxa are phylotypes that have at least one extant isolate.
Uncultivated taxa are phylotypes known only from clone sequences.
A detailed presentation of the phylogenies of the 619 oral taxa can be seen in the phylogenetic trees shown in Fig. 1 to 7. The percentage of times a node was present in the resampling is shown only when it was greater than 50%. The name of each taxon is followed by its designated HOT number, clone or strain number, GenBank accession number, number of clones of the taxon observed in this study, and a symbol indicating each taxon's naming and cultivation status.
FIG. 1.
Neighbor-joining tree for human oral taxa in the phylum Tenericutes and the classes Bacilli and Erysipelotrichia of the phylum Firmicutes. The name of each taxon is followed by the oral taxon number, clone or strain number, GenBank accession number, the number of clones out of 34,879 that were identified as this taxon, and a symbol indicating that the taxon is a named species (•), an unnamed cultivated taxa (▪), or an uncultivated phylotype (□). The tree was constructed with MEGA 4.3 using the Jukes and Cantor correction neighbor-joining distance matrix. Comparisons with missing data were eliminated pairwise. The numbers to the left of the branches indicate the percentage of times the clade was recovered out of 1,000 bootstrap resamplings. Only bootstrap percentages greater than 50 are shown. Roman numerals in square brackets following a genus name indicate Collins' Clostridia cluster numbers (17). Major clades are marked as follows: encircled “1,” Bacilli; encircled “2,” Firmicutes; encircled “3,” Erysipelotrichia; and encircled “4,” Tenericutes (previously know as the class Mollicutes within Firmicutes).
Firmicutes and Tenericutes.
The phylum Tenericutes and the classes Bacilli and Erysipelotrichia of the phylum Firmicutes are shown in Fig. 1. The taxa belonging to the Firmicutes class Clostridia are shown in Fig. 2 and 3.
FIG. 2.
Neighbor-joining tree for human oral taxa in the class Clostridia of the phylum Firmicutes. Labeling and methods used are as described in Fig. 1. Major clades are marked as follows: encircled “1,” Collins XI; encircled “2,” Peptostreptococcaceae; encircled “3,” Collins XIII; encircled “4,” unnamed family F1; encircled “5,” Collins XV, Eubacteriaceae; encircled “6,” Collins XIVa, Lachnospiraceae; encircled “7,” Peptococcaceae; encircled “8,” Collins XIII, Syntrophomonadaceae; and encircled “9,” unnamed family F2.
FIG. 3.
Neighbor-joining tree for human oral taxa in the Veillonellaceae (previously Acidaminococcaceae) family of the class Clostridia of the phylum Firmicutes. Labeling and methods used are as described in Fig. 1.
The class Bacilli (Fig. 1, clade designated with an encircled “1”) contains 86 taxa. It includes the genus Streptococcus, whose members are the most abundant bacterial species in the mouth. The related, but less frequently studied, genera Abiotrophia, Gemella, and Granulicatella are also extremely common, and three species from these genera were among the 10 taxa most frequently detected in our clone libraries. Abiotrophia and Granulicatella require the addition of pyridoxal to grow on blood agar media and were formerly known as the nutritionally variant streptococci (12, 55).
The class Erysipelotrichia (Fig. 1, clade designated with an encircled “3”) contains the following four oral organisms: Bulleidia extructa, Solobacterium moorei, Erysipelothrix tonsillarum, and Lactobacillus [XVII] catenaformis. The meaning of the roman number in square brackets is explained below.
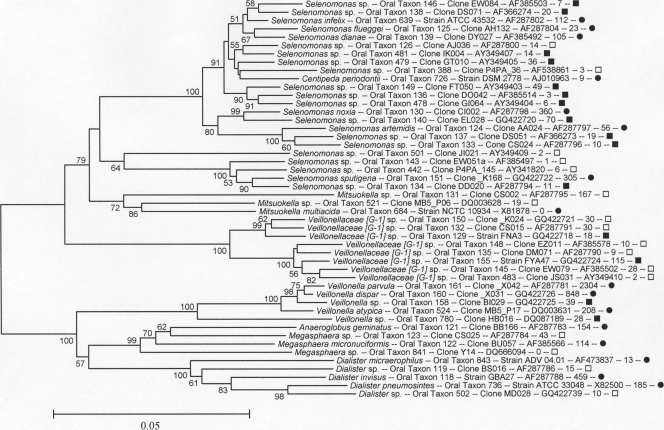
The Firmicutes class Clostridia is shown in Fig. 2 and 3. For the taxonomy of the Clostridia, a widely used classification is that described by Collins et al. (13). Therefore, in the Clostridia trees, Eubacterium infirmum is written Eubacterium [XI][G-1] infirmum to indicate that it is a misnamed Eubacterium sp. falling in Collins cluster XI and that it and neighboring taxa represent a novel genus, as yet unnamed, designated [G-1]. NCBI, and RDP taxonomies, for the most part, remain based on historic names rather than phylogenetic position, even when genera are known to be polyphyletic, thus creating illogical placements at the family and higher taxonomic levels. The major oral clades, corresponding roughly to family-level divisions, are marked in Fig. 2 as follows: Collins cluster XI (encircled “1”); the family Peptostreptococcaceae (encircled “2”); Collins cluster XIII (encircled “3”); a novel family level cluster, with no named species, designated F-1 (encircled “4”); Collins cluster XV, the Eubacteriaceae sensu stricto (encircled “5”); Collins cluster XIVa, the Lachnospiraceae (encircled “6”); the Peptococcaceae (encircled “7”); Collins cluster VIII, the Syntrophomonadaceae (encircled “8”); and a novel family-level cluster, with no named species, designated F-2 (encircled “9”). The largest family of the Clostridia is the Veillonellaceae (previously called Acidaminococcaceae), as shown in Fig. 3. The vast majority of human oral Clostridia fall in the families Lachnospiraceae, Peptostreptococcaceae, and Veillonellaceae.
All members of the Veillonellaceae, shown in Fig. 3, are Gram negative and include the genera Anaeroglobus, Centipeda, Dialister, Megasphaera, Selenomonas, and Veillonella. It is striking that a clade of Gram-negative organisms occurs in an otherwise Gram-positive phylum.
Tenericutes.
The phylum Tenericutes (Fig. 1, designated with an encircled “4”) has recently been created and was previously the class Mollicutes within the Firmicutes (38). Mycoplasma species have been detected in the saliva of 97% of individuals (68). Relatively few mycoplasmas have been detected in the clone libraries reported here, but representatives of Mycoplasma hominis, Mycoplasma salivarium, and Mycoplasma faucium were found. Tenericutes [G-1] sp. oral taxon 504 is very deeply branching and has tentatively been placed with the Tenericutes awaiting further information.
Actinobacteria.
The phyla Actinobacteria and Fusobacteria are shown in Fig. 4, and their clades are marked by an encircled “1” and an encircled “5,” respectively. Out of eight named Actinobacteria orders, oral taxa have thus far been found only in the orders Actinomycetales (Fig. 4, encircled “2”), Bifidobacteriales (encircled “3”), and Coriobacteriales (encircled “4”).
FIG. 4.
Neighbor-joining tree for human oral taxa in the phyla Actinobacteria and Fusobacteria. Labeling and methods used are as described in Fig. 1. Major clades are marked as follows: encircled “1,” phylum Actinobacteria; encircled “2,” order Actinomycetales; encircled “3,” order Bifidobacteriales; encircled “4,” order Coriobacteriales; and encircled “5,” phylum Fusobacteria.
Actinomycetales (Fig. 4, encircled “2”) include the genera Actinomyces, Rothia, Kocuria, Arsenicicoccus, Microbacterium, Propionibacterium, Mycobacterium, Dietzia, Turicella, and Corynebacterium. Bifidobacteriales (Fig. 4, encircled “3”) include the genera Bifidobacterium, Gardnerella, Scardovia, and Parascardovia. These genera are found primarily in dental caries and in denture plaque (40), except for Gardnerella, which is isolated primarily from the vagina. Coriobacteriales (Fig. 4, encircled “4”) include the following genera with oral members: Atopobium, Cryptobacterium, Eggerthella, Olsenella, and Slackia (17).
Fusobacteria.
The phylum Fusobacteria includes the following two genera frequently detected in the mouth: Fusobacterium and Leptotrichia. An unnamed and uncultivated genus, Fusobacteria [G-1], contains two taxa, with Fusobacteria [G-1] sp. oral taxon 220 being relative common with 47 clones detected.
Bacteroidetes.
The 107 Bacteroidetes taxa, shown in Fig. 5, fall into the genera Prevotella, Bacteroides, Porphyromonas, Tannerella, Bergeyella, Capnocytophaga, and eight that are unnamed.
FIG. 5.
Neighbor-joining tree for human oral taxa in the phylum Bacteroidetes. Labeling and methods used are as described in Fig. 1. Encircled “0” through “8” symbols refer to clades discussed in the text.
Prevotella is the largest genus, with approximately 50 species. The majority have been cultivated. The clade marked with an encircled “0” in Fig. 5, including Prevotella tannerae through Prevotella sp. oral taxon 308, warrants separate genus status, as it shares less than 80% 16S rRNA similarity with the main cluster of Prevotella (oral taxa 313 through 304).
In Fig. 5, eight currently unnamed genera (comprised of 1 to 5 phylotypes each) are marked by encircled “1” through encircled “8.” These taxa are deeply branching and have no closely related named species. Bacteroidaceae [G-1] sp. oral taxon 272 (encircled “1”) is both cultivable and common, with 210 clones observed. Three strains in the Moores' collection labeled Prevotella zoogleoformans have been identified as belonging to oral taxon 272. Bacteroidales [G-2] sp. oral taxon 274 (encircled “2”) is also cultivable and common, with 51 clones observed and an isolate designated “Bacteroides D59” identified from the Moores' collection. The six remaining unnamed genera, marked encircled “3” through encircled “8,” have, as yet, no cultivated isolates, but Bacteroidetes [G-3] sp. oral taxon 281 is common with 54 clones in our collection.
Bergeyella zoohelcum, the only named species of the genus, is a member of the canine oral microbiome and a human pathogen from dog bites (65). The two human oral Bergeyella taxa are uncultivated.
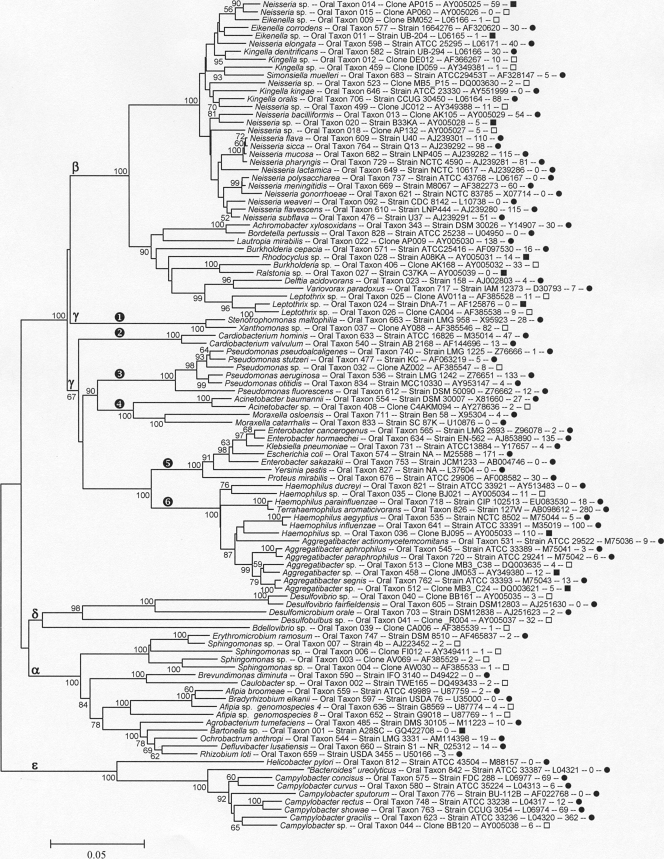
Proteobacteria.
All five classes of Proteobacteria, Alphaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Deltaproteobacteria, and Epsilonproteobacteria, contain taxa detected in the human oral cavity. These taxa are shown in Fig. 6, where the classes are labeled with their corresponding Greek letters.
FIG. 6.
Neighbor-joining tree for human oral taxa in the phylum Proteobacteria. Labeling and methods used are as described in Fig. 1. The Proteobacteria classes are marked with the corresponding Greek letters.
Betaproteobacteria include the genera Neisseria, Eikenella, Kingella, Simonsiella, Achromobacter, Bordetella, Lautropia, Burkholderia, Ralstonia, Delftia, Variovorax, and Leptothrix. Most of these genera are aerobes. Gammaproteobacteria include oral taxa in the following five families: Xanthomonadaceae (Fig. 6, encircled “1”), Cardiobacteriaceae (encircled “2”), Pseudomonadaceae (encircled “3”), Moraxellaceae (encircled “4”), Enterobacteriaceae (encircled “5”), and Pasteurellaceae (encircled “6”). The Xanthomonadaceae family (Fig. 6, encircled “1”) includes Stenotrophomonas maltophilia, which is an ubiquitous environmental organism that is an opportunistic pathogen which can cause nosocomial infections (57) but also can contaminate laboratory reagents and show up in cloning libraries as an artifact, as can many members of the Proteobacteria (63). BLAST interrogation of the GenBank database with Haemophilus parainfluenzae sequences frequently results in a match with accession no. AB098612, labeled “Terrahaemophilus aromaticivorans,” an isolate from petroleum sludge. This species has not been formally proposed and, in our opinion, was likely a human oral H. parainfluenzae contaminant in their sample.
Deltaproteobacteria include species in the genera Desulfovibrio, Desulfomicrobium, Desulfobulbus, and Bdellovibrio. Epsilonproteobacteria include the genera Campylobacter, Helicobacter, and the misnamed “Bacteroides ureolyticus,” which falls just inside or adjacent to the genus Campylobacter (66).
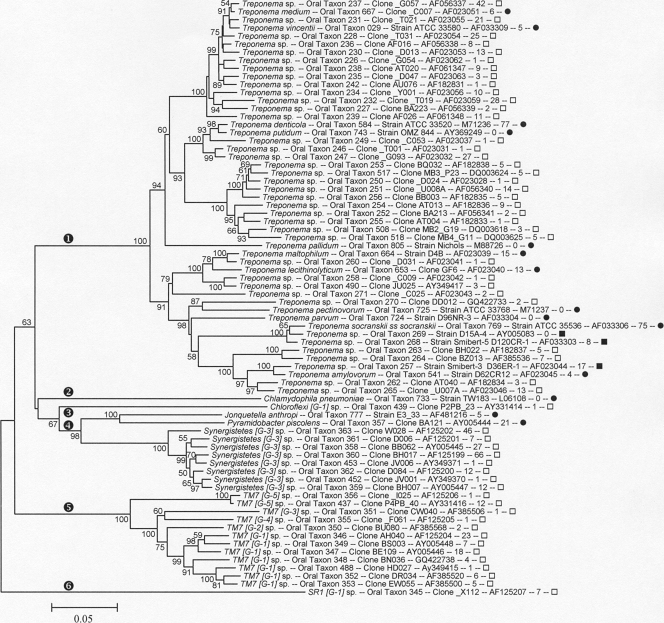
The Spirochaetes phylum is shown as the clade marked with an encircled “1” in Fig. 7. All human oral taxa identified to date in the phylum Spirochaetes are members of the genus Treponema. Over 70% of the 49 identified taxa have thus far resisted cultivation. The number of proposed taxa has decreased from 57 to 49 relative to our initial description of the oral treponemes (18), as some of the taxa with greater than 98.5% 16S rRNA sequence similarity were combined.
FIG. 7.
Neighbor-joining tree for human oral taxa in the phyla Spirochaetes, Chlamydiae, Chloroflexi, Synergistetes, TM7, and SR1. Labeling and methods used are as described in Fig. 1. The phyla are labeled as follows: encircled “1,” Spirochaetes; encircled “2,” Chlamydiae; encircled “3,” Chloroflexi; encircled “4,” Synergistetes; encircled “5,” TM7; and encircled “6,” SR1.
The Chlamydia phylum is marked with an encircled “2” in Fig. 7. Chlamydophila pneumoniae has been detected in dental plaque (39a) and is a recognized lung pathogen. Chlamydia were not observed in our cloning studies, perhaps because the common 9 to 27 (E. coli numbering) 16S rRNA PCR forward primer contains two mismatches with Chlamydia sequences (24). Use of the YM+3 forward primer set (24) may remedy this problem in future studies.
The Chloroflexi phylum is marked with an encircled “3” in Fig. 7. A single Chloroflexi phylotype has been identified. Chloroflexi are abundant in studies of wastewater treatment sludge (8). A closely related phylotype (>95% similarity) has been found in the canine oral cavity (F. E. Dewhirst, unpublished observation), suggesting there are multiple mammalian host-associated species in the phylum Chloroflexi.
Synergistetes taxa are shown in the clade marked with an encircled “4” in Fig. 7. Ten taxa in the phylum Synergistetes have been identified (34, 35). This recently described phylum includes a number of genera and phylotypes that have been previously misclassified as belonging to either the Deferribacteres or Firmicutes (28). Oral strains and clones fall into two main groups, with one which is readily cultivable and includes the named species Jonquetella anthropi (34) and Pyramidobacter piscolens (19). Most oral sequences fall into the second group (oral taxon 363 through 359), which until recently had no cultivable representatives. However, Vartoukian et al. (67) have successfully cultured a member of this group by a combination of coculture and colony hybridization-directed enrichment. Members of this phylum are selectively amplified and appear in significant numbers in 16S rRNA gene libraries generated with “Spirochaeta/Synergistetes” selective primer pair F24/M98 (see Table S2 in the supplemental material).
The TM7 phylum is marked with an encircled “5” in Fig. 7. There are 12 TM7 phylotypes of this phylum shown in Fig. 7. The name TM7 comes from Torf, mittlere Schicht, clone 7 (or peat, middle layer, clone 7), a study of organisms in a German peat bog (53). Organisms in this phylum are frequently detected in many environments (29), but despite the efforts of several laboratories, no members of this phylum have been cultivated, except as microcolonies (23).
The SR1 phylum is marked with an encircled “6” in Fig. 7. The Sulfur River 1 lineage is now recognized as a phylum distinct from OP11 (Obsidian Pool, candidate division 11), with which it was previously grouped (27). Earlier publications referred to SR1 sp. oral taxon 345 as clone X112 in phylum OP11 (47). Clones of oral taxon 345 have been obtained from several distinct individuals. This species is enriched in 16S rRNA gene libraries generated with “Bacteroidetes/TM7/SR1” selective primer pair F24/F01 (see Table S1 in the supplemental material). A closely related species (>95% similarity) has been found in the oral cavities of cats and dogs (F. E. Dewhirst, unpublished observation), indicating that there are multiple mammalian host-associated species in the SR1 phylum.
Web-based Human Oral Microbiome Database.
The taxonomic scheme and sequence database described above formed the basis for creating a publically accessible web-based Human Oral Microbiome Database (http://www.homd.org). It is a resource for phylogenetic, taxonomic, genomic, phenotypic and bibliographic data related to the human oral microbiome and is supported by a contract from the National Institute for Dental and Craniofacial Research to facilitate research by the oral microbiome community. The database was formally launched on 1 March 2008. The HOMD hardware, program architecture, and website navigation have been described in a separate publication (9). Briefly, the HOMD contains a taxon description page for each taxon with full taxonomy, its status as a named species, an unnamed isolate or an uncultivated phylotype, its type or reference strain number, and links to the literature. As a taxon moves from a phylotype, to an isolate, and to a named species, the database tracks the name and status of the bacterium and provides a stable link to the literature. The community can provide input to taxon descriptions and statuses using the HOMD interface. The HOMD contains a BLASTN tool for identifying the 16S rRNA sequences of isolates or clones. Hundreds of sequences can be submitted at one time, and the results can be downloaded as an Excel file showing the top four hits in the database for each query sequence. For some groups of taxa, 16S rRNA gene sequence analysis of partial sequences does not allow unequivocal identification at the species level. The HOMD BLASTN tool output allows easy detection of ambiguous identifications. It also contains visualization and analysis tools for examining the partial and completed genomes for any taxa of the human oral microbiome. It is likely that genomes for over 300 oral taxa will be available on the HOMD by the end of the Human Microbiome Project (51).
Clone analysis.
To validate and expand the species included in the HOMD and to better understand the diversity and taxon distribution of organisms in the human oral microbiome, 36,043 clones from 633 oral 16S rRNA gene libraries constructed and sequenced in our laboratories were analyzed. Following vector removal, screening for lengths of >300 bases, and chimera checking using multiple programs, 1,290 sequences were rejected, leaving 34,753 for analysis. The clones identified as potentially chimeric by the program Chimera Slayer are listed in Table S3 in the supplemental material. A total of 125 were validated as chimeras. Those not validated include intragenus, intraspecies potential chimeras. The majority of these nonvalidated chimeras were found multiple times, which also suggests that they are not chimeras. BLASTN analyses of the remaining clones against the HOMD Reference Sequence Set version 10 yielded identification of 89.1% of the clones in 525 oral taxa. Those not identified in the HOMD were analyzed by BLASTN comparison to the more than 70,000 16S rRNA sequences in RDP (11) and 302,066 sequences in the Greengenes prokMSA unaligned set (16). Using the RDP and Greengenes databases, an additional 8.7% of clones were matched to nonself sequences. The 2.2% of the clones which remained unidentified were clustered into 325 novel taxa, 220 of which were singletons. The clone analysis yielded a total of 654 additional taxa not included in the HOMD version 10. Because novel taxa based on singletons are suspect compared to those seen multiple times, a more conservative figure excluding the 220 singletons is 434 novel taxa. These additional taxa will be added to the HOMD when they meet the strict criteria described below.
A total of 94 of 619 taxa in the HOMD version 10 were not represented by any clone. The list of taxa not found is provided in Table S5 in the supplemental material. This group included most of the extrinsic pathogens we had added to the HOMD for completeness, such as Bordetella pertussis, Corynebacterium diphtheriae, Mycobacterium tuberculosis, and Neisseria gonorrhoeae. Other taxa not seen included those which would not have been expected to have been amplified with the “universal” primers used, such as Methanobrevibacter oralis (an archaeon) and Chlamydophila pneumoniae.
The phylogenetic distribution of clones in various phyla or divisions is shown in Table 2. The clones were found in 11 of 13 phyla included in the HOMD version 10. As the primers used do not amplify Chlamydiae or Archaea, the absence of clones representing these taxa was expected. Clones for the phyla Deinococcus (3 clones), Acidobacteria (1), and Cyanobacteria (1) were found. The clones for these phyla may represent transient exogenous bacteria. Nineteen clones representing four plants (chloroplast) were detected. As virtually all humans eat plants, it is not surprising that plant chloroplast 16S rRNA sequences should be detected in the oral cavity. Species identified included Triticum aestivum (wheat) and Manihot esculenta (cassava, source of tapioca). For each phylum, the percentage of taxa and clones that represent named species, unnamed taxa with cultivable isolates, and uncultivated phylotypes is presented in Table 2. When analyzed by clone distribution, 82% fall into cultivable named and unnamed taxa. When analyzed by taxa distribution, the percent cultivated drops to 32% (40% if singleton clone taxa are removed). This divergence in percent cultivated by clone or taxa count indicates that we have had great success culturing the most common species but have not yet identified isolates for the rarer taxa. The microbiologic community has cultured between 29% and 50% of the oral Firmicutes, Proteobacteria, Bacteroidetes, Actinobacteria, and Fusobacteria taxa. Less than 12% of taxa in the Spirochaetes and Synergistetes, however, have been cultured. TM7, SR1, and Chloroflexi have, as yet, no cultivated members (the phylum Chloroflexi has cultivated members but no cultivated human oral species). The rarer taxa, such as SR1, are present in the clone pool at level of only 1/5,000. This implies that even if the SR1 taxon is only moderately difficult to cultivate, it will still be challenging to find an isolate because of its rarity.
TABLE 2.
Phylogenetic distribution of 34,753 oral clones
| Phylum | No. of taxa (%) | % of taxa that represent: |
No. of clones (%) | % of clones that represent: |
||||
|---|---|---|---|---|---|---|---|---|
| Named species | Unnamed isolates | Uncultivated phylotypes | Named species | Unnamed isolates | Uncultivated Phylotypes | |||
| Firmicutes | 494 (41.9) | 20.6 | 8.7 | 70.6 | 23,354 (67.212) | 78.7 | 7.3 | 14.1 |
| Proteobacteria | 237 (20.1) | 33.3 | 3.0 | 63.7 | 3,909 (11.250) | 72.7 | 3.8 | 23.6 |
| Bacteroidetes | 153 (13.0) | 19.0 | 16.3 | 64.7 | 2,571 (7.403) | 58.6 | 24.1 | 17.3 |
| Actinobacteria | 133 (11.3) | 35.3 | 14.3 | 50.4 | 2,641 (7.572) | 55.7 | 20.3 | 24.0 |
| Spirochaetes | 73 (6.2) | 9.6 | 2.7 | 87.7 | 577 (1.671) | 33.8 | 4.3 | 61.9 |
| Fusobacteria | 42 (3.6) | 23.8 | 7.1 | 69.0 | 1,356 (3.893) | 76.6 | 4.0 | 19.4 |
| Synergistetes | 18 (1.5) | 11.1 | 0.0 | 88.9 | 209 (0.605) | 12.4 | 0.0 | 87.6 |
| TM7 | 15 (1.2) | 0.0 | 0.0 | 100.0 | 85 (0.244) | 0.0 | 0.0 | 100.0 |
| Tenericutes | 4 (0.3) | 75.0 | 0.0 | 25.0 | 10 (0.028) | 90.0 | 0.0 | 10.0 |
| Deinococcus | 3 (0.2) | 0.0 | 0.0 | 100.0 | 13 (0.037) | 0.0 | 0.0 | 100.0 |
| SR1 | 1 (0.1) | 0.0 | 0.0 | 100.0 | 7 (0.020) | 0.0 | 0.0 | 100.0 |
| Chloroflexi | 1 (0.1) | 0.0 | 0.0 | 100.0 | 1 (0.003) | 0.0 | 0.0 | 100.0 |
| Acidobacteria | 1 (0.1) | 0.0 | 0.0 | 100.0 | 1 (0.003) | 0.0 | 0.0 | 100.0 |
| Cyanobacteria | 1 (0.1) | 0.0 | 0.0 | 100.0 | 1 (0.003) | 0.0 | 0.0 | 100.0 |
| Plant chloroplast | 4 (0.3) | 100.0 | 0.0 | 0.0 | 19 (0.054) | 100.0 | 0.0 | 0.00 |
| Total | 1,179 (100) | 24.0 | 8.4 | 67.7 | 34,753 (100.000) | 73.3 | 8.8 | 17.8 |
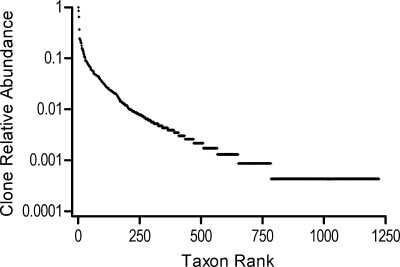
A plot of the relative abundance of clones in each of the 1,179 oral taxa is shown in Fig. 8. Veillonella parvula was the first ranked taxon, with 2,304 clones or 6.6% of the total clones observed. The complete listing of the rank abundance and relative abundance of each taxon is presented in Table S6 in the supplemental material. Because the 34,753 clones are from studies of different disease and health states, different oral sites, and different primer pairs, the data set cannot be used to infer the underlying population structure of the “oral cavity.” We therefore do not attempt to estimate the number of species in the oral cavity with these data. Furthermore, the question of “how many species are present in the human oral cavity?” is tricky, because the oral cavity is an open system where exogenous microorganisms from the environment are continually introduced by eating, drinking, and breathing. One answer to the question of how many microbial species would be found by exhaustive sampling of the oral cavities of the human population over time is all microbial species present in the earth's biosphere. A more straightforward question which can be answered for this data set is, “How many taxa are needed to account for the 90%, 95%, 98%, or 99% of the 34,753 clones sampled?” The answers are 259, 413, 655, and 875 taxa, respectively.
FIG. 8.
Rank abundance graph for 34,753 16S rRNA clones obtained from oral samples in 1,179 taxa. Clones were placed in taxa on the basis of 98% BLASTN identities. The first ranked taxon was Veillonella parvula, with 2,304 clones. Ranks 769 to 1,179 were singletons.
Transient versus endogenous species.
Because there is a constant stream of organisms introduced into the oral cavity from the environment, one needs to distinguish transient species from endogenous species, those that are part of what Theodor Rosebury called the indigenous microbes or “normal flora” (54). Unfortunately, the distinction between transient species and endogenous species cannot be deduced directly from human sampling studies. Rather, it has to come from comparing the human studies with environmental studies to determine the frequency with which clones of a particular genus are recovered as host associated or as environment associated. For example, species in the genus Prevotella have been found only in the microbiota of mammals, whereas species in the genus Sphingomonas are generally free-living species found in the environment in cloning studies of lake sediments, soils, etc. Table S7 in the supplemental material presents the categorization of the 169 genera currently included in the HOMD as strongly host associated, weakly host associated, or environmental based on a qualitative analysis of clone sequence sources. The degree of host association for various genera differs widely by phylogenetic position, as essentially all genera from the phylum Bacteroidetes are host associated, while nearly all genera from the Alphaproteobacteria are thought to be environmental transients.
Extended Reference Set for BLAST analyses.
Reference sequences for the 654 additional taxa identified in the clone analysis have been added to those for the 619 HOMD taxa to generate the Extended Reference Set (version 1), which is available for download or use in BLAST analysis at the HOMD website. Many sequences from the clone analysis are only 500 bases long, and thus, the Extended Reference Set, unlike the HOMD Reference Set 10, is composed of both full and partial sequences. The Extended Reference Set is useful for BLAST analysis of clone and other 5′ 500-base sequences but must be used with caution for analysis of full-length sequences.
Addition of taxa to the Human Oral Microbiome Database.
The 434 additional taxa identified in the clone analysis (excluding singletons) described above are candidates for addition to the HOMD. To be added, however, each taxon must meet the following criteria. (i) It must have been seen more than once, hence the exclusion of novel singletons. (ii) A near full-length (>1450-base) 16S rRNA gene sequence must be available for the taxon. (iii) The full sequence must be less than 98.5% similar to previously defined HOMD taxa. (iv) The full sequence must pass chimera and sequence quality screens. (v) If the sequence is for a named species, the sequence must greater than 98.5% similar to that of the type strain of the species. (vi) If there are multiple sequences in GenBank for a species, the longest and most accurate must be identified to represent the species. We expect that the majority of the taxa identified in the clone analysis will be validated and added in updates to the HOMD by January 2011.
The human oral microbiome has been extensively studied, is being examined as part of the Human Microbiome Project, and will continue to be examined in the future using ever more powerful sequencing technologies. We have created the Human Oral Microbiome Database taxonomic framework, with oral taxon numbers to facilitate communication between investigators exploring the diversity of the oral microbiome, as seen by 16S rRNA gene-based methods. It is critical that investigators can point to curated stably designated taxa rather than taxonomically undefined clone sequences in disseminating research findings. Investigators with oral strain or clone 16S RNA sequences can now definitively identify the vast majority of them by BLASTN analysis against the reference sets available at the HOMD website (www.homd.org). Analysis of approximately 35,000 clone sequences has allowed validation of the initial 619 species in the HOMD and identified more than 434 additional named and unnamed taxa to be added to the HOMD once full 16S rRNA sequences are obtained and the other criteria discussed above are met. The HOMD is the first example of a curated human body site-specific microbiome resource. Predominant members of the oral microbiome can also be found in fewer numbers at other body sites, such as the skin, gut, and vagina, and thus, the HOMD can be useful to the entire Human Microbiome Project and infectious disease communities. One can foresee the development of additional body site-specific curated microbiome resources based on the HOMD model or the framework of the HOMD to be expanded to include the entire human microbiome.
Supplementary Material
Acknowledgments
This work has been supported in part by contract U01 DE016937 and grants DE015847 and DE017106 from the National Institute of Dental and Craniofacial Research and a supplement to grant DE016937 from the American Recovery and Reinvestment Act of 2009.
We thank the following investigators for providing clinical samples or DNA for the construction of clone libraries: Ashraf F. Fouad, Ann L. Griffen, Anne D. Haffajee, Onir Leshem, Eugene J. Leys, Harlan J. Shiau, Sigmund S. Socransky, and Thomas R. Flynn. We thank Julia Downes for helpful discussions. We thank all of our colleagues who have deposited 16S rRNA gene sequences for oral strains in public databases.
Footnotes
Published ahead of print on 23 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1.Aas, J. A., S. M. Barbuto, T. Alpagot, I. Olsen, F. E. Dewhirst, and B. J. Paster. 2007. Subgingival plaque microbiota in HIV positive patients. J. Clin. Periodontol. 34:189-195. [DOI] [PubMed] [Google Scholar]
- 2.Aas, J. A., B. J. Paster, L. N. Stokes, I. Olsen, and F. E. Dewhirst. 2005. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2006. New screening software shows that most recent large 16S rRNA gene clone libraries contain chimeras. Appl. Environ. Microbiol. 72:5734-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Awano, S., T. Ansai, Y. Takata, I. Soh, S. Akifusa, T. Hamasaki, A. Yoshida, K. Sonoki, K. Fujisawa, and T. Takehara. 2008. Oral health and mortality risk from pneumonia in the elderly. J. Dent. Res. 87:334-339. [DOI] [PubMed] [Google Scholar]
- 5.Bahrani-Mougeot, F. K., B. J. Paster, S. Coleman, S. Barbuto, M. T. Brennan, J. Noll, T. Kennedy, P. C. Fox, and P. B. Lockhart. 2007. Molecular analysis of oral and respiratory bacterial species associated with ventilator-associated pneumonia. J. Clin. Microbiol. 45:1588-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck, J. D., and S. Offenbacher. 2005. Systemic effects of periodontitis: epidemiology of periodontal disease and cardiovascular disease. J. Periodontol. 76:2089-2100. [DOI] [PubMed] [Google Scholar]
- 7.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bjornsson, L., P. Hugenholtz, G. W. Tyson, and L. L. Blackall. 2002. Filamentous Chloroflexi (green non-sulfur bacteria) are abundant in wastewater treatment processes with biological nutrient removal. Microbiology 148:2309-2318. [DOI] [PubMed] [Google Scholar]
- 9.Chen, T., W.-H. Yu, J. Izard, O. V. Baranova, A. Lakshmanan, and F. E. Dewhirst. 2010. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database 2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Göbel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole, J. R., Q. Wang, E. Cardenas, J. Fish, B. Chai, R. J. Farris, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, T. Marsh, G. M. Garrity, and J. M. Tiedje. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141-D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Collins, M. D., and P. A. Lawson. 2000. The genus Abiotrophia (Kawamura et al.) is not monophyletic: proposal of Granulicatella gen. nov., Granulicatella adiacens comb. nov., Granulicatella elegans comb. nov. and Granulicatella balaenopterae comb. nov. Int. J. Syst. Evol. Microbiol. 50(Pt. 1):365-369. [DOI] [PubMed] [Google Scholar]
- 13.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 14.Corby, P. M., W. A. Bretz, T. C. Hart, N. J. Schork, J. Wessel, J. Lyons-Weiler, and B. J. Paster. 2007. Heritability of oral microbial species in caries-active and caries-free twins. Twin Res. Hum. Genet. 10:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lillo, A., F. P. Ashley, R. M. Palmer, M. A. Munson, L. Kyriacou, A. J. Weightman, and W. G. Wade. 2006. Novel subgingival bacterial phylotypes detected using multiple universal polymerase chain reaction primer sets. Oral Microbiol. Immunol. 21:61-68. [DOI] [PubMed] [Google Scholar]
- 16.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dewhirst, F. E., B. J. Paster, N. Tzellas, B. Coleman, J. Downes, D. A. Spratt, and W. G. Wade. 2001. Characterization of novel human oral isolates and cloned 16S rDNA sequences that fall in the family Coriobacteriaceae: description of olsenella gen. nov., reclassification of Lactobacillus uli as Olsenella uli comb. nov. and description of Olsenella profusa sp. nov. Int. J. Syst. Evol. Microbiol. 51:1797-1804. [DOI] [PubMed] [Google Scholar]
- 18.Dewhirst, F. E., M. A. Tamer, R. E. Ericson, C. N. Lau, V. A. Levanos, S. K. Boches, J. L. Galvin, and B. J. Paster. 2000. The diversity of periodontal spirochetes by 16S rRNA analysis. Oral Microbiol. Immunol. 15:196-202. [DOI] [PubMed] [Google Scholar]
- 19.Downes, J., S. R. Vartoukian, F. E. Dewhirst, J. Izard, T. Chen, W. H. Yu, I. C. Sutcliffe, and W. G. Wade. 2009. Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int. J. Syst. Evol. Microbiol. 59:972-980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dzink, J. L., S. S. Socransky, and A. D. Haffajee. 1988. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 15:316-323. [DOI] [PubMed] [Google Scholar]
- 21.Dzink, J. L., A. C. Tanner, A. D. Haffajee, and S. S. Socransky. 1985. Gram negative species associated with active destructive periodontal lesions. J. Clin. Periodontol. 12:648-659. [DOI] [PubMed] [Google Scholar]
- 22.Faveri, M., M. P. Mayer, M. Feres, L. C. de Figueiredo, F. E. Dewhirst, and B. J. Paster. 2008. Microbiological diversity of generalized aggressive periodontitis by 16S rRNA clonal analysis. Oral Microbiol. Immunol. 23:112-118. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari, B. C., S. J. Binnerup, and M. Gillings. 2005. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl. Environ. Microbiol. 71:8714-8720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frank, J. A., C. I. Reich, S. Sharma, J. S. Weisbaum, B. A. Wilson, and G. J. Olsen. 2008. Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol. 74:2461-2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genco, R. J., S. G. Grossi, A. Ho, F. Nishimura, and Y. Murayama. 2005. A proposed model linking inflammation to obesity, diabetes, and periodontal infections. J. Periodontol. 76:2075-2084. [DOI] [PubMed] [Google Scholar]
- 26.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 27.Harris, J. K., S. T. Kelley, and N. R. Pace. 2004. New perspective on uncultured bacterial phylogenetic division OP11. Appl. Environ. Microbiol. 70:845-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hugenholtz, P., G. W. Tyson, R. I. Webb, A. M. Wagner, and L. L. Blackall. 2001. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl. Environ. Microbiol. 67:411-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenkinson, H. F., and R. J. Lamont. 2005. Oral microbial communities in sickness and in health. Trends Microbiol. 13:589-595. [DOI] [PubMed] [Google Scholar]
- 31.Joshipura, K. J., H. C. Hung, E. B. Rimm, W. C. Willett, and A. Ascherio. 2003. Periodontal disease, tooth loss, and incidence of ischemic stroke. Stroke 34:47-52. [DOI] [PubMed] [Google Scholar]
- 32.Joshipura, K. J., E. B. Rimm, C. W. Douglass, D. Trichopoulos, A. Ascherio, and W. C. Willett. 1996. Poor oral health and coronary heart disease. J. Dent. Res. 75:1631-1636. [DOI] [PubMed] [Google Scholar]
- 33.Jukes, T., and C. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, NY.
- 34.Jumas-Bilak, E., J. P. Carlier, H. Jean-Pierre, D. Citron, K. Bernard, A. Damay, B. Gay, C. Teyssier, J. Campos, and H. Marchandin. 2007. Jonquetella anthropi gen. nov., sp. nov., the first member of the candidate phylum ‘Synergistetes’ isolated from man. Int. J. Syst. Evol. Microbiol. 57:2743-2748. [DOI] [PubMed] [Google Scholar]
- 35.Jumas-Bilak, E., L. Roudiere, and H. Marchandin. 2009. Description of ‘Synergistetes’ phyl. nov. and emended description of the phylum ‘Deferribacteres’ and of the family Syntrophomonadaceae, phylum ‘Firmicutes’. Int. J. Syst. Evol. Microbiol. 59:1028-1035. [DOI] [PubMed] [Google Scholar]
- 36.Kazor, C. E., P. M. Mitchell, A. M. Lee, L. N. Stokes, W. J. Loesche, F. E. Dewhirst, and B. J. Paster. 2003. Diversity of bacterial populations on the tongue dorsa of patients with halitosis and healthy patients. J. Clin. Microbiol. 41:558-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lederberg, J., and A. T. Mccray. 2001. 'Ome sweet 'omics—a genealogical treasury of words. Scientist 15:8-10. [Google Scholar]
- 38.Ludwig, W., K. H. Schleifer, and W. B. Whitman. 2009. Revised road map to the phylum Firmicutes. In P. De Vos, G. Garrity, D. Jones, N. R. Krieg, W. Ludwig, and F. A. Rainey (ed.), Bergey's manual of systematic bacteriology, vol. 3. Springer, New York, NY.
- 39.Mager, D. L., L. A. Ximenez-Fyvie, A. D. Haffajee, and S. S. Socransky. 2003. Distribution of selected bacterial species on intraoral surfaces. J. Clin. Periodontol. 30:644-654. [DOI] [PubMed] [Google Scholar]
- 39a.Mäntylä, P., M. Stenman, M. Paldanius, P. Saikku, T. Sorsa, and J. H. Meurman. 2004. Chlamydia pneumoniae together with collagenase-2 (MMP-8) in periodontal lesions. Oral Dis. 10:32-35. [DOI] [PubMed] [Google Scholar]
- 40.Mantzourani, M., S. C. Gilbert, H. N. Sulong, E. C. Sheehy, S. Tank, M. Fenlon, and D. Beighton. 2009. The isolation of bifidobacteria from occlusal carious lesions in children and adults. Caries Res. 43:308-313. [DOI] [PubMed] [Google Scholar]
- 41.Moore, W. E. C., L. V. Holdeman, E. P. Cato, R. M. Smibert, J. A. Burmeister, and R. R. Ranney. 1983. Bacteriology of moderate (chronic) periodontitis in mature adult humans. Infect. Immun. 42:510-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, W. E. C., L. V. Holdeman, R. M. Smibert, D. E. Hash, J. A. Burmeister, and R. R. Ranney. 1982. Bacteriology of severe periodontitis in young adult humans. Infect. Immun. 38:1137-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore, W. E. C., and L. V. Moore. 1994. The bacteria of periodontal diseases. Periodontol. 2000 5:66-77. [DOI] [PubMed] [Google Scholar]
- 44.Munson, M. A., A. Banerjee, T. F. Watson, and W. G. Wade. 2004. Molecular analysis of the microflora associated with dental caries. J. Clin. Microbiol. 42:3023-3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munson, M. A., T. Pitt-Ford, B. Chong, A. Weightman, and W. G. Wade. 2002. Molecular and cultural analysis of the microflora associated with endodontic infections. J. Dent. Res. 81:761-766. [DOI] [PubMed] [Google Scholar]
- 46.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3:233-250. [DOI] [PubMed] [Google Scholar]
- 47.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of campylobacters, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 49.Paster, B. J., W. A. Falkler, Jr., C. O. Enwonwu, E. O. Idigbe, K. O. Savage, V. A. Levanos, M. A. Tamer, R. L. Ericson, C. N. Lau, and F. E. Dewhirst. 2002. Prevalent bacterial species and novel phylotypes in advanced noma lesions. J. Clin. Microbiol. 40:2187-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paster, B. J., M. K. Russell, T. Alpagot, A. M. Lee, S. K. Boches, J. L. Galvin, and F. E. Dewhirst. 2002. Bacterial diversity in necrotizing ulcerative periodontitis in HIV-positive subjects. Ann. Periodontol. 7:8-16. [DOI] [PubMed] [Google Scholar]
- 51.Peterson, J., S. Garges, M. Giovanni, P. McInnes, L. Wang, J. A. Schloss, V. Bonazzi, J. E. McEwen, K. A. Wetterstrand, C. Deal, C. C. Baker, V. Di Francesco, T. K. Howcroft, R. W. Karp, R. D. Lunsford, C. R. Wellington, T. Belachew, M. Wright, C. Giblin, H. David, M. Mills, R. Salomon, C. Mullins, B. Akolkar, L. Begg, C. Davis, L. Grandison, M. Humble, J. Khalsa, A. R. Little, H. Peavy, C. Pontzer, M. Portnoy, M. H. Sayre, P. Starke-Reed, S. Zakhari, J. Read, B. Watson, and M. Guyer. 2009. The NIH Human Microbiome Project. Genome Res. 19:2317-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Preza, D., I. Olsen, J. A. Aas, T. Willumsen, B. Grinde, and B. J. Paster. 2008. Bacterial profiles of root caries in elderly patients. J. Clin. Microbiol. 46:2015-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rheims, H., F. A. Rainey, and E. Stackebrandt. 1996. A molecular approach to search for diversity among bacteria in the environment. J. Ind. Microbiol. Biotechnol. 17:159-169. [Google Scholar]
- 54.Rosebury, T. 1962. Microorganisms indigenous to man. McGraw-Hill Book Company, Inc., New York, NY.
- 55.Ruoff, K. L. 1991. Nutritionally variant streptococci. Clin. Microbiol. Rev. 4:184-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 57.Senol, E. 2004. Stenotrophomonas maltophilia: the significance and role as a nosocomial pathogen. J. Hosp. Infect. 57:1-7. [DOI] [PubMed] [Google Scholar]
- 58.Seymour, G. J., P. J. Ford, M. P. Cullinan, S. Leishman, and K. Yamazaki. 2007. Relationship between periodontal infections and systemic disease. Clin. Microbiol. Infect. 13(Suppl. 4):3-10. [DOI] [PubMed] [Google Scholar]
- 59.Socransky, S. S., and A. D. Haffajee. 1994. Evidence of bacterial etiology: a historical perspective. Periodontol. 2000 5:7-25. [DOI] [PubMed] [Google Scholar]
- 60.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 61.Tanner, A., M. F. Maiden, P. J. Macuch, L. L. Murray, and R. L. Kent, Jr. 1998. Microbiota of health, gingivitis, and initial periodontitis. J. Clin. Periodontol. 25:85-98. [DOI] [PubMed] [Google Scholar]
- 62.Tanner, A. C., C. Haffer, G. T. Bratthall, R. A. Visconti, and S. S. Socransky. 1979. A study of the bacteria associated with advancing periodontitis in man. J. Clin. Periodontol. 6:278-307. [DOI] [PubMed] [Google Scholar]
- 63.Tanner, M. A., B. M. Goebel, M. A. Dojka, and N. R. Pace. 1998. Specific ribosomal DNA sequences from diverse environmental settings correlate with experimental contaminants. Appl. Environ. Microbiol. 64:3110-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Turnbaugh, P. J., R. E. Ley, M. Hamady, C. M. Fraser-Liggett, R. Knight, and J. I. Gordon. 2007. The human microbiome project. Nature 449:804-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vandamme, P., J. F. Bernardet, P. Segers, K. Kersters, and B. Holmes, 1994. New perspectives in the classification of the flavobacteria: description of Chryseobacterium gen. nov., Bergeyella gen. nov., and Empedobacter nom. rev. Int. J. Syst. Bacteriol. 44:827-831. [Google Scholar]
- 66.Vandamme, P., M. I. Daneshvar, F. E. Dewhirst, B. J. Paster, K. Kersters, H. Goossens, and C. W. Moss. 1995. Chemotaxonomic analyses of Bacteroides gracilis and Bacteroides ureolyticus and reclassification of B. gracilis as Campylobacter gracilis comb. nov. Int. J. Syst. Bacteriol. 45:145-152. [DOI] [PubMed] [Google Scholar]
- 67.Vartoukian, S. R., R. M. Palmer, and W. G. Wade. 2010. Cultivation of a Synergistetes strain representing a previously uncultivated lineage. Environ. Microbiol. 12:916-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Watanabe, T., M. Matsuura, and K. Seto. 1986. Enumeration, isolation, and species identification of mycoplasmas in saliva sampled from the normal and pathological human oral cavity and antibody response to an oral mycoplasma (Mycoplasma salivarium). J. Clin. Microbiol. 23:1034-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuger, J., H. Luthi-Schaller, and R. Gmur. 2007. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease-associated dental plaques. Microbiology 153:3809-3816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.