Abstract
Vibrio fischeri ES114, an isolate from the Euprymna scolopes light organ, produces little bioluminescence in culture but is ∼1,000-fold brighter when colonizing the host. Cell-density-dependent regulation alone cannot explain this phenomenon, because cells within colonies on solid medium are much dimmer than symbiotic cells despite their similar cell densities. To better understand this low luminescence in culture, we screened ∼20,000 mini-Tn5 mutants of ES114 for increased luminescence and identified 28 independent “luminescence-up” mutants with insertions in 14 loci. Mutations affecting the Pst phosphate uptake system led to the discovery that luminescence is upregulated under low-phosphate conditions by PhoB, and we also found that ainS, which encodes an autoinducer synthase, mediates repression of luminescence during growth on plates. Other novel luminescence-up mutants had insertions in acnB, topA, tfoY, phoQ, guaB, and two specific tRNA genes. Two loci, hns and lonA, were previously described as repressors of bioluminescence in transgenic Escherichia coli carrying the light-generating lux genes, and mutations in arcA and arcB were consistent with our report that Arc represses lux. Our results reveal a complex regulatory web governing luminescence and show how certain environmental conditions are integrated into regulation of the pheromone-dependent lux system.
Vibrio fischeri is a valuable model for examining bioluminescence, pheromone signaling, and symbiotic bacteria-animal interactions. Studies of V. fischeri's mutualistic interactions have gained momentum since the discovery that this bacterium's light organ symbiosis with the Hawaiian bobtail squid, Euprymna scolopes, can be reconstituted in the laboratory (54, 70, 83). Moreover, the bioluminescence induced by V. fischeri in the host light organ is a pheromone-mediated behavior, making this an attractive system for examining environmental influences on bacterial pheromone signaling in a natural infection. Largely because of interest in this symbiosis, strain ES114, which was isolated from the E. scolopes light organ, has become the experimental strain of choice for many studies of V. fischeri.
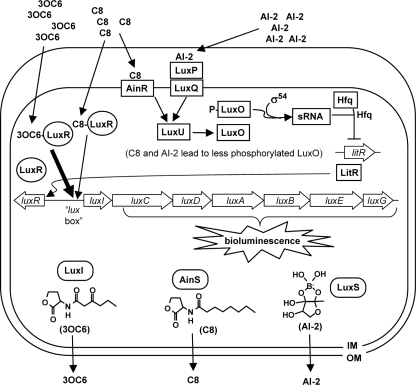
The genetic basis of bioluminescence in V. fischeri ES114 is fundamentally similar to that of other characterized V. fischeri strains (31, 32). The lux genes responsible for bioluminescence, luxABCDE and -G, are found together with the regulatory genes luxR and luxI and are arranged with luxR divergently transcribed from the luxICDABEG operon, as shown in Fig. 1 (24, 25, 56). Light is generated when luciferase, comprised of LuxA and LuxB, binds to FMNH2, O2, and an aliphatic aldehyde, and then converts these substrates to FMN, water, and an aliphatic acid (35, 76). LuxC, LuxD, LuxE, and LuxG (re)generate luciferase's aldehyde and FMNH2 substrates (12, 64).
FIG. 1.
Model of autoinducer-mediated regulation of bioluminescence in V. fischeri. Labeled open arrows correspond to genes, and the structures of three autoinducer molecules are presented with their respective synthases. The AI-2 structure is inferred from studies of V. harveyi (13) but has not been identified in V. fischeri. Interactions between autoinducers, proteins, genes, and small RNA (sRNA) are indicated. For simplicity, multimerization of proteins is not shown. This model is derived in part from experimental results but in some aspects relies on genomic predictions. For details see reference 73.
The remaining genes in this cluster, luxI and luxR, underlie a pheromone-mediated regulatory mechanism often referred to as quorum sensing. LuxI generates the membrane-permeable autoinducer pheromone N-3-oxohexananoyl-l-homoserine lactone (3-oxo-C6-HSL) (23, 41). As cell density increases, 3-oxo-C6-HSL accumulates until reaching a threshold concentration, whereupon it binds LuxR and together they activate transcription of luxICDABEG (24, 25, 77). As a result, the lux genes are most highly expressed at high cell densities, when the bacteria have reached a “quorum” (28). Like many bacterial quorum-sensing systems, the lux operon constitutes a positive feedback circuit, because the 3-oxo-C6-HSL produced by LuxI leads to increased transcription of luxICDABEG. As a result, environmental regulatory inputs to the lux system can be amplified and spread in a population.
V. fischeri generates two additional autoinducers: N-octanoyl-l-homoserine lactone (C8-HSL) produced by AinS (44) and AI-2 (46), which may be a furanosyl borate diester as it is in Vibrio harveyi (13). As Fig. 1 illustrates, AI-2 and C8-HSL presumably function through distinct receptors that both act via LuxU and LuxO, Hfq, and a small RNA to increase levels of LitR, which in turn increases luxR expression (26, 47, 58). C8-HSL can also activate LuxR directly (47). Although C8-HSL is a weaker activator of LuxR than 3-oxo-C6-HSL (67), ES114 produces more C8-HSL in broth cultures (73), and under these conditions it is the main activator of bioluminescence in ES114 (47).
Interestingly, despite conserved lux circuitry, ES114 and other isolates from E. scolopes are much dimmer in culture than previously studied V. fischeri strains (8). ES114 colonies on solid medium are not visibly luminescent, and cells in these colonies produce ∼1,000-fold less luminescence than do symbiotic ES114 cells, despite achieving similar high population densities. Thus, cell density alone cannot account for the dim luminescence of ES114 in culture, and environmentally responsive regulators must also play a critical role in regulating the luxICDABEG operon.
Relatively little is known about environmental influences on luxICDABEG expression in V. fischeri, particularly in ES114. CRP-mediated regulation of lux in response to glucose has been documented by using transgenic lux-containing Escherichia coli (18-20), although the response of V. fischeri to glucose is less clear and may be strain specific (8, 27). Similarly, the luminescence of strain MJ1 is inhibited by iron (36), but iron does not affect ES114 luminescence (8). We found that the redox-responsive ArcA/ArcB system repressses lux (10); however, as we report here, ArcA/ArcB does not account for aeration-dependent regulation of luminescence (73). Other genes affecting luminescence in V. fischeri ES114 have been reported (38, 79, 84), but to date no directed study has screened for regulators that could account for the relatively low lux expression by ES114 in culture or its induction/derepression upon entering the host.
Control of bacterial pheromone systems such as lux by environmentally responsive regulators is widespread and has important functional implications, yet the significance of this phenomenon remains obscure. Therefore, our goal is to elucidate regulators and environmental contingencies that control induction of the lux operon. In this study, we screened for “luminescence-up” mutants of V. fischeri ES114, which led to the identification of conditions and regulators that affect bioluminescence and expression of the lux operon.
MATERIALS AND METHODS
Bacteria and media.
V. fischeri strain ES114, originally isolated from an E. scolopes light organ (8), was the wild-type strain used throughout this study. V. fischeri mutants KV2801 (79), KV2850 (79), KV1651 (38), and KV2655, with disruptions in ptsI, crr, phoB, and phoU, respectively, were obtained from Karen Visick. E. coli strains DH5α (34) or DH5αλpir (21) were transformed by plasmids in the cloning steps outlined below. Transfer of plasmids to V. fischeri was accomplished by triparental matings using conjugative helper strain CC118λpir pEVS104 as previously described (72). E. coli was grown in LB medium (57) or brain heart infusion (BHI) medium (Bacto), and V. fischeri strains were grown in one of three medium types: (i) LBS, which contained, per liter of water, 10 g of tryptone, 5 g of yeast extract, 20 g of NaCl, and 50 mM Tris (pH 7.5); (ii) SWTO, which contained, per liter of total volume, 5 g of tryptone, 3 g of yeast extract, 3 ml of glycerol, 700 ml of Instant Ocean mixed to 36 ppt (Aquarium Systems, Mentor, OH), and an additional 170 mM NaCl; (iii) FMM, which contained, per liter, 950 ml of water, 378 μl of 1 M NaPO4 (pH 7.5), 50 ml of 1 M Tris (pH 7.5), 3 mg of FeSO4·7H2O, 13.6 g of MgSO4·7H2O, 0.59 g of NH4Cl, 0.83 g of KCl, 19.5 g of NaCl, 1.62 g of CaCl2·2H2O, 1 g of Casamino Acids, and 3 ml of glycerol. Solid media were prepared with 15 mg ml−1 agar for plating. For selection of E. coli, chloramphenicol (CAM) and kanamycin (KAN) were added to LB at final concentrations of 20 and 40 μg ml−1, respectively, and 150 μg ml−1 erythromycin (ERM) was added to BHI. For selection of V. fischeri on LBS, CAM, ERM, and KAN were used at concentrations of 2, 5, and 100 μg ml−1, respectively. 5-Bromo-4-chloro-3-indolyl-β-d-galactoside (X-Gal) and isopropyl-β-d-thiogalactoside (IPTG) were added to media at final concentrations of 60 and 50 μg ml−1, respectively.
Transposon mutagenesis.
Transposon mutants were generated as previously described (1, 48). Briefly, pEVS170 was transferred to ES114 by triparental matings (72). Following overnight incubation, mating spots were resuspended in LBS and dilution plated onto LBS supplemented with ERM. Plates were incubated overnight at ∼24°C, and images were generated using a Fluor-S Max-2 imager (Bio-Rad Laboratories, Hercules, CA) set to high-sensitivity chemiluminescence mode with a 20-min exposure. Colonies that were more luminescent than JRM100, an ERM-resistant derivative of ES114 (52), were patched onto LBS supplemented with ERM and imaged following overnight incubation to confirm their luminescence phenotype. To ensure that ermR colonies carried transposon insertions and were not harboring pEVS170, mutants were screened for kanamycin resistance, which is present on pEVS170 outside the transposon, and KAN-resistant mutants were discarded.
Genetic techniques and analyses.
Plasmids were generated using standard techniques. DNA ligase and restriction enzymes were obtained from New England Biolabs (Beverly, MA). Oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). PCR was performed with an iCycler (Bio-Rad Laboratories) using KOD DNA polymerase (Novagen, Madison, WI) or Phusion high-fidelity polymerase (Finnzymes, Finland). Plasmids used for cloning were purified using Qiagen miniprep kits (Valencia, CA) or the GenElute plasmid miniprep kit (Sigma-Aldrich, Inc., St. Louis, MO). For supercoiling analyses, plasmids were purified using the ChargeSwitch-ProDNA plasmid miniprep kit (Invitrogen, Carlsbad, CA). DNA was purified from PCR, digestion, and ligation reactions with the DNA Clean and Concentrator-5 kit (Zymo Research, Orange, CA). To clone PCR products into the pCR-BluntII-TOPO plasmid, we used the ZeroBlunt-TOPO PCR cloning kit (Invitrogen) and screened for white colonies on plates containing X-Gal. Cloned PCR products were sequenced at the University of Michigan DNA Sequencing Core Facility, and sequences were compared to the ES114 genomic database using Sequencher 4.1.2 (Gene Codes Corp., Ann Arbor, MI) to ensure that no unintended base pair changes were incorporated.
Plasmids pJLB52 (10), pJLB114, pAS5, pNL4, pNL18, pNL75, pNL6, pNL72, and pCL112 (47) were used to mobilize native copies of arcA, arcB, acnB, topA, lonA, pstA, hns, phoQ, and ainS, respectively, into mutants with transposon insertions in these genes. A description of new complementation plasmid construction follows. In each case the cloned gene was expressed by including ∼500 bp of upstream sequence and/or by a promoter(s) on the vector. For example, the pstA and phoQ genes appear to be toward the distal ends of operons and were cloned by themselves such that they would be expressed by the lacZ promoter on the vector. arcB was PCR amplified using primers ARC B1 (5′ CCG CCC TAG GCA GGG TAT GTT TAT GAA GCA GTT AA 3′) and ARC B2 (5′ GGG GTA CCG TGT TGC GGC AAA TAG TAC CTT CTT C 3′), and the blunt product was cloned into pCR-BluntII-TOPO to generate pJLB106. pJLB106 was linearized with AvrII and ligated to XbaI-digested pVSV105 (22), generating pJLB110, which was then digested with KpnI and self-ligated to excise the pCR-BluntII-TOPO vector, generating pJLB114. acnB was PCR amplified using primers JBACNB5 (5′ GCG CCA TAA GTC GTA TGT TGT TTG TTG TGG G 3′) and JBACNB7 (5′ CCA GCC CT TAA ATA AAA AAG CAG CCA ATT GCC 3′), and the blunt product was ligated directly into HpaI-digested pJLB103 to generate pJLB130. The vector pJLB103, which contains the R6K origin of replication and encodes KAN resistance, was derived from pVSV104 (22) by deleting the pES213 origin on a BamHI fragment. pJLB130 was digested with AvrII and KpnI, and the fragment containing acnB was ligated into XbaI- and KpnI-digested pVSV105 (22) to generate pAS5. topA was PCR amplified using primers pr_NL3 (5′ CAG CCT CAG AAA TGG ATT TTT TAT CGC TCA TAA G 3′) and pr_NL4 (5′ GAG CCG CAT TTC TGC AGC TCT TTC 3′), and the blunt product was cloned into pCR-BluntII-TOPO to generate pNL2. pNL2 was digested with EcoRV, and the topA-containing fragment was ligated into SmaI-digested pVSV105 (22) to generate pNL4. lonA was PCR amplified using primers pr_NL1 (5′ GGC CGT TTA CCT GTA ACA ACA ACG GAC 3′) and pr_NL2 (5′ GAG TGA CAA GTC ATT TCG ACT TGT CAG CCC 3′), and the blunt product was cloned into pCR-BluntII-TOPO to generate pNL3. pNL3 was digested with XbaI, and the lonA-containing fragment was ligated into SpeI-digested pVSV105 (22) to generate pNL18. hns was PCR amplified using primers pr_NL11 (5′ GCC AAA CCC AGA GCT ATA AGC GGG GGC 3′) and pr_NL12 (5′ TTT CGA GCA ATA ATA CGT TTC TAA ATG TAA TAA AAT GAA A 3′). pstA was PCR amplified using primers pr_NL13 (5′ GCG CTA GTT GTT GGC ATT GCA ATG GGA GCT GC 3′) and pr_NL14 (5′ AGA CAG GTG GTT AAC ATC CAT CGG TGA TAG 3′). phoQ was PCR amplified using primers pr_NL84 (5′ GCC TAA CGG TAC TAA AAA GCA TTC TGT ATG 3′) and pr_NL85 (5′ GAT GAA GAG CAT GAT TAT TAT TCT GAT GGA GAG ATA TTG G 3′). The blunt hns-, pstA-, and phoQ-containing products were cloned into SmaI-digested pVSV105 (22) to generate pNL6, pNL75, and pNL72, respectively.
The lacIq-PA1/34-luxC allele on pJLB101 was moved into the transposon mutants as described previously (11). Allelic exchange was confirmed by PCR and by IPTG inducibility of luminescence in the resulting strains. The lacIq-PA1/34-luxC allele from plasmid pJLB101 was crossed into the genomes of mutants EMH3, EMH5, EMH6, EMH7, EMH9, EMH12, EMH13, SLV4, SLV5, SLV10, SLV15, SLV16, SLV20, SLV29, SLV30, SLV32, SLV33, SLV41, SLV42, SLV43, NL1, NL3, NL4, NL6, and NL8 to generate strains NL18, NL19, NL20, NL21, NL22, NL23, NL24, NL25, NL26, NL27, NL28, NL29, NL30, NL31, NL32, NL33, NL34, NL35, NL36, NL37, NL38, NL39, NL40, NL41, and NL42, respectively. Similarly, the ΔlitR::kanR allele on plasmid pMF7 (26) was crossed into the genome of NL2 by using two-step allelic exchange to generate the mutant NL11.
To generate a ΔainR allele on plasmid pNL30, the 1,350-bp region upstream of ainR was PCR amplified using primers pr_NL27 (5′ GTA CTC ATA ACA CCA CTA CCT ATT TTT ACT ATA CTG 3′) and pr_NL28.3 (5′ GGG CCT AGG CAT TTA TAT AAA ACT CAC TGA TTT CGA AGT TT 3′), and the product was cloned into pCR-BluntII-TOPO plasmid to generate pNL28. The 1,480-bp region downstream of ES114 ainR was PCR amplified using primers pr_NL29 (5′ GGG GCC TAG GTA ACA CCG ATA AAA AAA TAG CCA GAA C 3′) and pr_NL30 (5′ CCC CAC TAG TCA TGA CTC TGT TGC GGG TCT TGA TGA AGC T 3′). AvrII and SpeI sites incorporated into the PCR product on primers pr_NL29 and pr_NL30 were digested with these enzymes, and this fragment was ligated into AvrII-digested pEVS118 (21) to generate pNL29. Plasmids pNL28 and pNL29 were linearized with AvrII and ligated together to generate pNL30, which contains upstream and downstream sequences fused at the ΔainR allele, with the ainR start and stop codons separated only by the 6-bp AvrII recognition sequence. The mutant strain NL43 (ΔainR) was generated by crossing the mutant ΔainR allele from plasmid pNL30 into ES114. Replacement of ainR with the ΔainR allele in NL43 was confirmed by PCR using primers pr_NL35 (5′ GAG TCC GTT AGC AAG GTC ACA CTT TGT TG 3′) and pr_NL36 (5′ ACC CAA AAC GTA AGA CCA TTG GTA TGC G 3′).
The ΔainSR allele was generated on plasmid pNL32 by PCR amplification of the 1,430-bp region upstream of ainS using primers pr_NL62 (5′ GGC GCT TTA CCG TTT GGT GAA AAC TTA CTT C 3′) and pr_NL63 (5′ GGG CCT AGG CTA CTC TTT TAT AAA TTC ATA TTG CAG GTT TT 3′). This product was cloned into pCR-BluntII-TOPO plasmid to generate pNL31. Plasmids pNL29 and pNL31 were linearized with AvrII and ligated together to generate pNL32, which contains the upstream and downstream sequences fused at the ΔainSR allele, with the ainS start and ainR stop codons separated by the 6-bp AvrII recognition sequence. Crossing the mutant ΔainSR allele from plasmid pNL32 into ES114 generated the mutant strain NL55 (ΔainSR). Replacement of ainSR with the ΔainSR allele was confirmed by PCR using primers DMC2 (5′ GGC GGT ACC AGA ACC AAG ACC TGC TCG TGC TAA 3′) and pr_NL36.
Luminescence and fluorescence measurements in culture.
Overnight V. fischeri cultures were diluted 1:1,000 in 50 ml of LBS or SWTO in 250-ml flasks and then incubated with shaking (200 rpm) at 24°C. At regular intervals, 500-μl samples were removed and the optical density at 595 nm (OD595) was measured with a BioPhotometer (Brinkman Instruments, Westbury, NY). Relative luminescence was measured with a TD-20/20 luminometer (Turner Designs, Sunnyvale, CA) immediately following shaking to aerate the sample. Specific luminescence was calculated as the luminescence per OD595 unit. Luminescence images of strains patched on plates and incubated overnight at ∼24°C were obtained with a Bio-Rad Fluor-S Max2 imager. Fluorescence of green fluorescent protein (GFP) expressed from reporter plasmids pJLB36 and pJBL38 (10) was measured with a TD-700 fluorometer (Turner Designs) with excitation and emission filters of 486 nm and >510 nm, respectively. Fluorescence values for strains carrying the promoterless vector pVSV33 (22) were subtracted as background. Unless otherwise indicated, the reported mean fluorescence for cultures was between OD595 values of 2.0 and 2.8, a range in which ES114-specific luminescence is approximately constant.
RESULTS
Isolation of luminescence-up mutants of V. fischeri ES114.
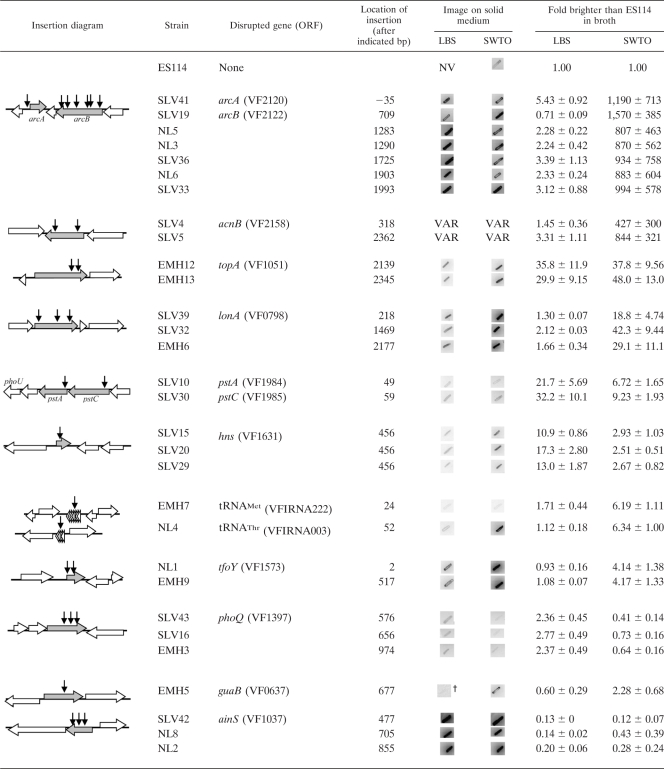
We screened ∼20,000 mini-Tn5 mutants of ES114 for increased luminescence and isolated 30 candidate luminescence-up clones that displayed consistent and significant increases in luminescence. The chromosomal location of the transposon insertions in these strains and their luminescence phenotypes are summarized in Table 1. Three of these mutants are likely siblings, because they have insertions in the same location in hns and originated from the same mating. Thus, our screen yielded 28 independent luminescence-up mutants of ES114 distributed over several loci (Table 1).
TABLE 1.
Analysis of luminescence-up transposon mutantsa
Horizontal arrows indicate disrupted genes (shaded) and flanking (open) open reading frames (ORFs). Vertical arrows correspond to locations of transposon insertions, and the insertion location is provided in bp relative to the A of the ATG start codon. All mutants were patched onto LBS and SWTO plates, and representative negative images of patches are shown, such that darker patches indicate greater bioluminescence. †, the contrast was increased to visualize the luminescence phenotype of mutant patches. NV, patches were not visible even with enhanced contrast. VAR, variable result as discussed in the text. All mutants were also grown in LBS and SWTO broth, and the average maximum luminescence/OD595 relative to ES114 ± standard error (n ≥ 2) is reported.
The mutants displayed a wide range of luminescence phenotypes that were from 2-fold to more than 1,000-fold brighter than their wild-type parent (Table 1). When grown in broth, all of the mutants still displayed cell-density-dependent regulation of luminescence (data not shown); however, with the exception of ainS mutants, they achieved brighter luminescence than the wild type in broth and some induce luminescence at lower cell densities (data not shown). Most mutants’ luminescence phenotypes varied depending on whether the cells were grown on plates or in broth and whether they were grown in LBS or SWTO. Both LBS and SWTO contain tryptone and yeast extract; however, LBS is buffered and supplemented with NaCl, whereas SWTO is unbuffered, supplemented with marine salts and glycerol, and is near marine osmolarity (71). Mutants with insertions in arcA, arcB, acnB, lonA, tfoY, guaB, and specific tRNA genes showed greater increases in luminescence relative to ES114 when grown in SWTO than in LBS (Table 1). In contrast, mutants with insertions in pstA, pstC, hns, and phoQ were comparatively brighter than ES114 when cultured in LBS (Table 1). Only the topA mutants showed similar luminescence-up phenotypes in both LBS and SWTO (Table 1). We found an unexpected result, discussed below, in that ainS mutants were much brighter than ES114 on plates (Table 1), despite being dimmer than ES114 in broth (Table 1), as was previously shown (47).
Comprehensiveness of the screen.
Based on previous work with mini-Tn5 derivatives (1, 39), we estimated that the ∼20,000 mutants screened should be approaching saturation of nonessential genes. Consistent with this prediction, multiple independent mutants were isolated at most of the loci identified, including all of the loci where dramatic and easily detected luminescence-up effects were observed (Table 1). On the other hand, some mutants of V. fischeri previously identified as having luminescence-up phenotypes in broth were not found in our study. These strains were patched onto LBS to test whether they would have been detected in our screen. Strains CL42 (47) and KV2801 (79), containing mutations within luxO and ptsI (E1), respectively, were dim on plates and would not have been identified in our screen (data not shown). A crr mutant (KV2850) yielded patches that were only marginally brighter on plates than ES114 (data not shown), and the phenotype was inconsistent; therefore, it is not surprising that a crr mutant was not isolated in our screen. A phoU mutant (KV2655) was brighter than ES114 on plates (data not shown), and this was missed in our screen; however, insertions were identified in pstA and pstC, which are upstream of phoU in what appears to be a phosphate transport operon. Taken together, these results suggest that continuing to screen additional transposon mutants under these conditions would yield relatively few additional new insights, although screening under different growth conditions might reveal novel mutants missed here.
Complementation of mutants.
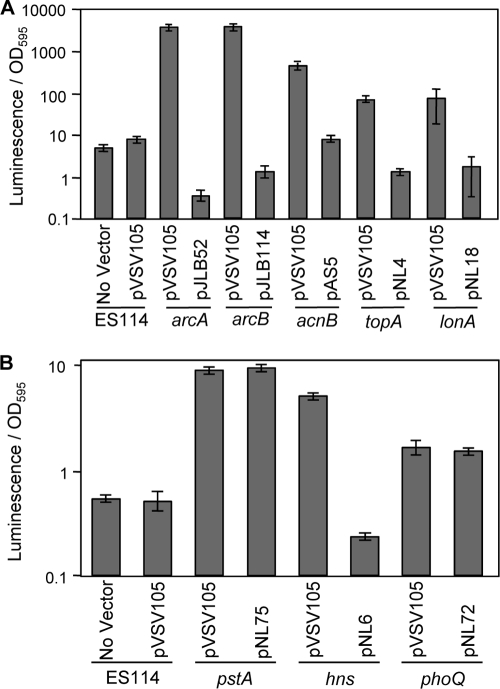
To confirm that the luminescence-up phenotypes resulted from the disruption of the genes identified, we genetically complemented the arcA, arcB, acnB, topA, lonA, pstA, phoQ, and hns mutants by providing the respective native genes in trans on low-copy-number shuttle plasmids (Fig. 2). In all strains except the pstA and phoQ mutants, wild-type luminescence was restored following reintroduction of the native gene. The pstA mutant maintained its luminescence-up phenotype (Fig. 2B), and this was likely due to polar effects on the downstream phoU. The relationship between this phosphate uptake system and luminescence is explored further below. Although the luminescence phenotype of the phoQ mutant was not complemented in LBS medium (Fig. 2B), it was complemented by phoQ in trans during growth in minimal medium (see below). There is no apparent cotranscribed gene downstream of phoQ (Table 1), making polar effects of this insertion unlikely. We further describe the effects of phoQ and Mg2+ on luminescence below. Because the ainS mutants only displayed a luminescence-up phenotype on plates, we evaluated genetic in trans complementation of ainS mutants in patches by using pCL112 (47), which restored wild-type-like dim luminescence (data not shown). We did not attempt to complement mutants with insertions in tfoY, guaB, or tRNA genes, which had relatively modest luminescence-up phenotypes. Given the apparent lack of cotranscribed genes downstream of tfoY (Table 1), it seems unlikely that the phenotype of this insertion mutant is caused by polar effects.
FIG. 2.
Genetic complementation of transposon mutants. Luminescence-up mutants were complemented with the respective complete gene carried on shuttle vector pVSV105. ES114 and mutants carrying the empty pVSV105 vector were included as controls. (A) Complementation of mutants with increased luminescence in SWTO. (B) Complementation of mutants with increased luminescence in LBS. Data for one representative mutant from each locus are shown, including SLV41 (arcA), NL3 (arcB), SLV4 (acnB), EMH12 (topA), SLV32 (lonA), SLV10 (pstA), SLV15 (hns), and SLV16 (phoQ). Error bars represent standard errors (n = 2).
Instability of acnB mutants.
The observable phenotypes of the luminescence-up mutants remained stable with the notable exception of mutants with insertions in acnB, which encodes the tricarboxylic acid cycle enzyme aconitase. There is no other aconitase gene apparent in the V. fischeri genome, and acnB mutants all grew relatively poorly on aerobic streak plates. Faster-growing suppressors arose frequently and had dim luminescence, like that of the wild type. We were able to confirm complementation of an acnB mutant in trans (described above) by curing the complementing plasmid with concomitant restoration of slow growth and bright luminescence. However, we found further genetic manipulation of these strains difficult due to the rapid appearance of suppressors. We have therefore omitted acnB mutants from the genetic analyses below and will describe the influence of acnB on luminescence in greater detail once the nature of the suppressors is more fully understood.
Dependence of luminescence-up phenotypes on the native luxICDABEG promoter.
We hypothesized that the luminescence-up phenotype of the mutants in Table 1 was due to regulatory effects on lux gene expression; however, the bright phenotypes of these mutants might alternatively be due to metabolic effects caused, for example, by increasing the availability of the FMNH2 or O2 substrates for LuxAB. To test this possibility, we placed a nonnative IPTG-inducible promoter construct, lacIq-PA1/34 between luxI and luxCDABEG (11), into the chromosomes of the mutant strains, with the exception of acnB mutants. Transposon insertions in arcA, arcB, lonA, pstA, pstC, hns, tfoY, phoQ, guaB, ainS, and the tRNAs had no effect on luminescence when luxCDABEG was expressed from this nonnative promoter, either with or without addition of IPTG (data not shown). Thus, the luminescence-up phenotype associated with most insertions was dependent on the native lux promoter and transcript.
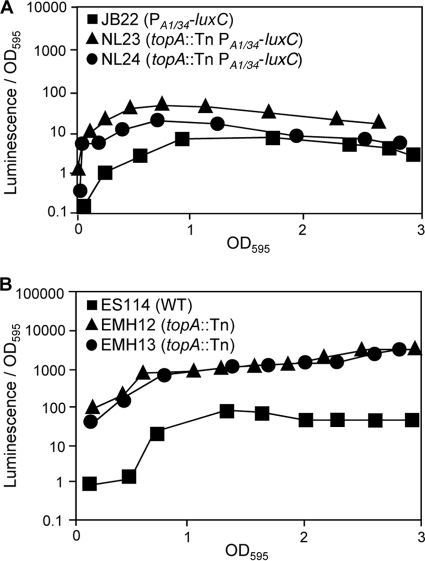
Only topA mutations yielded an increased level of luminescence in the lacIq-PA1/34-luxC background. topA mutations in the lacIq-PA1/34-luxC background led to enhanced luminescence at low ODs (Fig. 3A) but did so to a lesser extent than the ∼35- to 40-fold effect seen in the native lux promoter background (Table 1 and Fig. 3B). This suggests the topA mutants’ luminescence-up phenotype may be due to combined effects that are both dependent and independent of the native lux promoter.
FIG. 3.
Luminescence of topA mutants in SWTO. (A) Specific luminescence phenotype of JB22 as well as topA mutants NL23 and NL24, each of which have the luxCDABEG genes controlled by lacIq and the PA1/34 promoter. JB22 has the PA1/34- luxCDABEG allele in an otherwise-wild-type (ES114) background. No IPTG was added. (B) Specific luminescence of wild-type (WT) ES114 as well as topA mutants EMH12 and EMH13. Averages of two replicate flasks are shown.
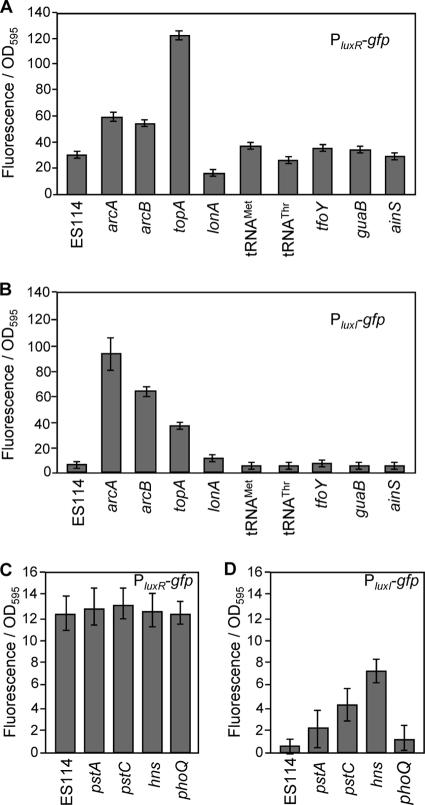
To test whether the luminescence-up mutations enhanced transcription from the luxI or luxR promoters, we moved reporter plasmids pJLB36 (PluxR-gfp) and pJLB38 (PluxI-gfp) into each mutant. Fluorescence data for these reporters were determined by growing the mutant strains in the medium (SWTO or LBS) that showed the greatest luminescence-up phenotype relative to the wild type. Consistent with our previous report (10), the PluxR-gfp and PluxI-gfp reporters yielded ∼2- and ∼15-fold greater fluorescence, respectively, in either the arcA or arcB mutants than in parental strain ES114 (Fig. 4A and B). These reporters also yielded higher fluorescence in topA mutants than in ES114, although the relative influence on the PluxR-gfp was greater than in the arcA or arcB backgrounds (Fig. 4A and B). The PluxI-gfp reporter yielded ∼4- and ∼8-fold greater fluorescence in the pstC and hns backgrounds, respectively, although these mutations had no effect on the PluxR-gfp reporter (Fig. 4C and D). None of the other transposon insertions, with the possible exception of those in unstable acnB mutants (data not shown), had a significant (P < 0.05) effect on either reporter under these conditions. This lack of effect in most mutants may reflect limitations of these reporters. It is worth noting that arc and topA mutants have much larger effects on luminescence (Table 1) than on gfp reporter activity (Fig. 4), and similar results were obtained with lacZ reporters (data not shown). These plasmid-borne reporters are maintained at ∼9.4 copies per genome (21), potentially titrating out regulators, although at least in the case of ArcA effects of similar magnitude were seen with lux-gfp reporters on a plasmid or on the chromosome in single copy (10). Taken together, it is perhaps not surprising that mutations with moderate effects on luminescence have no discernible influence on these reporters.
FIG. 4.
Effects of transposon mutations on luxR and luxI promoter-gfp reporters. Specific fluorescence generated from reporter plasmids pJLB36 (PluxR-gfp) (A and C) or pJLB38 (PluxI-gfp) (B and D) harbored in ES114 or in luminescence-up mutants is shown. Reporters were assayed in mutants grown in either SWTO (A and B) or LBS (C and D), depending on which medium yielded the greatest effect on luminescence. Fluorescence from ES114 harboring the promoterless parent vector pVSV33 was subtracted as background. Reporters were tested in each mutant described in Table 2, and data for one representative mutant from each locus are shown, including SLV41 (arcA), NL3 (arcB), EMH12 (topA), SLV32 (lonA), SLV10 (pstA), SLV30 (pstC), SLV15 (hns), EMH7 (tRNA-Met), NL4 (tRNA-Thr), NL1 (tfoY), SLV16 (phoQ), EMH5 (guaB), and NL2 (ainS). In all panels, data represent the average specific fluorescence when the culture OD595 was between 2.0 and 2.8, a range in which specific luminescence is constant for these strains. Averages and standard errors were calculated from 8 to 12 distinct samples taken from two independent replicate flasks of each examined strain. Mutants with insertions at the same locus were analyzed together in two separate experiments with similar results, and data for one representative mutant from one experiment are shown.
Role of ainS-mediated signaling in repression of luminescence.
The luminescence-up phenotype of ainS mutants on plates was unexpected given their previously reported diminished luminescence in broth (47), and we examined these mutants further. The AinS-produced pheromone C8-HSL is thought to act as a signal through two pathways (Fig. 1). In one pathway C8-HSL binds to LuxR and activates transcription, much like the LuxI product 3-oxo-C6-HSL. C8-HSL is a weaker activator of LuxR (67) and can apparently compete with the stronger activator 3-oxo-C6-HSL to dim luminescence (47). In the second pathway, C8-HSL is thought to be sensed by AinR (30), which as illustrated in Fig. 1 ultimately leads to an increase in the regulator LitR (26, 47, 58).
To explore which C8-HSL-responsive pathway is responsible for the luminescence-up phenotype of ainS mutants on plates, we used targeted mutants lacking components of these signaling pathways and compared their luminescence in patches on solid medium. Table 2 shows our results, which include the following: (i) the luminescence-up phenotype associated with loss of ainS was independent of ainR, indicating that this phenotype is not simply due to a lack of C8-HSL-AinR; (ii) the luminescence-up phenotype of the ainS mutant was dependent on luxI, indicating a key role for 3-oxo-C6-HSL in generating the bright luminescence; (iii) the luminescence-up phenotype of ainS mutants was also dependent on both luxS and litR, and such dependence could be relieved by inactivating luxO, even though a luxO mutation itself did not result in bright luminescence on plates (data not shown). When taken together and compared with the model of signaling presented in Fig. 1, these data are consistent with the luminescence-up phenotype of ainS mutants on plates resulting from the removal of competition for LuxR activation between C8-HSL and the stronger inducer 3-oxo-C6-HSL. Moreover, under these conditions it appears that LuxS and AI-2 are sufficient for the signaling through LuxO necessary to generate LitR, rendering AinS and C8-HSL dispensable in this regard.
TABLE 2.
Luminescence analysis of mutants involved in the AinS signaling pathways
| Strain | Reference | Genotype | Patches on LBS platesa |
|---|---|---|---|
| ES114 | 8 | Wild type | − |
| NL2 | This study | ES114 ainS::mini-Tn5-ermR | + |
| NL43 | This study | ES114 ΔainR | − |
| NL55 | This study | ES114 ΔainSR | + |
| CL24 | 47 | ES114 luxS::kanR | − |
| NL11 | This study | ES114 ainS::mini-Tn5-ermR litR::kanR | − |
| CL39 | 46 | ES114 luxI-ainS::catR | − |
| CL41 | 46 | ES114 ainS::catR luxS::kanR | − |
| CL64 | 47 | ES114 ainS::catR luxO::kanR | + |
| CL90 | 46 | ES114 luxS::kanR luxO::kanR | − |
| CL91 | 46 | ES114 ainS::catR luxS::kanR luxO::kanR | + |
All strains were patched onto LBS plates, and the results of the negative images are reported as follows: +, patches were bright; −, patches were not bright (compared to ainS mutant patches [Table 1]).
Roles of PhoB and phosphate in regulation of luminescence.
The luminescence-up phenotypes of the phoU, pstA, and pstC mutants (Table 1 and data not shown) suggested a role for inorganic phosphate (Pi) in the regulation of luminescence. In E. coli, the pst genes encode a high-affinity Pi transport system that is active under low-Pi conditions (86), and mutations in the pst genes result in increased expression of genes associated with the pho regulon in a mechanism dependent on the PhoR/PhoB two-component regulatory system (74). At low Pi, the sensor PhoR phosphorylates the response regulator PhoB, which then activates the transcription of specific genes within the pho regulon (49, 50). PhoU, which is encoded at the distal end of the pst operon, counteracts PhoB when [Pi] is high (59).
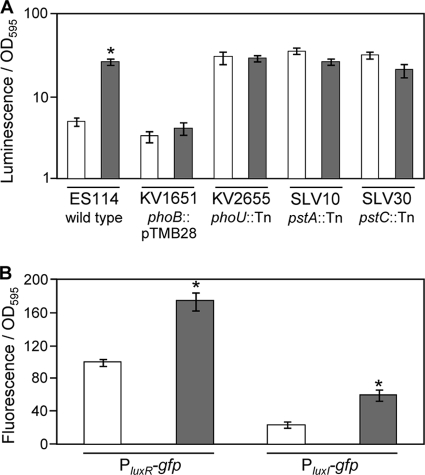
To test whether luminescence and lux regulation are tied to the pho regulon, luminescence was measured for ES114 as well as pst, phoU, and phoB transposon-insertion mutants cultured at relatively high and low Pi concentrations (Fig. 5A). In a medium with reduced Pi levels, the luminescence of ES114 was ∼10-fold higher at an OD595 of 0.5, and this response to low Pi was absent in a phoB mutant (Fig. 5A). Moreover, mutants with insertions in phoU, pstA, or pstC displayed constitutively high levels of luminescence regardless of [Pi]. These data suggest that luminescence is controlled in part by the PhoR/PhoB system in response to low Pi.
FIG. 5.
Effects of Pi concentration on luminescence and lux promoter-reporters. (A) Specific luminescence at OD595 of 0.5 for strains ES114 (wild type), KV1651 (phoB::pTMB28), KV2655 (phoU::mini-Tn5-ermR), SLV10 (pstA::mini-Tn5-ermR), and SLV30 (pstC::mini-Tn5-ermR) grown in FMM containing 378 μM (open bars) or 37.8 μM (gray bars) phosphate. (B) Specific fluorescence from PluxR-gfp and PluxI-gfp reporters on plasmids pJLB36 and pJLB38, respectively, harbored in ES114 grown in FMM containing 378 μM (open bars) or 37.8 μM (gray bars) phosphate. Fluorescence from ES114 harboring promoterless parent vector pVSV33 was subtracted as background. Data represent average specific fluorescence when culture OD595 values were ∼0.5. Bars indicate standard errors (n = 2). *, significant difference between high and low Pi conditions as determined with Student's t test (P ≤ 0.01).
To further examine the mechanism of Pi-mediated lux regulation, ES114 cells containing PluxR-gfp and PluxI-gfp reporter plasmids were grown in high- and low-Pi media (Fig. 5B). Under conditions with lower Pi levels, the activities of PluxR-gfp and PluxI-gfp each increased ∼2-fold. As a control we tested a constitutive promoter-gfp reporter and found no relative effect of high and low Pi (data not shown). Thus, low Pi leads to an increase in activity from the lux promoters, which may account for the luminescence-up phenotypes observed in phoU, pstA, and pstC mutant strains.
Effect of PhoQ and Mg2+ on luminescence.
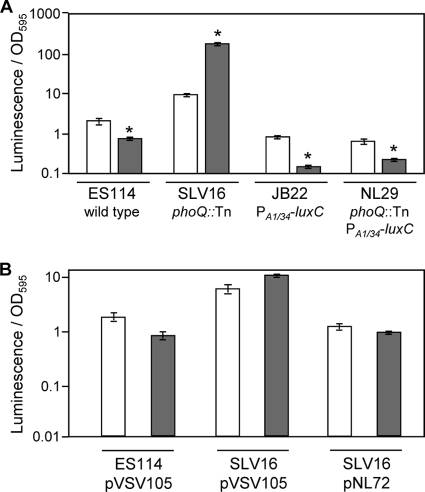
We also identified luminescence-up mutants with insertions in phoQ (Table 1). In other bacteria PhoQ together with the response regulator PhoP act as a two-component regulatory system that responds to low Mg2+ (42, 63, 78). Based on a simple model drawn from the luminescence-up phenotype of phoQ mutants and the function of PhoQ in other systems, we predicted a PhoQ-dependent repression of luminescence when Mg2+ was relatively low.
To further examine the effects of PhoP/PhoQ and Mg2+ on luminescence, we measured the luminescence of wild-type ES114, the constitutive lux-expressing strain JB22 (11), and their respective phoQ mutants, SLV16 and NL29, grown in a defined medium with differing concentrations of Mg2+. Consistent with our prediction, low Mg2+ led to decreased luminescence in ES114 but not in the phoQ mutant (Fig. 6A). Providing phoQ in trans restored low luminescence to the phoQ mutant (Fig. 6B). However, we also observed an unexpected increase in luminescence at low Mg2+ in the phoQ mutant (Fig. 6), suggesting a PhoQ-independent mechanism that functions counter to PhoQ to increase luminescence at low Mg2+. Moreover, low Mg2+ also decreased luminescence in both strains JB22 and NL29, in which luminescence is expressed from the constitutive nonnative PA1/34-luxC promoter (Fig. 6). These data indicate that luminescence is influenced by Mg2+ through multiple mechanisms that are both PhoQ dependent and PhoQ independent.
FIG. 6.
Effect of Mg2+ concentration on luminescence. (A) Specific luminescence of V. fischeri strains ES114 (wild type), SLV16 (phoQ::mini-Tn5-ermR), JB22 (ES114 lacIq-PA1/34-luxC), and NL29 (SLV16 lacIq-PA1/34-luxC). (B) Specific luminescence of ES114 and SLV16 carrying the empty pVSV105 vector and SLV16 carrying pNL72, which contains the native phoQ allele. Both panels report data from cultures grown in FMM, which contains 55 mM magnesium (open bars), or FMM with low (0.03 mM) Mg2+ (gray bars). *, significant difference in luminescence for that strain grown in low-Mg2+ medium relative to normal FMM as determined with Student's t test (P ≤ 0.05).
Effect of GuaB and guanine on luminescence.
GuaB and GuaA are required for the synthesis of GMP (55, 75), and we predicted that the insertion disrupting guaB would disrupt guanine synthesis. As predicted, in FMM medium a severe growth defect of the guaB mutant was observed that was recovered by adding 0.25 mM guanine (data not shown). Furthermore, luminescence of the guaB mutant was restored to dimmer, wild-type levels by adding 0.15 mM guanine to SWTO (data not shown).
Regulation of luminescence in response to aeration is topA dependent.
When ES114 is poorly aerated, its expression of luminescence is repressed (73); however, no mechanism for this regulation has yet been shown. The isolation of luminescence-up mutants with insertions in arcA, arcB, acnB, and topA (Table 1) suggested possible mechanisms for regulation of luminescence in response to culture aeration, because ArcA/ArcB, aconitase, and DNA supercoiling, which is mediated in part by TopA, have been implicated in redox-dependent control of gene expression in E. coli (2, 6, 33). Due to the problems with suppressors arising in cultures of acnB mutants as mentioned above, we limited our investigation to possible effects of ArcA/ArcB or TopA on aeration-dependent regulation of luminescence.
The redox-responsive ArcA/ArcB system represses luminescence presumably in response to reducing conditions (10). However, we found that ArcA cannot account for the repression of luminescence when cultures are grown in poorly aerated broth. In both the wild type and a ΔarcA mutant, the luminescence of cells grown in poorly aerated flasks began to diverge from that of well-aerated cells at an OD595 of ∼0.7 (Fig. 7A). Thus, although the ΔarcA mutant is brighter than ES114 under both conditions, the effect of poor aeration on luminescence is similar in both strains.
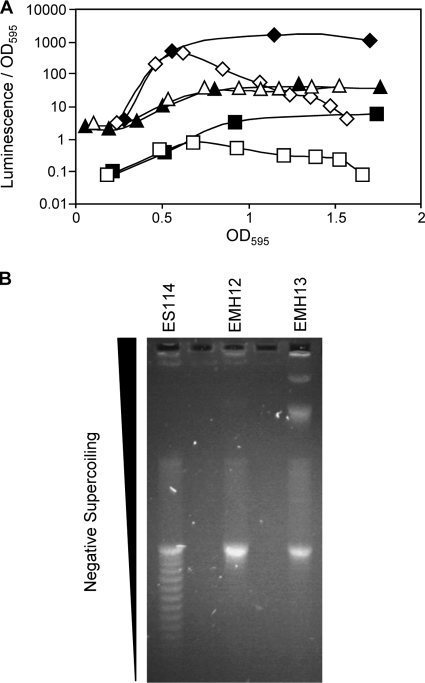
FIG. 7.
Effect of topA mutations on aeration-dependent regulation of luminescence and supercoiling. (A) Effect of topA mutation on aeration-mediated luminescence. Specific luminescence of wild-type ES114 (squares), topA mutant EMH12 (triangles), and arcA mutant AMJ2 (diamonds) in well-aerated (closed) or poorly aerated (open) cultures. Poor aeration conditions were created by growing cultures in 250-ml flasks containing 200 ml of SWTO, and well-aerated conditions consisted of 50 ml of SWTO in 250-ml flasks. (B) Supercoiling of plasmid DNA isolated from ES114 or topA mutants was assayed by electrophoresis through 0.8% agarose containing 20 μg/ml chloroquine. DNA that is less negatively supercoiled prior to loading will migrate more rapidly at this concentration of chloroquine (29). Laddering on the gel is indicative of topoisomers of the same plasmid that differ by one linking number. The gel was washed in distilled H2O for 2 h, stained in 1 μg/ml ethidium bromide for 1 h, and then imaged.
We next examined the topoisomerase I (topA) mutants. In E. coli, DNA is more negatively supercoiled in anaerobically grown cultures, leading to changes in gene expression (17, 37). Similarly, we found that plasmid supercoiling in V. fischeri is affected by culture aeration (data not shown). Topoisomerase I removes negative supercoils from DNA (43, 80), and we hypothesized that topA mutations might prevent cells from using genomic supercoiling as part of their global response to aeration. We confirmed the supercoiling phenotypes of topA mutants, assaying the level of negative supercoiling by isolating plasmid DNA from the wild type and the topA mutant strain and examining them in chloroquine gels. As predicted, supercoiling was affected in a topA mutant background (Fig. 7B). We also examined the luminescence phenotype of a topA mutant grown under different aeration conditions. In contrast to the wild type and arcA mutants, the luminescence of the topA mutant remained similar in both well-aerated and poorly aerated cultures (Fig. 7A). This suggests that a topA-dependent effect on the level of negative supercoiling may regulate luminescence in response to aeration.
DISCUSSION
In many bacteria, achieving a high cell density quorum is necessary but not sufficient to elicit full induction of pheromone-dependent behaviors. Therefore, understanding the biological significance of pheromone-mediated regulation in bacteria will require examining the environmentally responsive regulators that govern these systems. Only by elucidating the environmental contingencies required for induction of pheromone-controlled regulons will we be able to fully appreciate their functions in nature and contributions to bacterial fitness. In V. fischeri, environmental conditions clearly play an important role in modulating lux expression, and the ∼1,000-fold increase in luminescence of V. fischeri upon colonizing E. scolopes cannot be explained by cell density (8, 9). However, in contrast to a detailed understanding of pheromone-mediated activation of luminescence (Fig. 1), control of the luxICDABEG operon by environmentally responsive regulators has been less thoroughly explored.
In this study we initiated a systematic examination of the regulatory mechanisms accounting for the dim luminescence of V. fischeri ES114 in culture in order to elucidate the conditions that promote upregulation of luminescence and autoinducer synthesis. Using transposon mutagenesis, we isolated 28 independent luminescence-up mutants with insertions in 14 loci. Through the characterization of these mutants along with subsequent analyses of ES114 in different media, we have shown that luminescence in ES114 is responsive to a broad regulatory web influenced by specific environmental conditions. Given that the autoinducer synthase gene luxI is cotranscribed with the genes responsible for luminescence, these data support the idea that the quorum-sensing circuitry is regulated in response to the environment through multiple regulatory systems.
Environment-dependent lux regulation.
The importance of environmental context in luminescence regulation was underscored by simple analyses of the luminescence-up mutants. For example, in all mutants except topA strains, the luminescence-up phenotype was medium specific (Table 1). In some instances, these medium-dependent effects could be rationalized, given the genetic analysis of the mutants. For instance, mutants SLV16, SLV43, and EMH3 displayed a larger luminescence-up effect relative to ES114 when grown in LBS medium as opposed to SWTO (Table 1). These mutants have transposon insertions in phoQ, encoding the response regulator of the PhoPQ two-component regulatory system, which responds to environmental signals, including low [Mg2+] (42, 63, 78). Given the much lower levels of Mg2+ in LBS than in SWTO, the medium-dependent phenotype of these mutants was what one would predict if PhoQ repressed luminescence in response to low Mg2+. This model was tested further and validated by showing a phoQ-dependent repressive effect of Mg2+ on luminescence (Fig. 6). Although in other systems PhoPQ also responds to [Ca2+] (78), antimicrobial peptides (4), and pH (5), these factors did not show clear PhoQ-dependent effects on luminescence in V. fischeri (data not shown). Hussa et al. previously reported multiple homologs of phoP in V. fischeri (38), and the corresponding regulation may be more complex than in E. coli. Interestingly, our data indicate PhoQ-independent effects on luminescence, and taken together Mg2+ appears to influence luminescence through multiple mechanisms.
Similarly, analysis of luminescence-up mutants led to the identification of Pi as a key environmental factor governing luminescence. Mutations in pstA, pstC, or phoU led to increased luminescence (Table 1 and Fig. 5), and by analogy to E. coli, mutants in this Pi uptake operon (16, 69) would be expected to display the PhoB-dependent Pi starvation response (81). Consistent with this model, luminescence in ES114 is elevated in response to low Pi levels in a PhoB-dependent manner (Fig. 5). Interestingly, the “low Pi” concentrations utilized in this work appear to be much higher than levels found in the Hawaiian coastal waters inhabited by E. scolopes. Assays completed to determine the [Pi] (14) of seawater showed phosphate levels of <100 nM (data not shown), which is consistent with previous studies (3, 65, 68). However, this does not account for organic phosphate sources, which along with phosphate availability in the host light organ should be considered in future work.
Finally, the identification of luminescence-up mutations in topA, arcA, arcB, and acnB hinted at an important role for redox in luminescence regulation, as each of these loci has been connected to redox-dependent regulation in other systems. The degree of aeration influences regulation of lux expression in V. fischeri (62, 73), and here we show that this is dependent on topA, suggesting a role for supercoiling in this regulatory phenomenon (Fig. 7). Ongoing studies are aimed at understanding whether the luminescence-up mutations in arc or acnB reflect other environmental conditions that influence the cellular redox state.
Future experiments will also be aimed at understanding whether environmental conditions influence regulation mediated by genes such as ainS, lonA, hns, and tfoY (Table 1). Preliminary work suggests that at least ainS is subject to regulation in response to the environment, and the other genes may similarly have greater or lesser effects on luminescence in a context-dependent manner.
Mechanisms of luminescence regulation.
Understanding the mechanism(s) underlying the regulation of luminescence in luminescence-up mutants is important for at least two reasons. First, regulation of the native luxICDABEG promoter is likely to affect LuxI synthase expression and 3-oxo-C6-HSL synthesis in addition to affecting luminescence, leading to positive feedback and the potential for cell-cell signaling. Most mutations reported here did exert their effects on luminescence through mechanisms dependent on the native lux promoter, and therefore such regulation has the potential for population-wide 3-oxo-C6-HSL-mediated effects. Second, it will be important to ascertain whether regulators act directly on the lux promoter or indirectly, for example, by modulating one of the direct regulators. This distinction will be important for building robust predictive models of luminescence regulation. For example, interdependence of different regulators would imply that multiple environmental conditions must be sensed simultaneously to elicit a regulatory response.
Little is yet known about the specific mechanisms by which the regulators identified here control lux operon expression. It is thought that Lon targets LuxR (51), and we have previously shown that ArcA binds directly to the lux promoter (10), but mechanisms for the other regulators are less clear and await testing. For example, although luxR promoter activity is increased under low-Pi conditions, a search of the lux intergenic region did not reveal a clear pho box typical of a PhoB binding site, suggesting that it may be regulating luminescence indirectly via an unknown mechanism.
For some mutants, the underlying regulatory mechanism is perplexing. Notably, transposon insertions in tRNAMet and tRNAThr produced mutants with an ∼6-fold increase in luminescence during growth in SWTO; however, the significance of this finding is unclear. These interrupted tRNAs and those downstream from them are not unique in the V. fischeri genome. There are several loci for both tRNAMet and tRNAThr annotated within the genome with at least one additional locus having 100% identity to the mutated genes. Thus, although we cannot rule out a model of regulation by rare tRNAs, such a mechanism is not immediately apparent.
Other implications for pheromone signaling research.
Our discovery of regulators that respond to environmental stimuli and control lux expression has important implications. For example, autoinducer-mediated regulation in V. fischeri has often been used as a model system for mathematically describing a genetic regulatory circuit; however, the effects of environmental regulation on lux expression have been largely overlooked in these studies. Rather, many mathematical models of lux regulatory circuitry either omit environmental regulation entirely (40, 45, 53, 60, 61, 66, 82, 85) or consider only CRP-mediated regulation in response to glucose (7, 15). In the future, components of the environmentally responsive regulators connected to lux should be incorporated into the models of this regulatory circuit.
This work will also direct research aimed at understanding the environment experienced by V. fischeri inside its host, E. scolopes. With a better appreciation of the conditions and environmentally responsive regulators that lead to dim luminescence in culture, we can now develop clear hypotheses regarding the environmental cues that lead to lux induction during colonization. For example, the studies described above have prompted greater interest in examining redox, Mg2+, and Pi levels in the light organ. Our ability to reconstitute and observe this symbiosis offers a unique opportunity to assess the dynamics and regulation of pheromone-mediated signaling in a context that is ecologically relevant for the bacterium.
Acknowledgments
We thank Erica M. Hall, Susan L. Vescovi, and Alecia N. Septer for technical assistance and Karen Visick for providing strains.
This material is based upon work supported by the National Science Foundation (NSF) under grants CAREER MCB-0347317 and IOS-0841480, by the Army Research Office under grant 49549LSII, and by the National Institutes of Health under grant AI50661 to M. McFall-Ngai. A.N.S. was supported by a National Defense Science and Engineering Graduate Fellowship, 32 CFR 168a, under and awarded by DoD, Air Force Office of Scientific Research. S.L.V. was supported by an NSF Research Experience for Undergraduates site award (DBI-0453353). Genome information was provided by the Vibrio fischeri Genome Project supported by the W. M. Keck Foundation.
Footnotes
Published ahead of print on 6 August 2010.
REFERENCES
- 1.Adin, D. M., K. L. Visick, and E. V. Stabb. 2008. Identification of a cellobiose utilization gene cluster with cryptic beta-galactosidase activity in Vibrio fischeri. Appl. Environ. Microbiol. 74:4059-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeeva, S., K. J. Hellingwerf, and M. J. Teixeira de Mattos. 2003. Requirement of ArcA for redox regulation in Escherichia coli under microaerobic but not anaerobic or aerobic conditions. J. Bacteriol. 185:204-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armstrong, F. A. J., and H. W. Harvey. 1950. The cycle of phosphorus in the waters of the English Channel. J. Mar. Biol. Assoc. U. K. 29:145-162. [Google Scholar]
- 4.Bader, M. W., S. Sanowar, M. E. Daley, A. R. Schneider, U. Cho, W. Xu, R. E. Klevit, H. Le Moual, and S. I. Miller. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461-472. [DOI] [PubMed] [Google Scholar]
- 5.Bearson, B. L., L. Wilson, and J. W. Foster. 1998. A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180:2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bebbington, K. J., and H. D. Williams. 2001. A role for DNA supercoiling in the regulation of the cytochrome bd oxidase of Escherichia coli. Microbiology 147:591-598. [DOI] [PubMed] [Google Scholar]
- 7.Belta, C., J. Schug, T. Dang, V. Kumar, G. J. Pappas, H. Rubin, and P. Dunlap. 2001. Stability and reachability analysis of a hybrid model of luminescence in the marine bacterium Vibrio fischeri, p. 869-874. Proc. 40th IEEE Conf. on Decision Control 2001. IEEE, Piscataway, NJ.
- 8.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boettcher, K. J., and E. G. Ruby. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bose, J. L., U. Kim, W. Bartkowski, R. P. Gunsalus, A. M. Overley, N. L. Lyell, K. L. Visick, and E. V. Stabb. 2007. Bioluminescence in Vibrio fischeri is controlled by the redox-responsive regulator ArcA. Mol. Microbiol. 65:538-553. [DOI] [PubMed] [Google Scholar]
- 11.Bose, J. L., C. S. Rosenberg, and E. V. Stabb. 2008. Effects of luxCDABEG induction in Vibrio fischeri: enhancement of symbiotic colonization and conditional attenuation of growth in culture. Arch. Microbiol. 190:169-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boylan, M., C. Miyamoto, L. Wall, A. Graham, and E. Meighen. 1989. Lux C, D and E genes of the Vibrio fischeri luminescence operon code for the reductase, transferase, and synthetase enzymes involved in aldehyde biosynthesis. Photochem. Photobiol. 49:681-688. [DOI] [PubMed] [Google Scholar]
- 13.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 14.Cogan, E. B., G. B. Birrell, and O. H. Griffith. 1999. A robotics-based automated assay for inorganic and organic phosphates. Anal. Biochem. 271:29-35. [DOI] [PubMed] [Google Scholar]
- 15.Cox, C. D., G. D. Peterson, M. S. Allen, J. M. Lancaster, J. M. McCollum, D. Austin, L. Yan, G. S. Sayler, and M. L. Simpson. 2003. Analysis of noise in quorum sensing. OMICS 7:317-334. [DOI] [PubMed] [Google Scholar]
- 16.Cox, G. B., H. Rosenberg, J. A. Downie, and S. Silver. 1981. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J. Bacteriol. 148:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorman, C. J., G. C. Barr, N. N. Bhriain, and C. F. Higgins. 1988. DNA supercoiling and the anaerobic and growth phase regulation of tonB gene expression. J. Bacteriol. 170:2816-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunlap, P. V., and E. P. Greenberg. 1985. Control of Vibrio fischeri luminescence gene expression in Escherichia coli by cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 164:45-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunlap, P. V., and E. P. Greenberg. 1988. Control of Vibrio fischeri lux gene transcription by a cyclic AMP receptor protein-LuxR protein regulatory circuit. J. Bacteriol. 170:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunlap, P. V., and J. M. Ray. 1989. Requirement for autoinducer in transcriptional negative autoregulation of the Vibrio fischeri luxR gene in Escherichia coli. J. Bacteriol. 171:3549-3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 22.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 24.Engebrecht, J., K. Nealson, and M. Silverman. 1983. Bacterial bioluminescence: isolation and genetic analysis of functions from Vibrio fischeri. Cell 32:773-781. [DOI] [PubMed] [Google Scholar]
- 25.Engebrecht, J., and M. Silverman. 1984. Identification of genes and gene products necessary for bacterial bioluminescence. Proc. Natl. Acad. Sci. U. S. A. 81:4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fidopiastis, P. M., C. M. Miyamoto, M. G. Jobling, E. A. Meighen, and E. G. Ruby. 2002. LitR, a new transcriptional activator in Vibrio fischeri, regulates luminescence and symbiotic light organ colonization. Mol. Microbiol. 45:131-143. [DOI] [PubMed] [Google Scholar]
- 27.Friedrich, W. F., and E. P. Greenberg. 1983. Glucose repression of luminescence and luciferase in Vibrio fischeri. Arch. Microbiol. 134:87-91. [Google Scholar]
- 28.Fuqua, C., S. C. Winans, and E. P. Greenberg. 1996. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu. Rev. Microbiol. 50:727-751. [DOI] [PubMed] [Google Scholar]
- 29.Garner, M. M., G. Felsenfeld, M. H. O'Dea, and M. Gellert. 1987. Effects of DNA supercoiling on the topological properties of nucleosomes. Proc. Natl. Acad. Sci. U. S. A. 84:2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilson, L., A. Kuo, and P. V. Dunlap. 1995. AinS and a new family of autoinducer synthesis proteins. J. Bacteriol. 177:6946-6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gray, K. M., and E. P. Greenberg. 1992. Physical and functional maps of the luminescence gene cluster in an autoinducer-deficient Vibrio fischeri strain isolated from a squid light organ. J. Bacteriol. 174:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gray, K. M., and E. P. Greenberg. 1992. Sequencing and analysis of luxR and luxI, the luminescence regulatory genes from the squid light organ symbiont Vibrio fischeri ES114. Mol. Mar. Biol. Biotechnol. 1:414-419. [Google Scholar]
- 33.Gruer, M. J., and J. R. Guest. 1994. Two genetically-distinct and differentially-regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531-2541. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 35.Hastings, J. W., and K. H. Nealson. 1977. Bacterial bioluminescence. Annu. Rev. Microbiol. 31:549-595. [DOI] [PubMed] [Google Scholar]
- 36.Haygood, M. G., and K. H. Nealson. 1985. Mechanisms of iron regulation of luminescence in Vibrio fischeri. J. Bacteriol. 162:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsieh, L. S., R. M. Burger, and K. Drlica. 1991. Bacterial DNA supercoiling and [ATP]/[ADP]. Changes associated with a transition to anaerobic growth. J. Mol. Biol. 219:443-450. [DOI] [PubMed] [Google Scholar]
- 38.Hussa, E. A., T. M. O'Shea, C. L. Darnell, E. G. Ruby, and K. L. Visick. 2007. Two-component response regulators of Vibrio fischeri: identification, mutagenesis, and characterization. J. Bacteriol. 189:5825-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, L. Chun-Rong, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.James, S., P. Nilsson, G. James, S. Kjelleberg, and T. Fagerstrom. 2000. Luminescence control in the marine bacterium Vibrio fischeri: an analysis of the dynamics of lux regulation. J. Mol. Biol. 296:1127-1137. [DOI] [PubMed] [Google Scholar]
- 41.Kaplan, H. B., and E. P. Greenberg. 1985. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J. Bacteriol. 163:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kato, A., H. Tanabe, and R. Utsumi. 1999. Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181:5516-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kirkegaard, K., and J. C. Wang. 1985. Bacterial DNA topoisomerase I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 185:625-637. [DOI] [PubMed] [Google Scholar]
- 44.Kuo, A., N. V. Blough, and P. V. Dunlap. 1994. Multiple N-acyl-L-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol. 176:7558-7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuttler, C., and B. A. Hense. 2008. Interplay of two quorum sensing regulation systems of Vibrio fischeri. J. Theor. Biol. 251:167-180. [DOI] [PubMed] [Google Scholar]
- 46.Lupp, C., and E. G. Ruby. 2004. Vibrio fischeri LuxS and AinS: comparative study of two signal synthases. J. Bacteriol. 186:3873-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lupp, C., M. Urbanowski, E. P. Greenberg, and E. G. Ruby. 2003. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Mol. Microbiol. 50:319-331. [DOI] [PubMed] [Google Scholar]
- 48.Lyell, N. L., A. K. Dunn, J. L. Bose, S. L. Vescovi, and E. V. Stabb. 2008. Effective mutagenesis of Vibrio fischeri by using hyperactive mini-Tn5 derivatives. Appl. Environ. Microbiol. 74:7059-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makino, K., H. Shinagawa, M. Amemura, T. Kawamoto, M. Yamada, and A. Nakata. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551-559. [DOI] [PubMed] [Google Scholar]
- 50.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85-95. [DOI] [PubMed] [Google Scholar]
- 51.Manukhov, I. V., V. Kotova, and G. B. Zavil'genskii. 2006. Role of GroEL/GroES chaperonin system and Lon protease in regulation of expression Vibrio fischeri lux genes in Escherichia coli cells. Mol. Biol. (Mosk.) 40:277-283. [PubMed] [Google Scholar]
- 52.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCollum, J. M., G. D. Peterson, C. D. Cox, M. L. Simpson, and N. F. Samatova. 2006. The sorting direct method for stochastic simulation of biochemical systems with varying reaction execution behavior. Comput. Biol. Chem. 30:39-49. [DOI] [PubMed] [Google Scholar]
- 54.McFall-Ngai, M. J., and E. G. Ruby. 1991. Symbiont recognition and subsequent morphogenesis as early events in an animal-bacterial mutualism. Science 254:1491-1494. [DOI] [PubMed] [Google Scholar]
- 55.Mehra, R. K., and W. T. Drabble. 1981. Dual control of the gua operon of Escherichia coli K12 by adenine and guanine nucleotides. J. Gen. Microbiol. 123:27-37. [DOI] [PubMed] [Google Scholar]
- 56.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117-139. [DOI] [PubMed] [Google Scholar]
- 57.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Plainview, NY.
- 58.Miyamoto, C. M., Y. H. Lin, and E. A. Meighen. 2000. Control of bioluminescence in Vibrio fischeri by the LuxO signal response regulator. Mol. Microbiol. 36:594-607. [DOI] [PubMed] [Google Scholar]
- 59.Muda, M., N. N. Rao, and A. Torriani. 1992. Role of PhoU in phosphate transport and alkaline phosphatase regulation. J. Bacteriol. 174:8057-8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller, J., C. Kuttler, and B. A. Hense. 2008. Sensitivity of the quorum sensing system is achieved by low pass filtering. Biosystems 92:76-81. [DOI] [PubMed] [Google Scholar]
- 61.Muller, J., C. Kuttler, B. A. Hense, M. Rothballer, and A. Hartmann. 2006. Cell-cell communication by quorum sensing and dimension-reduction. J. Math. Biol. 53:672-702. [DOI] [PubMed] [Google Scholar]
- 62.Nealson, K. H., and J. W. Hastings. 1977. Low oxygen is optimal for luciferase synthesis in some bacteria. Ecological implications. Arch. Microbiol. 112:9-16. [DOI] [PubMed] [Google Scholar]
- 63.Newcombe, J., J. C. Jeynes, E. Mendoza, J. Hinds, G. L. Marsden, R. A. Stabler, M. Marti, and J. J. McFadden. 2005. Phenotypic and transcriptional characterization of the meningococcal PhoPQ system, a magnesium-sensing two-component regulatory system that controls genes involved in remodeling the meningococcal cell surface. J. Bacteriol. 187:4967-4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nijvipakul, S., J. Wongratana, C. Suadee, B. Entsch, D. P. Ballou, and P. Chaiyen. 2008. LuxG is a functioning flavin reductase for bacterial luminescence. J. Bacteriol. 190:1531-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Redfield, A. C., H. P. Smith, and B. H. Ketchum. 1937. The cycle of organic phosphorus in the Gulf of Maine. Biol. Bull. 73:421-443. [Google Scholar]
- 66.Romero-Campero, F. J., and M. J. Perez-Jimenez. 2008. A model of the quorum sensing system in Vibrio fischeri using P systems. Artif. Life 14:95-109. [DOI] [PubMed] [Google Scholar]
- 67.Schaefer, A. L., B. L. Hanzelka, A. Eberhard, and E. P. Greenberg. 1996. Quorum sensing in Vibrio fischeri: probing autoinducer-LuxR interactions with autoinducer analogs. J. Bacteriol. 178:2897-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Solorzano, L., and J. H. Sharp. 1980. Determination of total dissolved phosphorus and particulate phosphorus in natural waters. Limnol. Oceanogr. 25:754-758. [Google Scholar]
- 69.Sprague, G. F., Jr., R. M. Bell, and J. E. Cronan, Jr. 1975. A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol. Gen. Genet. 143:71-77. [DOI] [PubMed] [Google Scholar]
- 70.Stabb, E. V. (ed.). 2006. The Vibrio fischeri-Euprymna scolopes light organ symbiosis. ASM Press, Washington, DC.
- 71.Stabb, E. V., M. S. Butler, and D. M. Adin. 2004. Correlation between osmolarity and luminescence of symbiotic Vibrio fischeri strain ES114. J. Bacteriol. 186:2906-2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 73.Stabb, E. V., A. Schaefer, J. L. Bose, and E. G. Ruby. 2008. Quorum signaling and symbiosis in the marine luminous bacterium Vibrio fischeri, p. 233-250. In S. W. Winans and B. A. Bassler (ed.), Chemical communication among microbes. ASM Press, Washington, DC.
- 74.Steed, P. M., and B. L. Wanner. 1993. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J. Bacteriol. 175:6797-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tiedeman, A. A., and J. M. Smith. 1984. Isolation and characterization of regulatory mutations affecting the expression of the guaBA operon of Escherichia coli K-12. Mol. Gen. Genet. 195:77-82. [DOI] [PubMed] [Google Scholar]
- 76.Tu, S. C., and H. I. Mager. 1995. Biochemistry of bacterial bioluminescence. Photochem. Photobiol. 62:615-624. [DOI] [PubMed] [Google Scholar]
- 77.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Véscovi, E. G., F. C. Soncini, and E. A. Groisman. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165-174. [DOI] [PubMed] [Google Scholar]
- 79.Visick, K. L., T. M. O'Shea, A. H. Klein, K. Geszvain, and A. J. Wolfe. 2007. The sugar phosphotransferase system of Vibrio fischeri inhibits both motility and bioluminescence. J. Bacteriol. 189:2571-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang, J. C. 1971. Interaction between DNA and an Escherichia coli protein ω. J. Mol. Biol. 55:523-526. [DOI] [PubMed] [Google Scholar]
- 81.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 82.Ward, J. P., J. R. King, A. J. Koerber, P. Williams, J. M. Croft, and R. E. Sockett. 2001. Mathematical modelling of quorum sensing in bacteria. IMA J. Math Appl. Med. Biol. 18:263-292. [PubMed] [Google Scholar]
- 83.Wei, S. L., and R. E. Young. 1989. Development of symbiotic bacterial bioluminescence in a nearshore cephalopod, Euprymna scolopes. Mar. Biol. 103:541-546. [Google Scholar]
- 84.Whistler, C. A., and E. G. Ruby. 2003. GacA regulates symbiotic colonization traits of Vibrio fischeri and facilitates a beneficial association with an animal host. J. Bacteriol. 185:7202-7212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Williams, J. W., X. Cui, A. Levchenko, and A. M. Stevens. 2008. Robust and sensitive control of a quorum-sensing circuit by two interlocked feedback loops. Mol. Syst. Biol. 4:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Willsky, G. R., and M. H. Malamy. 1980. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J. Bacteriol. 144:356-365. [DOI] [PMC free article] [PubMed] [Google Scholar]