Abstract
The SCHIC domain of the B12-binding domain family present in the Rhodobacter sphaeroides AppA protein binds heme and senses oxygen. Here we show that the predicted SCHIC domain PpaA/AerR regulators also bind heme and respond to oxygen in vitro, despite their low sequence identity with AppA.
Cells monitor fluctuations in oxygen tension by means of oxygen-sensing proteins, many of which contain heme (10, 11). We recently described a new class of heme-binding protein domains designated SCHIC (sensor containing heme instead of cobalamin) (18, 23). SCHIC comprises approximately 120 amino acids and belongs to the vitamin B12-binding domain family (Pfam: PF02310 [9]).
The only SCHIC domain protein characterized thus far, AppA (14, 15), functions as a light- and oxygen-sensing antirepressor, which, in partnership with the repressor PpsR, controls photosynthesis gene expression in Rhodobacter sphaeroides (4, 12, 16, 20, 22). The SCHIC domain of AppA responds gradually to changes in oxygen concentrations, which allows the AppA-PpsR system to function as an oxygen-dependent transcriptional rheostat (23). The AppA proteins are found exclusively in the R. sphaeroides species. However, our sequence and phylogenetic analyses (23) predicted that aerobic regulators of photosynthesis gene expression of the PpaA/AerR family (7, 13, 21) also contained SCHIC domains. The PpaA proteins lack the light-sensing BLUF and the Cys-rich domains of AppA but often contain Arg-rich C termini (Fig. 1A). Because sequence identity among the SCHIC domains can be as low as 20% (23) (Fig. 1B), it has been unclear whether members of the PpaA/AerR family indeed bind heme instead of cobalamins.
FIG. 1.
(A) Domain structure of the R. sphaeroides AppA and PpaA proteins. SCHIC, sensor containing heme instead of cobalamin (23); BLUF, sensor of blue light using FAD (17); Cys, cysteine-rich domain; Arg, arginine-rich domain. Domains of unknown function are shown on a gray background. (B) Alignment of the SCHIC domains from anaerobic (top four) and aerobic anoxygenic phototrophic proteobacteria. Black shading indicates identities within the group; gray shading indicates similarities. The consensus is derived from a larger (58 sequences) alignment of the SCHIC domains.
To address this question, we characterized predicted SCHIC domain PpaA proteins from two groups of the anoxygenic photosynthetic proteobacteria—those that use photosynthesis only under anaerobic conditions (e.g., Rhodobacter) and those that require the presence of oxygen (e.g., Jannaschia). The PpaA proteins from R. sphaeroides 2.4.1 and Jannaschia sp. CCS1 were chosen because they share relatively little similarity to each other, i.e., 35% identity. Here we report that, in spite of low sequence similarity to each other and even lower similarity to the SCHIC domain from AppA (23% identity), the SCHIC domains of the PpaA proteins bind heme. Further, the SCHIC domain of R. sphaeroides PpaA, SCHICRs, responds to oxygen and CO in vitro in a manner that is remarkably similar to that of AppA.
The SCHIC domains of the PpaA proteins bind heme.
We cloned and overexpressed fragments of the R. sphaeroides PpaA (residues 55 to 249) and Jannaschia sp. CCS1 PpaA (residues 60 to 258) proteins encompassing predicted SCHIC domains (Fig. 1A) as C-terminal fusions to the maltose-binding protein (MBP). The R. sphaeroides ppaA gene fragment was PCR amplified using plasmid pSmNo (13) and the primers Ppa-TEPY-RI (5′ CGGAATTCCGCTCGATGGATCGCGCCC) and Ppa-TEPY-Spe (5′ GGACTAGTCACTGCAGCGCCGCCTCAATG) and cloned in vector pMAL-c2x (New England Biolabs). The Jannaschia sp. ppaA fragment was amplified from genomic DNA using the primers CCS1_F (5′GCCGCATGCCCTGTGAGGCGTTTCTGTCCGACAGC) and CCS1_R (5′CAGGAAGCTTTAATCCACGCCGGTCGCCTTCTTC) and cloned into a modified pMAL-c2x vector. E. coli DH5α cells containing the MBP-SCHIC plasmids were used for protein overexpression and purification. The fusion proteins were purified using affinity chromatography on amylose resin essentially as described previously (23).
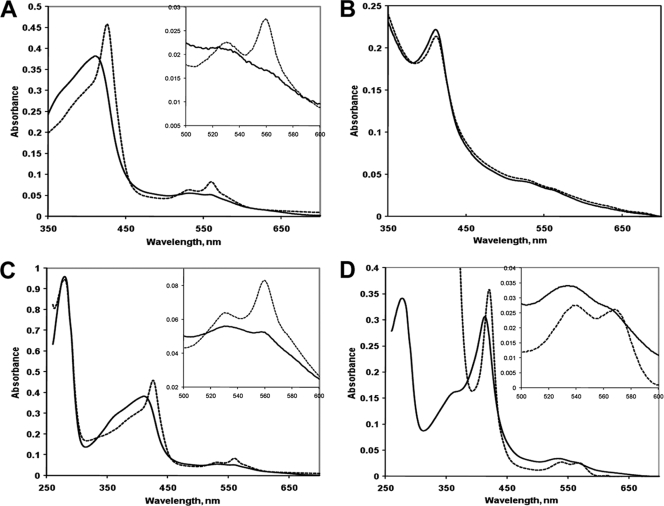
The purified SCHIC domain from R. sphaeroides PpaA, MBP-SCHICRs, demonstrated a Soret absorption peak at 413 nm, indicative of a bound tetrapyrrole (Fig. 2A); however, the amount of bound tetrapyrrole was relatively low (A413/A280 ratio, 0.11). To test for the chemical nature of the tetrapyrrole, we added a strong reductant, sodium dithionite, to the protein preparation. As a result, the Soret peak was shifted to 427 nm, and two additional prominent peaks, α (560 nm) and β (530 nm), appeared. These spectral changes are typical of the conversion of a hexacoordinated heme-protein complex from the met, Fe3+ form to the deoxy, Fe2+ form. Further, the positions of the Soret, α, and β peaks of the deoxy, Fe2+ form of MBP-SCHICRs were identical to those of the deoxy, Fe2+ form of the SCHIC domain from AppA (23).
FIG. 2.
(A) Electronic absorption spectra of the MBP fusion to the SCHIC domain of the R. sphaeroides PpaA protein, MBP-SCHICRs, as purified from E. coli. Solid trace, original spectrum; dashed trace, spectrum obtained after addition of dithionite. (B) Spectra of MBP-SCHICRs purified from anaerobic E. coli cells grown in the absence (solid trace) and presence (dotted trace) of 50 μM cyanocobalamin. (C) Spectra of MBP-SCHICRs after reconstitution with hemin in vitro. Solid trace, original spectrum; dashed trace, spectrum obtained after reduction with dithionite (and dithionite removal by size exclusion chromatography). (D) Spectra of MBP-SCHICJs after reconstitution with hemin in vitro (approximately 1:1 molar ratio). Solid trace, original spectrum; dashed trace, spectrum obtained after reduction with dithionite. Insets show magnified long-wavelength regions.
To compare the affinities of MBP-SCHICRs for heme and vitamin B12, we expressed MBP-SCHICRs in E. coli grown anaerobically in media supplemented with high (50 μM) concentration of vitamin B12 (cyanocobalamin; Sigma). Vitamin B12 is not synthesized by E. coli but can be taken up from the medium, as evidenced by the pinkish color of the anaerobically grown E. coli DH5α cells and presence of cobalamin in crude cell extracts (data not shown). We found that MBP-SCHICRs purified from these cells and cells not exposed to cyanocobalamin had identical spectra indicative of bound heme, not cyanocobalamin (Fig. 2B). This observation suggests that MBP-SCHICRs readily discriminates between heme and cobalamin, even when the latter compound is present in excess.
Next, we reconstituted MBP-SCHICRs with hemin in vitro, under the anaerobic conditions (anaerobic chamber), using methods described by us previously (23). The met, Fe3+ and deoxy, Fe2+ forms of the holoprotein showed the same absorbance peaks as those of MBP-SCHICRs purified from E. coli (Fig. 2C), which suggested that reconstitution in vitro had reproduced the heme-protein complex present in the purified MBP-SCHICRs.
The PpaA fragment from Jannaschia sp. CCS1, MBP-SCHICJs, as purified from E. coli, also contained relatively small amounts of bound heme, similar to MBP-SCHICRs (data not shown). We reconstituted MBP-SCHICJs with hemin in vitro (to approximately 1:1 heme-protein molar ratio). The Soret peak of MBP-SCHICJs in the met, Fe3+ form was 414 nm, i.e., within 1 nm of the SCHIC domains of R. sphaeroides AppA (415 nm) and PpaA (413 nm). Upon reduction with dithionite, the Soret peak was shifted to 420 nm, and the α and β peaks emerged at 538 and 567 nm, respectively (Fig. 2D). These peaks are 7 to 8 nm shifted compared to the SCHIC domain proteins from R. sphaeroides. While the observed spectral differences between the SCHIC domains suggest slightly different heme environments, it is clear that all studied SCHIC domains specifically bind heme and that heme environment is overall quite similar.
It is noteworthy that the SCHIC domain of AppA overexpressed in E. coli and purified on air contained only small amounts of heme (23), similar to the PpaA proteins described above. We do not fully understand the reasons for relatively low heme content in the proteins purified from aerobically grown E. coli. Several factors may contribute to incomplete heme saturation, e.g., insufficient heme supply in E. coli, instability of the heme-protein complexes under the aerobic conditions of protein expression and purification, and loss of heme during purification. Our attempts to date to overexpress the PpaA or AppA proteins in R. sphaeroides have been unsuccessful because of protein toxicity.
Interactions of the SCHIC domain from R. sphaeroides PpaA with gaseous ligands.
We further explored interactions of MBP-SCHICRs with gaseous ligands, carbon monoxide (CO) and molecular oxygen. The addition of CO to the deoxy, Fe2+ form of MBP-SCHICRs resulted in a characteristic carbonmonoxy heme spectrum with a sharp Soret peak at 421 nm, indicative of specific binding (data not shown). We determined kinetic parameters for binding of CO using laser flash photolysis measurements as described previously (23). We found that the CO on and off rate constants for MBP-SCHICRs were very similar to those of AppA (Table 1). Although many heme proteins bind CO, the kinetics of interactions with CO vary considerably among heme-binding proteins (1, 6, 8, 24). Similar kinetics suggests that the heme-binding pockets of the SCHIC domains from AppA and PpaA share key structural features and, by extension, that R. sphaeroides AppA and PpaA share similar functional capabilities.
TABLE 1.
Kinetics of CO binding and dissociation in heme-containing SCHIC domain AppA and PpaA proteins from R. sphaeroidesa
| Protein | Constant for CO binding |
Equilibrium dissociation constant (μM) | |
|---|---|---|---|
| On rate (μM−1 s−1) | Off rate (s−1) | ||
| AppA | 1.03 ± 0.02b | 0.77 ± 0.09 | 0.75 |
| PpaA | 0.95 ± 0.04 | 0.70 ± 0.09 | 0.74 |
MBP-AppA (data from reference 23) and MBP-SCHICRs. Data are means ± standard deviations, determined based on kinetic traces that were recorded at least four times using two independently reconstituted protein samples.
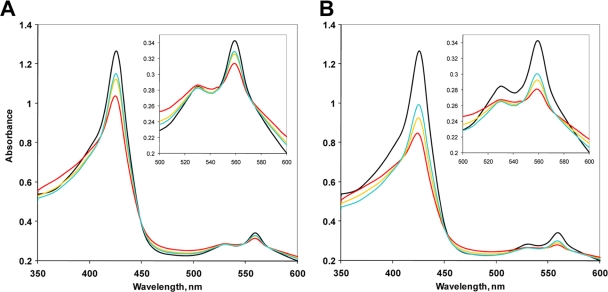
Next, we tested the interactions of the deoxy, Fe2+ form of MBP-SCHICRs with oxygen. Upon addition of oxygen, the amplitude of the Soret peak decreased and its position shifted to the blue region, while a shoulder at 380 nm characteristic of a loosely bound heme increased. The absorbance of the α and β peaks also decreased (Fig. 3, red traces). These changes indicate a loss of coordination (i.e., discoordination) of the heme iron from the axial position following oxygen binding. Importantly, these absorption changes were reversible. We observed restoration of the deoxy, Fe2+ form over time in the presence of the oxygen-scavenging system (a mixture of glucose, glucose oxidase, and catalase [3]) (Fig. 3). Also noteworthy is the observation that the magnitude of the spectral changes in MBP-SCHICRs was proportional to oxygen concentration. For example, addition of 100 μM oxygen decreased the Soret peak by approximately 0.25 units (α peak by 0.03), while addition of 200 μM oxygen decreased the Soret peak by approximately 0.5 units (α peak by 0.06) (Fig. 3A and B). This graded oxygen response of MBP-SCHICRs resembles the response of the SCHIC domain of AppA (23) and suggests that PpaA may function as “oxygen rheostat,” similar to AppA. However, the details of the sensory mechanism of R. sphaeroides PpaA and functional significance of this observation remain to be investigated.
FIG. 3.
Electronic absorption spectra of MBP-SCHICRs before and after exposure to 100 μM oxygen (final concentration) (A) and 200 μM oxygen (B). The medium contained 5 mM dithiothreitol and a glucose-glucose oxidase-catalase system of oxygen removal (3). Black traces, deoxy, Fe2+ protein; red traces, protein immediately after addition of oxygen; gold traces, protein after 3 min; cyan traces, protein after 15 min. Insets show magnified long-wavelength regions. The experimental details are given in reference 23.
The PpaA/AerR protein family.
The PpaA/AerR proteins are present in all sequenced genomes of the anoxygenic phototrophic proteobacteria (purple nonsulfur bacteria), which is in contrast to AppA, found exclusively in the R. sphaeroides species. The ppaA genes are located in the photosynthesis gene clusters of both anaerobic (Rhodobacter, Rhodopseudomonas, Rhodospirillum, etc.) and aerobic (Roseobacter, Erythrobacter, Jannaschia, etc.) anoxygenic photosynthetic proteobacteria. The representatives of the latter bacterial group are particularly abundant in marine habitats from coastal waters to deep-sea sediments, from the poles to the tropics, and are found either free living or associated with phytoplankton (2, 5). R. sphaeroides PpaA (13) and its counterparts from Rhodobacter capsulatus, AerR (7), and from Rhodocystis centenum, AerR (21), function as oxygen-dependent regulators of photosynthesis gene expression. The PpaA protein from R. sphaeroides and R. centenum function as aerobic activators of photosynthesis gene expression (13, 21), whereas AerR functions as an aerobic repressor (7). Presence of the heme-containing, oxygen-responsive SCHIC domain may explain the oxygen sensitivity of the PpaA/AerR family members that appear to be important for photosynthesis regulation on the global scale.
Acknowledgments
We are grateful to M. Morgan and C. Smith (University of Georgia) for Jannaschia sp. CCS1 genomic DNA.
This work was supported in part by National Institutes of Health National Center for Research Resources (COBRE) grant P20 RR15640 (M.G.), National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education and Extension Service, grant 2002-35318-14039 (M.A.G.G.), Welch Foundation grant I-1575, and National Science Foundation grant 620531 (M.A.G.G.).
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Arredondo-Peter, R., M. S. Hargrove, G. Sarath, J. E. Moran, J. Lohrman, J. S. Olson, and R. V. Klucas. 1997. Rice hemoglobins. Gene cloning, analysis, and O 2-binding kinetics of a recombinant protein synthesized in Escherichia coli. Plant Physiol. 115:1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 3.Bouin, J. C., M. T. Atallah, and H. O. Hultin. 1976. The glucose oxidase-catalase system. Methods Enzymol. 44:478-488. [DOI] [PubMed] [Google Scholar]
- 4.Braatsch, S., M. Gomelsky, S. Kuphal, and G. Klug. 2002. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol. Microbiol. 45:827-836. [DOI] [PubMed] [Google Scholar]
- 5.Brinkhoff, T., H. A. Giebel, and M. Simon. 2008. Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch. Microbiol. 189:531-539. [DOI] [PubMed] [Google Scholar]
- 6.Delgado-Nixon, V. M., G. Gonzalez, and M. A. Gilles-Gonzalez. 2000. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 39:2685-2691. [DOI] [PubMed] [Google Scholar]
- 7.Dong, C., S. Elsen, L. R. Swem, and C. E. Bauer. 2002. AerR, a second aerobic repressor of photosynthesis gene expression in Rhodobacter capsulatus. J. Bacteriol. 184:2805-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duff, S. M., J. B. Wittenberg, and R. D. Hill. 1997. Expression, purification, and properties of recombinant barley (Hordeum sp.) hemoglobin. Optical spectra and reactions with gaseous ligands. J. Biol. Chem. 272:16746-16752. [DOI] [PubMed] [Google Scholar]
- 9.Finn, R. D., J. Mistry, J. Tate, P. Coggill, A. Heger, J. E. Pollington, O. L. Gavin, P. Gunesekaran, G. Ceric, K. Forslund, L. Holm, E. L. Sonnhammer, S. R. Eddy, and A. Bateman. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilles-Gonzalez, M. A., G. S. Ditta, and D. R. Helinski. 1991. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350:170-172. [DOI] [PubMed] [Google Scholar]
- 11.Gilles-Gonzalez, M. A., and G. Gonzalez. 2005. Heme-based sensors: defining characteristics, recent developments, and regulatory networks. J. Inorg. Biochem. 99:1-22. [DOI] [PubMed] [Google Scholar]
- 12.Gomelsky, L., O. V. Moskvin, R. A. Stenzel, D. F. Jones, T. J. Donohue, and M. Gomelsky. 2008. Hierarchical regulation of photosynthesis gene expression by the oxygen-responsive PrrBA and AppA-PpsR systems of Rhodobacter sphaeroides. J. Bacteriol. 190:8106-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomelsky, L., J. Sram, O. V. Moskvin, I. M. Horne, H. N. Dodd, J. M. Pemberton, A. G. McEwan, S. Kaplan, and M. Gomelsky. 2003. Identification and in vivo characterization of PpaA, a regulator of photosystem formation in Rhodobacter sphaeroides. Microbiology 149:377-388. [DOI] [PubMed] [Google Scholar]
- 14.Gomelsky, M., and S. Kaplan. 1995. appA, a novel gene encoding a trans-acting factor involved in the regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 177:4609-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gomelsky, M., and S. Kaplan. 1998. AppA, a redox regulator of photosystem formation in Rhodobacter sphaeroides 2.4.1, is a flavoprotein. Identification of a novel FAD binding domain. J. Biol. Chem. 273:35319-35325. [DOI] [PubMed] [Google Scholar]
- 16.Gomelsky, M., and S. Kaplan. 1997. Molecular genetic analysis suggesting interactions between AppA and PpsR in regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 179:128-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gomelsky, M., and G. Klug. 2002. BLUF: a novel FAD-binding domain involved in sensory transduction in microorganisms. Trends Biochem. Sci. 27:497-500. [DOI] [PubMed] [Google Scholar]
- 18.Han, Y., M. H. Meyer, M. Keusgen, and G. Klug. 2007. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol. Microbiol. 64:1090-1104. [DOI] [PubMed] [Google Scholar]
- 19.Reference deleted.
- 20.Masuda, S., and C. E. Bauer. 2002. AppA is a blue light photoreceptor that antirepresses photosynthesis gene expression in Rhodobacter sphaeroides. Cell 110:613-623. [DOI] [PubMed] [Google Scholar]
- 21.Masuda, S., J. Berleman, B. M. Hasselbring, and C. E. Bauer. 2008. Regulation of aerobic photosystem synthesis in the purple bacterium Rhodospirillum centenum by CrtJ and AerR. Photochem. Photobiol. Sci. 7:1267-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskvin, O. V., L. Gomelsky, and M. Gomelsky. 2005. Transcriptome analysis of the Rhodobacter sphaeroides PpsR regulon: PpsR as master regulator of photosystem development. J. Bacteriol. 187:2148-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moskvin, O. V., S. Kaplan, M. A. Gilles-Gonzalez, and M. Gomelsky. 2007. Novel heme-based oxygen sensor with a revealing evolutionary history. J. Biol. Chem. 282:28740-28748. [DOI] [PubMed] [Google Scholar]
- 24.Quillin, M. L., R. M. Arduini, J. S. Oslon, and G. N. J. Phillips. 1993. High-resolution crystal structures of distal histidine mutants of sperm whale myoglobin. J. Mol. Biol. 234:140-155. [DOI] [PubMed] [Google Scholar]