Abstract
The diversity of the Escherichia coli species is in part due to the large number of mobile genetic elements that are exchanged between strains. We report here the identification of a new integrative and conjugative element (ICE) of the pKLC102/PAGI-2 family located downstream of the tRNA gene pheU in the E. coli strain BEN374. Indeed, this new region, which we called ICEEc2, can be transferred by conjugation from strain BEN374 to the E. coli strain C600. We were also able to transfer this region into a Salmonella enterica serovar Typhimurium strain and into a Yersinia pseudotuberculosis strain. This transfer was then followed by the integration of ICEEc2 into the host chromosome downstream of a phe tRNA gene. Our data indicated that this transfer involved a set of three genes encoding DNA mobility enzymes and a type IV pilus encoded by genes present on ICEEc2. Given the wide distribution of members of this family, these mobile genetic elements are likely to play an important role in the diversification of bacteria.
The fantastic diversity of the Escherichia coli species has been known for a long time. With modern sequencing strategies, the molecular bases of this diversity are now being unraveled (49). Analyzing the genome of 20 E. coli strains, Touchon et al. recently showed that only a minority of genes, approximately 1,900 genes, were shared by all E. coli strains and constituted the core genome of the E. coli species (50). Additionally, the total number of genes found in all E. coli strains, the pan-genome, is an order of magnitude larger than this core genome (50). The non-core genome of a strain, also called flexible gene pool, is therefore made of a wide diversity of genes. This genetic diversity of the E. coli species translates into a diversity of phenotypic properties. While most E. coli strains are commensal of the gastrointestinal tract of humans and warm-blooded animals, a significant number are responsible for different diseases in humans and animals (22), including extraintestinal infections in chickens; strains isolated from such cases are designated by the term APEC for avian pathogenic E. coli (10).
This diversity arises from frequent horizontal gene transfers of mobile genetic elements such as transposons, plasmids, phages, genomic islands, or integrative and conjugative elements (ICEs) (11, 21, 34). Among these mobile genetic elements, ICEs have a particular place as they share properties with both plasmids, genomic islands, and transposons; they can be defined as elements that encode all the necessary machineries that allow their excision from the chromosome, their transfer to a recipient strain, and their integration into the recipient strain's genome (5, 6, 46, 54). Well-known representatives of this class of genetic elements include Tn916 discovered in Enterococcus faecalis, the conjugative transposon CTnDOT in Bacteroides thetaiotaomicron, ICEKp1 in Klebsiella pneumoniae, SXT/R391-related elements, PFGI-1 in Pseudomonas fluorescens, and the clc element in Pseudomonas sp. strain B13 as well as ICEBs1 in Bacillus subtilis and ICEEc1 in the E. coli strain ECOR31 (1, 39, 44, 46, 54). Typically, ICEs contain at least three modules that are required for key steps in the ICE's life cycle: an excision/integration module, a transfer module, and a regulation module (54). Besides these, ICEs often contain cargo regions that confer on their host a diverse array of properties, such as virulence properties (ICEEc1), antibiotic resistance (SXT), or degradation of chemical compounds (clc). Because of their self-transfer abilities and their diverse accessory gene repertoires, ICEs are very likely to play a major role in bacteria evolution (46).
A new family of ICEs has recently gained interest and was named the pKLC102/PAGI-2 family. The first element of this family, the clc element, was discovered in Pseudomonas sp. strain B13 and confers on the bacteria the possibility to degrade aromatic compounds (42). The transfer of this element was discovered long before its complete sequence was characterized (16). Other members of this family include several elements present in Pseudomonas strains such as PAGI-1 and PAGI-2 as well as the pKLC102 element first considered to be a plasmid but later on shown to be an ICE because of its ability to integrate into the chromosome of its host (23, 52). pKLC102/PAGI-2 elements share a set of core genes (33) and, like most ICEs and genomic islands, are all integrated downstream of tRNA genes (26, 52). The transfer between strains has been demonstrated, albeit with different frequencies, for only a few members, such as the clc element, Pseudomonas aeruginosa pathogenicity island 1 (PAPI-1), and ICEHin1056 from Haemophilus influenzae (20, 37, 41); this transfer involves the type IV pilus (20), the integrase (40), and in some cases the formation of a circular intermediate of the excised ICE (24).
In order to identify new accessory genes of APEC strains, we previously described tRNA loci in the E. coli genome that could represent potential insertion sites for new genomic islands (18). We had already used this strategy to characterize the AGI-3 region that is involved in the virulence of an avian pathogenic E. coli strain and that confers the ability to grow on fructooligosaccharides (7, 43). During this tRNA screening, we showed that genomic islands might potentially be present downstream of the tRNA genes argW, leuX, pheU, pheV, selC, serU, and thrW in several APEC strains.
In this report, we describe the identification of a new genomic island located downstream of pheU in the APEC strain BEN374. This region, which we named ICEEc2, was fully sequenced, and its properties were analyzed in detail; ICEEc2 is a new ICE found in E. coli and belongs to the pKLC102/PAGI-2 family described above.
MATERIALS AND METHODS
Strains and growth conditions.
Bacterial strains used in this study are described in Table 1. E. coli strain BEN374 (O18:K1) was isolated from the organs of a chicken showing characteristic lesions of avian colibacillosis in 1992 in Spain (47). Bacteria were routinely grown in LB Lennox broth at 37°C except when indicated below (32). Ampicillin ([Amp] 100 μg·ml−1), kanamycin (50 μg·ml−1), nalidixic acid (30 μg·ml−1), trimethoprim (40 μg·ml−1), or streptomycin (50 μg·ml−1) was used when necessary.
TABLE 1.
Strains and cosmids used in this study
| Strain or cosmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| BEN79, BEN277, BEN278, BEN1588, BEN2908 | APEC strains | 47 |
| BEN374 | APEC O18:K1; Tmpr Strr Spcr | 18 |
| BEN374 ΔORF1-ORF3 | BEN374 with a deletion of the region of ORF1 to ORF3 | This study |
| BEN374 ΔORF14 | BEN374 with a deletion of ORF14 (pilS) | This study |
| C600 | E. coli K-12 | Coli Genetic Stock Center (CGSC 3004) |
| C600 Nalr | Spontaneous Nalr derivative of strain C600 | This study |
| S. enterica serovar Typhimurium DT104 | ||
| BN 9181 ΔacrB::Kanr | Kanr | 2 |
| Y. pseudotuberculosis | ||
| IP32953 O-Ag−b | Kanr | 36 |
| Cosmids | ||
| p9A11 | Ampr; contains region from 1 to 33182 of ICEEc2 | This study |
| p5E6 | Ampr; contains region from 27170 to 61545 of ICEEc2 | This study |
| p10A11 | Ampr; contains region from 55969 to 92215 of ICEEc2 | This study |
Nalr, nalidixic acid resistant; Strr, streptomycin resistant; Spcr, spectinomycin resistant; Ampr, ampicillin resistant; Kanr, kanamycin resistant; Tmpr, trimethoprim resistant.
O-Ag−, O-antigen negative.
Molecular biology techniques.
Restriction endonucleases and modification enzymes (New England Biolabs) were used according to the manufacturer's instructions. DNA fragments were purified from agarose gels using a Nucleospin Extract II purification kit (Macherey-Nagel).
Primers used in this study are described in Table 2. PCRs were performed with an Applied Biosystems 9700 apparatus using 1 U of Taq DNA polymerase from New England Biolabs in 1× buffer, a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 0.8 μM concentration of each primer, and 10 ng of chromosomal DNA in a 50-μl reaction volume. For PCR on single colonies, bacteria were resuspended in 50 μl of water, boiled for 10 min, and centrifuged at 10,000 × g for 5 min. Two microliters of supernatant was then used instead of chromosomal DNA. Cycling conditions were as follows: 1 cycle of 5 min/kb at 95°C, followed by 30 cycles of 10 s at 95°C, 10 s at 52°C, and 1 min/kb at 72°C, with a final extension of 5 min at 72°C. PCR products were separated on 1% agarose gels for 1 h at 10 V/cm of gel.
TABLE 2.
Primers used in this study
| Primer target or use | Primer name | Primer sequence |
|---|---|---|
| Tn7 | pheU374-Tn7F | AAGAGCAGTTGCGAGTAGC |
| pheU374-Tn7R | CGACTACGTTCCGTCAGATT | |
| iha | PG393 | CCCGTCTGGAAGTAATCACC |
| PG394 | TACAAACAGCGGAAAGGC | |
| 3′ region of ICEEc2 | pheU374-ilot-crD1 | CCGTTGTCTCAACAAAAGGTGG |
| pheU374-ilot-crD2 | GGATAACTGAGGTCAGCCGTGC | |
| 5′ region of ICEEc2 | pheU4-374 | GGTTCTCACTCCTGACCAGTGGC |
| Cosmid sequencing | PG122 | AGGGTTTTCCCAGTCACGAC |
| PG123 | CATAATACGACTCACTATAG | |
| ORF1-ORF3 mutant | PG381 | TCCTGAGAAGGCAGAGTGCGCTGACATTTCGTGAACGGAGGAATGCCATGGTGTAGGCTGGAGCTGCTTC |
| PG382 | ATTGCAACGGTTTTTCATGTTGTCAACAACTGTAAGCAATCACCTGATTAATGGGAATTAGCCATGGTCC | |
| ORF14 (pilS) mutant | PG383 | CCGCATACAGAATATTTCTGAAACATCCCATAAACCAGGAGTGATATATGGTGTAGGCTGGAGCTGCTTC |
| PG384 | CGGCAGTGACAGCATCCTCTGGTATTTAAGGATGCTGTCCGGAAACATTAATGGGAATTAGCCATGGTCC | |
| pheU_BEN374 | PG156 | GTGGTGCATTGACCTGACAGAAACACAG |
| PG158 | TGATGTGGGGAGAATCTGGTTGAGTTCG | |
| pheV_BEN374 | PG160 | GCCTGGTTTGCCTGACAATGCGTGC |
| PG162 | CTGGCAGCGGTGGTGTTCCTGTTTAGC | |
| pheU_Salmonella | pheU1-Sal-LT2 | CTGAAAAGCAGGCAGTCAGC |
| pheU2-Sal-LT2 | TCTGGCCTTCTCGTTATAGC | |
| pheV_Salmonella | pheV1-DT104 | GATAGATTGTGCAGTCTACG |
| pheV2-DT104 | TCTAATGGAGATCATATGGC | |
| pheU_Yersinia | PG402 | TGGAGTTGATTCTGGAGGGT |
| PG403 | ACCTGACCCTAAGCGACATTTCT | |
| pheV_Yersinia | PG404 | GAGAGCAGTTGATCGCCTTTATA |
| PG405 | CTAAAGCTAAACCACCCACT | |
| Circular intermediate | pheU374-circF | GACAACAACTTGACATGCCC |
| pheU374-circR | TCTGGCAAGTACTCTGATGG |
Deletions were obtained as described by Datsenko and Wanner using primers PG381/PG382 for deletion of ORF1 to ORF3 (ORF1-ORF3)and primers PG383/PG384 for deletion of ORF14 (8). The Kanr cassette was then removed using plasmid pCP20, leaving a scar of 80 bp. Deletions were then confirmed by PCR using primers flanking both regions.
Arbitrarily primed PCR was used to determine the DNA sequence of the region downstream of tRNA genes. Reactions were performed as described by O'Toole et al. (35). PCR products obtained were then sequenced (Cogenics).
Cosmid library.
Cosmid libraries were constructed as described in the SuperCos 1 Cosmid Vector Kit (Stratagene) except that the pWEB-TNC vector (Epicentre) was used instead of the SuperCos vector. Chromosomal DNA was partially digested by Sau3AI and ligated into the pWEB-TNC vector previously opened with BamHI and dephosphorylated with calf intestinal phosphatase (New England Biolabs). Cloned inserts were packaged into phage particles using Gigapack III XL packaging extracts (Stratagene). E. coli strain EPI100 was then incubated with phage particles, and the resulting clones were selected onto LB plates supplemented with Amp (LB-Amp). Plates were then incubated at room temperature for at least 48 h, and individual colonies were seeded in 96-well plates.
Screening of the cosmid library.
Primers were designed to amplify the junction between the pheU tRNA and the DNA region inserted downstream of this tRNA in strain BEN374. PCRs were then performed on each of the individual cosmid clones to detect the ones containing the target sequence. The ends of the cosmids identified in this way were sequenced using primers PG122 and PG123. The new sequence was then used for a new screening of the library; by repeating this strategy three times, three cosmids were obtained that covered the entire DNA region downstream of the pheU tRNA of strain BEN374. The DNA sequences of the selected cosmids were determined by shotgun sequencing (Cogenics).
Conjugation experiments.
Conjugation experiments were carried out in LB broth. The donor strain and the recipient strain (E. coli, Salmonella, or Yersinia) were mixed together with a donor-to-recipient ratio of approximately 4:1. This mix was incubated overnight at 37°C (30°C for conjugation with Yersinia as a recipient) without shaking. The next day, bacteria were plated on appropriate selective LB agar plates. Strain BEN374 is resistant to trimethoprim and streptomycin, two resistance phenotypes that are linked to ICEEc2 (see Results section). The E. coli K-12 recipient strain C600 Nalr is resistant to nalidixic acid (Nal); the Salmonella enterica serovar Typhimurium and Yersinia recipient strains are resistant to kanamycin. Nalidixic acid (30 μg ml−1) or kanamycin (50 μg ml−1) was used to select against E. coli BEN374 donor cells, and trimethoprim (50 μg ml−1) or streptomycin (40 μg ml−1) was used to select against unmated recipient cells. The transfer frequency of ICEEc2 was determined by dividing the number of ICEEc2 transconjugants by the number of donor cells.
Transfer of the entire ICEEc2 region was confirmed by performing PCRs on 20 conjugants of each conjugation to detect different regions of ICEEc2: the Tn7 transposon (primers Tn7F/Tn7R), the iha gene (PG393/PG394), and the 3′ end of ICEEc2 (crD1/crD2).
Sequence analysis and annotation.
Coding sequences were predicted using the AMIGene (annotation of microbial genomes) software (3) and then submitted to automatic functional annotation using the set of tools listed in Vallenet et al. (51). Manual validation of automatic annotations was performed in a relational database called PkGDB using the MaGe (magnifying genomes) web interface. This interface allows graphic visualization of the annotations enhanced by a synchronized representation of synteny groups in other genomes chosen for comparison, as described by Vallenet et al. (51). DNA repeats were identified using GenAlyzer (http://www.genomes.de/). Regions similar to ICEEc2 were identified in GenBank using tBLASTx to search for regions encoding proteins similar to the products of genes ORF58-ORF62 or ORF1-ORF3. Comparisons of chromosomal regions with ICEEc2 were performed using the Artemis Comparison Tool (ACT).
Phylogenetic analysis of ORF1 integrase.
The 50 proteins most similar to open reading frame 1 (ORF1) were retrieved from Uniref100 by using BlastP (www.ebi.ac.uk). The integrases used for phylogenetic analyses were the following (Uniprot identification [ID] number): A1JPX0, A3L3X2, A3LKM0, A6N596, A6V9Q0, A6V9Z5, A7FN12, A7ZRC9, A8G8R9, A8GJ53, A8GJC2, A9MES8, A9N3Y8, B0UVF5, B4EXE8, B5F495, B7LDP4, C1DRE8, C1HR63, C2AZI0, C2LLF7, C4UAJ4, C5BDJ6, C6ANK9, D0FNR2, D0KGZ7, Q1W561, Q4K7E9, Q4LBF7, Q4ZWK0, Q5NDA1, Q6D9K0, Q6EVN9, Q6X305, Q7N7I3, Q7N7S9, Q7WY55, Q7WZ26, Q8KRZ2, Q8Z1C4, and Q9F771. Integrases from other pKLC102/PAGI-2 elements were also selected: Q8GQA9, Q3BT84, Q8GPZ9, A9C2W6, C6BE38, and C4X1L7. Representative sequences of different subgroups of IntG and IntP integrases were selected based on the publication of Boyd et al. (4). Selected sequences were the following: A0KZG0, A2W8F3, A3EU83, A4WDI5, A6AQ85, A7JWT4, A8ANF3, C3NPT3, Q0HJK5, Q0I4F8, Q0TBE3, Q167M4, Q2N8A1, Q31YZ6, Q7MZ43, Q8FAM8, Q8XAK1, Q8XCM9, Q9KR84, A8CG97, O22009, P27077, Q6HA01, Q77WA5, Q859D2, Q9T205, YP_001700534, and YP_001718763.
A multiple alignment was performed using MUSCLE (12), and the alignment obtained was manually curated using Bioedit (www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenic analysis was then done by maximum likelihood (PhyML), neighbor-joining (BioNJ), and maximum parsimony (TNT) at www.phylogeny.fr (9). A consensus tree was then built using Consense and using the majority rule (www.bioweb.pasteur.fr).
Nucleotide sequence accession number.
The sequence of the ICEEc2 region was deposited in the GenBank under accession number GU725392.
RESULTS
A new genomic island is located downstream of pheU in strain BEN374.
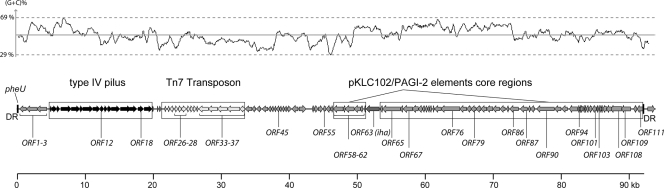
Considering the results of the tRNA screening previously performed (18), we used arbitrarily primed PCR to characterize the sequences present downstream of tRNA genes most frequently targeted by insertion of supernumerary DNA regions (argW, leuX, pheU, pheV, selC, serU, and thrW) in six different APEC strains. Translation of the 3′ part of the sequence located downstream of pheU (GenBank accession number DQ023207) in the APEC strain BEN374 showed homologies with several phage integrases. We therefore decided to further characterize the region located downstream of pheU in this strain. A cosmid library of strain BEN374 was screened by PCR, and three cosmids were isolated covering the entire region. This region, depicted in Fig. 1, is 92,215 bp long and was named ICEEc2 following the nomenclature of Burrus et al. for ICEs (see below) (5). Overall, the G+C content of ICEEc2 is identical to that of the E. coli K-12 chromosome (50.8%). Yet it is very variable. In particular, a region between positions 50000 and 73000 shows a higher G+C content (58%); such variations suggest that ICEEc2 was assembled from different origins.
FIG. 1.
Schematic representation of the ICEEc2 genomic island. The upper part represents the percent G+C content of ICEEc2. The different genes present in ICEEc2 are depicted. The 50-bp direct repeats (DR) flanking ICEEc2 are represented. Regions of interest mentioned in the text are indicated below the graph. The bottom part of the graphic is a scale in kb.
ICEEc2 is predicted to contain 111 genes (see Table S1 in the supplemental material). Several of these genes encode proteins with similarities to proteins of known function (Table 3). The complete annotation of ICEEc2 is available through the MaGe platform at www.genoscope.cns.fr/agc/mage. Most interesting is the presence in ICEEc2 of genes related to conjugative transfer. As described by Llosa et al., a conjugation machinery can be seen as the association of two processes, DNA replication and DNA secretion, through a coupling protein (29). ICEEc2 contains several genes encoding proteins potentially involved in DNA replication and in formation of a type IV pilus and a potential coupling protein (Table 3).
TABLE 3.
Functional classification of ICEEc2 genes encoding proteins with putative functions
| Cellular process | Gene | Putative product | Reference or source |
|---|---|---|---|
| DNA processing enzymes | |||
| DNA replication | ORF87 | Single-stranded binding protein | Swiss-Prot P25762 |
| ORF90 | DNA topoisomerase IA | Swiss-Prot P14294 | |
| ORF111 | CbiA-like chromosome partitioning protein | TrEMBL A7ZRE7 | |
| DNA recombination | ORF1 | XerC-like recombinase | Swiss-Prot Q9F771 |
| ORF18 | Rci recombinase | Swiss-Prot P16470 | |
| DNA transposition | ORF33-ORF37 (tnsABCDE) | Tn7 transposition proteins | Swiss-Prot P13988, P13989, P05846, P13991, P05845 |
| ORF86 | YhgA-like transposase | Swiss-Prot P31667 | |
| DNA unwinding | ORF2 | Putative helicase | TrEMBL A7ZRD0 |
| ORF3 | UvrD-like helicase | Swiss-Prot Q8K2I9 | |
| ORF109 | DnaB-type helicase | Swiss-Prot P0ACB0 | |
| DNA nuclease | ORF108 | ParB nuclease | Swiss-Prot Q92JI0 |
| DNA cleavage | ORF79 | Type IV restriction enzyme | Swiss-Prot P24202 |
| Conjugative transfer | |||
| Formation of type IV pilus | ORF6-ORF17 (pil genes) | Type IV pilus proteins | 28 |
| Coupling | ORF76 | TraD-like coupling protein | TrEMBL C2LLJ7 |
| Mating pair stabilization | ORF58 | TraG-like protein | Swiss-Prot P33790 |
| Other | ORF65 | TraC-like protein | TrEMBL C2LLI5 |
| ORF67 | TrbI protein | Swiss-Prot P05359 | |
| Xenobiotic resistance/adaptation to the environment | |||
| Antibiotic resistance | ORF26 (dfrA1) | Trimethoprim resistance | Swiss-Prot P00382 |
| ORF27 (sat2) | Streptothricin resistance | Swiss-Prot P13018 | |
| ORF28 (aadA1) | Streptomycin/spectinomycin resistance | Swiss-Prot P0AG06 | |
| Iron acquisition | ORF63 (iha) | Catecholate receptor/potential adhesin | TrEMBL Q6EZC5 |
| Miscellaneous | ORF45 | Antirestriction protein | TrEMBL B4TL18 |
| ORF55 | L31 ribosomal protein | Swiss-Prot B7L427 | |
| ORF94 | Sb34 phage protein | TrEMBL Q8HAA4 | |
| ORF101 | Ea22 phage protein | Swiss-Prot P03756 | |
| ORF103 | Ea22 phage protein | Swiss-Prot P03756 |
A Tn7 transposon is also present within ICEEc2 (ORF23 to ORF37) with a class II integron at its 5′ end; this integron contains three antibiotic resistance genes, ORF26 (dfrA1), ORF27 (sat2), and ORF28 (aadA1), which confer on strain BEN374 resistance to trimethoprim, streptothricin, and streptomycin/spectinomycin, respectively. Accordingly, upon transfer into the E. coli strain EPI100 (Epicentre) of the cosmid p9A11 containing genes ORF26-ORF28, the recipient strain became resistant to these three antibiotics (data not shown). In addition, ICEEc2 carries iha, a putative virulence gene that has been involved in the adhesive properties of different E. coli strains and in iron acquisition (27, 48).
ICEEc2 also contains a number of repeats that are described in Table 4. Among these, we shall mention the 50-bp imperfect repeats that flank ICEEc2, corresponding to the last 50 nucleotides of the pheU tRNA gene. Also, six 20-bp repeats (GTGCCAATCCGGTgtgTGGA; nucleotides representing exact matches are shown in uppercase), in either the direct or indirect orientation, are present at the end of the ORF17 (pilV) gene. A region starting at position 80038 is also characteristic, showing 17 highly conserved direct repeats of the motif aCTGTTGCCACTGGCAACgCCGgACacTTTTTAAcC that contains a 15-bp palindrome (underlined) and a T-rich region (bold).
TABLE 4.
DNA repeats present in ICEEc2
| Repeat no. | Typea | Size (bp) | Localization (nt)b | No. of repeats | Relevant characteristic(s) |
|---|---|---|---|---|---|
| 1 | DR | 50 | 2 | 2 | End of pheU tRNA; flanks ICEEc2 |
| 2 | DR, IR | 20 | 15329 | 6 | Sequence, GTGCCAATCCGGTGTGTGGA; putative targets of the Rci recombinase; could modify the C-terminal sequence of ORF17 (PilV) |
| 3 | DR | 249 | 42853 | 3 | Two of these repeats are flanking a putative transposase (ORF50) |
| 4 | DR | 67 | 54588 | 2 | Flanks the iha gene |
| 5 | DR | 36 | 80049 | 17 | Each repeat contains the palindromic sequence GTTGCCACTGGCAAC |
DR, direct repeat; IR, inverted repeat.
The localization of the first repeat is indicated. nt, nucleotide.
ICEEc2 is a member of the pKLC102/PAGI-2 family of mobile genetic elements.
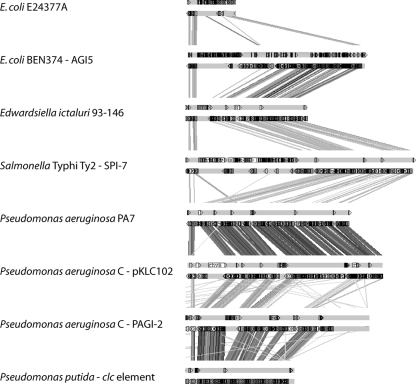
A search for regions similar to ICEEc2 was performed, and it revealed that ICEEc2 belongs to the pKLC102/PAGI-2 family of mobile genetic elements (33). ICEEc2 contains 33 genes whose products are homologous to those of genes present in pKLC102 (see Table S1 in the supplemental material). The region containing ORF58 to ORF62 (corresponding to CP87 to CP91 of pKLC102) is one of the most conserved regions in the different elements that we analyzed and that was also described by Mohd-Zain et al. (33). We thus used this region as a bait to retrieve regions that belong to the pKLC102/PAGI-2 family. Similarities were most often found at the protein level (Fig. 2), while similarities at the nucleotide level were restricted to regions present in other E. coli strains. Indeed, a region similar to ICEEc2 was found in the commensal E. coli strain ED1a, and remnants of such a region were present in the enterotoxigenic E. coli (ETEC) strain E24377A. In strain ED1a, while iha is present, the Tn7 transposon and the region downstream of ORF89 are absent. Additionally, the gene encoding the XerC-like protein is truncated. In the case of strain E24377A, the first three and the last four ORFs of ICEEc2 are present, suggesting that a region similar to ICEEc2 was present at some time in the history of this strain but has since been deleted.
FIG. 2.
tBLASTx comparisons between ICEEc2 and other elements of the pKLC102/PAGI-2 family. tBLASTx comparisons were performed using DoubleAct (http://www.hpa-bioinfotools.org.uk/pise/double_act.html) and analyzed using the Artemis Comparison Tool (ACT). For each genetic element analyzed, the upper line indicates ORFs encoded on the direct strand, and the bottom line indicates ORFs encoded on the complementary strand. The threshold for ACT visualization was set at 200.
ICEEc2 belongs to a subfamily of pKLC102/PAGI-2 elements with XerC-like integrases.
Analysis of the members of the pKLC102/PAGI-2 family revealed that two groups can be distinguished in this family based on the type of integrase they possess: either an integrase of the P4 family, such as in the PAGI-3 region of P. aeruginosa SG17M (33), or one of the XerC family, such as ORF1 in ICEEc2.
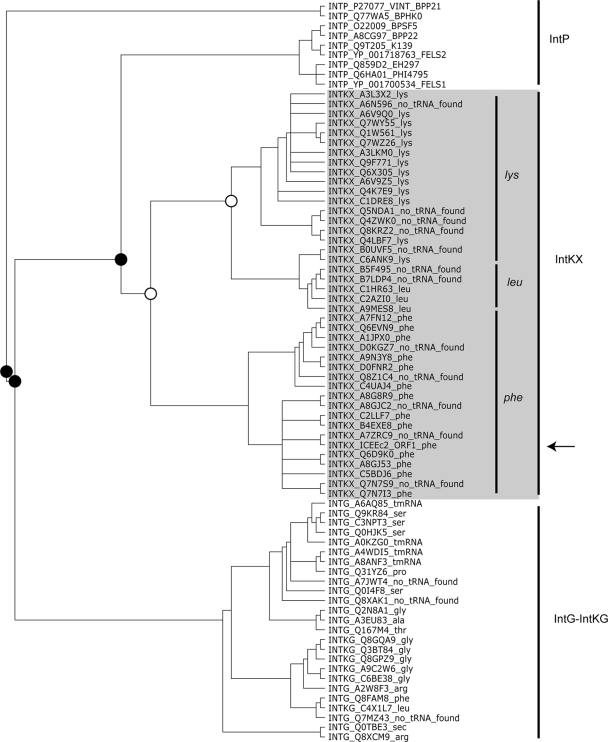
Interestingly, when a multiple alignment was performed with proteins most similar to ORF1, a cluster of recombinases was identified, and these recombinases were all located within regions similar to ICEEc2 or within remnants of ICEEc2-like elements such as in E. coli E24377A. These recombinases (designated as IntKX for integrase of pKLC102-PAGI-2 elements similar to XerC) form a cluster of sequences that are phylogenetically closer to integrases of prophages (IntP group) than to integrases of genomic islands (IntG group), two groups that were identified by Boyd et al. (4) (Fig. 3). In contrast, integrases retrieved from pKLC102/PAGI-2 members with a P4-like integrase (designated as IntKG for integrase of pKLC102-PAGI-2 elements similar to genomic island integrases) clustered with IntG sequences.
FIG. 3.
Phylogenetic analysis of the pKLC102/PAGI-2 element integrases. Sequences homologous to ORF1 from pKLC102-PAGI-2 elements and integrases of the IntP and IntG family were aligned using MUSCLE. The alignment was then manually curated, taking into account the different boxes in integrases described by Esposito et al. (13). The region upstream of the genes encoding integrases of pKLC102/PAGI-2 elements were analyzed; when a tRNA gene was found immediately upstream of this gene, its name was in included on the figure. The absence of a tRNA or the fact that it was not annotated is also indicated (no tRNA found). Analysis was then performed by neighbor joining, maximum likelihood, and parsimony. The tree represented in this figure is the one generated by Consense and is rooted using IntP_Q77WA5_BPHK0 and IntP_P27077_VINT_BPP21 as outgroups. Nodes relevant for our analysis and found in all three analyses are indicated by filled circles while those supported by only two analyses are indicated by open circles. The arrow indicates the position of ORF1.
The most variable regions of integrases are located in their N-terminal regions which contain the DNA binding region that determines their specificity. These analyses were thus performed using only the catalytic C-terminal part of the integrases containing the different boxes identified in tyrosine recombinases (13). Yet although the N-terminal DNA binding regions were removed, IntKXs were clustered according to their insertion sites; integrases located downstream of phe tRNA were well separated from those present downstream of lys or leu tRNA. This last result suggests that part of the specificities of these integrases may also be linked to their C-terminal regions and points to a coevolution between these integrases and the insertion sites of pKLC102/PAGI-like elements possessing an IntKX integrase. Interestingly, while elements possessing an IntKX integrase were most often located downstream of phe, lys, or leu tRNA, most pKLC102/PAGI-2-like elements with an IntKG integrase were inserted downstream of gly tRNA.
The ICEEc2 island is transferable by conjugation.
Among the members of the pKLC102/PAGI-2 family, some of them such as PAPI-1 or the clc element have been shown to be transferable by conjugation. Additionally, the detailed analysis of genes present in ICEEc2 indicated that some of them encode proteins that are very likely to fulfill functions involved in conjugative DNA transfer: DNA replication, type IV pilus formation, and coupling (TraD protein). Furthermore, ICEEc2 is flanked by 50-bp direct repeats and possesses a gene encoding an integrase that could act on these repeats to form an excised form typical of ICEs. Given these data, we hypothesized that ICEEc2 was also an ICE that could be transferred by conjugation.
We thus followed the transfer of ICEEc2 using the trimethoprim or streptomycin resistances linked to Tn7. Because Tn7 can spontaneously excise itself from ICEEc2 and insert into the host chromosome in a TnsD- or TnsE-dependent pathway (53), trimethoprim or streptomycin resistance does not necessarily reflect the transfer of the complete ICEEc2 element. We therefore confirmed the complete transfer of ICEEc2 in the conjugants by PCR amplification of regions within the iha gene, the Tn7 transposon, and ORF111. We tested the intraspecies transfer of ICEEc2 into E. coli K-12 strain C600 as well as the interspecies transfer using an S. Typhimurium strain and the Yersinia pseudotuberculosis strain IP32953 as recipient hosts. Transfer frequencies are indicated in Table 5. These results indicated that transfer of ICEEc2 occurred at high frequency in E. coli C600 (4.1 × 10−3) and in Salmonella strains (2.6 × 10−4). We were also able to transfer ICEEc2 into a Y. pseudotuberculosis IP32953 strain, but in this case the frequency was much lower (4.4 × 10−8). Furthermore, when Y. pseudotuberculosis IP32953 was the recipient, PCR analysis of 20 transconjugants showed that the entire ICEEc2 region had been transferred in only two conjugants. In the 18 other conjugants, only the Tn7 transposon had been transferred.
TABLE 5.
Conjugation frequency
| E. coli donor strain | Recipient strain | Transfer frequencya |
|---|---|---|
| BEN374 | E. coli C600 Nalr | 4.10 × 10−3 |
| S. Typhimurium DT104 BN 9181 ΔacrB::Kanr | 2.59 × 10−4 | |
| Y. pseudotuberculosis IP32953 O-Ag−, Kanrb | 4.4 × 10−8 | |
| BEN374 ΔORF1-ORF3 | E. coli C600 Nalr | ND |
| BEN374 ΔORF14 | E. coli C600 Nalr | 1.58 × 10−5 |
Calculated by dividing the number of ICEEc2 transconjugants by the number of donor cells. ND, not detected.
O-Ag−, O-antigen negative.
ICEEc2 integrates into the host's chromosome after conjugation.
The transfer of ICEs by conjugation is followed by their integration into the host's chromosome. We thus investigated if transfer of ICEEc2 into E. coli C600, Salmonella, or Yersinia strains was followed by its integration into the chromosome of the recipient strains.
Because our analysis indicated that elements sharing an integrase similar to ORF1 were all located downstream of phe tRNA (Fig. 3), we investigated integration of ICEEc2 only downstream of phe tRNA genes. E. coli, Salmonella, and Yersinia strains actually contain two phe tRNA genes identical in sequence, termed pheU and pheV in the case of E. coli. We thus used PCR primers located upstream of pheU or pheV to investigate the integration downstream of each of these two tRNA genes.
DNA was prepared directly from colonies obtained after conjugation. In all conjugants analyzed, obtained from either E. coli, Salmonella, or Yersinia recipients and containing a complete copy of ICEEc2, PCR amplification was obtained for reactions used to probe the integration both downstream of pheU and downstream of pheV (data not shown). PCR amplification from these same conjugants was also observed using primers located on both sides of pheU or pheV, indicating intact phe regions (data not shown). Altogether, these results indicate the existence, within the same colonies, of a mixed population of bacteria, some of them with ICEEc2 integrated downstream of pheU and others with ICEEc2 downstream of pheV.
Transfer of ICEEc2 requires the type IV pilus and formation of a circular intermediate.
Based on the current knowledge of the transfer of pKLC102/PAGI-2 family members, we hypothesized that the transfer of ICEEc2 required the type IV pilus and that the first three ORFs of ICEEc2 were involved in the formation of a circular intermediate necessary for the transfer of ICEEc2. Such a circular intermediate has already been detected in the case of pKLC102 (24). To address these questions, derivatives of strain BEN374 with a deletion of either ORF14 (pilS) or the ORF1-ORF3 region were constructed using the method described by Datsenko and Wanner (8). We then tested the transfer efficiency of these mutated ICEEc2 regions. Transfer efficiency from BEN374 ΔORF14 in the E. coli C600 strain was reduced by more than 2 orders of magnitude, and no transfer at all was observed when BEN374 ΔORF1-ORF3 was used as the donor. These results indicate that these regions play a major role in the transfer of ICEEc2 (Table 5).
We then analyzed by PCR the possible formation of a circular intermediate of ICEEc2 in strain BEN374. By using PCR primers pheU374_circF and pheU374_circR, located at the ends of ICEEc2 and orientated outward of ICEEc2, a circular form of ICEEc2 was detected (Fig. 4). This circular form was also detected in the ΔORF14 (pilS) mutant but was absent in the ΔORF1-ORF3 mutant (Fig. 4). This suggests that the formation of the circular form of ICEEc2 depends on genes ORF1-ORF3 and is required for efficient transfer.
FIG. 4.
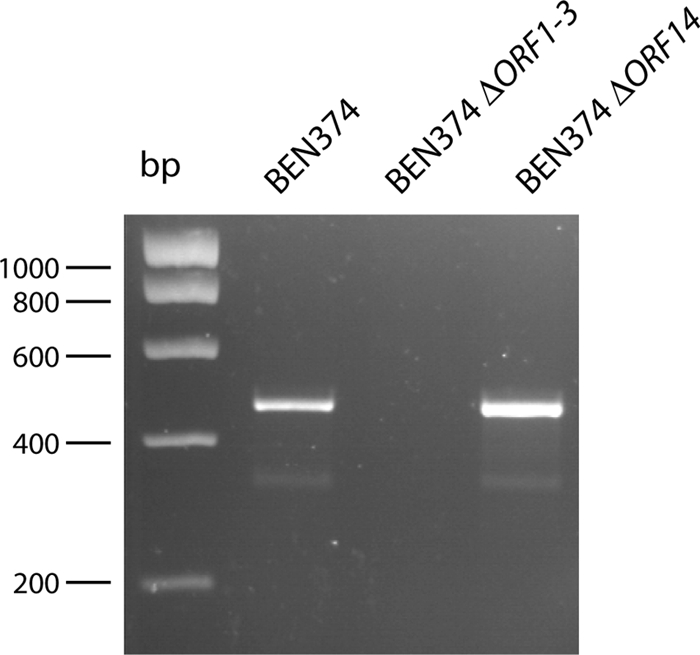
Formation of a circular intermediate is dependent upon the ORF1-ORF3 region. Formation of a circular intermediate was demonstrated by PCR using primers pheU374-circF and pheU374-circR and genomic DNA from strains BEN374, BEN374 ΔORF1-ORF3, and BEN374 ΔORF14.
DISCUSSION
In this report, we describe the identification of a new genomic island present downstream of the pheU tRNA gene in the extraintestinal pathogenic E. coli (ExPEC) strain BEN374 and demonstrated that this region could be transferred to other bacteria by conjugation.
ICEEc2 is a new ICE belonging to the pKLC102/PAGI-2 family.
Detailed analysis of the DNA sequence of ICEEc2 indicated that ICEEc2 belongs to the pKLC102/PAGI-2 family of genetic elements (24, 33). Members of this family have been categorized as integrative and conjugative elements that in some cases, in addition to having transfer and integration capacities, appear to be present as circular intermediates (6, 24, 46).
We thus investigated whether ICEEc2 could be a new ICE of the E. coli species by characterizing its transfer by conjugation and its integration into the host chromosome. Our results clearly establish that ICEEc2 can be transferred at high frequency from strain BEN374 to another E. coli strain or to a Salmonella strain and, at a much lower frequency, to a Yersinia strain. ICEEc2, similar to the clc element, is thus another example of a pKLC102/PAGI-2 element that can be transferred between strains of different species. A possible explanation for the low transfer efficiency in the Yersinia strain tested is the presence of genomic islands downstream of pheU and pheV in this strain (GenBank accession number BX936398). While all Yersinia conjugants obtained had the Tn7 transposon, the integration of ICEEc2 occurred in only two conjugants. DNA transfer has therefore occurred in a much higher percentage of recipients. We hypothesize that the presence of genomic islands in the Yersinia strain tested prior to the arrival of ICEEc2 might have disrupted the local organization around the phe tRNAs and thus prevented integration of ICEEc2.
The transfer of the entire ICEEc2 region was followed by integration downstream of a phe tRNA gene, as evidenced by PCR amplifications from single colonies. Yet PCR results indicated that within the same colony, amplification also occurred when using primers specific for phe tRNA genes without any downstream integration. It is thus possible that within a single colony some bacteria have integrated ICEEc2 downstream of pheU while in others ICEEc2 integrated downstream of pheV. Alternative hypotheses are that ICEEc2 shuttles within a single bacteria between an integrated form and an episomal form, as suggested for pKLC102 and PAPI-1 (24), or that multiple transfers occurred in the colony. Multiple insertions downstream of both pheU and pheV are also possible (for example, multiple pKLC102/PAGI-2 elements are present in Serratia proteamaculans strain 568), but putative ICE exclusion mechanisms are likely to restrict this possibility (54).
Together, these findings demonstrate that ICEEc2 is a new ICE that belongs to the pKLC102/PAGI-2 ICE family (6, 46). ICEEc2 is thus the second ICE characterized in E. coli, after ICEEc1 found in strain ECOR31. Even though ICEEc1 is considered the precursor of the high-pathogenicity island (HPI) present in E. coli and Yersinia strains, conjugative transfer of the complete ICEEc1 has never been demonstrated (44). In contrast, it is clear from our results that intra- and interspecies transfer of ICEEc2 occurs. Analyses of the different E. coli genomes available so far indicate that ICEEc2-related elements (or remnants of such elements) are found in other E. coli strains such as the commensal strain ED1a (50) or the intestinal pathogenic strain E24377A (38). It is thus likely that ICEEc2-related elements are present in a significant number of E. coli strains.
The transfer of ICEEc2 involves the formation of a circular intermediate.
The present work along with previous reports highlighted some common features concerning the transfer of pKLC102/PAGI-2 elements (20, 37, 41, 42). The first one is the presence of a circular intermediate (reference 24 and the present report). Our work in addition showed that, in the case of ICEEc2, the transfer occurs mainly through the type IV pilus and requires the formation of this circular intermediate as deletion of the first three ORFs strongly reduced its formation and the transfer efficiency. Another observation is the fact that transfer was possible between species. This property is shared by other members of this family such as the clc element, which was shown to be transferable to bacterial species different from the donor strain (42). A possible explanation for this broad range of possible recipients is that specificity is likely to be determined by the type IV pilus and more specifically by the PilV subunit. The type IV pilus genes are most similar to those of the type IV pilus from the plasmid pO113 found in the enterohemorrhagic E. coli (EHEC) strain EH41 (28). Within the region containing the pilV gene, we identified DNA repeats characteristic of shufflon regions on which the Rci recombinase (ORF18) could act to change the C-terminal sequence of the PilV protein. This, in turn, could influence conjugation specificity. This phenomenon has already been described for other type IV pili such as those encoded by the R64 plasmid (19, 25). Additionally, the antirestriction protein ORF45 might play a role in the broad specificity observed, as already observed in the case of Tn916 (31).
pKLC102/PAGI-2 members share a set of genes and, like other ICEs, have a modular structure, with each module contributing an essential function of the ICE life cycle: excision/integration, conjugation, and regulation (33, 54). Yet differences from other ICEs concerning the replication machinery and the regulation module exist. Data reported here and by others suggest that these elements can be present in the host both as episomal and chromosomally integrated forms. The question then arises as to whether the excised form is produced continuously by excision or by replication. In particular, genes responsible for this behavior remain to be characterized. The maintenance of ICEEc2 is likely to involve ORF111, a gene similar to the soj gene shown to be essential for the maintenance of PAPI-1 in P. aeruginosa (37). Because the region containing the ORF58 to ORF62 genes is one of the most conserved among pKLC102/PAGI-2 members, we speculated that it would be important for one of the steps of the life cycle of ICEEc2. Yet our attempts to address this question were unsuccessful.
Another important issue is to characterize the conjugation machinery of these elements, which seems to be quite different from that of already described conjugative plasmids. We could not find within ICEEc2 a gene encoding a classical relaxase, an essential enzyme responsible for the formation of the relaxosome that replicates DNA prior to transfer (17). It is tempting to speculate that the two activities of these relaxases, namely, DNA nicking and DNA unwinding activities (30), are fulfilled by two independent proteins such as the ParB protein (ORF108) and the DnaB helicase (ORF109). Other genes that potentially contribute to the conjugation process include those that are related to plasmid replication, such as ssb, topB, and other DNA-related enzymes. The role of repeat region 5 in the conjugation of ICEEc2 is also worth investigating: this region is reminiscent of the direct repeats described in the putative replication origin of pKLC102 by Klockgether et al. (23). Moreover, the central region of the palindrome is conserved between the two types of motifs (TGCCACTGGCA). We therefore suggest that the repeat 5 regions are likely to be involved in the life cycle of pKLC102/PAGI-2 elements.
ICEEc2 belongs to a group of pKLC102/PAGI-2 elements with XerC-like integrases.
Two types of elements in the pKLC102/PAGI-2 family can actually be distinguished based on the type of tyrosine recombinase they encode, either a XerC-like tyrosine recombinase (IntKX) or a P4-related tyrosine recombinase (IntKG). It is interesting that IntKX integrases are phylogenetically closer to prophage integrases (IntP) while IntKG integrases are closer to integrases present in genomic islands (IntG). Interestingly, Boyd et al. have recently shown that IntG and IntP are clearly different from a phylogenetic perspective (4).
In addition to this clear phylogenetic difference between IntKX and IntKP integrases, several points are also worth mentioning. First, the transfer frequencies of elements with IntKX integrases seems to be higher than those of elements with IntKG integrases. Transfer efficiencies as high as 10−2 have been observed for ICEHin1056, an IntKX pKLC102/PAGI-2 element, and we observed a transfer efficiency of 4.1 × 10−3 for ICEEc2 (33). In contrast, the transfer efficiency of the clc element of Pseudomonas sp. strain B13, a member of the IntKG subgroup, is only about 3.5 × 10−8 (41). In addition, Klockgether et al. (24) did not observe any circular forms of PAGI-2 and PAGI-3, two members of the IntKG subgroup, while PAPI-1 and pKLC102, members of the IntKX subgroup, formed circular intermediates. Furthermore, pKLC102/PAGI-2 regions with an IntKX integrase have long repeats flanking them (approximately 50 bp), suggesting that these long repeats are a key determinant in the mobility of these elements. In contrast, members of the IntKG subgroup are flanked by shorter repeats (approximately 20 bp). Finally, mobility of pKLC102/PAGI-2 elements is also likely to be regulated by the expression of these integrases, as has been shown for the clc element (45). Whether the IntKG and IntKX integrases are regulated by different mechanisms remains to be tested.
These different points suggest that pKLC102/PAGI-2 elements with an IntKG integrase have a lower mobility than those with an IntKX integrase. An interesting hypothesis is that ICEs with an IntKG integrase are in the process of stabilization into the chromosome of their host and are engaged in their transformation into a genomic island.
Cargo regions of pKLC102/PAGI-2 members and their potential role in virulence and fitness.
ICEEc2 combines two characteristics that deserve to be highlighted: it carries three antibiotic resistance genes and the iha gene that may contribute to the virulence properties of strain BEN374. Whether ICEEc2 really contributes to the virulence of strain BEN374 remains to be studied, but it is likely that it contributes at least to its survival in iron-depleted environments. Indeed, it has been demonstrated that Iha is a catecholate siderophore receptor (27). Iha was also described as involved in adhesion of an enterohemorrhagic E. coli strain to eukaryotic cells; ICEEc2 is thus likely to be involved in the adhesion of strain BEN374 to eukaryotic cells (48).
The fact that ICEEc2 combines features that may increase its distribution, i.e., an antibiotic resistance cluster, an iron acquisition protein, and a potential adhesin, highlights the amazing diversity of functions that are encoded by cargo regions of pKLC102/PAGI-2 elements: antibiotic resistance (33) and aromatic compound degradation (41, 42), as well as fitness of the recipient strain (15). Recently, ICEPm1, a member of the pKLC102/PAGI-2 family present in Proteus mirabilis strains, was shown to be more prevalent in isolates obtained from urine samples than in those from other body sites, suggesting a potential role in virulence (14). Salmonella pathogenicity island 7 (SPI-7) from S. enterica serovar Typhi, on which are located the Vi antigen genes, is another element of this family carrying genes involved in the virulence of its host.
Because of this diversity of function and of their ubiquitous character, pKLC102/PAGI-2 genetic elements could be major players in the diversification of proteobacteria in terms of adaptation of bacteria to their environment (new hosts, contaminated environments, etc.). Yet many unanswered questions remain concerning the biology and the life cycle of these elements. Addressing these questions would bring new clues concerning what seems to be an important modality in the evolution of bacterial genomes.
Supplementary Material
Acknowledgments
We thank Axel Cloeckaert for providing us with Salmonella strains and Elizabeth Carniel for the Yersinia strain.
This project was supported by the European Community under FP5 (COLIRISK, grant number QLK2-CT-2002-00944) and by the AIP Genome et Séquençage 2007 of the INRA.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Auchtung, J. M., C. A. Lee, R. E. Monson, A. P. Lehman, and A. D. Grossman. 2005. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 102:12554-12559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baucheron, S., S. Tyler, D. Boyd, M. R. Mulvey, E. Chaslus-Dancla, and A. Cloeckaert. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bocs, S., S. Cruveiller, D. Vallenet, G. Nuel, and C. Medigue. 2003. AMIGene: annotation of microbial genes. Nucleic Acids Res. 31:3723-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., S. Almagro-Moreno, and M. A. Parent. 2009. Genomic islands are dynamic, ancient integrative elements in bacterial evolution. Trends Microbiol. 17:47-53. [DOI] [PubMed] [Google Scholar]
- 5.Burrus, V., G. Pavlovic, B. Decaris, and G. Guedon. 2002. Conjugative transposons: the tip of the iceberg. Mol. Microbiol. 46:601-610. [DOI] [PubMed] [Google Scholar]
- 6.Burrus, V., and M. K. Waldor. 2004. Shaping bacterial genomes with integrative and conjugative elements. Res. Microbiol. 155:376-386. [DOI] [PubMed] [Google Scholar]
- 7.Chouikha, I., P. Germon, A. Brée, P. Gilot, M. Moulin-Schouleur, and C. Schouler. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J. F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J. M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465-W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 11.Dobrindt, U., B. Hochhut, U. Hentschel, and J. Hacker. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat. Rev. Microbiol. 2:414-424. [DOI] [PubMed] [Google Scholar]
- 12.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito, D., and R. Craigie. 1998. Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein-DNA interaction. EMBO J. 17:5832-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flannery, E. L., L. Mody, and H. L. Mobley. 2009. Identification of a modular pathogenicity island that is widespread among urease-producing uropathogens and shares features with a diverse group of mobile elements. Infect. Immun. 77:4887-4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaillard, M., N. Pernet, C. Vogne, O. Hagenbuchle, and J. R. van der Meer. 2008. Host and invader impact of transfer of the clc genomic island into Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U. S. A. 105:7058-7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaillard, M., T. Vallaeys, F. J. Vorholter, M. Minoia, C. Werlen, V. Sentchilo, A. Puhler, and J. R. van der Meer. 2006. The clc element of Pseudomonas sp. strain B13, a genomic island with various catabolic properties. J. Bacteriol. 188:1999-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcillan-Barcia, M. P., M. V. Francia, and F. de la Cruz. 2009. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33:657-687. [DOI] [PubMed] [Google Scholar]
- 18.Germon, P., D. Roche, S. Melo, S. Mignon-Grasteau, U. Dobrindt, J. Hacker, C. Schouler, and M. Moulin-Schouleur. 2007. tDNA locus polymorphism and ecto-chromosomal DNA insertion hot-spots are related to the phylogenetic group of Escherichia coli strains. Microbiology 153:826-837. [DOI] [PubMed] [Google Scholar]
- 19.Gyohda, A., and T. Komano. 2000. Purification and characterization of the R64 shufflon-specific recombinase. J. Bacteriol. 182:2787-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Juhas, M., D. W. Crook, I. D. Dimopoulou, G. Lunter, R. M. Harding, D. J. Ferguson, and D. W. Hood. 2007. Novel type IV secretion system involved in propagation of genomic islands. J. Bacteriol. 189:761-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Juhas, M., J. R. van der Meer, M. Gaillard, R. M. Harding, D. W. Hood, and D. W. Crook. 2009. Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33:376-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaper, J. B., J. P. Nataro, and H. L. Mobley. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123-140. [DOI] [PubMed] [Google Scholar]
- 23.Klockgether, J., O. Reva, K. Larbig, and B. Tummler. 2004. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J. Bacteriol. 186:518-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klockgether, J., D. Wurdemann, O. Reva, L. Wiehlmann, and B. Tummler. 2007. Diversity of the abundant pKLC102/PAGI-2 family of genomic islands in Pseudomonas aeruginosa. J. Bacteriol. 189:2443-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komano, T., A. Kubo, and T. Nisioka. 1987. Shufflon: multi-inversion of four contiguous DNA segments of plasmid R64 creates seven different open reading frames. Nucleic Acids Res. 15:1165-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leveille, S., M. Caza, J. R. Johnson, C. Clabots, M. Sabri, and C. M. Dozois. 2006. Iha from an Escherichia coli urinary tract infection outbreak clonal group A strain is expressed in vivo in the mouse urinary tract and functions as a catecholate siderophore receptor. Infect. Immun. 74:3427-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leyton, D. L., J. Sloan, R. E. Hill, S. Doughty, and E. L. Hartland. 2003. Transfer region of pO113 from enterohemorrhagic Escherichia coli: similarity with R64 and identification of a novel plasmid-encoded autotransporter, EpeA. Infect. Immun. 71:6307-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llosa, M., F. X. Gomis-Ruth, M. Coll, and F. de la Cruz. 2002. Bacterial conjugation: a two-step mechanism for DNA transport. Mol. Microbiol. 45:1-8. [DOI] [PubMed] [Google Scholar]
- 30.Lucas, M., B. Gonzalez-Perez, M. Cabezas, G. Moncalian, G. Rivas, and F. de la Cruz. Relaxase DNA binding and cleavage are two distinguishable steps in conjugative DNA processing that involve different sequence elements of the nic site. J. Biol. Chem. 285:8918-8926. [DOI] [PMC free article] [PubMed]
- 31.McMahon, S. A., G. A. Roberts, K. A. Johnson, L. P. Cooper, H. Liu, J. H. White, L. G. Carter, B. Sanghvi, M. Oke, M. D. Walkinshaw, G. W. Blakely, J. H. Naismith, and D. T. Dryden. 2009. Extensive DNA mimicry by the ArdA anti-restriction protein and its role in the spread of antibiotic resistance. Nucleic Acids Res. 37:4887-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 33.Mohd-Zain, Z., S. L. Turner, A. M. Cerdeno-Tarraga, A. K. Lilley, T. J. Inzana, A. J. Duncan, R. M. Harding, D. W. Hood, T. E. Peto, and D. W. Crook. 2004. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J. Bacteriol. 186:8114-8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Osborn, A. M., and D. Boltner. 2002. When phage, plasmids, and transposons collide: genomic islands, and conjugative- and mobilizable-transposons as a mosaic continuum. Plasmid 48:202-212. [DOI] [PubMed] [Google Scholar]
- 35.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 36.Pouillot, F., A. Derbise, M. Kukkonen, J. Foulon, T. K. Korhonen, and E. Carniel. 2005. Evaluation of O-antigen inactivation on Pla activity and virulence of Yersinia pseudotuberculosis harbouring the pPla plasmid. Microbiology 151:3759-3768. [DOI] [PubMed] [Google Scholar]
- 37.Qiu, X., A. U. Gurkar, and S. Lory. 2006. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 103:19830-19835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasko, D. A., J. A. Phillips, X. Li, and H. L. Mobley. 2001. Identification of DNA sequences from a second pathogenicity island of uropathogenic Escherichia coli CFT073: probes specific for uropathogenic populations. J. Infect. Dis. 184:1041-1049. [DOI] [PubMed] [Google Scholar]
- 39.Ravatn, R., S. Studer, D. Springael, A. J. Zehnder, and J. R. van der Meer. 1998. Chromosomal integration, tandem amplification, and deamplification in Pseudomonas putida F1 of a 105-kilobase genetic element containing the chlorocatechol degradative genes from Pseudomonas sp. strain B13. J. Bacteriol. 180:4360-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ravatn, R., S. Studer, A. J. Zehnder, and J. R. van der Meer. 1998. Int-B13, an unusual site-specific recombinase of the bacteriophage P4 integrase family, is responsible for chromosomal insertion of the 105-kilobase clc element of Pseudomonas sp. strain B13. J. Bacteriol. 180:5505-5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravatn, R., A. J. Zehnder, and J. R. van der Meer. 1998. Low-frequency horizontal transfer of an element containing the chlorocatechol degradation genes from Pseudomonas sp. strain B13 to Pseudomonas putida F1 and to indigenous bacteria in laboratory-scale activated-sludge microcosms. Appl. Environ. Microbiol. 64:2126-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reineke, W., S. W. Wessels, M. A. Rubio, J. Latorre, U. Schwien, E. Schmidt, M. Schlömann, and H.-J. Knackmuss. 1982. Degradation of monochlorinated aromatics following transfer of genes encoding chlorocatechol catabolism. FEMS Microbiol. Lett. 14:291-294. [Google Scholar]
- 43.Schouler, C., A. Taki, I. Chouikha, M. Moulin-Schouleur, and P. Gilot. 2009. A genomic island of an extraintestinal pathogenic Escherichia coli strain enables the metabolism of fructooligosaccharides, which improves intestinal colonization. J. Bacteriol. 191:388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schubert, S., S. Dufke, J. Sorsa, and J. Heesemann. 2004. A novel integrative and conjugative element (ICE) of Escherichia coli: the putative progenitor of the Yersinia high-pathogenicity island. Mol. Microbiol. 51:837-848. [DOI] [PubMed] [Google Scholar]
- 45.Sentchilo, V., A. J. Zehnder, and J. R. van der Meer. 2003. Characterization of two alternative promoters for integrase expression in the clc genomic island of Pseudomonas sp. strain B13. Mol. Microbiol. 49:93-104. [DOI] [PubMed] [Google Scholar]
- 46.Seth-Smith, H., and N. J. Croucher. 2009. Genome watch: breaking the ICE. Nat. Rev. Microbiol. 7:328-329. [DOI] [PubMed] [Google Scholar]
- 47.Stordeur, P., D. Marlier, J. Blanco, E. Oswald, F. Biet, M. Dho-Moulin, and J. Mainil. 2002. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet. Microbiol. 84:231-241. [DOI] [PubMed] [Google Scholar]
- 48.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenaillon, O., D. Skurnik, B. Picard, and E. Denamur. 2010. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8:207-217. [DOI] [PubMed] [Google Scholar]
- 50.Touchon, M., C. Hoede, O. Tenaillon, V. Barbe, S. Baeriswyl, P. Bidet, E. Bingen, S. Bonacorsi, C. Bouchier, O. Bouvet, A. Calteau, H. Chiapello, O. Clermont, S. Cruveiller, A. Danchin, M. Diard, C. Dossat, M. E. Karoui, E. Frapy, L. Garry, J. M. Ghigo, A. M. Gilles, J. Johnson, C. Le Bouguenec, M. Lescat, S. Mangenot, V. Martinez-Jehanne, I. Matic, X. Nassif, S. Oztas, M. A. Petit, C. Pichon, Z. Rouy, C. S. Ruf, D. Schneider, J. Tourret, B. Vacherie, D. Vallenet, C. Medigue, E. P. Rocha, and E. Denamur. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet. 5:e1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vallenet, D., L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli, and C. Medigue. 2006. MaGe: a microbial genome annotation system supported by synteny results. Nucleic Acids Res. 34:53-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van der Meer, J. R., and V. Sentchilo. 2003. Genomic islands and the evolution of catabolic pathways in bacteria. Curr. Opin. Biotechnol. 14:248-254. [DOI] [PubMed] [Google Scholar]
- 53.Wolkow, C. A., R. T. DeBoy, and N. L. Craig. 1996. Conjugating plasmids are preferred targets for Tn7. Genes Dev. 10:2145-2157. [DOI] [PubMed] [Google Scholar]
- 54.Wozniak, R. A., and M. K. Waldor. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat. Rev. Microbiol. 8:552-563. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.