Abstract
In this study, we identified and characterized the enzymatic properties of MG_186, a calcium-dependent Mycoplasma genitalium nuclease. MG_186 displays the hallmarks of nucleases, as indicated by its amino acid sequence similarity to other nucleases. We cloned, UGA corrected, expressed, purified, and demonstrated that recombinant MG_186 (rMG_186) exhibits nuclease activity similar to that of typical sugar-nonspecific endonucleases and exonucleases. Biochemical characterization indicated that Ca2+ alone enhances its activity, which was inhibited by divalent cations, such as Zn2+ and Mn2+. Chelating agents EGTA and EDTA also inhibited nuclease activity. Mycoplasma membrane fractionation and Triton X-114 phase separation showed that MG_186 was a membrane-associated lipoprotein, and electron microscopy revealed its surface membrane location. Incubation of purified human endometrial cell nuclei with rMG_186 resulted in DNA degradation and morphological changes typical of apoptosis. Further, immunofluorescence analysis of rMG_186-treated nuclei indicated that morphological changes were linked to the disintegration of lamin and the internalization of rMG_186. Since M. genitalium has the capacity to invade eukaryotic cells and localize to the perinuclear and nuclear region of parasitized target cells, MG_186 has the potential to provide M. genitalium, which possesses the smallest genome of any self-replicating cell, with the ability to degrade host nucleic acids both as a source of nucleotide precursors for growth and for pathogenic purposes.
Mycoplasma genitalium was first identified as a urogenital tract pathogen in men and subsequently implicated in a range of women pathologies, including pelvic inflammatory diseases, cervicitis, endometritis, salpingitis, and tubal factor infertility (5, 37, 40). In addition to its urogenital niche, M. genitalium has been detected in synovial and respiratory tract specimens (3, 39). M. genitalium DNA sequencing revealed a reduced genome size of 580 kb and a low GC content, along with 482 protein-encoding genes, of which 76 were categorized as hypothetical proteins (18). The streamlined genome of M. genitalium results in gene deficits that dramatically limit its biosynthetic capabilities, leading to a complete dependence on the host for metabolic precursors, such as nucleotides, amino acids, fatty acids, and sterols.
Since M. genitalium, like most mollicutes, is unable to synthesize de novo purine and pyrimidine bases (27), it must scavenge nucleotides from the host in order to replicate and persist. Only Mycoplasma penetrans has an orotate-related pathway for converting carbamoyl-phosphate to uridine-5′-monophosphate (34). The importance of nucleases in the life cycle of mycoplasmas is reinforced by their detection in at least 20 Mycoplasma species (26). Purification of membrane-associated Ca2+/Mg2+-dependent M. penetrans and Mycoplasma hyorhinis nucleases and their relation to mycoplasma survival and pathogenesis have been reported (7, 8, 29, 30). Also, a membrane nuclease gene, mnuA, was identified and cloned from Mycoplasma pulmonis (20, 25). mnuA orthologous sequences were found in M. penetrans, Mycoplasma pneumoniae, Mycoplasma hyopneumoniae, Mycoplasma gallisepticum, and Ureaplasma urealyticum but not in M. genitalium. However, recent nuclease studies with M. hyopneumoniae (nuclease gene designated mhp379) revealed the existence of orthologous sequences in M. genitalium as well as in M. pneumoniae, M. pulmonis, M. gallisepticum, and Mycoplasma synoviae (35).
M. genitalium was initially described as an extracellular pathogen. Subsequently, we reported that M. genitalium can be observed in the cytoplasmic and perinuclear regions of infected mammalian cells and can persist long-term within these compartments (4, 13, 24). The latter supports the contention that M. genitalium is capable of intracellular replication and survival. Furthermore, our recent evidence suggests that M. genitalium and its protein products are capable of intranuclear localization within infected endometrial cells (41). Therefore, understanding how M. genitalium overcomes its biosynthetic deficiencies and successfully parasitizes host tissues may provide insights into its biological uniqueness as the smallest pathogen capable of “independent” growth. In this report, we characterized a putative lipoprotein, MG_186, that retains the thermostable nuclease motif found in other bacterial nucleases. The gene encoding MG_186 was cloned and expressed in Escherichia coli, and the biochemical properties of purified recombinant MG_186 (rMG_186) nuclease protein were examined along with its impact on intact nuclei isolated from endometrial cells.
MATERIALS AND METHODS
Bacterial strains and cell culture conditions.
M. genitalium (G37) cells were grown to mid- to late log phase in SP-4 medium at 37°C in 150-cm2 tissue culture flasks. Surface-attached mycoplasmas were harvested by being washed three times with phosphate-buffered saline (PBS; pH 7.4), scraped, and pelleted at 12,500 × g for 15 min at 4°C. Escherichia coli TOP10 (Invitrogen) and E. coli BL21(DE3) (Stratagene) were grown in Luria-Bertani (LB) broth.
Human endometrial cell line EM42, which originated from benign proliferative endometrium (15), was grown in RPMI 1640 medium supplemented with 5% (vol/vol) fetal bovine serum and 2 mM l-glutamine (Invitrogen). All cell cultures were grown under air-5% CO2 at 37°C and routinely certified to be free of mycoplasma contamination (MycoProbe Mycoplasma detection kit [R&D Systems]). Nuclei from EM42 cells were isolated as detailed below, and DNA was purified using the Easy DNA isolation kit (Invitrogen). Total RNA was purified using the RNeasy RNA purification kit (Qiagen).
Cloning, expression, and purification of rMG_186.
M. genitalium chromosomal DNA was isolated using Easy DNA isolation kits (Invitrogen). Plasmid DNA was purified using the QIAprep spin protocol according to the manufacturer's instructions (Qiagen). Based on the published genome sequence, the MG_186 gene was amplified by PCR, using M. genitalium strain G37 chromosomal DNA as a template. PCR amplification and UGA corrections were performed as described previously for the M. pneumoniae community-acquired respiratory distress syndrome (CARDS) toxin (21). Primers were designed without the N-terminal signal peptide sequence to facilitate the expression of soluble recombinant protein. Specific mg186 oligonucleotide primers are given in Table 1.
TABLE 1.
Primers used to amplify and UGA correct the mg186 gene
| Primer | Primer sequence (5′ to 3′)a | Position (size in bp)b |
|---|---|---|
| MG186F | CATATGTGCATTGCTGAAAAACCAGTTAAC | 73-96 (30) |
| MG186R | GGATCCTTAGCCATTGTTTAGTTTCAATTCATAGATG | 723-753 (37) |
| MG186F1 | GCAAGGGTTAACCACTGGAGAGATGGGGATAC | 142-173 (32) |
| MG186R1 | GTATCCCCATCTCTCCAGTGGTTAACCCTTGC | 142-173 (32) |
| MG186F2 | GTAGTGAGGTGTGGATCTGGCCACTAAATAGCTATAG | 332-368 (37) |
| MG186R2 | CTATAGCTATTTAGTGGCCAGATCCACACCTCACTAC | 332-368 (37) |
| MG186F3 | GTTGACCAAAGTTGGACAAGGTATTTAGCTC | 649-679 (31) |
| MG186R3 | GAGCTAAATACCTTGTCCAACTTTGGTCAAC | 649-679 (31) |
Underlining indicates introduced restriction enzyme sites. Bolding indicates TGA-to-TGG changes of the codon to permit expression of tryptophan in E. coli.
The position of nucleotides indicates their location within the encoding ORF.
The resulting PCR product was cloned into pCR2.1, generating a plasmid designated pCR-MG_186, which was subsequently digested with NdeI and BamHI and ligated into pET19b to yield pET-MG_186. This plasmid was transformed into competent E. coli BL21(DE3), and recombinant colonies were screened for resistance to ampicillin and expression of rMG_186 protein. Verification of UGA-corrected pET-MG_186 was achieved by complete DNA sequencing (Department of Microbiology and Immunology Nucleic Acids Core Facility, University of Texas Health Science Center at San Antonio). Induction of recombinant protein synthesis in E. coli was accomplished by the addition of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG; Sigma-Aldrich), and bacteria were incubated for 3 h at 37°C under aeration at 220 rpm. Fusion proteins were purified by nickel affinity chromatography under native conditions (Qiagen). rMG_186 was desalted in 50 mM Tris-HCl buffer (pH 8.0) plus 5% glycerol using PD-10 columns, and protein purity was analyzed by SDS-PAGE.
Quantification of protein and nucleic acids.
Protein concentrations were estimated by the bicinchoninic acid method (Pierce, Rockford, IL) with bovine serum albumin (BSA) as a standard. DNA, RNA, and purified plasmid sample concentrations were assayed at 260 nm using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific).
Assays for nuclease activity.
Nuclease activity of rMG_186 was analyzed by agarose gel electrophoresis and zymogram as described previously (29). Approximately 0.5 to 1 μg of rMG_186 was incubated at 37°C in 50 μl of nuclease reaction buffer (100 mM Tris-HCl [pH 8.3] plus 10 mM CaCl2) containing 1 to 5 μg of nucleic acid substrate (EM42 chromosomal DNA [double stranded], EM42 RNA, or M13 phage DNA [single stranded; New England BioLabs]). To measure endonuclease activity, we used closed circular plasmid DNA (pET-MG_186) as the substrate. Aliquots (10 μl) were removed at different time intervals, and reactions were arrested by adding EDTA. Reaction products were visualized by 1% agarose gel electrophoresis, and nucleic acid degradation was monitored by staining with ethidium bromide. To study the effect of divalent cations or salts, each was added individually or in the presence of CaCl2, and assays were performed as described above. To quantify and compare native and rMG_186 nuclease activities, total M. genitalium proteins or rMG_186 were resolved using 12% SDS-PAGE gels containing 160 μg/ml herring sperm DNA. Nuclease activity was initiated by soaking gels in 100 mM Tris-HCl (pH 8.3), 10 mM CaCl2, and 10 mM MgCl2. Then, DNA was stained with ethidium bromide (0.5 mg/ml) and visualized with UV light, and proteins were stained with Coomassie brilliant blue.
Preparation of antisera against rMG_186.
New Zealand white rabbits were immunized subcutaneously with 100 to 200 μg of rMG_186 protein suspended in complete Freund's adjuvant. Individual rabbits were boosted three times with the same amounts of antigen in incomplete Freund's adjuvant at 21-day intervals. Serum samples were collected and used for immunological characterization and neutralization assays as described below.
M. genitalium membrane purification and Triton X-114 phase separation.
Mid- to late log phase cultures (400 ml) of M. genitalium cells were pelleted and subjected to membrane isolation. Membranes were purified by sucrose gradient centrifugation as previously described (14), and protein concentrations in total cell and membrane fractions were estimated by the bicinchoninic acid method (Pierce, Rockford, IL). Equal amounts of each fraction were further separated by 4 to 12% NuPAGE gel electrophoresis and then transferred to nitrocellulose membranes. Membrane phase separation was carried out as described previously (16, 17). In brief, pelleted mycoplasma cells were washed three times with PBS and solubilized by adding Triton X-114 to a final concentration of 1%. Following incubation at 4°C for 2.5 h, phase separation was induced by incubating Triton X-114-solubilized proteins at 37°C for 10 min. After centrifugation at 2,000 × g for 5 min, the aqueous phase and the pellet were subjected to gel electrophoresis.
Immunoblot analysis.
Equal amounts of M. genitalium whole-cell lysate and membrane-, cytoplasmic-, and Triton X-114-treated fractions were separated on 4 to 12% NuPAGE gels (Invitrogen) and transferred to nitrocellulose membranes. Immunoblotting was performed using rabbit antiserum reactive against rMG_186 (1:2,000), elongation factor G (EF-G) (1:2,000; a kind gift from R. Herrmann), or P140 adhesin (1:2,000) (24) plus goat anti-rabbit alkaline phosphatase (1:2,000) antibodies.
Immunogold electron microscopy.
Immunogold labeling of M. genitalium was performed as described earlier (2). M. genitalium cells attached on Formvar-coated nickel grids were fixed with 1% glutaraldehyde-4% formaldehyde for 20 min at room temperature (RT). Mycoplasma cells on fixed grids were then incubated with rabbit IgG-purified antiserum raised against rMG_186 or with prebleed serum (1:100) followed by goat anti-rabbit IgG gold particles (20 nm; 1:20) in PBS (pH 7.4) containing 1% bovine serum albumin. Individual grids were examined by JEOL 1230 transmission electron microscopy at an 80-kV accelerating voltage after being stained with 7% uranyl acetate followed by Reynolds' lead citrate.
Neutralization assays for nuclease activity.
To avoid endogenous nucleases present in test antisera, IgG fractions were purified by protein G column chromatography before use in the following neutralization test. rMG_186 (0.5 μg) was preincubated with different concentrations of specific IgG (0.5 to 5 μg) fractions from rabbit serum reactive against rMG_186 or recombinant CARDS (rCARDS) toxin (22) at 37°C for 10 min, and nuclease activity was measured as described above using EM42 chromosomal DNA.
Immunoreactivity of M. genitalium-infected patient sera against rMG_186.
M. genitalium-positive patient sera (20 individuals) collected from sexually transmitted disease clinics throughout San Antonio, TX (34), were evaluated for MG_186 antibodies by Western blot analysis using rMG_186. Human sera were diluted at 1:100 in 3% milk in Tris-buffered saline-Tween 20 (TBST), and secondary goat anti-human (Zymed) was diluted in TBST at a 1:2,000 dilution.
Impact of rMG_186 on EM42 nuclei and DNA.
EM42 nuclei were purified as described previously (29). Purified nuclei were resuspended in 50 mM Tris-HCl (pH 8.0), 5 mM MgCl2, 0.1 mM EDTA, and 40% glycerol and stored on ice or at −80°C. To study the effect of rMG_186 on EM42 nuclei, DNA degradation was assessed by agarose gel electrophoresis and morphological changes of nuclei. For analysis by agarose gel electrophoresis, nuclei were pelleted by centrifugation and resuspended in 200 μl of 100 mM Tris-HCl, pH 8.3, containing 10 mM EDTA or CaCl2. After the addition of rMG_186, test preparations were incubated at 37°C and 10-μl aliquots (containing 1 μg of rMG_186) were collected at specific intervals. DNA was precipitated with isopropanol, washed with ethanol, dried, and separated in 1% agarose gels.
To study morphological changes, nuclei were stained with SYTOX green nucleic acid stain (Invitrogen) as described previously (29) and analyzed by light microscopy. In brief, for SYTOX staining, nuclei were diluted in two volumes of 10 mM HEPES (pH 7.0), 40 mM β-glycerophosphate, 50 mM NaCl, and 10 mM CaCl2. After the addition of 5 μl of 20 mM Tris-HCl (pH 8.3) (negative control) or 5 μl rMG_186 nuclease (1 ng to 1 μg), nuclear preparations were incubated for 1 h to overnight at RT. Nuclei were fixed in 2% formaldehyde and applied to coverslips to dry for at least 2 h. Nuclear DNA was stained with 50 nM SYTOX in PBS for 15 min, and excess dye was removed by three washes with PBS. Finally, nuclei were mounted in Vectashield (Vector Laboratories) and examined using inverted fluorescence microscopy (Carl Zeiss Cell Observer Z1). The SYTOX/DNA complex has excitation and emission maxima of 504 nm and 523 nm, respectively.
For indirect immunofluorescence, rMG_186-treated and untreated nuclei were fixed in ice-cold 2% paraformaldehyde in PBS for 15 min. Nuclei were washed three times with 0.2% goat serum in PBS and incubated with anti-rMG_186 rabbit polyclonal antibodies (1:1,000) and/or anti-lamin A mouse monoclonal antibody (Abcam; 1:1,000) for 1 h at 4°C. After nuclei were washed three times with 0.2% goat serum in PBS, nuclei were incubated for 1 h at 4°C with goat anti-rabbit Alexa Fluor 633 (Invitrogen; 1:500) to identify anti-rMG_186 antibodies or with goat anti-mouse Alexa Fluor 488 (Invitrogen; 1:500) to detect anti-lamin A antibodies. Individual samples were washed again with 0.2% goat serum in PBS and stained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 min. Nuclear preparations were mounted on glass slides using Vectashield mounting medium and examined using inverted fluorescence microscopy (Carl Zeiss Cell Observer Z1 with AxioVision 4.8 software). Nuclear morphology was analyzed, and images were captured at ×400 magnification. Serially produced sections (0.5 μm; z-series) were obtained through fluorescent specimens by combining a series of x-y scans taken along the z axis.
Computer-assisted analysis.
Amino acid identity matches were performed using the National Center for Biotechnology Information's sequence similarity search tool designed to support analysis of nucleotide and protein databases at http://www.ncbi.nlm.nih.gov/blast/. All M. genitalium sequence data used in this study were downloaded from the comprehensive microbial resource database at http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=gmg. Protein domains, families, and functional sites were analyzed using http://expasy.org/prosite/.
RESULTS
Sequence similarities of MG_186 to calcium-dependent nucleases.
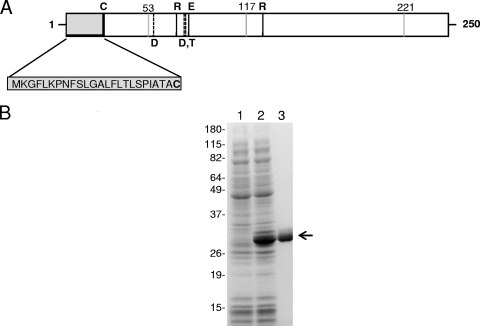
The MG_186 gene encodes a protein which consists of 250 amino acid residues with an estimated molecular mass of 28.28 kDa and an isoelectric point of 7.8. Analysis of the MG_186 amino acid sequence revealed the presence of a classical lipoprotein signal peptide consisting of approximately 25 amino acid residues with a typical cysteine cleavage site at residue 25 (Fig. 1 A). BLASTP (1) sequence comparisons of MG_186 identified more than 50 bacterial homologues in both Gram-negative and Gram-positive species. Amino acid sequence 1 to 245 of MG_186 is 39% (113/288) identical to that of M. pneumoniae MPN133, and amino acids 48 to 231 share 31% (62/197) identity with those of the recently reported M. hyopneumoniae mhp379 nuclease (35). Prosite analysis reveals that MG_186 contains key and invariant amino acid motifs, as in thermonuclease (TNASE_3), that mediate binding of calcium metal ions and active site conformation (Fig. 1A). These motifs were first identified in the nuclease of Staphylococcus aureus (11, 23, 36). Thus, sequence similarity and conservation of the essential catalytic residues between M. genitalium MG_186 and other nuclease sequences suggest that MG_186 is a Ca2+-dependent nuclease.
FIG. 1.
MG_186 organization, cloning, expression, and purification. (A) Schematic of MG_186. MG_186 is comprised of 250 amino acids and contains a hydrophobic amino-terminal signal sequence and prokaryotic lipoprotein cleavage site (indicated by the gray box and the letter C, respectively). Amino acids 44 through 200 have significant identity and similarity with the thermonuclease domain profile first described for the Nuc thermonuclease of Staphylococcus aureus (11, 23, 36). Conserved amino acid residues involved in binding of calcium ions (dashed vertical lines) are aspartate (D57, D77) and tyrosine (T78). Amino acid residues arginine (R72, R126) and glutamate (E80) are also conserved and comprise the active catalytic site (black vertical lines). Positions of UGA-encoded tryptophans (53, 117, and 221) are indicated by gray vertical lines. (B) Expression and purification of rMG_186. The MG_186 gene without its signal peptide was cloned, expressed, and purified as a His-tagged protein as detailed in Materials and Methods. Protein fractions were resolved on 4 to 12% NuPAGE gradient gels. Lane 1, uninduced E. coli pET19b-MG186 total cell lysate; lane 2, induced E. coli pET19b-MG186 total cell lysate; and lane 3, Ni-NTA column-purified rMG_186. The arrow indicates rMG_186. Molecular weight markers are indicated on the left.
Cloning, site-directed mutagenesis, expression, and purification of rMG_186.
To determine whether mg186 encoded a functional nuclease, the open reading frame (ORF) corresponding to the MG_186 protein was analyzed for UGA-encoded tryptophans, and three residues at positions 53, 117, and 221 were identified. To express MG_186 in E. coli, the three tryptophan codons were changed from UGA to UGG by site-directed mutagenesis, and the region lacking the signal peptide (Fig. 1A and Table 1) of the complete ORF was PCR amplified, confirmed by sequencing, and cloned into expression vector pET19b to generate N-terminal His-tagged protein. Upon IPTG induction, E. coli BL21(DE3) cells, which were transformed with plasmid pET-MG_186, expressed a soluble protein that migrated with an expected molecular mass of 29 kDa (Fig. 1B, lane 2). Uninduced cells lacked a strong protein band in that region (Fig. 1B, lane 1). The overexpressed His-tagged rMG_186 protein was purified to homogeneity by nickel nitrilotriacetic acid (Ni-NTA) chromatography (Fig. 1B, lane 3), and its mass was identical to the theoretical molecular mass of rMG_186 (28.56 kDa).
Nuclease activity of rMG_186.
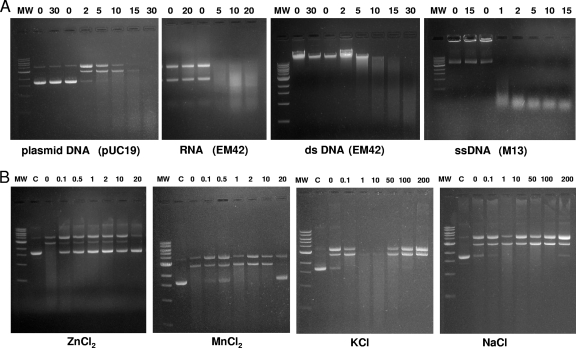
Since the MG_186 thermonuclease motif is predicted to require divalent cation Ca2+ (Fig. 1A), we initially monitored rMG_186 nuclease activity in the presence of 10 mM CaCl2 using plasmid DNA as the substrate. Under these conditions, plasmid DNA was digested completely within 30 min (Fig. 2 A). To further examine rMG_186 nuclease activity, we used different nucleic acids as substrates and determined enzymatic activity by analyzing patterns of degradation at different time points, as described in Materials and Methods. With CaCl2, single-strand DNA and RNA were degraded within 5 min, in contrast to plasmid DNA or double-strand DNA, which required a longer time frame, suggesting that MG_186 displays a preference for single-strand nucleic acids (Fig. 2A). However, in the absence of Ca2+, MG_186 did not exhibit nuclease activity. To determine whether other divalent cations were effective in activating rMG_186 nuclease activity, assays were performed in the presence of increasing concentrations of MnCl2 and ZnCl2. In contrast to Ca2+, the addition of Mn2+ or Zn2+ inhibited nuclease activity (Fig. 2B). Mg2+, a common divalent cation required by many nucleases, barely stimulated rMG_186 nuclease activity (data not shown). Thus, with plasmid DNA as the substrate, MG_186 shows a strong preference for Ca2+ over other divalent cations. As expected, Ca2+-stimulated nuclease activity was also inhibited by EGTA or EDTA (data not shown). It has been reported that salt concentrations in the reaction mixture can influence nuclease activity (7). Strikingly, we found that the Ca2+-mediated rMG_186 nuclease activity was stimulated to higher levels in the presence of 1 mM and 10 mM KCl (Fig. 2B), while KCl at 0.1 mM or above 50 mM did not enhance nuclease activity. A wide range of NaCl concentrations showed modest or no effect (Fig. 2B). Since MG_186 possesses the thermonuclease motif, the thermostablility of MG_186 was analyzed by performing nuclease assays at different temperatures (37 to 65°C) in the presence of Ca2+. Highest enzymatic activities were observed between 37°C to 55°C. Temperatures at 65°C and above completely abolished activity.
FIG. 2.
Nuclease activity of rMG_186. (A) Nuclease activity of rMG_186 on different nucleic acid substrates. The far left lane of each panel includes molecular weight markers (MW). The next two lanes of each panel represent nucleic acid samples (1 μg/lane) that were not treated with rMG_186. The subsequent lanes include rMG_186 in the presence of 10 mM CaCl2. Specific nucleic acid substrates (1 μg/lane) and incubation times in minutes are indicated. Individual sample reactions were arrested by adding EDTA followed by incubation at 70°C for 10 min. (B) Effect of other divalent cations and salts on nuclease activity of rMG_186. Plasmid DNA (1 μg) was treated with different concentrations (mM) of salts and divalent cations (indicated above each lane) in the presence of rMG_186 and 10 mM CaCl2. Reactions were stopped as described above. Test samples were resolved on 1% agarose gel along with untreated plasmid DNA (lanes designated C).
MG_186 expression and nuclease activity in M. genitalium.
To confirm that MG_186 was synthesized during M. genitalium growth in SP-4 medium, we examined mycoplasma cell lysates during log (24 h) to stationary (120 h) phases for nuclease activity. DNA-impregnated SDS-PAGE gels exhibited a clear and intense zone in all fractions around 28 kDa, suggesting that MG_186 is expressed at all mycoplasma growth points. No other region of the gel exhibited similar nuclease activity. As a representative of mycoplasma-associated nuclease activity, 72-h culture cell lysates are shown in Fig. 3 (lane 2). Interestingly, Coomassie blue staining of the same gel did not identify a prominent band around the clear zone region (Fig. 3, lane 1, arrow), suggesting that MG_186 is active, even at low levels of expression. To further demonstrate the enzymatic capacity of rMG_186, we examined a concentration range of purified rMG_186 (5 ng to 500 ng) and showed that even at 5 ng, strong nuclease activity was detected (Fig. 3, lane 3).
FIG. 3.
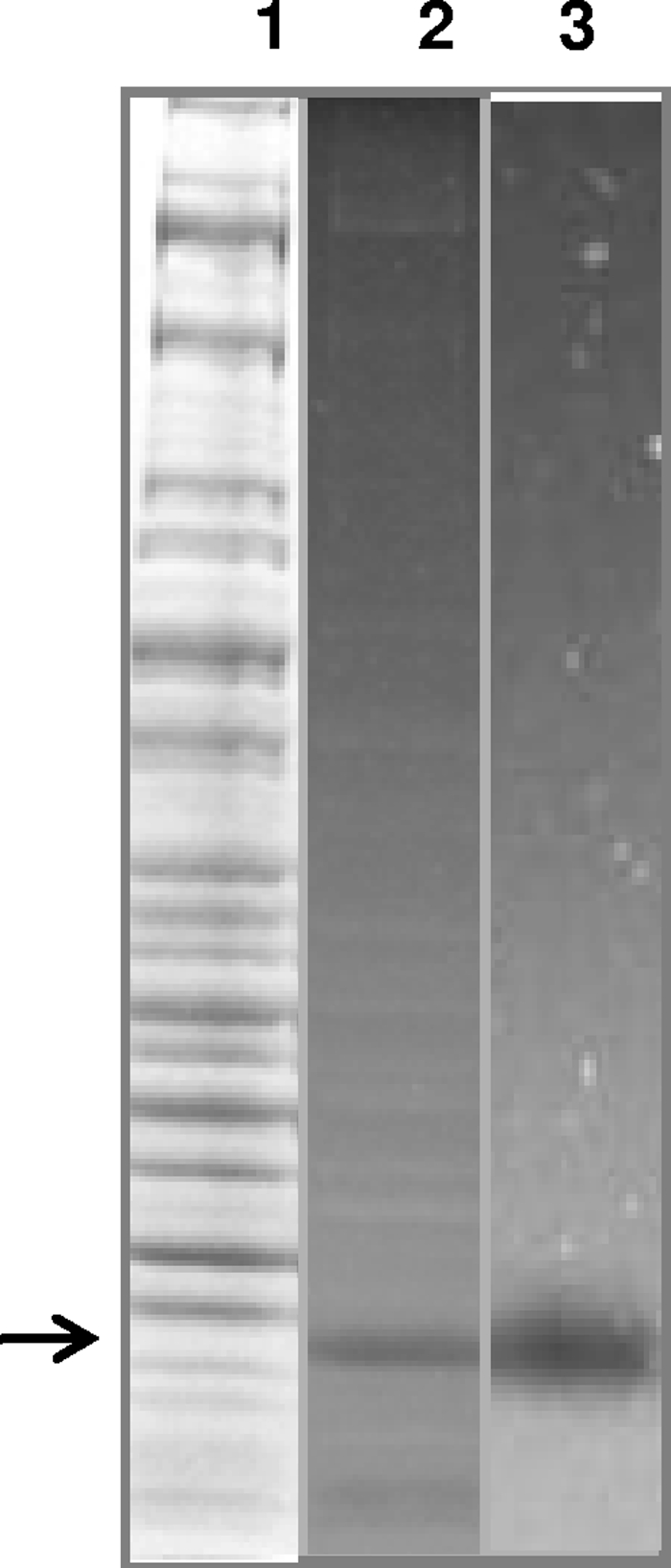
Zymogram analysis of nuclease activity of MG_186. Whole-cell lysates of M. genitalium were prepared by sonication, and samples were run on 10% SDS-PAGE gels impregnated with herring sperm DNA. Nuclease activity was detected as detailed in Materials and Methods. Purified rMG_186 was also analyzed for nuclease activity by zymogram analysis. Lane 1, Coomassie blue staining of whole-cell lysates of M. genitalium; lane 2, zymogram analysis of lane 1 before Coomassie blue staining; and lane 3, zymogram analysis of purified rMG_186.
Membrane-associated surface-exposed lipoprotein properties of MG_186.
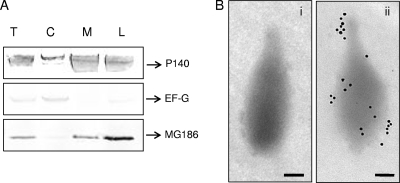
Prosite analysis predicted MG_186 to be a possible lipoprotein due to its signal peptide. To examine the distribution of MG_186 in M. genitalium, membrane fractions and lipid-associated membrane protein (LAMP) extracts were prepared and analyzed for the presence of MG_186. Using rabbit anti-rMG_186 serum, we showed by immunoblot analysis that MG_186 is membrane associated, like the major adhesin protein P140 of M. genitalium (Fig. 4 A). Although LAMP preparations showed slight contamination of elongation factor G, it was clear that MG_186 is enriched in the LAMP fraction, reinforcing the lipoprotein nature of MG_186. To further confirm the membrane location of MG_186, intact M. genitalium cells were treated with IgG-purified rabbit antiserum reactive against rMG_186 or with prebleed serum, followed by immunogold-labeled secondary antibodies and electron microscopy. As shown in Fig. 4B, panel ii, MG_186 is surface exposed and localized to the tip organelle and other regions of M. genitalium. Prebleed serum revealed no surface-labeling pattern. Overall, these data implicate MG_186 as a surface-associated lipoprotein.
FIG. 4.
MG_186 as a membrane-associated lipoprotein. (A) Localization of MG_186 in M. genitalium by immunoblotting membrane and LAMP fractions. Total cell lysates (T), cytoplasmic fractions (C), membrane fractions (M), and LAMP extracts (L) were resolved on 4 to 12% NuPAGE gradient gels and transferred to nitrocellulose membranes, and filter sections were cut into three pieces based on the expected protein sizes before exposure to specific antibodies (rabbit anti-P140, anti-EF-G, and anti-rMG_186). Adhesin P140 (140 kDa), the major surface-associated lipoprotein, served as the positive membrane control, and EF-G (77 kDa), a cytoplasmic protein, served as the internal cytosolic control. (B) Surface localization of MG_186 on intact M. genitalium cells by immunogold labeling. Intact M. genitalium cells were treated with prebleed rabbit serum (i) and anti-rabbit rMG_186 antiserum (ii) at a 1:100 dilution, followed by goat anti-rabbit IgG gold particles (20 nm; 1:20). Bars, 1 μm.
Neutralization of MG_186 nuclease activity by anti-MG_186 antibodies.
To investigate whether nuclease activity of rMG_186 can be neutralized by antibodies raised against rMG_186, rabbit IgG-purified anti-rMG_186 antibodies were preincubated with rMG_186, or as a negative control, with IgG-purified antibodies reactive against the M. pneumoniae CARDS toxin. Clearly, rMG_186 nuclease activity was negated by anti-MG_186 antibodies (Fig. 5, lane 4) and not by anti-CARDS toxin antibodies.
FIG. 5.
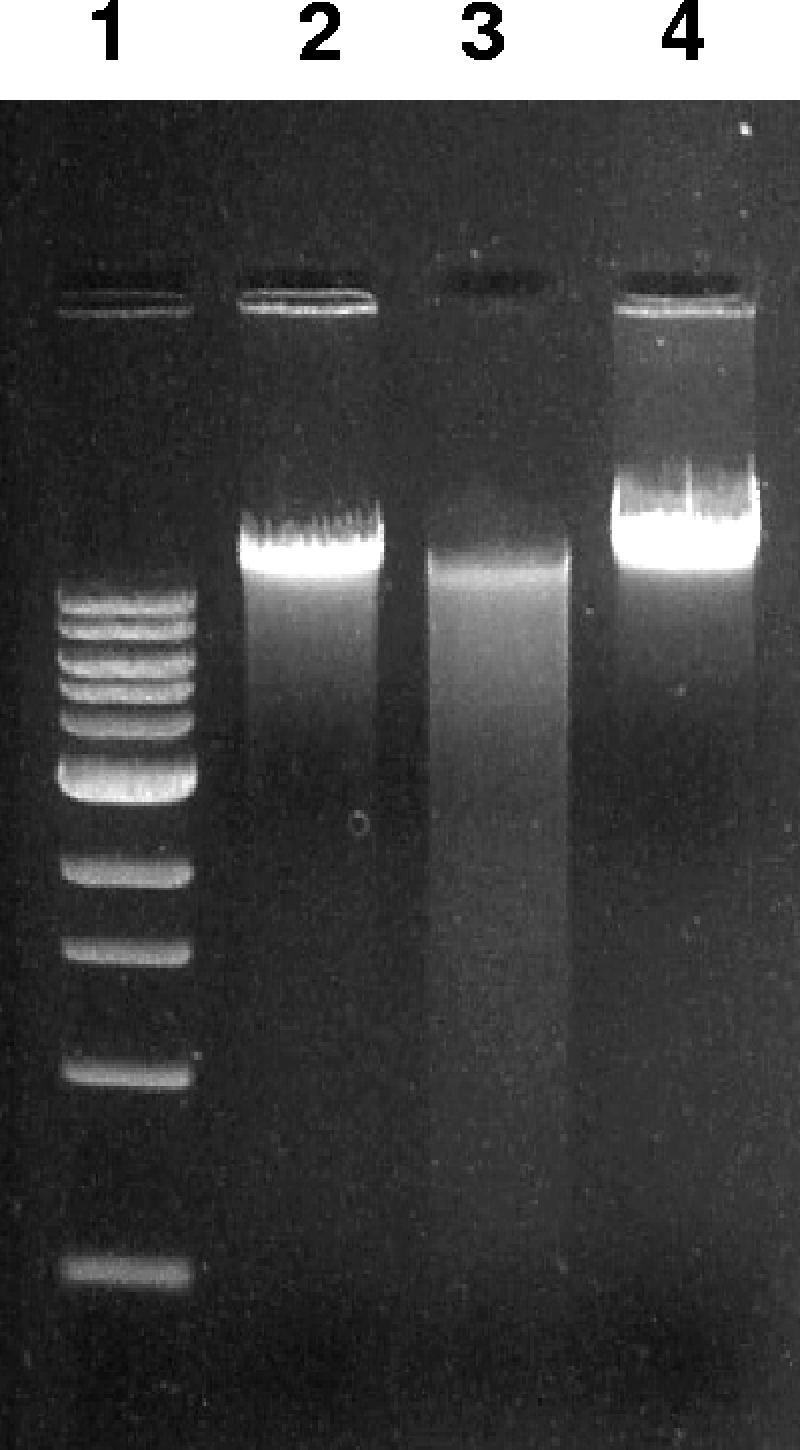
Neutralization of nuclease activity of rMG_186. rMG_186 was preincubated with anti-rMG_186 IgG or the irrelevant control (M. pneumoniae anti-IgG rCARDS toxin) at 37°C for 20 min. Then, 1 μg EM42 chromosomal DNA was added for 30 min, and nuclease activity was analyzed on agarose gels. Lane 1, molecular weight markers; lane 2, untreated EM42 DNA; lane 3, EM42 DNA after incubation with rMG_186 plus anti-rCARDS toxin IgG; and lane 4, EM42 DNA after incubation with rMG_186 plus anti-rMG_186 IgG.
Since MG_186 is surface associated and antibodies against rMG_186 neutralize its nuclease activity, we analyzed M. genitalium-infected patient sera for immunoreactivity against rMG_186. Infected patient sera exhibited weak to no response to rMG_186 (data not shown).
Induction of morphological and structural changes in the EM42 nucleus by rMG_186.
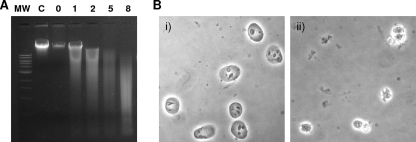
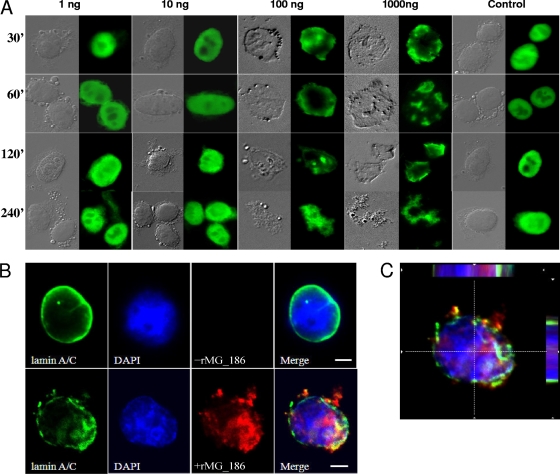
Since it was reported that mycoplasma nuclease activity leads to apoptosis and internucleosomal DNA fragmentation in contaminated mammalian cells (8, 29, 30), we evaluated the impact of rMG_186 on DNA degradation and the integrity of isolated and intact nuclei from EM42 cells. After 1 h of incubation at 37°C in the presence of rMG_186 and CaCl2, DNA degradation was apparent, which increased dramatically over time as high-molecular-weight DNA was fragmented to smaller sizes (Fig. 6 A). Ultimately, complete degradation of chromosomal DNA occurred. Using phase-contrast microscopy, we observed dramatic nuclear shrinkage in contrast to nuclei treated with buffer alone (Fig. 6B). We also stained MG_186-treated nuclei with the DNA binding fluorescence dye SYTOX and analyzed nuclear integrity by fluorescence microscopy. At low concentrations of rMG_186 (1 to 10 ng), we detected no differences between treated and untreated samples. However, rMG_186 concentrations between 100 and 1,000 ng revealed substantial variations in nuclear morphology. Further, the DNA appeared condensed, with plaque formation at the inner nuclear membrane or completely digested with resultant ghost nuclei (Fig. 7 A). In order to further characterize the structural changes in the morphology of EM42 nuclei treated with rMG_186, we evaluated the integrity of the nuclear membrane using lamin as a marker. As presented in Fig. 7B, rMG_186-treated nuclei showed disintegration of lamin compared to untreated controls. Interestingly, antibodies reactive against rMG_186 clearly indicate intranuclear localization of rMG_186, which is consistent with the observed nuclear DNA degradation (Fig. 7C and 6A).
FIG. 6.
Effect of rMG_186 on intact EM42 nuclei. Nuclei from EM42 mammalian cells were isolated, and the intactness of nuclear preparations was confirmed by phase contrast microscopy. (A) Agarose gel electrophoresis analysis of nuclear DNA after treatment with rMG_186. EM42 nuclei were incubated with 1 μg of rMG_186 in the presence of 10 mM CaCl2 and collected at different intervals (designated 1, 2, 5, or 8 [in hours] above each lane). DNA was isolated from control and treated samples and examined on agarose gels. Nuclei incubated with carrier buffer plus CaCl2 served as controls (C represents the 8-h control sample). (B) Morphological changes of untreated control and rMG_186-treated intact nuclei after 3 h. Phase contrast microscopy was used to examine nuclei treated with buffer alone (i) and nuclei treated with rMG_186 (ii).
FIG. 7.
Effect of rMG_186 on morphological changes in nuclei, DNA, and lamin organization. (A) Intact EM42 nuclei were incubated with 1 ng to 1 μg of rMG_186 for various time intervals (30 to 240 min). Differences in morphology of rMG_186-treated and untreated nuclei and their parallel SYTOX green staining are readily observed over time. (B) Lamin nuclear patterns after incubation with rMG_186. EM42 nuclei were treated with (bottom) or without (top) 100 ng of rMG_186 for 30 min and subjected to immunofluorescence using rabbit anti-rMG_186 and mouse anti-lamin antibodies. The upper panels (untreated control nuclei) indicate the normal organization of lamin in the intact nucleus (goat anti-mouse Alexa Flour 488 [green]). Also, DAPI staining (blue) reveals the undisturbed nuclear DNA. The lower panels (rMG_186-treated nuclei) indicate the disintegration of lamin topography and fragmented DNA (DAPI staining), also revealing chromatin condensation probably due to nuclease activity of rMG_186 (anti-rabbit Alexa Flour 633 [red]). (C) Intranuclear localization of rMG_186. A magnified optical cross section of the bottom merged panel of B clearly reveals the distribution of rMG_186 (red) at the nuclear membrane along with lamin (green) and within DNA (blue). Bars, 5 μm.
DISCUSSION
Nucleases are ubiquitous and implicated in a range of cellular functions, including replication, recombination, and DNA degradation and repair. For microbial pathogens, especially mycoplasmas, nuclease-mediated degradation of host nucleic acids is essential as a source of nucleotides for biosynthetic and survival purposes. Nucleases have been detected in numerous Mycoplasma species and are either membrane associated or secreted (7, 12, 26, 29, 31, 33). However, no functional M. genitalium nuclease has yet been identified, although it was recently reported that a homolog of M. hyopneumoniae, designated mhp379 (35), which is closely related to a family of bacterial thermostable nucleases, exists in M. genitalium. Our interest in the characterization of M. genitalium nuclease activity results from its intracellular behavior, streamlined genome, and absolute parasitic dependence. Earlier, we reported that human pathogenic mycoplasmas, including M. genitalium, are capable of establishing an intracellular niche and displaying a distinct tropism for the perinuclear region of human target cells (4, 13, 24, 41). This unusual cellular distribution was also reinforced in the clinical setting, where we detected intracellular M. genitalium cells located within vaginal cells of patients infected with M. genitalium (9). Remarkably, M. genitalium appears capable of intranuclear penetration and possibly even residence (41). How this unique and curious relationship between M. genitalium and host nuclei/DNA could have evolved is unknown, although it would require unusual biological and biochemical subtleties.
Our current study shows that MG_186 represents the newest member of a group of mycoplasma nucleases that share similar domain organizations and surface protein localizations. Also, the genomic organization of MG_186 indicates that several transporter genes are located immediately downstream of MG_186, similar to that of M. hyopneumoniae and other mycoplasmas (35). It is possible that these transporters are involved in nucleic acid precursor import (31). By combining genomic analysis with measurements of rMG_186 enzymatic activity, we reinforced nuclease properties of MG_186 (Fig. 2). Further analysis of the entire M. genitalium genome and nuclease activity of total M. genitalium cell lysates demonstrated the presence of only one nuclease, MG_186 (Fig. 3), in contrast to other mycoplasmas that possess several functional nucleases (26). Interestingly, the expression of MG_186 in M. genitalium cells is low, as measured by Coomassie blue-stained total protein gels, which is consistent with evidence that patients infected with M. genitalium do not mount a strong immune response to MG_186.
Like other mycoplasma nucleases (7, 29), MG_186 requires Ca2+ for optimal activity, while Mn2+ and Zn2+ inhibit activity (Fig. 2B). Further, Mg2+ does not stimulate MG_186 enzymatic activity in contrast to the major nuclease of M. penetrans (7). Cytosolic Ca2+ signals can be evoked in the host by bacterial infection, especially by toxins or chemical stimuli, such as hormones and growth factors and environmental changes (i.e., pH or temperature shifts). Depending on the host target cell and the nature and extent of the environmental stimulus, Ca2+ signals can be transient, oscillatory, or sustained (38) and can occur as cellular or subcellular events (10). It has been previously shown that treatment of animals with progesterone, which also leads to increases in intracellular calcium, is a prerequisite for the establishment of genital tract colonization by M. genitalium (19). Also, estradiol, which is known to reduce intracellular calcium, is ineffective in facilitating M. genitalium colonization (6, 19). Therefore, the impact of infection(s), hormonal changes, and other environmental stressors in the host will likely influence calcium levels and M. genitalium nuclease activity.
The detection of MG_186 in the Triton fraction of M. genitalium cell lysate (Fig. 4A), its surface-exposed distribution (Fig. 4B), and signal peptide prediction (Fig. 1) suggest that MG_186 functions as a surface-active protein, like the major adhesin P140. Since intracellular M. genitalium cells preferentially localize around the perinuclear region, it seems apparent that host DNA and RNA offer potential and essential nucleotide pools to intracellular mycoplasmas. As a consequence, M. genitalium competes with the host for important metabolic precursors. Therefore, the potent membrane nucleases of mycoplasmas, combined with other mycoplasma determinants, may be responsible for pathogenic effects and chromosomal aberrations observed in infected eukaryotic cells. For example, the morphological changes in the nuclei elicited by rMG_186 (Fig. 6B and 7A) are substantial, which prompted us to examine lamin distribution and stability. Lamins are nuclear intermediate filament proteins that maintain nuclear shape and mediate chromatin-nuclear membrane interactions. Lamin disintegration and its impact on the morphological changes of nuclei during apoptosis have been described (32). The abnormal ultrastructure and morphology of rMG_186-treated nuclei and the accompanying aberrant lamin appearance are consistent with nuclear events associated with condensation of chromatin and DNA degradation (Fig. 6 and 7). These observations correspond to reports that mycoplasma infection of mammalian cells or organ cultures or xenografts can lead to alterations in host nucleic acid metabolism and resultant chromosomal aberrations. Recently, M. genitalium infection has also been linked to malignant transformation of benign human prostate cells (28).
Consequently, it appears that MG_186 is a critical pathogenic contributor to M. genitalium colonization and persistence by providing essential nucleotide precursors for mycoplasma biosynthetic functions and replication while also competing with the host for nucleotide pools. These events likely trigger pathways to host cell death and subsequent pathological consequences. Therefore, therapeutic targeting of MG_186 could result in the interruption or prevention of mycoplasma-mediated inflammatory diseases and other complications in the urogenital tract.
Acknowledgments
This study was supported by award number U19AI045429 from the National Institute of Allergy and Infectious Diseases.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
We thank Rose Garza for her assistance in assembling the manuscript.
Footnotes
Published ahead of print on 16 July 2010.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian, S., T. R. Kannan, P. J. Hart, and J. B. Baseman. 2009. Amino acid changes in elongation factor Tu of Mycoplasma pneumoniae and Mycoplasma genitalium influence fibronectin binding. Infect. Immun. 77:3533-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baseman, J. B., S. F. Dallo, J. G. Tully, and D. L. Rose. 1988. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J. Clin. Microbiol. 26:2266-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baseman, J. B., M. Lange, N. L. Criscimagna, J. A. Giron, and C. A. Thomas. 1995. Interplay between mycoplasmas and host target cells. Microb. Pathog. 19:105-116. [DOI] [PubMed] [Google Scholar]
- 5.Baseman, J. B., and J. G. Tully. 1997. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg. Infect. Dis. 3:21-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beagley, K. W., and C. M. Gockel. 2003. Regulation of innate and adaptive immunity by the female sex hormones oestradiol and progesterone. FEMS Immunol. Med. Microbiol. 38:13-22. [DOI] [PubMed] [Google Scholar]
- 7.Bendjennat, M., A. Blanchard, M. Loutfi, L. Montagnier, and E. Bahraoui. 1997. Purification and characterization of Mycoplasma penetrans Ca2+/Mg2+-dependent endonuclease. J. Bacteriol. 179:2210-2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendjennat, M., A. Blanchard, M. Loutfi, L. Montagnier, and E. Bahraoui. 1999. Role of Mycoplasma penetrans endonuclease P40 as a potential pathogenic determinant. Infect. Immun. 67:4456-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blaylock, M. W., O. Musatovova, J. G. Baseman, and J. B. Baseman. 2004. Determination of infectious load of Mycoplasma genitalium in clinical samples of human vaginal cells. J. Clin. Microbiol. 42:746-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bootman, M. D., T. J. Collins, C. M. Peppiatt, L. S. Prothero, L. MacKenzie, P. De Smet, M. Travers, S. C. Tovey, J. T. Seo, M. J. Berridge, F. Ciccolini, and P. Lipp. 2001. Calcium signalling—an overview. Semin. Cell Dev. Biol. 12:3-10. [DOI] [PubMed] [Google Scholar]
- 11.Cotton, F. A., E. E. Hazen, Jr., and M. J. Legg. 1979. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3′,5′-bisphosphate-calcium ion complex at 1.5-A resolution. Proc. Natl. Acad. Sci. U. S. A. 76:2551-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowen, B. S., and S. C. Smith. 1972. Nuclease activities of Mycoplasma gallisepticum as a function of culture age in different media. J. Bacteriol. 109:21-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallo, S. F., and J. B. Baseman. 2000. Intracellular DNA replication and long-term survival of pathogenic mycoplasmas. Microb. Pathog. 29:301-309. [DOI] [PubMed] [Google Scholar]
- 14.Dallo, S. F., T. R. Kannan, M. W. Blaylock, and J. B. Baseman. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 46:1041-1051. [DOI] [PubMed] [Google Scholar]
- 15.Desai, N. N., E. A. Kennard, D. A. Kniss, and C. I. Friedman. 1994. Novel human endometrial cell line promotes blastocyst development. Fertil. Steril. 61:760-766. [PubMed] [Google Scholar]
- 16.Feng, S. H., and S. C. Lo. 1994. Induced mouse spleen B-cell proliferation and secretion of immunoglobulin by lipid-associated membrane proteins of Mycoplasma fermentans incognitus and Mycoplasma penetrans. Infect. Immun. 62:3916-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feng, S. H., and S. C. Lo. 1999. Lipid extract of Mycoplasma penetrans proteinase K-digested lipid-associated membrane proteins rapidly activates NF-κB and activator protein 1. Infect. Immun. 67:2951-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraser, C. M., J. D. Gocayne, O. White, M. D. Adams, R. A. Clayton, R. D. Fleischmann, C. J. Bult, A. R. Kerlavage, G. Sutton, J. M. Kelley, R. D. Fritchman, J. F. Weidman, K. V. Small, M. Sandusky, J. Fuhrmann, D. Nguyen, T. R. Utterback, D. M. Saudek, C. A. Phillips, J. M. Merrick, J. F. Tomb, B. A. Dougherty, K. F. Bott, P. C. Hu, T. S. Lucier, S. N. Peterson, H. O. Smith, C. A. Hutchison III, and J. C. Venter. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270:397-403. [DOI] [PubMed] [Google Scholar]
- 19.Furr, P. M., and D. Taylor-Robinson. 1993. Factors influencing the ability of different mycoplasmas to colonize the genital tract of hormone-treated female mice. Int. J. Exp. Pathol. 74:97-101. [PMC free article] [PubMed] [Google Scholar]
- 20.Jarvill-Taylor, K. J., C. VanDyk, and F. C. Minion. 1999. Cloning of mnuA, a membrane nuclease gene of Mycoplasma pulmonis, and analysis of its expression in Escherichia coli. J. Bacteriol. 181:1853-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kannan, T. R., and J. B. Baseman. 2006. ADP-ribosylating and vacuolating cytotoxin of Mycoplasma pneumoniae represents unique virulence determinant among bacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 103:6724-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kannan, T. R., D. Provenzano, J. R. Wright, and J. B. Baseman. 2005. Identification and characterization of human surfactant protein A binding protein of Mycoplasma pneumoniae. Infect. Immun. 73:2828-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Loll, P. J., and E. E. Lattman. 1989. The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+, and the inhibitor pdTp, refined at 1.65 A. Proteins 5:183-201. [DOI] [PubMed] [Google Scholar]
- 24.Mernaugh, G. R., S. F. Dallo, S. C. Holt, and J. B. Baseman. 1993. Properties of adhering and nonadhering populations of Mycoplasma genitalium. Clin. Infect. Dis. 17(Suppl. 1):S69-S78. [DOI] [PubMed] [Google Scholar]
- 25.Minion, F. C., and J. D. Goguen. 1986. Identification and preliminary characterization of external membrane-bound nuclease activities in Mycoplasma pulmonis. Infect. Immun. 51:352-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Minion, F. C., K. J. Jarvill-Taylor, D. E. Billings, and E. Tigges. 1993. Membrane-associated nuclease activities in mycoplasmas. J. Bacteriol. 175:7842-7847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell, A., and L. R. Finch. 1977. Pathways of nucleotide biosynthesis in Mycoplasma mycoides subsp. mycoides. J. Bacteriol. 130:1047-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Namiki, K., S. Goodison, S. Porvasnik, R. W. Allan, K. A. Iczkowski, C. Urbanek, L. Reyes, N. Sakamoto, and C. J. Rosser. 2009. Persistent exposure to Mycoplasma induces malignant transformation of human prostate cells. PLoS One 4:e6872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paddenberg, R., A. Weber, S. Wulf, and H. G. Mannherz. 1998. Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases. Cell Death Differ. 5:517-528. [DOI] [PubMed] [Google Scholar]
- 30.Paddenberg, R., S. Wulf, A. Weber, P. Heimann, L. A. Beck, and H. G. Mannherz. 1996. Internucleosomal DNA fragmentation in cultured cells under conditions reported to induce apoptosis may be caused by mycoplasma endonucleases. Eur. J. Cell Biol. 71:105-119. [PubMed] [Google Scholar]
- 31.Pollack, J. D., and P. J. Hoffmann. 1982. Properties of the nucleases of mollicutes. J. Bacteriol. 152:538-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rao, L., D. Perez, and E. White. 1996. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 135:1441-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Razin, S., A. Knyszynski, and Y. Lifshitz. 1964. Nucleases of Mycoplasma. J. Gen. Microbiol. 36:323-332. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki, Y., J. Ishikawa, A. Yamashita, K. Oshima, T. Kenri, K. Furuya, C. Yoshino, A. Horino, T. Shiba, T. Sasaki, and M. Hattori. 2002. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 30:5293-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt, J. A., G. F. Browning, and P. F. Markham. 2007. Mycoplasma hyopneumoniae mhp379 is a Ca2+-dependent, sugar-nonspecific exonuclease exposed on the cell surface. J. Bacteriol. 189:3414-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taniuchi, H., C. B. Anfinsen, and A. Sodja. 1967. The amino acid sequence of an extracellular nuclease of Staphylococcus aureus. 3. Complete amino acid sequence. J. Biol. Chem. 242:4752-4758. [PubMed] [Google Scholar]
- 37.Taylor-Robinson, D. 2002. Mycoplasma genitalium—an update. Int. J. STD AIDS. 13:145-151. [DOI] [PubMed] [Google Scholar]
- 38.Thomas, A. P., G. S. Bird, G. Hajnoczky, L. D. Robb-Gaspers, and J. W. Putney, Jr. 1996. Spatial and temporal aspects of cellular calcium signaling. FASEB J. 10:1505-1517. [PubMed] [Google Scholar]
- 39.Tully, J. G., D. L. Rose, J. B. Baseman, S. F. Dallo, A. L. Lazzell, and C. P. Davis. 1995. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J. Clin. Microbiol. 33:1851-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tully, J. G., D. Taylor-Robinson, R. M. Cole, and D. L. Rose. 1981. A newly discovered mycoplasma in the human urogenital tract. Lancet 1:1288-1291. [DOI] [PubMed] [Google Scholar]
- 41.Ueno, P. M., J. Timenetsky, V. E. Centonze, J. J. Wewer, M. Cagle, M. A. Stein, M. Krishnan, and J. B. Baseman. 2008. Interaction of Mycoplasma genitalium with host cells: evidence for nuclear localization. Microbiology 154:3033-3041. [DOI] [PubMed] [Google Scholar]