Abstract
In the diazotrophic filaments of heterocyst-forming cyanobacteria, an exchange of metabolites takes place between vegetative cells and heterocysts that results in a net transfer of reduced carbon to the heterocysts and of fixed nitrogen to the vegetative cells. Open reading frame alr2355 of the genome of Anabaena sp. strain PCC 7120 is the ald gene encoding alanine dehydrogenase. A strain carrying a green fluorescent protein (GFP) fusion to the N terminus of Ald (Ald-N-GFP) showed that the ald gene is expressed in differentiating and mature heterocysts. Inactivation of ald resulted in a lack of alanine dehydrogenase activity, a substantially decreased nitrogenase activity, and a 50% reduction in the rate of diazotrophic growth. Whereas production of alanine was not affected in the ald mutant, in vivo labeling with [14C]alanine (in whole filaments and isolated heterocysts) or [14C]pyruvate (in whole filaments) showed that alanine catabolism was hampered. Thus, alanine catabolism in the heterocysts is needed for normal diazotrophic growth. Our results extend the significance of a previous work that suggested that alanine is transported from vegetative cells into heterocysts in the diazotrophic Anabaena filament.
Cyanobacteria such as those of the genera Anabaena and Nostoc grow as filaments of cells (trichomes) that, when incubated in the absence of a source of combined nitrogen, present two cell types: vegetative cells that perform oxygenic photosynthesis and heterocysts that perform N2 fixation. Heterocysts carry the oxygen-labile enzyme nitrogenase, and, thus, compartmentalization is the way these organisms separate the incompatible activities of N2 fixation and O2-evolving photosynthesis (9). In Anabaena and Nostoc, heterocysts are spaced along the filament so that approximately 1 in 10 to 15 cells is a heterocyst. Heterocysts differentiate from vegetative cells in a process that involves execution of a specific program of gene expression (12, 15, 39). In the N2-fixing filament, the heterocysts provide the vegetative cells with fixed nitrogen, and the vegetative cells provide the heterocysts with photosynthate (38). Two important aspects of the diazotrophic physiology of heterocyst-forming cyanobacteria that are still under investigation include the actual metabolites that are transferred intercellularly and the mechanism(s) of transfer (10).
Because the ammonium produced by nitrogenase is incorporated into glutamate to produce glutamine in the heterocyst and because the heterocyst lacks the main glutamate-synthesizing enzyme, glutamine(amide):2-oxoglutarate amino transferase (GOGAT; also known as glutamate synthase), a physiological exchange of glutamine and glutamate resulting in a net transfer of nitrogen from the heterocysts to the vegetative cells has been suggested (21, 36, 37). On the other hand, a sugar is supposed to be transferred from vegetative cells to heterocysts. Because high invertase activity levels are found in the heterocysts (34) and because overexpression of sucrose-degrading sucrose synthase in Anabaena sp. impairs diazotrophic growth (4), it is possible that sucrose is a transferred carbon source. Indeed, determination of 14C-labeled metabolites in heterocysts isolated from filaments incubated for short periods of time with [14C]bicarbonate identified sugars and glutamate as possible compounds transferred from vegetative cells to heterocysts (13). However, this study also identified alanine as a metabolite possibly transported from vegetative cells to heterocysts.
The cyanobacteria bear a Gram-negative type of cell envelope, carrying an outer membrane (OM) outside the cytoplasmic membrane (CM) and the peptidoglycan layer (9, 15). In filamentous cyanobacteria, whereas the CM and peptidoglycan layer surround each cell, the OM is continuous along the filament, defining a continuous periplasmic space (10, 19). In Anabaena sp. strain PCC 7120, the OM is a permeability barrier for metabolites such as glutamate and sucrose (27). Two possible pathways for intercellular molecular exchange in heterocyst-forming cyanobacteria have been discussed: the periplasm (10, 19) and cell-to-cell-joining proteinaceous structures (11, 22, 25). Whereas the latter would mediate direct transfer of metabolites between the cytoplasm of adjacent cells, the former would require specific CM permeases to mediate metabolite transfer between the periplasm and the cytoplasm of each cell type (10).
In Anabaena sp. strain PCC 7120, two ABC-type amino acid transporters have been identified that are specifically required for diazotrophic growth (29, 30). The N-I transporter (NatABCDE), which shows preference for neutral hydrophobic amino acids, is present exclusively in vegetative cells (30). The N-II transporter (NatFGH-BgtA), which shows preference for acidic and neutral polar amino acids, is present in both vegetative cells and heterocysts (29). A general phenotype of mutants of neutral amino acid transporters in cyanobacteria is release into the culture medium of some hydrophobic amino acids, especially alanine (16, 23, 24), which is accumulated at higher levels in the extracellular medium of cultures incubated in the absence than in the presence of a source of combined nitrogen (30).
Thus, alanine is a conspicuous metabolite in the diazotrophic physiology of heterocyst-forming cyanobacteria, and the possibility that it moves in either direction between heterocysts and vegetative cells has been discussed (13, 29, 30). Alanine dehydrogenase, which catalyzes the reversible reductive amination of pyruvate, has been detected in several cyanobacteria (8). In Anabaena spp., alanine dehydrogenase has been found at higher levels or exclusively in diazotrophic cultures (26), and in the diazotrophic filaments of Anabaena cylindrica it is present at higher levels in heterocysts than in vegetative cells (33). Open reading frame (ORF) alr2355 of the Anabaena sp. strain PCC 7120 genome is predicted to encode an alanine dehydrogenase (14). In this work we addressed the expression and inactivation of alr2355, identifying it as the Anabaena ald gene and defining an important catabolic role for alanine dehydrogenase in diazotrophy.
MATERIALS AND METHODS
Strains and growth conditions.
Anabaena sp. strain PCC 7120 (also known as Nostoc sp. strain PCC 7120) was grown in BG11 medium (containing ferric citrate instead of ferric ammonium citrate [32] and NaNO3 as the nitrogen source), BG110 medium (free of combined nitrogen), or BG110 medium with ammonium (BG110 medium containing 3 to 5 mM NH4Cl and, respectively, 6 to 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer, pH 7.5) at 30°C in the light (25 μE m−2 s−1) in shaken (100 rpm) liquid cultures or in medium solidified with 1% Difco agar. Alternatively, cultures (referred to as bubbled cultures and denoted BG11C or BG110C) were supplemented with 10 mM NaHCO3 and bubbled with a mixture of air and 1% (vol/vol) CO2 in the light (75 μE m−2 s−1). In this case, the ammonium-containing medium was supplemented with 8 mM NH4Cl and 16 mM TES-NaOH buffer (pH 7.5). For the mutants described below, antibiotics were used at the following concentrations: 2 to 5 μg ml−1 streptomycin (Sm), 2 to 5 μg ml−1 spectinomycin (Sp), and 10 to 20 μg ml−1 neomycin (Nm) for liquid cultures; and 5 μg ml−1 Sm, 5 μg ml−1 Sp, and 40 μg ml−1 Nm for solid cultures. DNA was isolated from Anabaena sp. by the method of Cai and Wolk (2).
Escherichia coli DH5α was used for plasmid constructions. This strain and strains HB101 and ED8654, used for conjugations with Anabaena sp., were grown in LB medium, supplemented when appropriate with antibiotics at standard concentrations (1).
Strain construction.
To produce a green fluorescent protein (GFP) fusion to the N terminus of the Ald protein (Ald-N-GFP), a 460-bp fragment from the 5′ and upstream regions of alr2355 was amplified by PCR using oligodeoxynucleotide primers alr2355-7120-5 and alr2355-7120-6 (which contain a ClaI and an EcoRV restriction site, respectively) (Table 1) and DNA from strain PCC 7120 as a template. This fragment was cloned in vector pMBL-T (Dominion MBL) and transferred as a ClaI/EcoRV-ended fragment (resulting from a partial EcoRV restriction reaction) to ClaI/EcoRV-digested pCSEL21 (28), producing a fusion of the gfp-mut2 gene (3) to the first codon of alr2355. The resulting fusion was finally transferred as an EcoRI-ended fragment to EcoRI-digested pCSV3, producing pCSR100 (Smr Spr). To inactivate the ald gene, an internal 545-bp fragment of alr2355 was amplified by PCR using primers alr2355-7120-1 and alr2355-7120-2 (which contain BamHI restriction sites in their 5′ ends) and DNA from strain PCC 7120 as a template. The amplified fragment was cloned into vector pMBL-T and transferred to BamHI-digested pCSV3 (30) producing pCSR88 (Smr/Spr) or to pRL424 (7) producing pCSR84 (Nmr).
TABLE 1.
Oligodeoxynucleotide primers used in this worka
| Primer | Sequence (5′→ 3′) |
|---|---|
| alr1004-7120-3 | CCT CTA GAA GTT CCC TCC |
| alr1004-7120-4 | GTC AGG GTT GGT AGT CTG |
| alr2355-7120-1 | GGA TCC GGA GTC GAG AGT TAG TAG |
| alr2355-7120-2 | GGA TCC CTC TGC GTC CTA GTA CC |
| alr2355-7120-3 | CCA CAA TAC GGA AGT GTC |
| alr2355-7120-4 | GCG TGT AAG GTT TCC AC |
| alr2355-7120-5 | GGT GGC ATA ATC GAT GAC TAC C |
| alr2355-7120-6 | GGA ACG CCG ATA TCC ATT TAA CGC |
| alr2355-7120-gfp | GGA GGT AAG AGG CTG GAG AG |
| gfp-4 | CAA GAA TTG GGA CAA CTC C |
Introduced restriction enzyme cutting sites are indicated in boldface.
Conjugation of Anabaena sp. strain PCC 7120 or CSX60 (Smr/Spr) (30) with E. coli HB101 carrying pCSR84, pCSR88, or pCSR100 with helper and methylation plasmid pRL623 was effected by the conjugative plasmid pRL443, carried in E. coli ED8654, and performed as described previously (6) with selection for resistance to Sm/Sp or Nm. The genetic structure of selected clones was studied by PCR with DNA from these clones and the primer pair alr2355-7120-3 and alr2355-7120-4 for the insertion mutants (strains CSR24 and CSX60-R24) and, for the Ald-N-GFP fusion strain (CSR30), the pair alr2355-7120-gfp and alr2355-7120-4 and the pair alr2355-7120-gfp and gfp-4.
Confocal microscopy.
Samples from cultures of Anabaena sp. set atop solidified medium were visualized using a Leica HCX Plan-Apo 63× (1.4 numerical aperture [NA]) oil immersion objective attached to a Leica TCS SP2 confocal laser scanning microscope. GFP was excited using 488-nm irradiation from an argon ion laser. Fluorescent emission was monitored by collection across windows of 500 to 538 nm (GFP imaging) and 630 to 700 nm (cyanobacterial autofluorescence).
Growth rates and nitrogenase activity.
The growth rate constant (μ = ln2/td, where td is the doubling time) was calculated from the increase of protein content, determined in 0.2-ml samples of shaken liquid cultures (23). Protein concentration was determined by a modified Lowry procedure (20). Chlorophyll a (Chl) content of cultures was determined by the method of Mackinney (18). Nitrogenase activity was determined under oxic conditions by the acetylene reduction assay as described previously (23). For determination of both the growth rate and nitrogenase activity, filaments grown in BG11 medium (in the presence of antibiotics for the mutant) were washed with BG110 medium and used to inoculate BG110 medium without antibiotics. An amount of filaments corresponding to 0.2 and 1 μg of Chl ml−1 was used to inoculate the cultures for the growth rate and nitrogenase determinations, respectively.
Alanine dehydrogenase activity.
Filaments (corresponding to about 5 to 10 mg of Chl) from 48-h bubbled BG110C cultures were harvested and resuspended in 50 mM Tris-HCl (pH 7.5), 10 mM EDTA, 5 mM β-mercaptoethanol, and a 1 mM concentration of the protease inhibitor phenylmethylsulfonyl fluoride (from Sigma). The suspension was then passed three times through a French pressure cell at 20,000 lb/in2 and centrifuged at 35,000 × g for 20 min at 4°C. The obtained supernatant was used to determine alanine dehydrogenase according to the method of Rowell and Stewart (33). The reaction mixture contained the following in a 1-ml volume: 100 mM Tris-HCl (pH 8), 1 mM Na-pyruvate, 133 mM NH4Cl, 100 μM NADH, and cell extract containing 0.1 to 0.5 mg of protein. The reaction was started by addition of NADH, and the mixture was incubated at room temperature (about 25°C). Oxidation of NADH was monitored following the decrease in A340 for 5 min. Activity is expressed as nmol of NADH oxidized per mg of protein per min.
Determination of extracellular amino acids.
Extracellular amino acids were determined in the medium of bubbled BG110C cultures incubated for 24 h and 48 h at 30°C in the light (75 to 100 μE m−2 s−1). Culture samples of 10 ml were filtered, the filtrates were lyophilized, and the resulting powder was resuspended in 0.25 to 0.5 ml of 0.1 M HCl. Samples from these preparations were used for high-performance liquid chromatography (HPLC) analysis as previously described (29). The amino acids detected with this method were Asp, Glu, Gln, Asn, Ser, Gly, His, Thr, Ala, Arg, Pro, Tyr, Val, Met, Cys, Ile, Leu, Phe, and Lys.
Alanine and pyruvate metabolism.
Filaments grown for 48 h in bubbled BG110C cultures were harvested, washed, and suspended in 25 mM N-tris(hydroxymethyl)-methylglycine (Tricine)-NaOH buffer (pH 8.1) for uptake assays. For isolation of heterocysts, filaments incubated for 48 h in bubbled BG110C cultures were harvested, washed, and resuspended in buffer 1 containing 50 mM imidazole and 0.5 mM EDTA (pH 7.5). The filaments were then broken by passage through a French pressure cell at 3,000 lb/in2. Samples were enriched in heterocysts after successive steps of centrifugation (200 × g for 10 min at room temperature) and washing with buffer 1. Purity of heterocyst preparations was assessed by microscopy, and the heterocysts were finally suspended in Tricine buffer. No differences between the heterocysts isolated from strains PCC 7120 and CSR24 were apparent. The uptake assays, with an amount of filaments corresponding to about 10 μg of Chl ml−1 or heterocysts corresponding to about 100 μg of protein ml−1, were carried out at 30°C in the light (200 μE m−2 s−1) with 3 μM l-[U-14C]alanine or 10 μM [1-14C]pyruvate (both from GE Healthcare, United Kingdom) for 10 or 60 min, as indicated below.
The filaments or isolated heterocysts from a 1-ml sample of uptake assay suspension were collected by filtration and washed with 5 to 10 ml of Tricine buffer. The filter was immediately subjected to boiling in 2 ml of H2O for 5 min, and the resulting suspension was centrifuged for 5 min at 1,400 × g to sediment the cell debris. A 1.5-ml sample of the supernatant (water-soluble extract) was lyophilized, and the resulting material was resuspended in 20 μl of H2O. Samples (3 to 5 μl) from this concentrated material were subjected to thin-layer chromatography (TLC) in 0.1-mm cellulose plates (20 by 20 cm; Merck). The TLC was run with the following solvents: first dimension, n-butanol-acetone-ammonium hydroxide-water (20:20:10:4, vol/vol/vol/vol); second dimension, isopropanol-formic acid-water (20:1:5, vol/vol/vol). Radioactive spots in the plates were visualized and quantified with a Cyclone storage phosphor system (Packard). Cochromatography with stable amino acids, which were visualized with ninhydrin (31) or with labeled pyruvate, was used to identify spots.
RESULTS
Expression of Ald-N-GFP.
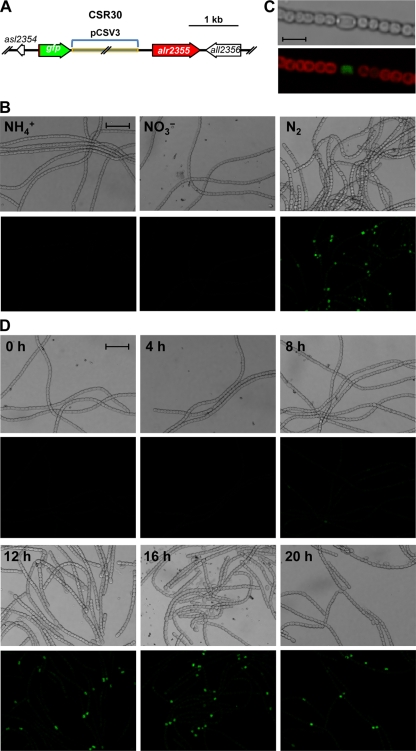
To investigate the culture conditions and cell specificity of expression of the ald gene in the filaments of Anabaena sp. strain PCC 7120, a strain (CSR30) carrying a fusion of the gfp-mut2 gene to the 5′ terminus of ald was prepared (Fig. 1A) (see Materials and Methods). In this construct, the gfp-mut2 is expressed from the transcription and translation signals of ald. Whereas in filaments of strain CSR30 grown with ammonium or incubated for 24 h with nitrate as the nitrogen source the GFP fluorescence was undetectable, in filaments incubated for 24 h without a source of combined nitrogen, the GFP signal was clearly seen localized to particular cells (Fig. 1B) which, as shown by a further magnification, correspond to heterocysts (Fig. 1C).
FIG. 1.
Expression of an Ald-N-GFP fusion protein. (A) Schematic of the ald (alr2355) locus in strain CSR30 carrying the ald-gfp-mut2 fusion. Note that the fusion gene was incorporated into the chromosome through single recombination between strain PCC 7120 DNA cloned in the plasmid and the corresponding chromosomal region (i.e., the promoter region of the gene), and therefore an intact copy of the ald gene remains. (B) Bright-field (top panels) and GFP fluorescence (bottom panels) micrographs of filaments of strain CSR30 grown with ammonium (NH4+) or grown with ammonium and incubated with nitrate (NO3−) or no source of combined nitrogen (N2) for 24 h. Scale bar, 20 μm. (C) Increased magnification bright-field (top) and cyanobacterial autofluorescence/GFP fluorescence overlay (bottom) micrographs of strain CSR30 incubated without added nitrogen as above. Scale bar, 5 μm. (D) Bright-field (top panels) and GFP fluorescence (bottom panels) micrographs of filaments of strain CSR30 grown with ammonium and incubated without a source of combined nitrogen for the time periods indicated. Scale bar, 20 μm.
To study ald expression during differentiation, ammonium-grown filaments of strain CSR30 were subjected to N step-down, and the GFP fluorescence was checked every 4 h. Whereas some GFP signal could be observed at 8 h, strong signals localized to presumptive proheterocysts were observed at 12 h and later times (Fig. 1D). Because under our culture conditions heterocyst differentiation is complete 20 to 24 h after N step-down, ald expression appears to be activated at an intermediate step of the differentiation process.
Construction and phenotype of Anabaena ald mutants.
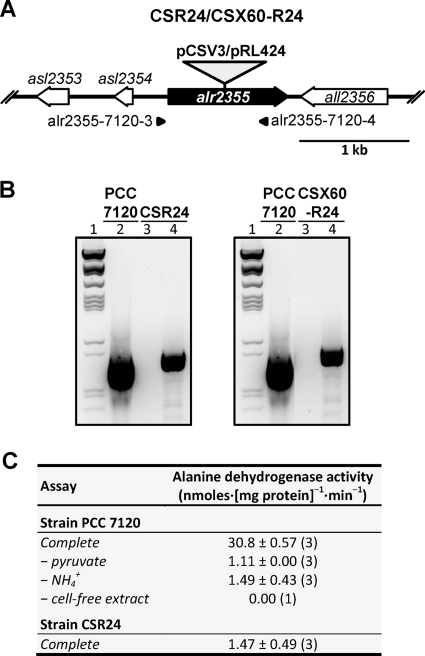
To investigate the role of alanine dehydrogenase in the heterocysts and in the diazotrophic physiology of Anabaena sp. strain PCC 7120, an ald mutant was constructed by insertion of a plasmid into the gene (Fig. 2A) (see Materials and Methods). The ald::pCSV3 mutant, strain CSR24, was homozygous for mutant chromosomes (Fig. 2B) and did not show alanine dehydrogenase activity (Fig. 2C), confirming the identity of alr2355 as the Anabaena ald gene.
FIG. 2.
Insertional inactivation of the ald gene. (A) Schematic of the ald (alr2355) locus with indication of the plasmids inserted to produce strains CSR24 and CSX60-R24. The approximate positions of the primers used to check segregation of the mutant chromosomes by PCR are indicated. (B) PCR analysis of the genetic structure in the ald region in strains PCC 7120, CSR24, and CSX60-R24. Electrophoresis gels show the following: lanes 1, λ DNA digested with ClaI; lanes 2, products of PCR with DNA from strain PCC 7120 and primers alr2355-7120-3 and alr2355-7120-4; lanes 3, products of PCR with DNA from strain CSR24 (left panel) or CSX60-R24 (right panel) and primers alr2355-7120-3 and alr2355-7120-4; lanes 4, products of control PCR with DNA from strain CSR24 (left panel) or CSX60-R24 (right panel) and primers alr1004-7120-3 and alr1004-7120-4, which amplify an unrelated chromosomal region. The expected amplification products are as follows: for alr2355-7120-3 with alr2355-7120-4, 910 bp (PCC 7120), 4,957 bp (CSR24), and 4,017 bp (CSX60-R24); for alr1004-7120-3 with alr1004-7120-4, 907 bp (note that the large fragments of 4,957 and 4,017 bp were not produced in the PCRs performed). (C) Alanine dehydrogenase activity in cell extracts of strains PCC 7120 and CSR24, determined as described in Materials and Methods (values are means and standard deviations; the number of assays is in parentheses). The background level of NADH oxidation observed with the extract of the mutant is similar to the level observed with the wild-type extract without pyruvate or ammonium and, therefore, should correspond to activity of other NADH oxidases present in the extracts.
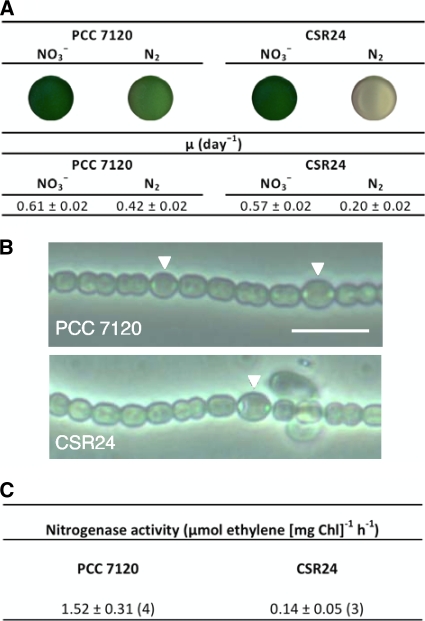
Growth of strain CSR24 was significantly impaired specifically under diazotrophic conditions. Whereas in nitrate-containing medium the mutant showed a growth rate constant similar to that of the wild type, in BG110 medium (lacking any source of combined nitrogen) the growth rate constant was about 50% for the mutant compared to that of the wild type (Fig. 3A). This result indicates that the heterocyst-expressed ald gene has a specific role in diazotrophy. Nonetheless, diazotrophic filaments of strain CSR24 contained heterocysts with a normal appearance, including the presence of cyanophycin-containing polar granules (Fig. 3B). These filaments showed a low but consistent nitrogenase activity (determined by the acetylene reduction assay) of about 10% of that observed in the wild type (Fig. 3C).
FIG. 3.
Phenotype of the ald mutant. (A) Growth characteristics. Filament suspensions (2 ml) of strains PCC 7120 and CSR24 grown for 9 days in BG11 (NO3−) or BG110 (N2) medium were loaded into 3-ml wells and photographed to show the appearance of the cultures (top). The growth rate constants (μ) of strains PCC 7120 and CSR24 in BG11 (NO3−) or BG110 (N2) medium are shown (bottom). Values correspond to the means and standard deviations of the means of the data from four independent experiments. (B) Phase-contrast micrographs of filaments of strains PCC 7120 and CSR24 incubated in BG110 medium for 44 h. Triangles point to heterocysts, in which polar granules are evident. Scale bar, 10 μm. (C) Nitrogenase activity determined by the acetylene reduction assay in suspensions of filaments of strains PCC 7120 and CSR24 incubated in BG110 medium for 44 h. Values are the means and standard deviations of the means of the number of determinations (with independent cultures) indicated in parentheses.
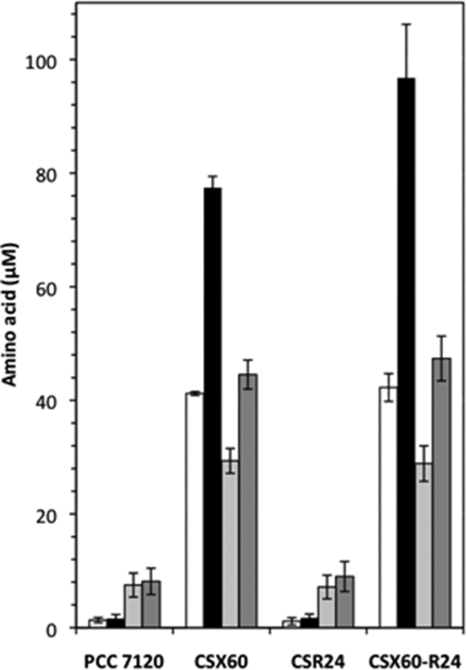
As mentioned above, cyanobacterial mutants of the neutral amino acid transporters characteristically release hydrophobic amino acids that accumulate in the culture medium, with alanine accumulating at the highest levels. To test whether alanine dehydrogenase has any role in this production of alanine, an ald mutant was constructed on an natA background. (The natA gene encodes an ATPase subunit of the N-I system [30]). For this, an ald::pRL424 construct was transferred into strain CSX60 (natA::C.S3) producing strain CSX60-R24 (Fig. 2A) (see Materials and Methods), which was homozygous for the mutant chromosomes (Fig. 2B). Figure 4 shows an analysis of the accumulation of amino acids in medium without a source of combined nitrogen. Strain CSX60-R24 produced hydrophobic amino acids at levels similar to those produced by strain CSX60, and specifically production of alanine was about 25% higher in the double mutant at 48 h of incubation. Therefore, alanine dehydrogenase is not required for the synthesis of alanine that is released by the natA mutant.
FIG. 4.
Amino acids in the culture medium of Anabaena sp. strains PCC 7120 (wild type), CSX60 (natA::C.S3), CSR24 (ald::pCSV3), and CSX60-R24 (natA::C.S3 ald::pRL424). Filaments grown in bubbled BG11C medium (with antibiotics for the mutants) were inoculated at 5 μg of Chl ml−1 and incubated in bubbled cultures of BG110C medium without antibiotics. Amino acids in the extracellular medium were analyzed by HPLC as described in Materials and Methods. White and black bars, alanine concentrations after 24 h and 48 h of incubation, respectively; light gray and dark gray bars, concentrations of Ile, Leu, Phe, Tyr, and Val combined after 24 h and 48 h of incubation, respectively. Amino acids other than those shown in the figure or Pro, which reached a concentration of 5.5 and 8.3 μM in strains CSX60 and CSX60-R24, respectively, after 48 h of incubation, were found at concentrations of <2 μM. Values are the means and standard deviations of the means (error lines) of data from three independent experiments.
Alanine and pyruvate metabolism.
Filaments and isolated heterocysts of Anabaena sp. strain PCC 7120 take up alanine efficiently (13, 29, 30). Metabolism of alanine was studied in the wild type and in strain CSR24 by means of TLC analysis of radioactive compounds produced in filaments or isolated heterocysts incubated in the presence of l-[U-14C]alanine. The most highly labeled compounds were amino acids, on which we centered our analysis (Table 2). In wild-type whole filaments, alanine accounted for 27.9% of total label after 10 min of incubation, showing that a significant fraction of the alanine taken up was metabolized, and most label in metabolic products accumulated in glutamate (27.5%) and aspartate (6.7%). After 60 min of incubation, only 8.5% of the label remained as alanine and 39.7% was found as glutamate. In whole filaments of the ald mutant, strain CSR24, alanine accounted for 81% of the label after 10 min of incubation, with only 6.6% of the label accumulating in glutamate (Table 2). This result indicates that inactivation of ald largely reduces alanine metabolism, suggesting a catabolic role for alanine dehydrogenase under the tested conditions. Nonetheless, in the ald mutant, the amount of label found as alanine after 60 min of incubation decreased to 67.2%, suggesting the presence of alternative catabolic possibilities.
TABLE 2.
Metabolism of [14C]alanine and [14C]pyruvate in Anabaena sp. strains PCC 7120 (wild type) and CSR24 (ald::pCSV3)
| Labeled substrate, sample type, and labeled product | Distribution (%) of radioactivity by incubation period and straina |
|||
|---|---|---|---|---|
| 10 min |
60 min |
|||
| PCC 7120 | CSR24 | PCC 7120 | CSR24 | |
| l-[U-14C]alanine | ||||
| Filaments | ||||
| Ala | 27.9 | 81.0 | 8.5 | 67.2 |
| Asp | 6.7 | 0.6 | 3.1 | 0.4 |
| Gln | 3.2 | 0.4 | 2.2 | 0.6 |
| Glu | 27.5 | 6.6 | 39.7 | 7.3 |
| Pyr | 1.3 | 0.4 | 8.0 | 2.2 |
| Origin | 14.1 | 4.3 | 11.5 | 5.9 |
| Heterocysts | ||||
| Ala | 70.2 | 93.7 | 24.1 | 83.4 |
| Asp | 3.4 | 0.3 | 4.1 | 0.4 |
| Gln | 6.8 | 0.8 | 14.8 | 2.4 |
| Glu | 4.2 | 0.6 | 8.4 | 1.5 |
| Pyr | 0.0 | 0.0 | 0.5 | 0.0 |
| Origin | 8.2 | 2.4 | 18.0 | 4.7 |
| [1-14C]pyruvate | ||||
| Filaments | ||||
| Ala | 5.1 | 29.3 | 1.9 | 15.5 |
| Asp | 11.6 | 3.4 | 5.5 | 1.5 |
| Gln | 0.2 | 0.1 | 1.3 | 0.7 |
| Glu | 3.3 | 0.8 | 12.5 | 3.0 |
| Pyr | 17.5 | 7.5 | 21.7 | 16.7 |
| Origin | 15.6 | 12.6 | 35.4 | 24.9 |
Values correspond to the percentage of radioactivity associated to the indicated compound (100 % corresponds to the sum of radioactivity in spots visualized in the TLC plates). The percentage of radioactivity in pyruvate may be underestimated because this compound is somewhat volatile (13). For filaments of CSR24 incubated with [1-14C]pyruvate for 60 min, significant label was also found in Ser (4.1 %) and Gly (2.0 %). One unidentified spot accounting for about 6 % of the radioactivity was observed for both time points in whole filaments of either strain incubated with l-[U-14C]alanine, and two other unidentified spots accounting together for about 9 % of the radioactivity were observed in wild-type filaments incubated with l -[U-14C]alanine for 60 min. Lack of reaction with ninhydrin indicated that none of these unidentified spots was an amino acid. All other observed spots were less than 2 % of the total radioactivity in any of the analyzed TLC plates.
In isolated heterocysts of the wild type, alanine was also subjected to catabolism, decreasing its labeling from 70.2% at 10 min incubation to 24.1% at 60 min of incubation (Table 2), and glutamine was the most abundantly labeled metabolic product. A low but still significant production of glutamate was deleted, whereas aspartate was labeled to a similar extent as in the whole filaments. In the heterocysts isolated from the ald mutant, alanine accounted for 93.7% and 83.4% of the label after 10 min and 60 min of incubation, respectively, indicating that alanine metabolism was largely hampered and suggesting, again, an important catabolic role of alanine dehydrogenase.
In contrast to alanine, pyruvate is taken up efficiently by whole filaments but not by isolated heterocysts of Anabaena sp. strain PCC 7120 (13; also our unpublished results). We therefore could check pyruvate metabolism reliably only in whole filaments. Label from [1-14C]pyruvate was more widely distributed than label from l-[U-14C]alanine, but other than a spot that could correspond to pyruvate itself (which accounted for 7.5 to 21.7% of the label) and the radioactivity that remained in the origin of the chromatography (that was more than that observed with l-[U-14C]alanine), significant label was again mainly found in amino acids. In the wild type, after both 10 min and 60 min of incubation, labeled alanine, glutamate, and aspartate were clearly observed (Table 2). In the ald mutant, the label significantly increased in alanine and decreased in glutamate and aspartate compared to levels in the wild type. This result indicates that, in addition to being subject to catabolism, pyruvate is used as a substrate for the biosynthesis of alanine, the catabolism of which is impaired in the ald mutant, as described above.
DISCUSSION
ORF alr2355 is the ald gene encoding alanine dehydrogenase in Anabaena sp. strain PCC 7120. The gene is not essential for growth, but an ald insertional mutant is impaired specifically in diazotrophic growth. Because, as shown with a GFP fusion, the gene is expressed mainly in differentiating and mature heterocysts, a specific role of alanine dehydrogenase in these differentiated cells can be suggested. Induction of this gene in response to nitrogen deprivation has also been observed in microarray analysis of gene expression in Anabaena sp. strain PCC 7120, but in this analysis an earlier induction time was detected (5, 39). Because it is possible that a certain amount of GFP has to accumulate to give an appreciable fluorescence signal, the difference in detection times for mRNA and GFP fluorescence may indicate a relatively low expression level of ald.
Although biosynthesis of alanine that could be mediated by alanine dehydrogenase has been observed in Anabaena filaments incubated with l-methionine-dl-sulfoximine, an inhibitor of glutamine synthetase that promotes accumulation of ammonium in the cells (37), our results indicate a catabolic role for alanine dehydrogenase that is consistent with the known catalytic properties of the enzyme. In A. cylindrica, alanine dehydrogenase exhibits a very high (nonphysiological) Km for ammonium, whereas its Km for alanine is 0.4 mM (33), which is around the physiological concentration of this amino acid in the cyanobacterial cytoplasm (30). A role in alanine degradation has also been found for alanine dehydrogenase in cells of the unicellular cyanobacterium Synechococcus elongatus subjected to nitrogen deprivation, conditions under which protein degradation can produce alanine (17). Alanine dehydrogenase could operate in the catabolism of alanine resulting from protein degradation during heterocyst differentiation (38, 39). Additionally, a catabolic alanine dehydrogenase could have a role in the heterocysts in the metabolism of alanine that, as suggested by Jüttner (13), could be received from the vegetative cells.
The main catabolic products of l-[U-14C]alanine in whole filaments of Anabaena sp. strain PCC 7120 are aspartate and glutamate (Table 2). These results indicate incorporation of the labeled carbon skeleton from alanine in central metabolism reaching the reductive and oxidative branches of the incomplete tricarboxylic acid (TCA) cycle that is present in cyanobacteria (35). First, alanine-derived pyruvate can be transformed via phosphoenolpyruvate into oxaloacetate, which can be transaminated, producing aspartate. Second, oxaloacetate and acetyl-coenzyme A (also produced from alanine-derived pyruvate) can make citrate that will be oxidized down to 2-oxoglutarate, which is a substrate of the glutamine synthetase (GS)-GOGAT pathway producing glutamine and glutamate. In our experiments with whole filaments, most labeling accumulated in glutamate, which is the final product of the pathway. In isolated heterocysts, however, more labeling accumulated in glutamine than in glutamate, which is consistent with lack of GOGAT in the heterocysts (21, 36, 37). Nonetheless, the observed labeling of glutamate and glutamine would require some synthesis of glutamate in the isolated heterocysts. (Because of the low pyruvate uptake observed with our heterocyst preparations, significant contamination of such preparations with vegetative cells is unlikely.) Aspartate-2-oxoglutarate and glutamate-2-oxoglutarate transaminase activities have been detected in isolated heterocysts of A. cylindrica (36). If an enzyme catalyzing one or both of these transaminase activities were present in the heterocysts of Anabaena sp. strain PCC 7120, it could produce labeled glutamate using labeled 2-oxoglutarate as substrate.
The experiments performed with [1-14C]pyruvate in whole filaments rendered results consistent with those described above, with significant accumulation of labeling in aspartate and glutamate. It should be noted, however, that in this case most labeling should proceed via oxaloacetate, since the C-1 is removed in oxidative decarboxylation of pyruvate. Nonetheless, some radioactivity could be recovered through fixation of the released 14CO2. The other major product of [1-14C]pyruvate is alanine (Table 2), which could result from a transamination reaction. ORF alr4853 of the Anabaena genome encodes an aspartate transaminase (14) that is also the predicted Anabaena protein most similar to alanine transaminase (glutamate-pyruvate transaminase). A homozygous insertional mutant of this gene, however, could not be obtained in this work (results not shown).
Oxidation of alanine provides reducing equivalents that can be used in the metabolism of the heterocysts, which includes the nitrogenase reaction. The low nitrogenase activity exhibited by strain CSR24 (about 10% of the wild-type value) is consistent with such a role of alanine and could account for the observed impairment of the mutant in diazotrophic growth (a growth rate constant about 50% of that exhibited by the wild type). The fact that a 90% decrease in nitrogenase activity determined with the artificial substrate acetylene results in only a 50% decrease in growth rate suggests that in the wild type this activity is substantially above the growth-limiting activity.
We have previously shown that mutants of the N-I transporter or of both the N-I and N-II transporters of Anabaena sp. strain PCC 7120 release hydrophobic amino acids (mainly alanine) to the growth medium and are impaired in diazotrophic growth (29, 30). Production of alanine in an natA genetic background is somewhat enhanced by the ald mutation (Fig. 4), which is consistent with a catabolic role of alanine dehydrogenase as discussed above. If alanine is transferred from vegetative cells to heterocysts (13) and, as shown in this work, if catabolism of alanine in these differentiated cells is important in the diazotrophic physiology, leakage of alanine from the filaments would be detrimental for diazotrophic growth as observed in the N-I and N-II transport mutants (29, 30). Thus, we concur with Jüttner's proposal (13) of alanine transfer from vegetative cells to heterocysts. In this scenario, a role of the N-I and N-II transporters in diazotrophy could be the recovery of amino acids leaked out from the cytoplasm.
In summary, as a result of the expression of the ald gene in the differentiating and mature heterocysts, alanine dehydrogenase is compartmentalized in the diazotrophic Anabaena filament mediating catabolism of alanine in the heterocysts. Sources of alanine can be protein degradation during heterocyst differentiation (39) and intercellular transport of alanine from vegetative cells into heterocysts (13). Alanine is not the only identified nitrogen-containing compound that appears to be transferred from vegetative cells to heterocysts since, as mentioned above, glutamate also appears to be imported into heterocysts. In the heterocysts, whereas glutamate would be used as the carbon skeleton for the biosynthesis of glutamine (37), alanine appears to contribute to production of the carbon skeleton for the biosynthesis of aspartate (which is conspicuously incorporated into cyanophycin [9]) and to be oxidized, as discussed above, producing reducing equivalents. If alanine and glutamate are transferred from vegetative cells to heterocysts at significant levels, much of a nitrogen-rich amino acid such as glutamine should move in the opposite orientation to result in a net transfer of nitrogen from heterocysts to vegetative cells.
Acknowledgments
We thank M. Burnat, J. E. Frías, and V. Mariscal for useful help and A. Orea and C. Parejo for skillful technical assistance. Use of DNA sequences from the Kazusa DNA Research Institute (Japan) database is acknowledged.
This work was supported by grant number BFU2008-03811 from the Ministerio de Ciencia y Tecnología (Madrid, Spain), cofinanced by FEDER.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2010. Current protocols in molecular biology. Wiley Interscience, New York, NY.
- 2.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172:3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cormack, B. P., R. H. Valdivia, and S. Falkow. 1996. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 173:33-38. [DOI] [PubMed] [Google Scholar]
- 4.Curatti, L., E. Flores, and G. Salerno. 2002. Sucrose is involved in the diazotrophic metabolism of the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 513:175-178. [DOI] [PubMed] [Google Scholar]
- 5.Ehira, S., and M. Ohmori. 2006. NrrA, a nitrogen-responsive response regulator facilitates heterocyst development in the cyanobacterium Anabaena sp. strain PCC 7120. Mol. Microbiol. 59:1692-1703. [DOI] [PubMed] [Google Scholar]
- 6.Elhai, J., A. Vepritskiy, A. M. Muro-Pastor, E. Flores, and C. P. Wolk. 1997. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 179:1998-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68:119-138. [DOI] [PubMed] [Google Scholar]
- 8.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 9.Flores, E., and A. Herrero. 2010. Compartmentalized function through cell differentiation in filamentous cyanobacteria. Nat. Rev. Microbiol. 8:39-50. [DOI] [PubMed] [Google Scholar]
- 10.Flores, E., A. Herrero, C. P. Wolk, and I. Maldener. 2006. Is the periplasm continuous in filamentous multicellular cyanobacteria? Trends Microbiol. 14:439-443. [DOI] [PubMed] [Google Scholar]
- 11.Flores, E., R. Pernil, A. M. Muro-Pastor, V. Mariscal, I. Maldener, S. Lechno-Yossef, Q. Fan, C. P. Wolk, and A. Herrero. 2007. Septum-localized protein required for filament integrity and diazotrophy in the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 189:3884-3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herrero, A., A. M. Muro-Pastor, A. Valladares, and E. Flores. 2004. Cellular differentiation and the NtcA transcription factor in filamentous cyanobacteria. FEMS Microbiol. Rev. 28:469-487. [DOI] [PubMed] [Google Scholar]
- 13.Jüttner, F. 1983. 14C-labeled metabolites in heterocysts and vegetative cells of Anabaena cylindrica filaments and their presumptive function as transport vehicles of organic carbon and nitrogen. J. Bacteriol. 155:628-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaneko, T., Y. Nakamura, C. P. Wolk, T. Kuritz, S. Sasamoto, A. Watanabe, M. Iriguchi, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, M. Kohara, M. Matsumoto, A. Matsuno, A. Muraki, N. Nakazaki, S. Shimpo, M. Sugimoto, M. Takazawa, M. Yamada, M. Yasuda, and S. Tabata. 2001. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 8:205-213. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, K., R. A. Mella-Herrera, and J. W. Golden. 2010. Cyanobacterial heterocysts. Cold Spring Harb. Perspect. Biol. 2:a000315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labarre, J., P. Thuriaux, and F. Chauvat. 1987. Genetic analysis of amino acid transport in the facultatively heterotrophic cyanobacterium Synechocystis sp. strain 6803. J. Bacteriol. 169:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lahmi, R., E. Sendersky, A. Perelman, M. Hagemann, K. Forchhammer, and R. Schwarz. 2006. Alanine dehydrogenase activity is required for adequate progression of phycobilisome degradation during nitrogen starvation in Synechococcus elongatus PCC 7942. J. Bacteriol. 188:5258-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:109-112. [Google Scholar]
- 19.Mariscal, V., A. Herrero, and E. Flores. 2007. Continuous periplasm in a filamentous, heterocyst-forming cyanobacterium. Mol. Microbiol. 65:1139-1145. [DOI] [PubMed] [Google Scholar]
- 20.Markwell, M. A. K., S. M. Hass, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 21.Martín-Figueroa, E., F. Navarro, and F. J. Florencio. 2000. The GS-GOGAT pathway is not operative in the heterocysts. Cloning and expression of glsF gene from the cyanobacterium Anabaena sp. PCC 7120. FEBS Lett. 76:282-286. [DOI] [PubMed] [Google Scholar]
- 22.Merino-Puerto, V., V. Mariscal, C. W. Mullineaux, A. Herrero, and E. Flores. 2010. Fra proteins influencing filament integrity, diazotrophy and localization of septal protein SepJ in the heterocyst-forming cyanobacterium Anabaena sp. Mol. Microbiol. 75:1159-1170. [DOI] [PubMed] [Google Scholar]
- 23.Montesinos, M. L., A. Herrero, and E. Flores. 1995. Amino acid transport systems required for diazotrophic growth in the cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 177:3150-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montesinos, M. L., A. Herrero, and E. Flores. 1997. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 179:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mullineaux, C. W., V. Mariscal, A. Nenninger, H. Khanum, A. Herrero, E. Flores, and D. G. Adams. 2008. Mechanism of intercellular molecular exchange in heterocyst-forming cyanobacteria. EMBO J. 27:1299-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neilson, A. H., and M. Doudoroff. 1973. Ammonia assimilation in blue-green algae. Arch. Mikrobiol. 89:15-22. [DOI] [PubMed] [Google Scholar]
- 27.Nicolaisen, K., V. Mariscal, R. Bredemeier, R. Pernil, S. Moslavac, R. López-Igual, I. Maldener, A. Herrero, E. Schleiff, and E. Flores. 2009. The outer membrane of a heterocyst-forming cyanobacterium is a permeability barrier for uptake of metabolites that are exchanged between cells. Mol. Microbiol. 74:58-70. [DOI] [PubMed] [Google Scholar]
- 28.Olmedo-Verd, E., A. M. Muro-Pastor, E. Flores, and A. Herrero. 2006. Localized induction of the ntcA regulatory gene in developing heterocysts of Anabaena sp. strain PCC 7120. J. Bacteriol. 188:6694-6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pernil, R., S. Picossi, V. Mariscal, A. Herrero, and E. Flores. 2008. ABC-type amino acid uptake transporters Bgt and N-II of Anabaena sp. strain PCC 7120 share an ATPase subunit and are expressed in vegetative cells and heterocysts. Mol. Microbiol. 67:1067-1080. [DOI] [PubMed] [Google Scholar]
- 30.Picossi, S., M. L. Montesinos, R. Pernil, C. Lichtlé, A. Herrero, and E. Flores. 2005. ABC-type neutral amino acid permease N-I is required for optimal diazotrophic growth and is repressed in the heterocysts of Anabaena sp. strain PCC 7120. Mol. Microbiol. 57:1582-1592. [DOI] [PubMed] [Google Scholar]
- 31.Quintero, M. J., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2000. Arginine catabolism in the cyanobacterium Synechocystis sp. strain PCC 6803 involves the urea cycle and arginase pathway. J. Bacteriol. 182:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111:1-61. [Google Scholar]
- 33.Rowell, P., and W. D. P. Stewart. 1975. Alanine dehydrogenase of the N2-fixing blue-green alga, Anabaena cylindrica. Arch. Microbiol. 107:115-124. [DOI] [PubMed] [Google Scholar]
- 34.Schilling, N., and K. Ehrnsperger. 1985. Cellular differentiation of sucrose metabolism in Anabaena variabilis. Z. Naturforsch. 40c:776-779. [Google Scholar]
- 35.Smith, A. J., J. London, and R. Y. Stanier. 1967. Biochemical basis of obligate autotrophy in blue-green algae and thiobacilli. J. Bacteriol. 94:972-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas, J., J. C. Meeks, C. P. Wolk, P. W. Shaffer, and S. M. Austin. 1977. Formation of glutamine from [13N]ammonia, [13N]dinitrogen, and [14C]glutamate by heterocysts isolated from Anabaena cylindrica. J. Bacteriol. 129:1545-1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolk, C. P., J. Thomas, P. W. Shaffer, S. M. Austin, and A. Galonsky. 1976. Pathway of nitrogen metabolism after fixation of 13N-labeled nitrogen gas by the cyanobacterium, Anabaena cylindrica. J. Biol. Chem. 251:5027-5034. [PubMed] [Google Scholar]
- 38.Wolk, C. P., A. Ernst, and J. Elhai. 1994. Heterocyst metabolism and development, p. 769-823. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 39.Xu, X., J. Elhai, and C. P. Wolk. 2008. Transcriptional and developmental responses by Anabaena to deprivation of fixed nitrogen, p. 383-422. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution. Caister Academic Press, Norfolk, United Kingdom.