Abstract
Oxoglutarate dehydrogenase (ODH) and pyruvate dehydrogenase (PDH) complexes catalyze key reactions in central metabolism, and in Corynebacterium glutamicum there is indication of an unusual supercomplex consisting of AceE (E1), AceF (E2), and Lpd (E3) together with OdhA. OdhA is a fusion protein of additional E1 and E2 domains, and odhA orthologs are present in all Corynebacterineae, including, for instance, Mycobacterium tuberculosis. Here we show that deletion of any of the individual domains of OdhA in C. glutamicum resulted in loss of ODH activity, whereas PDH was still functional. On the other hand, deletion of AceF disabled both PDH activity and ODH activity as well, although isolated AceF protein had solely transacetylase activity and no transsuccinylase activity. Surprisingly, the isolated OdhA protein was inactive with 2-oxoglutarate as the substrate, but it gained transsuccinylase activity upon addition of dihydrolipoamide. Further enzymatic analysis of mutant proteins and mutant cells revealed that OdhA specifically catalyzes the E1 and E2 reaction to convert 2-oxoglutarate to succinyl-coenzyme A (CoA) but fully relies on the lipoyl residues provided by AceF involved in the reactions to convert pyruvate to acetyl-CoA. It therefore appears that in the putative supercomplex in C. glutamicum, in addition to dihydrolipoyl dehydrogenase E3, lipoyl domains are also shared, thus confirming the unique evolutionary position of bacteria such as C. glutamicum and M. tuberculosis.
Pyruvate dehydrogenase (PDH) and 2-oxoglutarate dehydrogenase (ODH) activities catalyze key reactions in central metabolism. They exist as huge enzyme complexes of up to 11 MDa to convert a 2-oxoacid to an acyl-coenzyme A (CoA) derivative, which is acetyl- or succinyl-CoA, respectively (for reviews, see references 28 and 29 and references therein). The reaction requires distinct enzyme activities and involves the sequential actions of thiamine-pyrophosphate-dependent oxidative decarboxylation (E1, EC 1.2.4.2), with the concomitant transfer of the respective acyl group to a lipoamide residue. This is followed by the acyl group transfer to CoA, catalyzed by dihydrolipoyl transacylase activity (E2, EC 2.3.1.6), and, finally, the last step is dihydrolipoamide reoxidation to lipoamide by an FAD-dependent dihydrolipoyl dehydrogenase (E3, EC 1.8.1.4), thus enabling the initiation of a new catalytic cycle. As a result, the energy of the C1-C2 bond of an α-oxoacid is preserved in acetyl-CoA and succinyl-CoA, respectively, and NADH.
PDH and ODH are structurally closely related assemblies. Structural data for the three-dimensional organization of PDH of Bacillus stearothermophilus have culminated in the current view that the complex consists of an E2 core, to which E1 and E3 are flexibly tethered (20-22). This has similarly been disclosed for the PDH of Escherichia coli (23), as well as for components of ODH (6, 8, 18, 37). The PDH possesses specific E1p and E2p proteins, and ODH possesses specific E1o and E2o proteins, whereas the dihydrolipoyl dehydrogenase component E3 is shared by the two multienzyme complexes (28, 29). Thus, PDH and ODH complexes share one identical polypeptide plus very similar polypeptides, and they also have a similar overall quaternary structure (21, 23).
Within the Gram-positives, the Corynebacterineae, such as Mycobacterium tuberculosis and Corynebacterium glutamicum, have a number of distinctive features. This includes the synthesis of mycolic acids enabling the formation of a periplasmic space as in Gram-negatives (15) and the possession of unusual glycans and lipoylated glycans in their cell wall (1). It now has become clear that also the PDH and ODH of these organisms have unique properties, with respect to their protein components, three-dimensional organization, and regulation (25, 36). There is only one E2 protein present and with the isolated protein, it is shown to reconstitute PDH activity together with E1 and E3 proteins (35). An E2 protein specific to ODH is absent in M. tuberculosis, as is the case with C. glutamicum as well. Instead, Corynebacterineae possess one large fusion protein, termed OdhA in C. glutamicum and Kgd in M. tuberculosis, consisting of an E2 domain plus an E1 domain (36). However, as a lipoylated protein in Mycobacterium, only the E2 protein, which confers PDH activity in the reconstitution assay, is known, and no ODH activity is detectable in M. tuberculosis (35). A further remarkable feature found for C. glutamicum is the formation of a mixed 2-oxoacid dehydrogenase complex, since tagged OdhA copurified with the E2, E3, and E1p proteins, and vice versa, tagged E1p copurified with the E2 and E3 proteins together with OdhA (25). Another conspicuous feature shared by the OdhA and Kgd proteins is their interaction with a small regulatory protein which contains a phosphopeptide recognition domain (FHA domain) well characterized for many eukaryotic regulatory proteins. The protein is termed OdhI for C. glutamicum and GarA for M. tuberculosis (4, 25), and the structure of OdhI has recently been resolved (3). These proteins themselves are phosphorylated by one or several serine/threonine protein kinases present in the Corynebacterineae (25, 32), and they interact in their unphosphorylated form with OdhA or Kgd, respectively, to inhibit the activity of these proteins (25, 26).
Due to these remarkable features of activities and structures enabling pyruvate and 2-oxoglutarate conversion in the Corynebacterineae, we decided to study PDH and ODH as well as features of their constituent polypeptides in C. glutamicum in somewhat more detail, leading to the detection of the unprecedented structural and functional organization of these important enzyme complexes within central metabolism.
MATERIALS AND METHODS
Strains and cultivations.
The wild type (WT) of Corynebacterium glutamicum ATCC 13032 or its recombinant derivatives were used throughout. All strains utilized are listed in Table 1. Growth of strains was at 30°C on brain heart infusion (BHI; Difco Laboratories) or on the defined salt medium CGXII with 4% (wt/vol) glucose as a substrate (7). For recombinant work, E. coli DH5α was used and was grown at 30°C on Luria-Bertani medium.
TABLE 1.
C. glutamicum strains, relevant plasmids, and plasmids used for overexpression together with relevant primers
| Strain designation | Description or primer sequencea |
|---|---|
| Strains | |
| ATCC 13032 | Wild type (WT) |
| WTΔodhA | Chromosomal deletion of nt 1174001-1177720 |
| WTΔodhAE1 | Chromosomal deletion of nt 1174001-1176601 |
| WTΔodhAE2 | Chromosomal deletion of nt 1176641-1177330 |
| WTΔodhAE1L | Chromosomal deletion of nt 1177361-1177624 |
| WTΔaceF | Chromosomal deletion of nt 2310962-2312939 |
| WTΔaceE | Chromosomal deletion of aceE, obtained from reference 31 |
| WT aceF(K43Q-K162Q) | Chromosomal point mutations to result in AceF-K43Q-K162Q |
| WT aceF(K162Q-K278Q) | Chromosomal point mutations to result in AceF-K162Q-K278Q |
| Plasmids | |
| pK19mobsacB-K43Q-K162Q | pK19mobsacB containing custom-synthesized 1,240-bp fragment of aceF |
| pK19mobsacB-K162Q-K278Q | pK19mobsacB containing custom-synthesized 1,200-bp fragment of aceF |
| pEKEx5-H6-AceE | AATGGCCGATCAAGCAAAACTTG |
| TTATTCCTCAGGAGCGTTTGGAT | |
| pEKEx5-H6-AceF | CGCGGATCCGCGATGGCGTTCTCCGTAGAGATGC |
| CGCGGATCCGCGGAGCTGCAGATCGCCTTCGAAG | |
| pEKEx2-odh′-H6 | CTGAGCTAGGATCCCTCTACGACCCCAATGAAGG |
| TAGCTCAGGAATTCATTGTGATGGTGATGGTGATGGGCAGCCTTTGGTGCTGGTT | |
| pEKEx5-odhA-T294A | CGTTACCGTTTCCTTGGCCAACCCAGGTGGCATC |
| GATGCCACCTGGGTTGGCCAAGGAAACGGTAACG | |
| pEKEx5-odhA-H352C | CACCTCCACCTACGATTGCCGCGTGATCCAGGGTG |
| CACCCTGGATCACGCGGCAATCGTAGGTGGAGGTG | |
| pEKEx5-odhA-Q356D | CTACGATCACCGCGTGATCGATGGTGCTGTGTCCGGTG |
| CACCGGACACAGCACCATCGATCACGCGGTGATCGTAG | |
| pEKEx5-odhA-H352C-Q-356D | CTCCACCTACGATTGCCGCGTGATCGATGGTGCTGTGTCCGGTGA |
| TCACCGGACACAGCACCATCGATCACGCGGCAATCGTAGGTGGAG |
The nucleotide positions refer to the genome sequence of NC_006958.1. Primer sequences are given in direction 5′ to 3′. The primers given for the pEKEx5 plasmids show those with mutations in odhA with the mutated nucleotides in italics. nt, nucleotides.
Mutant generation and recombinant work.
To introduce the chromosomal mutations into the WT in order to derive WTΔodhA, WTΔodhAE1, WTΔodhAE2, WTΔodhAE1L, WTΔaceF2, WT aceF(K43Q-K162Q), and WT aceF(K162Q-K278Q), nonreplicative vectors based on pK19mobsacB (30) were first made carrying the deletion, or mutation, together with adjacent chromosomal sequences. These vectors, together with the primers used to construct them by overlap extension PCR, are given in Table 1. All vectors were confirmed by sequencing and utilized in two rounds of positive selection to obtain the desired allelic exchange, which was verified by PCR.
Vector constructions.
To construct pEKEx3, the shuttle vector pEKEx2 (8) was restricted with XhoI and StuI, blunted, and dephosphorylated to delete the kanamycin resistance gene. The spectinomycin resistance gene was derived as a 1,176-bp BglII fragment from pEC-S18mob2 (17) and ligated with the pEKEx2 backbone to yield pEKEx3. To construct the pEKEx4 vector, pEKEx2 was digested with SbfI and EcRI to delete the multiple cloning site (MCS). To construct the pEKEx5 vector, pEKEx3 was digested with SbfI and EcRI to delete the MCS. In the remaining backbones, the 252-bp MunI and NheI fragments of pQE30 (Qiagen), providing a ribosome binding site (RBS), a deca-His tag, MCS, and terminator, were ligated to yield pEKEx4 and pEKEx5. pEKEx4 provides kanamycin (Kan) resistance (50 μg ml−1 for both C. glutamicum and E. coli), whereas vectors pEKEx3 and pEKEx5 provide spectinomycin (Spe) resistance (250 μg ml−1 for C. glutamicum and 100 μg ml−1 for E. coli). Vectors pEKEx4 and pEKEx5 provide an amino-terminal His tag. All vectors allow IPTG (isopropyl-β-d-thiogalactopyranoside) (0.5 mM)-inducible expression.
To enable OdhA and AceF isolation, the corresponding genes were amplified with the primers given in Table 1 and cloned in pEKEx5. To generate OdhA mutant proteins, odhA was first cloned in pUC18, mutated by site-directed mutagenesis, and cloned as a BamHI fragment in pEKEx5 to yield pEKEx5-odhA-T294A, pEKEx5-odhA-H352C, pEKEx5-odhA-Q356D, and pEKEx5-odhA-H352C-Q-356D. All vector inserts were confirmed by sequencing.
In order to determine the amino-terminal end of OdhA, vector pEKEx-odh′ was constructed. A 930-bp fragment encompassing the suggested translational start was amplified using primers enabling the synthesis and fusion of a hexa-His tag at position 1178338 of the genome sequence of NC_006958.1. The resulting fragment was ligated with pEKEx2.
Protein isolation and analysis.
Proteins were isolated from the C. glutamicum WT carrying the respective pEKEx derivative. For protein isolation, cells were grown in LB-2% glucose, induced at an optical density (OD) of 1 (measured at 600 nm) and harvested 2 to 4 h later. Cells were disrupted by sonication, with subsequent centrifugation yielding crude extract. For some applications, and always in the case of ODH and PDH activity determinations, this was followed by an ultracentrifugation step (7°C, 50,000 rpm, model TI70.1, 1 h, Optima L80-XP ultracentrifuge; Beckman Coulter). Proteins were isolated by Ni-nitrilotriacetic acid (NTA) chromatography.
For determination of the amino-terminal end of OdhA, the C. glutamicum WT transformed with pEKEx-odh′ was grown on BHI with 25 μg/ml kanamycin up to the late exponential phase, and crude extract applied to a Ni-NTA column. Protein eluted with 250 mM imidazole and 300 mM NaCl was separated by PAGE. It yielded one band with an expected size of about 14 kDa. The protein was blotted on a polyvinylidene difluoride (PVDF) membrane and subjected to Edman degradation (Proteome Factory AG, D-12489 Berlin, Germany).
Enzyme assays.
For ODH and PDH assays, only fresh extracts were used, prepared in 0.1 M TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid] (pH 7.2), 10 mM MgCl2, 3 mM cysteine, 10% glycerol. The assay mixture (1 ml) was 50 mM TES (pH 7.7), 10 mM MgCl2, 3 mM cysteine, 2 mM NAD, 0.9 mM thiamine diphosphate, 0.05 mM chlorpromazine, and 1.5 mM 2-oxoglutarate or 6 mM pyruvate. The reaction was started with 0.2 mM CoA, and extinction followed at 340 nm.
The dehydrogenase/decarboxylase activity, E1, was assayed by measuring the reduction of 2,6-dichlorophenolindophenol (DCPIP) at 600 nm using an extinction coefficient of 21 mM−1 cm−1 in the following system (1 ml): 80 mM potassium phosphate (pH 7.5), 3 mM KCN, 0.23 mM DCPIP, 2.2 mM phenazine methosulfate, 1 mM MgCl2, 0.2 mM thiamine pyrophosphate, and 1 mM 2-oxoglutarate or pyruvate.
The transacylase activity, E2, was determined in the following system (1 ml): 50 mM Tris-HCl (pH 7.2), 5 mM MgCl2, 0.5 mM EDTA, 0.2 mM NADH, 0.2 mM lipoamide, 5 U dihydrodilipoamide dehydrogenase (Sigma), which was allowed to react for 10 min to achieve equilibrium. Then protein was added, and the reaction started by the addition of 0.15 mM acyl-CoA. At several points in time, 100-μl samples were withdrawn, the reaction was stopped by the addition of trichloroacetic acid, and after centrifugation, supernatants were used for CoA and acyl-CoA quantification via high-pressure liquid chromatography (HPLC). Separation was achieved on a LiChrospher 100 RP 18EC-5μ column using a gradient consisting of NaH2PO4 and acetonitrile on an Agilent 1100 analysis system.
Kinetic data were analyzed using the Origin7G software (Northampton) and nonlinear regression. Regression to the Michaelis-Menten equation yielded in all cases a ratio of chi-square to degrees of freedom of <2.1 × 10−4.
RESULTS
Consequences of odhA deletions.
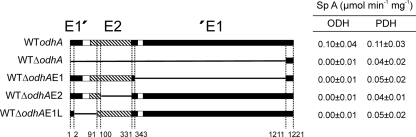
OdhA is a fusion protein with two major domains exhibiting structural features of an E1 and E2 protein, and a short sequence stretch of E1 localized at the N terminus, which is connected by a linker region of 69 amino acids (aa) to the rest of the protein (Fig. 1). We made four plasmid-encoded odhA mutant alleles with individual parts of odhA deleted and used them to delete the corresponding parts in the chromosome of the C. glutamicum WT. The resultant strains were confirmed to have the appropriate deletion via PCR. Growth of these mutants was studied with mineral salt medium CGXII with 4% glucose as the substrate (7). Whereas growth of the wild type was completed within 20 h and an OD of 49 was reached, even after 40 h, only very weak growth to an OD of at best 5 was obtained with each of the deletion mutants (data not shown). An organic acid analysis revealed that mutants accumulated up to 39 mM 2-oxoglutarate and 61 mM acetate in the medium, whereas with the wild type, these metabolites did not accumulate. These data indicate that all parts of odhA are required for a functional tricarboxylic acid cycle in C. glutamicum, thus enabling growth, and that, specifically, the conversion of 2-oxoglutarate is limited.
FIG. 1.
Organization of OdhA in domains and effects of chromosomal deletions on ODH and PDH activities. The organization of OdhA, as present in the wild type of C. glutamicum, is shown at the top, together with the location of domains and specific amino acid positions. The chromosomal deletions in the WTΔodhA, WTΔodhAE1, WTΔodhAE2, and WTΔodhAE1L mutants are shown below. Numbers give the locations still present in the deletions introduced, and the bars the approximate location of domains which are shaded as follows: black, E1; hatched, E2; white, linker regions. On the right, the resulting ODH (2-oxoglutarate dehydrogenase) and PDH (pyruvate dehydrogenase) specific activities of the deletion mutants compared to those of the wild type are listed. Enzyme activity determinations were done from independently grown cultures several times, and the result of one typical experiment is given.
For an enzymatic characterization, the mutants were grown on complex BHI medium and their ODH and PDH activities determined. Both ODH and PDH exhibited a high specific activity of about 0.1 μmol min−1 (mg protein)−1 in the wild type (Fig. 1), but all mutants had lost their ODH activity. This was also the case for the mutant termed WTΔodhAE1L, in which only aa 2 to 91 specific to the N-terminal extension were deleted, suggesting also that this part, which is strictly conserved within OdhA proteins of the Corynebacterineae (see Discussion and Fig. 6), has structural or functional significance. The significantly reduced PDH activity as a consequence of each OdhA mutation could indicate that the overall structure of the supercomplex is disturbed, also influencing slightly the domain interaction required for full PDH activity.
AceF is required for both ODH and PDH activity.
In addition to the fused E1 and E2 domains within OdhA, C. glutamicum possesses two further individual E1 and E2 domains. Absence of the E1-encoding aceE gene (NCgl2167) was shown to result in loss of PDH activity (31). We deleted the E2-encoding aceF gene (NCgl2126) in the C. glutamicum WT to result in WTΔaceF and used this strain together with WTΔaceE to determine ODH and PDH activities. With extracts of WTΔaceE no PDH activity was found, thus confirming the prior analysis (31), whereas with this strain, the ODH activity was 0.09 ± 0.12 μmol min−1 (mg protein)−1, which is almost indistinguishable from that of the wild type, with an activity of 0.10 ± 0.04 μmol min−1 (mg protein)−1. However, in WTΔaceF both ODH and PDH activities were virtually absent (≤0.01 ± 0.01 μmol min−1 [mg protein]−1). Consequently, AceE is specific to PDH, representing E1p, whereas AceF (E2) is essential for both PDH and ODH activities, although OdhA has an E2 domain. This might indicate that AceF is either unspecific or contributes in a still unknown manner to the E2 reaction of both the ODH and PDH activities.
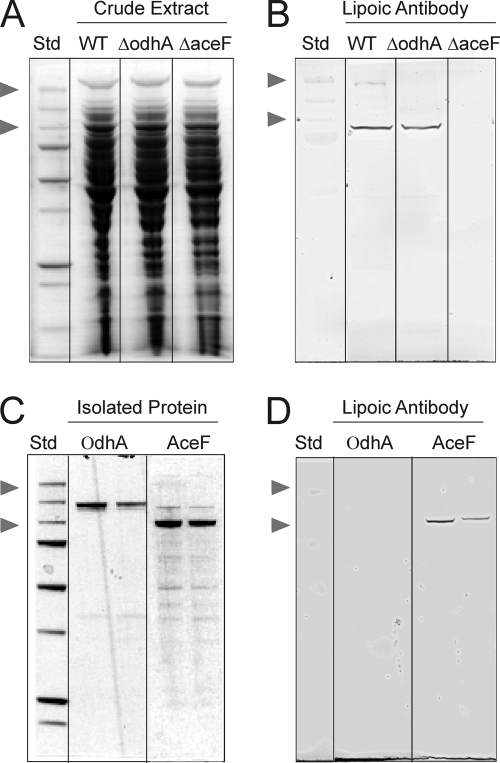
Lipoylated proteins in C. glutamicum.
The E2-acyltransferase reaction requires lipoyl residues as an essential cofactor. We therefore probed extracts of the C. glutamicum WT with an anti-lipoic acid antibody. As shown in Fig. 2, one intensive band was produced in immunoblots from WT or WTΔodhA. This band migrates at a size of about 72 kDa and, as expected for AceF, is absent with WTΔaceF, therefore suggesting that AceF is the prominent lipoylated protein in C. glutamicum, as is similarly the case in M. tuberculosis and Mycobacterium smegmatis (35), and that OdhA is nonlipoylated. For further confirmation and to exclude the possibility that small amounts of OdhA in crude extracts limit their immunological detection, the OdhA and AceF proteins were isolated from C. glutamicum. Their immunological analysis is shown at the bottom of Fig. 2 and confirms that OdhA has no detectable lipoyl residues.
FIG. 2.
AceF is lipoylated, but OdhA is not. (A) SDS gel of crude extract obtained from WT, WTΔodhA, and WTΔaceF. (B) The corresponding immunoblot using anti-lipoic acid antibody. (C) SDS gel of isolated OdhA and AceF protein. (D) The corresponding immunoblot. For panel A, 10 μg crude extract was applied in each case, whereas for the isolated proteins (panel C), 0.4 μg and 0.2 μg were applied side by side. The two arrowheads label the position of standards of 250 and 100 kDa, respectively.
Acyltransferase activities of AceF and OdhA.
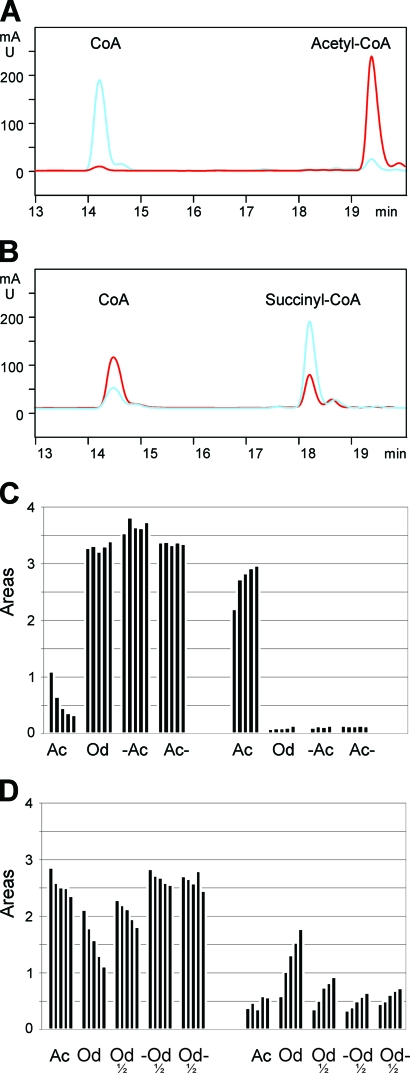
Since AceF is the only lipoylated protein and its absence results in the loss of PDH and ODH activity, it might be argued that this enzyme has broader substrate specificity and acts on pyruvate and 2-oxoglutarate as well. The orthologous protein in M. tuberculosis is SucB (Rv2215, alias DlaT). In M. tuberculosis this is also a constituent protein of PDH (35), but in addition it is proposed as a component of a second complex which has NAD(P)H-dependent peroxidase and peroxynitrite reductase activity (5), but acyltransferase activity has not been demonstrated. We assayed the AceF and OdhA proteins of C. glutamicum with either acetyl- or succinyl-CoA as a substrate. The proteins used were isolated from C. glutamicum to ensure posttranslational modifications. The assay system was that described by Hall and Weizman (12), in which dihydrolipoamide is continuously regenerated by dihydrolipoamide dehydrogenase from lipoamide and NADH, and acyltransferase activity converts the added acyl-CoA to free CoA. However, in contrast to this procedure in which NADH oxidation is followed, we directly followed acyl-CoA and CoA concentrations by HPLC since this gave more-reliable data. When a reaction assay was analyzed 15 min after incubation with either 50 μg AceF or 50 μg OdhA and acyl-CoA as a substrate (Fig. 3A), AceF fully converted acetyl-CoA to CoA (blue), whereas OdhA did not react with acetyl-CoA (red). Thus, AceF has high transacylase activity with acetyl-CoA. However, when the assay contained succinyl-CoA (Fig. 3B), OdhA catalyzed succinyl-CoA conversion to CoA (red), whereas with AceF succinyl-CoA was hardly utilized. This indicates that without covalently linked lipoyl domains, OdhA is able to catalyze the E2 reaction as part of ODH activity.
FIG. 3.
Activities of AceF and OdhA with acetyl- and succinyl-CoA as substrate. HPLC chromatograms with acetyl-CoA (A) or succinyl-CoA (B) as substrate for AceF (blue) or OdhA (red) with 50 μg each after 15-min reaction time. The positions of acyl-CoA and CoA in the chromatograms are marked accordingly. (C) Time-dependent activities with acetyl-CoA as substrate. The left 4-column set shows acetyl-CoA utilization, and the right 4-column set CoA liberation, where each column is the amount of substrate consumed and product formed, respectively, after 3, 6, 9, 12, and 15 min (from left to right). The assays are marked as follows: Ac, 50 μg AceF; Od, 50 μg OdhA; −Ac, minus enzyme; Ac−, minus dihydrolipoamide. (D) Time-dependent activities with succinyl-CoA as substrate. Assays are as described in the legend to panel C, except that in addition, 25 μg OdhA (Od) was used. The bars represent the areas (10−3), and the initial concentration for acetyl- and succinyl-CoA added was 150 μM.
A more detailed analysis in which reactions were observed over time, together with appropriate controls, showed that with AceF, even after just 3 min of reaction time, the major fraction of acetyl-CoA was converted to CoA (Fig. 3C). The protein has a specific catalytic activity of 628 nmol min−1 mg−1. In assays with OdhA or controls without AceF or without dihydrolipoamide, no CoA was formed when acetyl-CoA was the substrate. With succinyl-CoA the situation was slightly different (Fig. 3D). Some CoA formation also occurred in controls without OdhA or without dihydrolipoamide. This partial conversion of succinyl-CoA is due to the high rate of chemical hydrolysis at the neutral pH in the assay (19). However, with 50 μg and 25 μg OdhA, a time-dependent succinyl-CoA conversion with concomitant CoA release in a dihydrolipoamide-dependent manner was also apparent. The OdhA-specific catalytic activity was 112 nmol min−1 mg−1.
Dehydrogenase activities of AceE and OdhA.
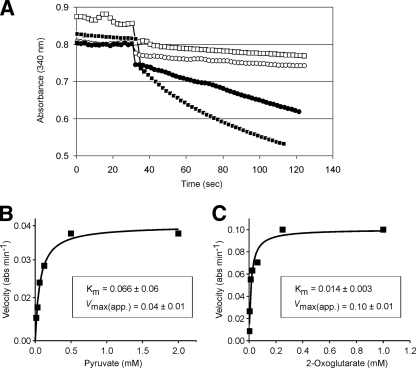
Following the unexpected outcome that OdhA can apparently utilize free dihydrodilipoamide to perform the E2 reaction specifically with succinyl-CoA, we investigated the activity and specificity of the E1 domain within OdhA and that of the separate E1 protein AceE. Proteins isolated from C. glutamicum were used to determine activities by monitoring the reduction of 2,6-DCPIP (24). As shown in Fig. 4, AceE has reductase activity with pyruvate but did not react with 2-oxoglutarate, even at the high concentration of 1 mM substrate used. The opposite was the case for OdhA, which exhibited activity with 2-oxoglutarate but did not react with pyruvate. The specific activity for AceE was 250 nmol min−1 mg−1, and that for OdhA was 110 nmol min−1 mg−1, respectively.
FIG. 4.
Dehydrogenase activities of AceE and OdhA. (A) Isolated proteins, each 50 μg, were used to follow the 2,6-DCPIP-dependent reduction at 600 nm due to the presence of either 1 mM 2-oxoglutarate or 1 mM pyruvate added after 30 s. ○, OdhA with pyruvate; •, OdhA with 2-oxoglutarate; ▪, AceE with pyruvate; □, AceE with 2-oxoglutarate. (B) Kinetic characterization of pyruvate dehydrogenase. (C) Kinetic characterization of 2-oxoglutarate dehydrogenase.
Recording the initial reaction velocities with different substrate concentrations and nonlinear regression yielded a good fit to the Michaelis-Menten equation (Fig. 4). From this analysis for OdhA and 2-oxoglutararate, a Km of 0.014 ± 0.003 mM and for AceE and pyruvate a Km value of 0.066 ± 0.06 mM were derived. The value for AceE is close to that known for the E1 reaction of the E. coli PDH, for which the Km is 0.110 mM (16), or that of human E1 protein, for which it is 0.026 mM (38).
Mutations in the lipoyl domains.
Since OdhA and AceF apparently share the AceF-bound lipoyl residues, the question arose of whether the accessibility of these residues for the two catalytic centers is different. To address this question, enzyme complexes were created with fewer than three lipoyl domains per AceF chain. The lipoyl groups are bound via an amide linkage to the N6 amino groups of conserved lysine residues (27). The corresponding lysine codons were replaced in C. glutamicum WT by glutamine codons in analogy to identical mutations made in E2p of E. coli (2). As a result, C. glutamicum WT aceF(K43Q-K162Q) and WT aceF(K162Q-K278Q) were created, and in them, only the third or the first lipoylatable lysine residue within the three tandemly repeated lipoyl domains was retained. As shown in Table 2, both mutant strains still yielded ODH and PDH activities, demonstrating efficient active site coupling by the formation and transfer of acyl groups by the respective dehydrogenase and acyltransferase domains. The detailed kinetic characterization for the WT aceF(K43Q-K162Q) mutant did not reveal any loss of catalytic efficiency (Vmax/Km) for either pyruvate or 2-oxoglutarate compared to the WT. However, with the WT aceF(K162Q-K278Q) mutant a strong reduction in the catalytic efficiency with 2-oxoglutarate was produced, and also the efficiency with pyruvate was slightly influenced. This might indicate a significantly hindered access of the lipoyl domain attached to K43 of AceF to the active site within the E2 domain of OdhA.
TABLE 2.
Summary of effects of AceF mutant proteins on ODH and PDH activitiesa
| Strain | ODH |
PDH |
||||
|---|---|---|---|---|---|---|
| Km (mM) | Vmax (nmol min−1 mg−1) | Vmax/Km (μl min−1 mg−1) | Km (mM) | Vmax (nmol min−1 mg−1) | Vmax/Km (μl min−1 mg−1) | |
| WT | 0.13 | 134 | 1,048 | 1.69 | 148 | 87 |
| WT aceF(K43Q-K162Q) | 0.17 | 176 | 1,067 | 2.12 | 208 | 98 |
| WT aceF(K162Q-K278Q) | 0.32 | 78 | 244 | 2.24 | 154 | 69 |
Kinetic parameters for ODH and PDH were derived as described under Materials and Methods. In the WT aceF(K43Q-K162Q) mutant only the third lipoylatable lysine residue was retained, and in the WT aceF(K162Q-K278Q) mutant only the first. Assays were done in duplicate, and deviations were always below 5%.
Acyltransferase and dehydrogenase activities of OdhA muteins.
In order to obtain further evidence for succinyl-transferase activity of OdhA, we sought to locate putative residues involved in catalysis. A comparison with known acyltransferase structures yielded the highest similarities, with respect to sequence and structure, with E2p of E. coli, E2o of Bos taurus, E2o of Homo sapiens, and E2p of Azotobacter vinelandii (33). Within the A. vinelandii enzyme, S558, H610, and N614 are involved in proton activation and transfer (13), and these residues, or their functional groups, are fully retained in T294, H352, and Q356 of the OdhA protein (data not shown). We therefore generated the proteins OdhA-T294A, OdhA-H352C, and OdhA-Q356D with single amino acid substitutions and OdhA-H352C-Q356D with two of these mutations combined. The proteins were expressed and isolated from the corresponding recombinant wild type of C. glutamicum.
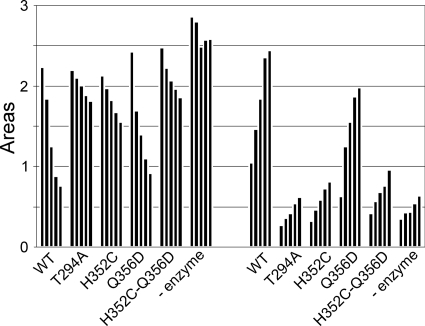
Absence of the imidazole side chain in OdhA-H352C and in OdhA-H352C-Q356D fully prevented succinyltransferase activity (Fig. 5). This agrees with the notion that this residue is part of the catalytic cycle in E2p of A. vinelandii by abstracting the hydrogen atom from the reactive sulfhydryl group of CoA. With the OdhA-Q356D protein, the activity was slightly reduced, which is also in complete agreement with the effect of the corresponding mutation in the A. vinelandii enzyme, since this residue is involved in the stabilization and increasing nucleophilicity of imidazole rather than directly participating in the catalytic cycle (13). Almost no activity was observed for OdhA-T294A, as was also found for the corresponding mutation in E2p of A. vinelandii. Overall, these data confirm the E2 activity of OdhA with the specific residues involved in catalysis, as is known from the A. vinelandii E2p enzyme, and agree with the notion that the E2 part of OdhA adopts a fully functional E2 domain.
FIG. 5.
Substrate specificity of OdhA muteins. The left 6-column set shows succinyl-CoA utilization, and the right 6-column set CoA liberation, where each column indicates the amount of substrate consumed and product formed, respectively, after 3, 6, 9, 12, and 15 min (from left to right). The assays included 50 μg OdhA protein either from unmutated OdhA (WT), OdhA protein with the point mutation(s) as indicated, or no protein (− enzyme). The bars represent the areas (10−3), and the initial concentration for succinyl-CoA added was 150 μM.
To assess possible effects of the mutations in the E2 domain of OdhA on the E1 reaction, proteins were also assayed for dehydrogenase activities. With mutein OdhA-T294A the E1 activity was 97 ± 6 nmol min−1 mg−1, which is almost identical to the activity of wild-type OdhA with its activity of 110 ± 10 nmol min−1 mg−1 (see above). With mutein OdhA-Q356D, the E1 activity was slightly reduced to 86 ± 10 nmol min−1 mg−1, and with muteins OdhA-H352C and OdhA-H352C-Q356D, the dehydrogenase activities were significantly reduced to 51 ± 12 and 36 ± 6 nmol min−1 mg−1, respectively. This strong effect of the mutations in the E2 domains on the E1 activity was unexpected. However, since OdhA-T294A does not have E2 activity but fully retains E1 activity, this illustrates that in principle the OdhA domains operate independently.
Amino-terminal end of OdhA.
OdhA has a stretch of about 40 amino acids at its N terminus resembling the N terminus of E1o (SucA) of E. coli (Fig. 6), and we noticed that the N-terminal region of the Kgd protein of M. tuberculosis as used by Tian et al. (35) is within a predicted α-helix. Furthermore, translation of upstream sequences of both kgd and odhA revealed further possibly conserved regions (data not shown). To solve this apparent inconsistency, we constructed plasmid pEKEx-odh′ with a 930-bp fragment covering a suggested promoter region together with the beginning of odhA and six His codons at the 3′ end. Plasmid pEKEx-odh′ was introduced into the C. glutamicum WT, and after growth in liquid culture, crude extracts were prepared and applied to a Ni-NTA column. One major protein was eluted. Sequencing its N-terminal end by Edman degradation revealed the sequence SSASTFG, which is thus in accordance with the NCgl1084 annotation of OdhA, except for the apparent posttranslational removal of the initiating methionine. Based on this determination and the strong conservation, Kgd is proposed to start 17 aa upstream of the start, as presently annotated and used in Rv1248c. As mentioned, the N-terminal region of OdhA shows similarity with the N-terminal region of E1o of E. coli, which has recently been demonstrated to enable subunit interaction within ODH (34). This part might therefore be of considerable structural relevance for OdhA or Kgd proteins of Corynebacterineae within enzyme complexes. This proposal agrees with our observation that deletion of aa 2 to 91 of OdhA causes absence of ODH activity (Fig. 1).
FIG. 6.
Start and structure of OdhA. The N-terminal ends are shown for OdhA of C. glutamicum (OdhA-Cg), Mycobacterium leprae (OdhA-Ml), Kgd of M. tuberculosis (Kgd-Mt), OdhA of Corynebacterium diphtheriae (OdhA-Cd), and SucA, E1o of E. coli (SucA-Ec). The annotated initiating methionines are given in boldface, and the sequence of OdhA-Cg determined by Edman degradation is underlined. The sequences were aligned using Clustal. The structural predictions as obtained for OdhA of C. glutamicum are given at the top, and that for SucA of E. coli at the bottom. The prediction was made with PredictProtein, and “rel sec” is the reliability index (0 = low and 9 = high), for either loop, L, or helix, H, given in “sub sec.” Asterisks, periods, and colons denote the degree of similarity of the amino acids.
DISCUSSION
The first evidence for a supercomplex of ODH and PDH proteins in C. glutamicum came from copurification experiments (25). The current information reveals even more intimate connections between protein components of ODH and PDH. As shown by the aceF deletion, the encoded AceF protein is required for both PDH and ODH activities. Whereas this could be interpreted as showing that AceF has both transacetylase and also transsuccinylase activity or even merely a structural function, this is ruled out by the enzyme assays with the isolated proteins. AceF has exclusively transacetylase activity, and if lipoic acid is present, then OdhA has only transsuccinylase activity. Free lipoic acid is the substrate for E2 and E3 of E. coli, although lipoylated domains would represent a much better substrate (10). We assume that the lipoyl residues required for ODH activity are contributed by AceF. High flexibility of lipoyl domains in 2-oxoacid dehydrogenases is an inherent characteristic to enable shuttling of intermediates between the different reactive centers required for the overall enzyme reaction. It is also possibly a reason for the variety of structural modifications of enzymes involving dihydrolipoamide or dehydrolipoyl dehydrogenase. Thus, for instance, the dehydrolipoyl dehydrogenase of Streptococcus pneumoniae has its own lipoic acid covalently attached (11), and in eukaryotic PDHs the E3 binding protein (referred to variously as E3BP, or protein X) possesses one lipoyl domain and is a constituent of the E2 core.
More experimental evidence of lipoyl domain sharing within the supercomplex comes from the AceF-K43Q-K162Q mutant, which has only the third lipoyl residue retained, or the AceF-K162Q-K278Q mutant, with only the first lipoyl residue present. First of all, it was found that with the AceF-K43Q-K162Q mutant the overall catalytic activity for both PDH and ODH was not significantly affected by the loss of the first two lipoyl domains. This indicates that the lipoyl residue attached to K278 can substitute in vivo for the first two lipoyl domains and agrees with the general view on the redundancy of lipoyl domains and their large structural flexibility, as concluded for a number of artificial lipoyl domain variations of E2p of E. coli, which also has three naturally occurring lipoyl domains (2). Also, visualizations of lipoyl domains in PDH and ODH structures of E. coli (23) and in the PDH of B. stearothermophilus as well (20-22) strengthen this view. Interestingly, with the AceF-K162Q-K278Q mutant, in which only the first lipoyl domain is retained, the activity of both ODH and PDH is influenced. This is a direct indication that this specific lipoyl residue can be shared by the partial reactions involving the lipoyl residue for both the transacetylase and the transsuccinylase cycle. Furthermore, the especially reduced efficiency with 2-oxoglutarate hints at structural constraints within the entire supercomplex regarding the accessibility of the E1 or E2 domain of OdhA to this lipoyl residue as part of the first two catalytic steps within the cycle of 2-oxoglutarate decarboxylation.
As demonstrated by a large number of studies and also most impressively by electron microscope techniques for the PDH of B. stearothermophilus (22) and E. coli (23), 2-oxoacid dehydrogenases represent very large flexible structures. The complex consists of a core, around which at a distance of about 70 to 90 Å there is a shell consisting of E1 and E3 domains. The E2 protein is essential for the whole structure. With its catalytic domain, it represents the core, and it has three additional sections which are located in the gap between the core and the surrounding shell of the E1 and E3 domains. In the E2 of C. glutamicum, all the domains, including the linker regions (aa 314 to 369 and aa 411 to 440) and the peripheric subunit binding domain (PSBD; aa 372 to 405), are clearly defined. It is in fact the case that E2 from C. glutamicum contains all the structural elements necessary to construct a complex consisting of E1, E2, and E3. This is confirmed by the fact that the PDH activity is only moderately influenced in the absence of the OdhA protein (Fig. 1). Although it is too early to speculate on the arrangement of OdhA within the proposed complex (25), it is very likely that the short N-terminal E1′ part of OdhA has an important structural function. An indication is the extremely strong conservation of this region in the OdhA proteins of the Corynebacterineae, including the incorrectly annotated Kgd protein of M. tuberculosis (Fig. 6). Moreover, there is indication that the E1o terminus of E. coli interacts with E2 (8, 9), and this region displays considerable structural similarities to the N terminus of OdhA (Fig. 6). In the same way, in the case of the E1o of Azotobacter vinelandii, the N terminus is considered to be responsible for interaction with E2 (14). Therefore, E1′ is proposed to carry a key function to enable anchoring of OdhA within the suggested PDH-ODH supercomplex.
Acknowledgments
We appreciate the continuous interest of H. Sahm in this work.
The work was supported by BMBF 0313704 (SysMap).
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Alderwick, L. J., H. L. Birch, A. K. Mishra, L. Eggeling, and G. S. Besra. 2007. Structure, function and biosynthesis of the Mycobacterium tuberculosis cell wall: arabinogalactan and lipoarabinomannan assembly with a view to discovering new drug targets. Biochem. Soc. Trans. 35:1325-1328. [DOI] [PubMed] [Google Scholar]
- 2.Allen, A. G., R. N. Perham, N. Allison, J. S. Miles, and J. R. Guest. 1989. Reductive acetylation of tandemly repeated lipoyl domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli is random order. J. Mol. Biol. 208:623-633. [DOI] [PubMed] [Google Scholar]
- 3.Barthe, P., C. Roumestand, M. J. Canova, L. Kremer, C. Hurard, V. Molle, and M. Cohen-Gonsaud. 2009. Dynamic and structural characterization of a bacterial FHA protein reveals a new autoinhibition mechanism. Structure 17:568-578. [DOI] [PubMed] [Google Scholar]
- 4.Belanger, A. E., and G. F. Hatfull. 1999. Exponential-phase glycogen recycling is essential for growth of Mycobacterium smegmatis. J. Bacteriol. 181:6670-6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 6.Bunik, V. I., and D. Degtyarev. 2008. Structure-function relationships in the 2-oxo acid dehydrogenase family: substrate-specific signatures and functional predictions for the 2-oxoglutarate dehydrogenase-like proteins. Proteins 71:874-890. [DOI] [PubMed] [Google Scholar]
- 7.Eggeling, L. and O. Reyes. 2005. Experiments, p. 535-566. In L. Eggeling and M. Bott (ed.), Handbook of Corynebacterium glutamicum. Taylor and Francis, Boca Raton, FL.
- 8.Eikmanns, B. J., E. Kleinertz, W. Liebl, and H. Sahm. 1991. A family of Corynebacterium glutamicum/Escherichia coli shuttle vectors for cloning, controlled gene expression, and promoter probing. Gene 102:93-98. [DOI] [PubMed] [Google Scholar]
- 9.Frank, R. A., A. J. Price, F. D. Northrop, R. N. Perham, and B. F. Luisi. 2007. Crystal structure of the E1 component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 368:639-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, L. D., L. C. Packman, and R. N. Perham. 1989. Kinetics and specificity of reductive acylation of lipoyl domains from 2-oxo acid dehydrogenase multienzyme complexes. Biochemistry 28:1574-1581. [DOI] [PubMed] [Google Scholar]
- 11.Håkansson, A. P., and A. W. Smith. 2007. Enzymatic characterization of dihydrolipoamide dehydrogenase from Streptococcus pneumoniae harboring its own substrate. J. Biol. Chem. 282:29521-29530. [DOI] [PubMed] [Google Scholar]
- 12.Hall, E. R., and P. D. Weitzman. 1974. A continuous spectrophotometric assay for the trans-acylase (E2) component of pyruvate and alpha-oxoglutarate dehydrogenase enzyme complexes. Anal. Biochem. 62:286-290. [DOI] [PubMed] [Google Scholar]
- 13.Hendle, J., A. Mattevi, A. H. Westphal, J. Spee, A. de Kok, A. Teplyakov, and W. G. Hol. 1995. Crystallographic and enzymatic investigations on the role of Ser558, His610, and Asn614 in the catalytic mechanism of Azotobacter vinelandii dihydrolipoamide acetyltransferase (E2p). Biochemistry 34:4287-4298. [DOI] [PubMed] [Google Scholar]
- 14.Hengeveld, A. F., C. P. van Mierlo, H. W. van den Hooven, A. J. Visser, and A. de Kok. 2002. Functional and structural characterization of a synthetic peptide representing the N-terminal domain of prokaryotic pyruvate dehydrogenase. Biochemistry 41:7490-7500. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmann, C., A. Leis, M. Niederweis, J. M. Plitzko, and H. Engelhardt. 2008. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. U. S. A. 105:3963-3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeng, J., A. T. Kallarakal, S. F. Kim, K. M. Popov, and B. J. Song. 1998. Pyruvate dehydrogenase E1 alpha isoform in rat testis: cDNA cloning, characterization, and biochemical comparison of the recombinant testis and liver enzymes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 120:205-216. [DOI] [PubMed] [Google Scholar]
- 17.Kirchner, O., and A. Tauch. 2003. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 104:287-299. [DOI] [PubMed] [Google Scholar]
- 18.Knapp, J. E., D. T. Mitchell, M. A. Yazdi, S. R. Ernst, L. J. Reed, and M. L. Hackert. 1998. Crystal structure of the truncated cubic core component of the Escherichia coli 2-oxoglutarate dehydrogenase multienzyme complex. J. Mol. Biol. 280:655-668. [DOI] [PubMed] [Google Scholar]
- 19.Lambeth, D. O., and W. W. Muhonen. 1992. The direct assay of kinases and acyl-CoA synthetases by HPLC: application to nucleoside diphosphate kinase and succinyl-CoA synthetase. Anal. Biochem. 6209:192-198. [DOI] [PubMed] [Google Scholar]
- 20.Lengyel, J. S., K. M. Stott, X. Wu, B. R. Brooks, A. Balbo, P. Schuck, R. N. Perham, S. Subramaniam, and J. L. Milne. 2008. Extended polypeptide linkers establish the spatial architecture of a pyruvate dehydrogenase multienzyme complex. Structure 16:93-103. [DOI] [PubMed] [Google Scholar]
- 21.Milne, J. L., D. Shi, P. B. Rosenthal, J. S. Sunshine, G. J. Domingo, X. Wu, B. R. Brooks, R. N. Perham, R. Henderson, and S. Subramaniam. 2002. 1. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 21:5587-5598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Milne, J. L., X. Wu, M. J. Borgnia, J. S. Lengyel, B. R. Brooks, D. Shi, R. N. Perham, and S. Subramaniam. 2006. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J. Biol. Chem. 281:4364-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murphy, G. E., and G. J. Jensen. 2005. Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. Structure 13:1765-1773. [DOI] [PubMed] [Google Scholar]
- 24.Nemeria, N., Y. Yan, Z. Zhang, A. M. Brown, P. Arjunan, W. Furey, J. R. Guest, and F. Jordan. 2001. Inhibition of the Escherichia coli pyruvate dehydrogenase complex E1 subunit and its tyrosine 177 variants by thiamin 2-thiazolone and thiamin 2-thiothiazolone diphosphates. Evidence for reversible tight-binding inhibition. J. Biol. Chem. 276:45969-45978. [DOI] [PubMed] [Google Scholar]
- 25.Niebisch, A., A. Kabus, C. Schultz, B. Weil, and M. Bott. 2006. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J. Biol. Chem. 281:12300-12307. [DOI] [PubMed] [Google Scholar]
- 26.O'Hare, H. M., R. Durán, C. Cerveñansky, M. Bellinzoni, A. M. Wehenkel, O. Pritsch, G. Obal, J. Baumgartner, J. Vialaret, K. Johnsson, and P. M. Alzari. 2008. Regulation of glutamate metabolism by protein kinases in mycobacteria. Mol. Microbiol. 70:1408-1423. [DOI] [PubMed] [Google Scholar]
- 27.Packman, L. C., G. Hale, and R. N. Perham. 1984. Repeating functional domains in the pyruvate dehydrogenase multienzyme complex of Escherichia coli. EMBO J. 3:1315-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perham, N. R. 2000. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu. Rev. Biochem. 69:961-1004. [DOI] [PubMed] [Google Scholar]
- 29.Reed, L. J., F. H. Pettit, M. H. Eley, L. Hamilton, J. H. Collins, and R. M. Oliver. 1975. Reconstitution of the Escherichia coli pyruvate dehydrogenase complex. Proc. Natl. Acad. Sci. U. S. A. 72:3068-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 31.Schreiner, M. E., D. Fiur, J. Holátko, M. Pátek, and B. J. Eikmanns. 2005. E1 enzyme of the pyruvate dehydrogenase complex in Corynebacterium glutamicum: molecular analysis of the gene and phylogenetic aspects. J. Bacteriol. 187:6005-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schultz, C., A. Niebisch, A. Schwaiger, U. Viets, S. Metzger, M. Bramkamp, and M. Bott. 2009. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol. Microbiol. 74:724-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Söding, J., A. Biegert, and A. N. Lupas. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244-W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, J., Y. H. Park, N. S. Nemeria, S. Kale, L. Kakalis, and F. Jordan. 2010. Nuclear magnetic resonance evidence for the role of the flexible regions of the E1 component of the pyruvate dehydrogenase complex from Gram-negative bacteria. J. Biol. Chem. 285:4680-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian, J., R. Bryk, S. Shi, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2005. Mycobacterium tuberculosis appears to lack alpha-ketoglutarate dehydrogenase and encodes pyruvate dehydrogenase in widely separated genes. Mol. Microbiol. 57:859-868. [DOI] [PubMed] [Google Scholar]
- 36.Usuda, Y., N. Tujimoto, C. Abe, Y. Asakura, E. Kimura, Y. Kawahara, O. Kurahashi, and H. Matsui. 1996. Molecular cloning of the Corynebacterium glutamicum (′Brevibacterium lactofermentum' AJ12036) odhA gene encoding a novel type of 2-oxoglutarate dehydrogenase. Microbiology 142:3347-3354. [DOI] [PubMed] [Google Scholar]
- 37.Wagenknecht, T., R. Grassucchi, and D. Schaak. 1990. Cryoelectron microscopy of frozen-hydrated alpha-ketoacid dehydrogenase complexes from Escherichia coli. J. Biol. Chem. 265:22402-22408. [PubMed] [Google Scholar]
- 38.Yi, J., N. Nemeria, A. McNally, F. Jordan, R. S. Machado, and J. R. Guest. 1996. Effect of substitutions in the thiamin diphosphate-magnesium fold on the activation of the pyruvate dehydrogenase complex from Escherichia coli by cofactors and substrate. J. Biol. Chem. 271:33192-331200. [DOI] [PubMed] [Google Scholar]