Abstract
Staphylococci contain a class Ib NrdEF ribonucleotide reductase (RNR) that is responsible, under aerobic conditions, for the synthesis of deoxyribonucleotide precursors for DNA synthesis and repair. The genes encoding that RNR are contained in an operon consisting of three genes, nrdIEF, whereas many other class Ib RNR operons contain a fourth gene, nrdH, that determines a thiol redoxin protein, NrdH. We identified a 77-amino-acid open reading frame in Staphylococcus aureus that resembles NrdH proteins. However, S. aureus NrdH differs significantly from the canonical NrdH both in its redox-active site, C-P-P-C instead of C-M/V-Q-C, and in the absence of the C-terminal [WF]SGFRP[DE] structural motif. We show that S. aureus NrdH is a thiol redox protein. It is not essential for aerobic or anaerobic growth and appears to have a marginal role in protection against oxidative stress. In vitro, S. aureus NrdH was found to be an efficient reductant of disulfide bonds in low-molecular-weight substrates and proteins using dithiothreitol as the source of reducing power and an effective reductant for the homologous class Ib RNR employing thioredoxin reductase and NADPH as the source of the reducing power. Its ability to reduce NrdEF is comparable to that of thioredoxin-thioredoxin reductase. Hence, S. aureus contains two alternative thiol redox proteins, NrdH and thioredoxin, with both proteins being able to function in vitro with thioredoxin reductase as the immediate hydrogen donors for the class Ib RNR. It remains to be clarified under which in vivo physiological conditions the two systems are used.
Ribonucleotide reductases (RNRs) are essential enzymes in all living cells, providing the only known de novo pathway for the biosynthesis of deoxyribonucleotides, the immediate precursors of DNA synthesis and repair. RNRs catalyze the controlled reduction of all four ribonucleotides to maintain a balanced pool of deoxyribonucleotides during the cell cycle (29). Three main classes of RNRs are known. Class I RNRs are oxygen-dependent enzymes, class II RNRs are oxygen-independent enzymes, and class III RNRs are oxygen-sensitive enzymes. Class I RNRs are divided into two subclasses, subclasses Ia and Ib.
Staphylococcus aureus is a Gram-positive facultative aerobe and a major human pathogen (24). S. aureus contains class Ib and class III RNRs, which are essential for aerobic and anaerobic growth, respectively (26). The class Ib NrdEF RNR is encoded by the nrdE and nrdF genes: NrdE contains the substrate binding and allosteric binding sites, and NrdF contains the catalytic site for ribonucleotide reduction. The S. aureus nrdEF genes form an operon containing a third gene, nrdI, the product of which, NrdI, is a flavodoxin (5, 33). Many other bacteria such as Escherichia coli (16), Lactobacillus lactis (17), and Mycobacterium and Corynebacterium spp. possess class Ib RNR operons that contain a fourth gene, nrdH (30, 44, 50), whose product, NrdH, is a thiol-disulfide redoxin (16, 17, 40, 43, 49). More-complex situations are found for some bacteria, where the class Ib RNR operon may be duplicated and one or more of the nrdI and nrdH genes may be missing or located in another part of the chromosome (29).
NrdH proteins are glutaredoxin-like protein disulfide oxidoreductases that alter the redox state of target proteins via the reversible oxidation of their active-site dithiol proteins. NrdH proteins function with high specificity as electron donors for class I RNRs (9, 16-18). They are widespread in bacteria, particularly in those bacteria that lack glutathione (GSH), where they function as a hydrogen donor for the class Ib RNR (12, 16, 17). In E. coli, which possesses class Ia and class Ib RNRs, NrdH functions in vivo as the primary electron donor for the class Ib RNR, whereas thioredoxin or glutaredoxin is used by the class Ia NrdAB RNR (12, 17). NrdH redoxins contain a conserved CXXC motif, have a low redox potential, and can reduce insulin disulfides. NrdH proteins possess an amino acid sequence similar to that of glutaredoxins but behave functionally more like thioredoxins. NrdH proteins are reduced by thioredoxin reductase but not by GSH and lack those residues in glutaredoxin that bind GSH and the GSH binding cleft (39, 40). The structures of the E. coli and Corynebacterium ammoniagenes NrdH redoxins reveal the presence of a wide hydrophobic pocket at the surface, like that in thioredoxin, that is presumed to be involved in binding to thioredoxin reductase (39, 40). NrdI proteins are flavodoxin proteins that function as electron donors for class Ib RNRs and are involved in the maintenance of the NrdF diferric tyrosyl radical (5, 33). In Streptococcus pyogenes, NrdI is essential for the activity of the NrdHEF system in a heterologous E. coli in vivo complementation assay (33). Class Ib RNRs are proposed to depend on two specific electron donors, NrdH, which provides reducing power to the NrdE subunit, and NrdI, which supplies electrons to the NrdF subunit (33).
In this report we identify an open reading frame (ORF) in S. aureus encoding an NrdH-like protein with partial sequence relatedness to the E. coli, Salmonella enterica serovar Typhimurium, L. lactis, and C. ammoniagenes NrdH proteins. In contrast to these bacteria, the S. aureus nrdH gene does not form part of the class Ib RNR operon. The S. aureus NrdH protein differs in its structure from the canonical NrdH in its redox-active site, C-P-P-C instead of C-M/V-Q-C, and in the absence of the C-terminal [WF]SGFRP[DE] structural motif. We show that in vitro, S. aureus NrdH reduces protein disulfides and is an electron donor for the homologous class Ib NrdEF ribonucleotide reductase.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. aureus strains were grown at 37°C under aerobic and anaerobic conditions in tryptone soy broth (TSB; Difco) as described previously (26). When appropriate, kanamycin (200 μg ml−1) or erythromycin (12 μg ml−1) was supplemented. Phage transductions were carried out with φ11 as described previously (26). E. coli cells were grown in Luria-Bertani (LB) medium with the addition of ampicillin (100 μg ml−1) or kanamycin (50 μg ml−1) as needed. The effect of the oxidative stress compounds diamide, menadione, hydrogen peroxide, and τ-butyl hydroperoxide on the growth of S. aureus strains was determined as previously described (46).
TABLE 1.
Bacteria, phage, and plasmids used in this study
| Strain, phage, or plasmid | Relevant characteristic(s)a | Source and/or reference(s) |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqlacZΔM15 Tn10(Tcr)] | Laboratory stock |
| HB101 | supE44 hsdS20(rB− mB−) recA ara-14 proA2 lacY1 galK2 rspL20 xyl-5 mtl-1 | Laboratory stock |
| BL21(DF3) | Laboratory stock | |
| S. aureus | ||
| 8325-4 | Wild-type strain UV cured of prophages; spontaneous mutant with an 11-bp deletion in the rsbU gene (σB negative) | Laboratory stock; 10, 19 |
| SH1000 | Derivative of 8325-4 carrying the intact rsbU gene (σB positive); rsbU+ | Gift of S. Foster; 15 |
| RN4220 | Restriction mutant of 8325-4 used as a primary recipient for plasmids propagated in E. coli | Laboratory stock |
| RN4220 ΔnrdH | RN4220 in which the arcR gene was deleted and replaced by the Ωkm-2 kanamycin cassette; Kmr | This study |
| SH1000 ΔnrdH | Φ11/RN4220 ΔnrdH × SH1000b; Kmr | This study |
| Phage φ11 | Transducing phage that can infect S. aureus 8325-4 | Laboratory stock |
| Plasmids | ||
| pGEM-T Easy | EcoRV-linearized pGEM vector with added 3′-terminal T bases; Apr | Promega |
| pBR322::Ωkm-2 | pBR322 carrying the omega-Km2 cassette; Apr Kmr | 15 |
| pMUTIN-4 | pUC18-based suicidal vector for Gram-positive bacteria; Pspac-lacZ PpenP-lacI Apr Emr | 47 |
| pUP | PacI-BamHI-digested pMUTIN-4 containing ∼1.2 kb PCR amplified upstream of the nrdH region; Apr Emr | This study |
| pUP-DW | BamHI-Bsp120I-digested pUP and ∼1.2-kb PCR-amplified 3′ end downstream of the nrdH region; Apr Emr | This study |
| pUP_Km_DW (pKm1) | BamHI-digested pUP-DW containing the BamHI-flanked omega-Km2 cassette; Apr Emr Kmr | This study |
| pET-28a(+) and pET-30a(+) | T7-based protein expression vectors carrying a hexahistidine tag (His tag); Kmr | Novagen |
| pET-nrdH | pET-30a(+)::nrdH (C-terminal His tag); Kmr | This study |
| pET-nrdE | pET-28a(+)::nrdE (N-terminal His tag); Kmr | This study |
| pET-nrdF | pET-28a(+)::nrdF (N-terminal His tag); Kmr | This study |
| pOUA | pET-14b(+)::trxA (N-terminal His tag); Apr | O. Uziel et al., unpublished data |
| pOUB | pET-14b(+)::trxB (N-terminal His tag); Apr | Uziel et al., unpublished |
Apr, Emr, Kmr, and Tcr, resistance to ampicillin, erythromycin-lincomycin, kanamycin, and tetracycline, respectively.
Transduction with phage φ11 from donor × recipient.
DNA manipulations.
For E. coli, the preparation of plasmids, DNA manipulations, and transformation of competent cells were performed as described previously (35). For S. aureus, Southern analysis and the transformation of competent cells were performed as described previously (26). Standard procedures were employed for restriction enzyme digestion, ligation, and PCR (35). Oligodeoxynucleotide primers used in this study are listed in Table S3 in the supplemental material.
Construction of the nrdH deletion mutant.
A gene replacement strategy was used to create an nrdH knockout in S. aureus RN4220. The ∼1.2-kb DNA region upstream of nrdH was amplified by PCR using primers (Up_For and Dw_For [see Table S3 in the supplemental material]) containing PacI and BamHI restriction sites and S. aureus NCTC 8325-4 genomic DNA as a template. The purified fragment was cut with PacI and BamHI and inserted into pMUTIN-4 (47) cut with the same enzymes to give pUP. A similar-size BamHI-Bsp120I PCR fragment (primers Dw_For and Dw_Rev) containing the DNA downstream region of nrdH was ligated into pUP cut with the same enzymes to yield pUP-DW. The pUP-DW construct was digested with BamHI, treated with an alkaline phosphatase enzyme to avoid self-ligation, and then ligated with the 2.24-kb BamHI-BamHI omega-Km2 cassette, encoding kanamycin resistance (31), to yield pUP_Km_DW, with two possible orientations of the cassette. Restriction and PCR analyses identified a clone (pKm1) in which the nrdH gene and the omega-Km2 cassette possessed the same orientation. pKm1 was propagated in E. coli HB101 (Dam−) cells and then introduced into S. aureus RN4220 cells by electroporation. Transformants were selected on TSB plates containing erythromycin and kanamycin. The expected single integration event resulting from recombination between the pUP_Km_DW construct (pKm1) and the host chromosomal region was confirmed by PCR and Southern analysis (data not shown). To select for spontaneous segregation of the wild-type nrdH allele, one transformant was propagated for ∼100 generations in TSB liquid medium containing kanamycin but lacking erythromycin, and colonies were screened for a loss of the erythromycin marker on TSB plates containing kanamycin. One kanamycin-resistant, erythromycin-sensitive clone was chosen for further analysis by PCR (primers For_del_nrdH and Rev_del_nrdH [Table S3]) and DNA sequencing to confirm the desired deletion-substitution in the nrdH gene. Transduction with phage φ11 was used to introduce the deletion-substitution mutation into S. aureus SH1000 (rsbU+) as described previously (26).
Cloning and expression.
The S. aureus nrdH, nrdE, and nrdF genes were amplified from genomic DNA by PCR using the primers listed in Table S3 in the supplemental material. DNA fragments were purified with the QIA extraction kit (Qiagen), ligated into the pGEM-T Easy vector (Promega), and electroporated into E. coli XL1-Blue cells. Transformants were selected on LB plates containing ampicillin, isopropyl-β-d-thiogalactopyranoside (IPTG) (0.5 mM), and X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (80 μg ml−1) to allow blue-white screening. The verified DNA inserts of positive clones were digested with either NdeI and EcoRI or NdeI and SalI and ligated into either pET28a(+) or pET30a(+) (Novagen) linearized with the corresponding enzymes to yield pET-nrdH, pET-nrdE, and pET-nrdF. The resulting constructs were introduced into E. coli XL1-Blue cells. The plasmid constructs were verified by sequencing and introduced into E. coli BL21(DE3) for protein expression. Cultures of E. coli BL21(DE3) containing pET-nrdH, pET-nrdE, and pET-nrdF as well as pOUA and pOUB, carrying genes encoding the S. aureus thioredoxin and thioredoxin reductase, respectively (46), were grown overnight at 37°C in LB medium containing kanamycin, diluted to an absorbance at 600 nm (A600) of 0.1, and shaken vigorously at 37°C. At an A600 of 0.6, IPTG was added to a final concentration of 0.4 to 1 mM. The cells were incubated for 4 h at 37°C or overnight at 18°C for thioredoxin reductase expression, harvested by centrifugation at 4,000 × g for 15 min at 4°C, and washed once with 50 mM Tris-HCl (pH 8.0), and the supernatant was discarded. The cell pellet was stored at −20°C. Frozen cells were thawed and suspended in 50 mM Tris-HCl (pH 8.0), phenylmethylsulfonyl fluoride (PMSF) was added to 1 mM, and the mixture was sonicated (Misonics). After centrifugation at 10,000 × g for 45 min at 4°C, the clear supernatant was loaded onto a His Trap HP column (GE Amersham Biosciences) using the AKTA Prime system equilibrated with a solution containing 50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 1 mM PMSF. The column was washed with 160 ml of the same buffer containing increasing amounts of imidazole. Bound His-tagged recombinant proteins were eluted with buffer containing 100 mM imidazole. Protein samples after Ni2+ affinity purification were dialyzed against 300 mM NaCl and 50 mM Tris-HCl (pH 8.0). The recovery of recombinant proteins was monitored by use of the Bradford assay (2). Purified recombinant proteins were stored at −70°C.
Assay of ribonucleotide reductase activity.
RNR activity was assayed by the conversion of CDP to dCDP (42). Assay mixtures of 50 μl were incubated for 20 min at 30°C and contained 50 mM Tris-HCl (pH 7.6), 5 mM MgCl, 10 mM dithiothreitol (DTT), 0.2 mM dATP, 0.56 mM [3H]CDP (28,000 cpm nmol−1), 20 μg of NrdE, and 10 μg of NrdF. The amount of dCDP formed was determined by a standard method as previously described (42). One unit of enzyme corresponds to 1 nmol of dCDP formed min−1, and the specific activity is expressed as units/mg of total protein.
Insulin, DTNB, and glutaredoxin assays.
Insulin assays were performed according to a method described previously by Holmgren (14), DTNB [5,5′-dithiobis-(2-nitrobenzoic acid)] assays were performed according to a method described previously by Jordan et al. (16), and GSH-disulfide oxidoreductase assays were performed according to a method described previously by Luthman and Holmgren (25).
Bioinformatics analysis.
Open reading frame (ORF) searches were performed with the National Center for Biotechnology Information (NCBI) ORF Finder server (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Sequence entries and primary analyses of DNA and protein sequences were performed with the Clone Manager 7 program (Scientific & Educational Software, Durham, NC). Primary sequences of S. aureus NrdH-like proteins were identified in databases of the University of Oklahoma's Advanced Center for Genome Technology (http://www.genome.ou.edu/), the Comprehensive Microbial Resource of the J. Craig Venter Institute (http://cmr.jcvi.org/tigr-scripts/CMR/CmrHomePage.cgi), the Staphylococcus aureus Sequencing Group at the Sanger Centre (http://www.sanger.ac.uk/sequencing/Staphylococcus/aureus/), and the NCBI Microbial Genomes server (http://www.ncbi.nlm.nih.gov/sutils/genom_table.cgi?organism=microb) by use of the tBLASTn algorithm (1). Pairwise and multiple-sequence alignments were performed with the ClustalW program, version 1.84 (13); with the EMBL ClustalW2 server (http://www.ebi.ac.uk/Tools/clustalw2/index.html); and with the Network Protein Sequence Analysis server (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_clustalw.html). Position-specific iterated (PSI) BLAST searches (1) were performed to find close homologs of S. aureus NrdH. The searches included three iterations on the UniProt database of proteins sequences (http://www.uniprot.org/). A multiple-sequence alignment was built by using MUSCLE (8) with 200 homologous proteins found by PSI BLAST and the two Protein Data Bank (PDB) templates. The aligned sequences of the S. aureus, C. ammoniagenes, and E. coli NrdH proteins were extracted and used to build structure models of S. aureus NrdH. Phylogenetic and molecular evolutionary analyses were conducted by using MEGA 4 (41).
Structure modeling.
Appropriate three-dimensional (3D) templates suitable for the modeling of S. aureus NrdH were obtained from the FFAS03 server (http://ffas.ljcrf.edu/ffas-cgi/cgi/ffas.pl) (34). The C. ammoniagenes NrdH X-ray structure (PDB accession number 1R7H) (39) was the best result, followed by the Brucella melitensis glutaredoxin nuclear magnetic resonance (NMR) structure (PDB accession number 2KHP) and the E. coli NrdH X-ray structure (PDB accession number 1H75) (40). The two X-ray structures were used to model the S. aureus NrdH structure, employing residues 2 to 77; two models were built, one based on a monomer template and the second based on a dimer template.
Model building and validation.
The first model was based on the monomeric-state X-ray structure of E. coli (PDB accession number 1H75) (40). The model was built by using the SCWRL4 side-chain conformation prediction software (20). The sequence identity between the S. aureus NrdH and E. coli NrdH proteins is 22%. Since the alignment between the sequences of the two proteins contains only one gap, which is located prior to the second alpha helix (α2), the sequence of S. aureus NrdH (between residues 2 and 77) was threaded onto the structure of E. coli NrdH, and the conformation of the side chains (rotamers) was modeled by using SCWRL4 (20). The second model was based on the domain-swapped dimer X-ray structure of C. ammoniagenes (PDB accession number 1R7H) (39). The model was built by using NEST (32). The sequence identity between residues 2 and 77 of S. aureus NrdH and C. ammoniagenes NrdH is 16.8%. Here the gap is located inside α2 of the template structure, so a method that models the side chains only is not sufficient in this case. The models were further assessed by using the MolProbity server (http://molprobity.biochem.duke.edu/) (6, 23), which examines ψ and φ angles, Cβ deviations, atom clashes, and rotamers. The conservation score calculated by ConSurf (21), based on the multiple-sequence alignment described above, was projected onto the two structure models in order to check whether the conserved amino acids are in the core of the protein structure and the variable amino acids are on its periphery. The three structures were superimposed by using the MultiProt server (http://bioinfo3d.cs.tau.ac.il/MultiProt/) (38). The root mean square deviation (RMSD) between S. aureus NrdH and C. ammoniagenes NrdH is 0.54 Å, and the RMSD between S. aureus NrdH and E. coli NrdH is 0.47 Å.
Nucleotide sequence accession number.
The nucleotide sequence of the DNA region containing the S. aureus strain RN4220 nrdH gene has been deposited in the EMBL/GenBank/DDBJ database under accession no. AJ416021.
RESULTS AND DISCUSSION
Staphylococci contain an NrdH-like protein.
We previously reported that S. aureus, unlike E. coli and L. lactis, lacks an nrdH gene in the class Ib RNR operon (26, 45). An identical operon structure is found in other staphylococci and in certain bacilli belonging to the Bacillus cereus group. We assumed that these bacteria, all of which contain a single aerobic class Ib RNR and lack a glutathione thiol redox system, are likely to possess an NrdH redox protein. Initial attempts to identify an nrdH gene in S. aureus strains COL (11) and NCTC 8325 were performed in BLAST searches using the E. coli (16), L. lactis (17), and C. ammoniagenes (9) NrdH amino acid sequences as queries but were unsuccessful. A more refined PSI-BLAST search using a single L. lactis NrdH protein sequence as a query and an E value threshold of 0.02 led to the identification of a remote nrdH homolog, which contained a characteristic C-X-X-C redox motif. The nrdH-like gene is located between, and oriented in the opposite direction of, the cbdAB genes, encoding subunit I of the bd-type ubiquinol oxidase, and the ptsI gene, encoding phosphoenolpyruvate-dependent phosphotransferase, and is far removed from the nrdIEF gene cluster (26). A single promoter containing well-defined −10 and −35 recognition elements is positioned upstream of the nrdH structural gene (data not shown). We identified nrdH-like orthologs in the genomes of other Staphylococcus species, including S. epidermidis, S. saprophyticus, S. haemolyticus, and S. carnosus and in species of the Bacillus cereus group, including Bacillus anthracis and Bacillus thuringiensis (26, 45). All of these species contain a C-P-P-C redox-active-site motif. Figure 1 shows a multiple-sequence alignment of the S. aureus strain COL NrdH-like protein with ORFs exhibiting the highest BLAST scores, together with the E. coli, L. lactis, and C. ammoniagenes NrdH proteins. Most of these ORFs are annotated in the databases as conserved hypothetical proteins, NrdH or NrdH-like proteins, or putative glutaredoxin-like proteins. They are assumed to be members of the NrdH redoxin family of proteins (http://www.ncbi.nlm.nih.gov/Structure/cdd/cddsrv.cgi?ascbin=8&maxaln=10&seltype=2&uid=48525), which are characterized by a glutaredoxin-like amino acid sequence and a thioredoxin-like activity profile (39). Alignment of 15 NrdH-like ORFs extracted from genomes of several geographically distinct strains of S. aureus and other Staphylococcus species revealed that they (i) contain a totally conserved C-P-P-C motif, (ii) share significant amino acid sequence similarity, and (iii) possess highly conserved N-and C-terminal regions that are separated by divergent regions (see Fig. S1 in the supplemental material).
FIG. 1.
ClustalW multiple-sequence alignment of NrdH and NrdH-like bacterial proteins. Experimentally characterized S. aureus (SAU), E. coli (ECO), and C. ammoniagenes (CAM) NrdH orthologs are indicated in white boldface in black boxes. Redox-active residues are shown in gray boxes. Identical residues are indicated by asterisks, highly similar residues are indicated by colons, and weakly similar residues are indicated by dots (all shown in boldface type). SEP, Staphylococcus epidermidis; SHA, S. haemolyticus; SWA, Staphylococcus warneri; BAN, B. anthracis; BWE, Bacillus weihenstephanensis; BCO, Bacillus coahuilensis; Bsp, Bacillus sp. NRRL B-14911; STY, S. Typhimurium; DZE, Dickeya zeae; AGR, Agrobacterium tumefaciens; BME, Brucella melitensis; BLO, Bifidobacterium longum; MTU, M. tuberculosis; AOD, Actinomyces odontolyticus; LAC, Lactococcus lactis; SPY, S. pyogenes; SPN, Streptococcus pneumoniae; SAG, Streptococcus agalactiae; EFA, Enterococcus faecalis; EFE, Enterococcus faecium; LPL, Lactobacillus plantarum; LSA, Lactobacillus sakei. GenBank accession numbers of the NrdH proteins shown are listed in Table S1 in the supplemental material.
The S. aureus NrdH-like protein has a molecular mass of ∼9.2 kDa and a relatively low isoelectric point (3.93 to 4.10 in different strains). Comparative sequence analysis of S. aureus NrdH shows that it is related to the well-characterized NrdH redoxin (NrdH) and glutaredoxin (Grx) families of proteins. For example, it shares 22% and 27% identities with E. coli NrdH and L. lactis NrdH, respectively, and 12% and 25% identities with E. coli glutoredoxin-1 (GrxA) and glutoredoxin-3 (GrxC), respectively. In the N-terminal portion of the protein, it contains a motif containing two cysteines, C-P-P-C, that resembles the canonical redox-active sites of NrdH (C-M/V-Q-C), thioredoxin (C-G-P-C), and glutaredoxin (C-P-Y-C). S. aureus NrdH is most similar to the putative B. cereus NrdH-like protein, which possesses the identical redox-active motif. However, S. aureus NrdH differs from the E. coli and L. lactis NrdH proteins (which are encoded in class Ib RNR operons) in that it lacks a unique sequence present in the C-terminal portion of the canonical NrdH redoxin, termed the [WF]SGFRP[DE] motif. The latter is considered to be specific for the NrdH subclass of proteins of the thioredoxin superfamily and are thought to stabilize the protein structure via an extended network of H-bonded interactions (40). As shown in Fig. 1, the corresponding S. aureus and B. anthracis NrdH sequences are significantly different, MYHVDLD (GenBank accession number YP_185957) and VAGFQIE (accession number NP_846437), respectively.
Stehr and Lindqvist (39) previously proposed that NrdH proteins can be divided into three classes based on the sequence of the active site and the C-terminal motif: group 1 (C-V-Q-C; W-S-G-F-R-P-[ED]), group 2 (C-[MVI]-Q-C; F-SG-F-[RQ]-P), and group 3 (C-[mi]-Q-C; G-P-X-P). In terms of these features, the S. aureus and B. anthracis groups of proteins clearly form a separate and distinct subset of the NrdH proteins. Nevertheless, a comparison of the predicted secondary structure of S. aureus NrdH with that of E. coli NrdH shows that they possess very similar secondary structures (data not shown). In the following sections, we refer to SACOL1093 (NrdH-like protein in S. aureus strain COL) and its homologs as NrdH proteins. The phylogeny of NrdH protein sequences confirms that the staphylococcal and bacillus NrdH proteins are separated from the canonical NrdH redoxins as well as from the glutaredoxins (39, 44) (Fig. 2). NrdH-like proteins have also been reported for members of the Archaea (3). The Methanococcus jannaschii redoxin Mj0307 (3) shares about 20% sequence identity with the mesophilic NrdH redoxins and is similar in its sequence to glutaredoxins yet is structurally different from E. coli NrdH and, like S. aureus NrdH, lacks the C-terminal motif.
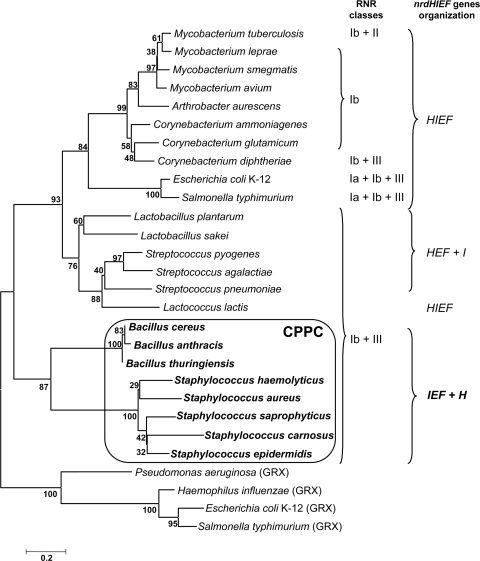
FIG. 2.
Molecular phylogeny of bacterial NrdH, NrdH-like, and glutaredoxin proteins. The CLUSTALW2 program (22) was used for the sequence alignment of NrdH, NrdH-like, and glutaredoxin (Grx) proteins. The neighbor-joining method was applied to estimate the relationships among the aligned sequences by using the MEGA 4 program (41). The right side schematically indicates the locations of the nrdH genes in different bacteria as being linked or unlinked to the class Ib RNR nrdEF gene cluster and the different RNR classes present in each organism. Complete names of bacteria and GenBank accession numbers of the NrdH and Grx proteins shown in this figure are listed in Table S2 in the supplemental material.
S. aureus NrdH structure prediction.
Homology modeling of the S. aureus NrdH structure, based on the known E. coli NrdH 3D structure, shows that their structures are closely related: each possesses a four-stranded β-sheet and three flanking helices typical of the thioredoxin superfamily (Fig. 3). Superposition of the S. aureus NrdH and E. coli NrdH structures resulted in an RMSD of 0.47 Å. The predicted S. aureus NrdH structure has an active site and surface topology similar to those of the E. coli redoxin but lacks those residues in glutaredoxins that are responsible for binding glutathione. NrdH proteins contain a highly conserved C-terminal sequence motif, 61WSGFRP(D/E)67, which in the E. coli NrdH structure links β4 to the α3 helix and creates an intricate network of H bonds involving the residue side chains and two water molecules. A salt bridge occurs between Arg65 and Asp67 (40). Conspicuously, the corresponding region in S. aureus NrdH 61MYHVDLD67 is quite different. Plausibly, it performs the same function as in E. coli NrdH in stabilizing the loop connecting β4 to α3. Note that in S. aureus NrdH, Asp65 replaces Arg65 in E. coli NrdH, which eliminates the salt bridge between Arg65 and Asp67. However, a new salt bridge may be formed between His63 and Asp65. The rim of the hydrophobic pocket in E. coli NrdH contains three arginines (Arg8, Arg44, and Arg49), which create a positively charged surface around the cavity; in the S. aureus NrdH structural model, the corresponding residues are Glu, Ile, and Phe. In contrast to E. coli NrdH, which has a deep hydrophobic pocket next to the active site where thioredoxin reductase is proposed to bind (40), the S. aureus NrdH cavity is shallower. The residues in S. aureus NrdH that correspond to those in E. coli NrdH that form the bottom of the pocket and surround it are similarly hydrophobic. However, the E. coli and S. aureus NrdH proteins differ noticeably in their electrostatic and hydrophobic profiles (see Fig. S2 in the supplemental material).
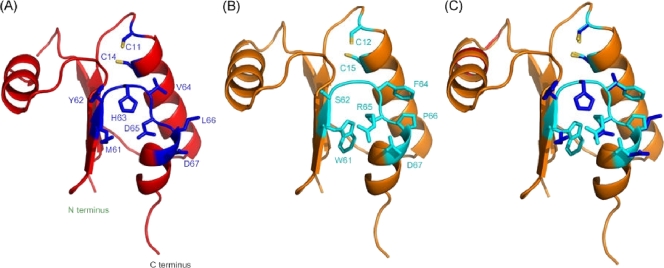
FIG. 3.
Homology structural model of S. aureus NrdH. (A and B) S. aureus NrdH model structure (see Materials and Methods) (A) and E. coli NrdH X-ray structure (PDB accession number 1H75) (B) (40) presented in cartoons. The two cysteines of the CXXC redox-active site are shown as sticks and are labeled. The residues of the loop connecting β4 to α3 are shown as sticks and are colored blue (A) and cyan (B). (C) Superposition of the two structures performed by using Multiprot (38). The RMSD between the two structures is 0.47 Å.
S. aureus nrdH is not essential for aerobic and anaerobic growth.
Preliminary experiments showed that S. aureus nrdH is transcribed under aerobic and anaerobic conditions (data not shown). To assess whether S. aureus nrdH is essential for growth, an nrdH deletion mutant was created by using a gene replacement strategy (see Materials and Methods). Plasmid pUP-Km-DW (Table 1), a derivative of pMUTIN-4 (47), was used to replace the chromosomal nrdH gene with the kanamycin resistance cassette. Strain SH1000 and the ΔnrdH deletion mutant were grown in liquid in TSB medium under aerobic and anaerobic conditions. No discernible difference was observed in the rate and extent of growth between the parent and mutant strains, establishing that NrdH is not essential under either growth condition. For aerobic growth, the generation time of both strains was ∼40 min, and maximal growth was reached, as judged by the absorbance, at the stationary phase (optical density at 600 nm [OD600] of ∼8). For anaerobic growth, the generation time of the two strains was ∼65 min, and maximal growth was reached in the stationary phase (OD600 of 1.8 to 1.9). These results establish that S. aureus NrdH is not essential for growth under the conditions employed.
To assess whether NrdH has a role in protection against oxidative stress, cultures of the wild type and the ΔnrdH mutant were grown in TSB medium and treated with hydrogen peroxide at concentrations of 0 to 25 mM, and the effect on growth was monitored as described previously (48). No significant difference was found for the growths of the parent and mutant strains (data not shown). Similarly, no significant difference was found when cells were treated with 2 to 8 mM diamide, a thiol-specific oxidant that promotes disulfide bond formation (data not shown). NrdH therefore does not appear to have a role in coping with oxidative stress under the conditions employed.
NrdH reduces protein disulfides.
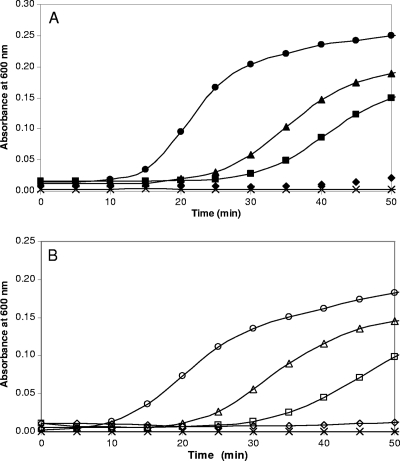
To assess whether S. aureus NrdH possesses thiol-disulfide redox activity, we tested the ability of recombinant NrdH to reduce protein disulfides. Insulin was used as the disulfide substrate in an assay previously developed for measuring thioredoxin redox activity (14). The chemical or enzymatic reduction of disulfide bonds results in the precipitation of insulin chains, which can be monitored by an increase in turbidity. E. coli thioredoxin was used as a positive control; DTT (dithiothreitol) at a concentration of 1 mM served as a negative control. Figure 4A shows that in the presence of the artificial disulfide reductant DTT (1 mM), the addition of NrdH to the standard reaction mix caused a marked increase in turbidity as measured by determining the OD600. Turbidity increased with increasing NrdH concentrations, 2 μM and 8 μM, and time of incubation. Figure 4B shows that the addition of 2 μM and 8 μM E. coli thioredoxin to the standard reaction mix caused a similar increase in turbidity, which further increased with time. In both cases, the reduction of insulin with 1 mM DTT alone is extremely low. The results demonstrate that S. aureus NrdH is as effective, if not more so, as E. coli thioredoxin in reducing insulin disulfides.
FIG. 4.
S. aureus NrdH and TrxA reduction of insulin disulfides. The assay was performed in the presence of 1 mM DTT and increasing concentrations of 2 μM (▪), 4 μM (▴), and 8 μM (•) the NrdH protein (A) and in the presence of 2 μM (□), 4 μM (▵), and 8 μM (○) TrxA (B). Controls were no DTT added (×), no NrdH added (⧫), and no TrxA added (⋄).
We next tested the ability of the S. aureus NrdH protein to transfer electrons from S. aureus thioredoxin reductase to reduce DTNB. Control reactions were performed with S. aureus and E. coli thioredoxins. Reactions were carried out in microplates as described in Materials and Methods. S. aureus NrdH was somewhat less effective in the reduction of DTNB than its cognate thioredoxin but was more effective than E. coli thioredoxin. Reduction rates measured by the ΔA405/min were 0.020 for S. aureus NrdH, 0.033 for S. aureus thioredoxin, and 0.013 for E. coli thioredoxin.
Although S. aureus lacks glutathione, we tested whether S. aureus NrdH possesses GSH-disulfide oxidoreductase activity. The experimental system employs NADPH as the source of reducing power, reduced glutathione, yeast glutathione reductase, and 2-hydroxyethyl disulfide (HED) as substrates (see Materials and Methods). No significant consumption of NADPH occurred upon the addition of NrdH to the standard reaction mix, as judged by the lack of a change in the OD340, indicating that NrdH, like thioredoxin, lacks GSH-disulfide oxidoreductase activity (data not shown).
NrdH is a hydrogen donor for the class Ib ribonucleotide reductase.
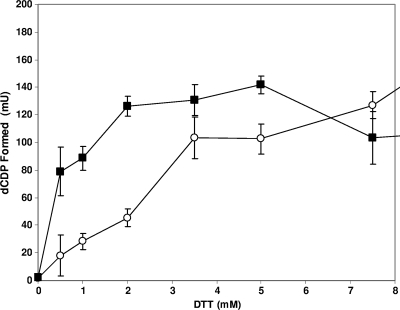
S. aureus (26) and many other Gram-positive bacteria, including C. ammoniagenes (44), Mycobacterium tuberculosis (7), and B. subtilis (37), contain a single aerobic class Ib RNR. Given that the S. aureus class Ib RNR is essential for aerobic growth (26) and lacks a glutathione-based redox system (27), we supposed that NrdH may play an important role in growth as a hydrogen donor for the class Ib RNR. To assess whether the S. aureus NrdH protein can function as an electron donor for the class Ib NrdEF RNR, we employed an in vitro assay to monitor the conversion of labeled CDP to dCDP (see Materials and Methods). His6-tagged S. aureus NrdE and NrdF proteins were expressed in E. coli and affinity purified on a nickel-chelating column (see Materials and Methods). The conversion of CDP to dCDP was dependent on the NrdE and NrdF proteins and a hydrogen donor system. Figure 5 shows NrdEF activity as a function of the DTT concentration in the standard reaction mixture with (top curve) and without (bottom curve) NrdH. In the presence of NrdH, activity was maximal at 2 mM DTT, corresponding to a level of stimulation about 2.5-fold higher that that without NrdH. The data have been adjusted to deduct the residual activity found in the absence of added DTT, which is presumably due to the presence of trace amounts of DTT in the NrdEF and NrdH protein preparations, which is difficult to remove even after exhaustive dialysis of the protein preparations. At higher DTT concentrations the relative stimulation decreases as the DTT contribution to activity increases. Thus, NrdH can serve as an electron donor for the class Ib RNR. As a control, hydroxyurea, an inhibitor of class I RNRs, abolished RNR activity in a concentration-dependent way (data not shown).
FIG. 5.
Effect of DTT on stimulating NrdH-dependent S. aureus NrdEF ribonucleotide reductase activity. NrdE (20 μg) and NrdF (10 μg) were incubated in the standard assay (see Materials and Methods) in the presence of 4.5 μg of the NrdH protein (▪) or in the absence of the NrdH protein (○).
To establish that S. aureus NrdH is able to reduce NrdEF via thioredoxin reductase, we replaced DTT in the activity assay mixture with NADPH and the cognate thioredoxin reductase. Table 2 shows that NrdH serves as a reductant for the class Ib RNR with NADPH and thioredoxin reductase as the hydrogen donor system. Reduction by NrdH was almost as effective as that by thioredoxin in transferring the reducing power from NADPH via thioredoxin reductase. These results are consistent with in vitro studies of the E. coli, L. lactis, S. Typhimurium, and C. ammoniagenes class Ib RNR systems and more-recent in vivo studies of E. coli by Gon and coworkers, which showed that NrdH specifically reduces class Ib and is recycled by thioredoxin reductase (12). Although S. aureus lacks glutathione, we asked whether NrdH was unable to mediate the transfer of electrons from NADPH to NrdEF when glutathione and glutathione reductase were used in place of NrdH and thioredoxin reductase. The results (Table 2) show that no activity was detected.
TABLE 2.
S. aureus thioredoxin TrxA and NrdH function with the thioredoxin reductase TrxB as hydrogen donors for the class Ib ribonucleotide reductase NrdEFa
| Hydrogen donor system | dCDP formed (mU) | Sp act (U/μg protein) |
|---|---|---|
| None | 9 | 0.4 |
| TrxB | 39 | 1.8 |
| TrxA + TrxB | 213 | 9.7 |
| NrdH + TrxB | 168 | 7.6 |
| NrdH + GSH + GR | ND | ND |
RNR assays were performed with [3H]CDP as a substrate and 15 μg NrdE, 7 μg NrdF, and 0.4 mM NADPH as the source of reducing power. The reaction mixtures contained 5 μg thioredoxin (TrxA) plus 1 μg thioredoxin reductase (TrxB), 4.5 μg of the NrdH protein plus 1 μg thioredoxin reductase, or 1 mM glutathione (GSH) plus 6 μg glutathione reductase (GR) as hydrogen donors. mU, milliunits; ND, not detectable.
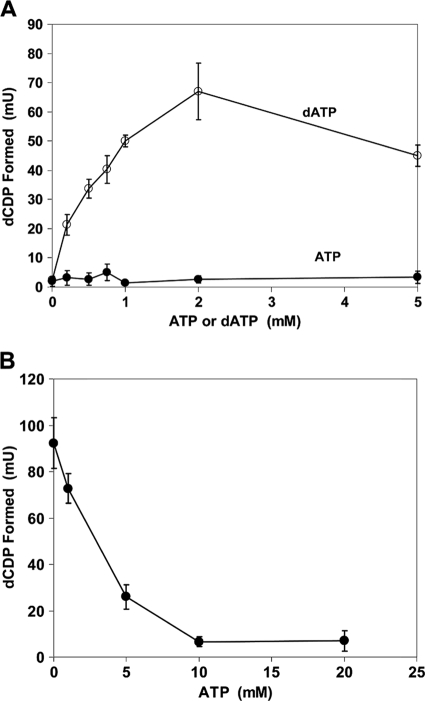
The effects of the allosteric effectors ATP and dATP on the class Ib RNR activity were examined. dATP was found to strongly stimulate CDP reduction with an optimal activity at 2.0 mM, whereas ATP had little or no effect on activity (Fig. 6A). When ATP was added to the standard reaction mixture containing 2.0 mM dATP, the reaction rate decreased with increasing ATP concentrations from 0 to 10 mM (Fig. 6B). These finding are similar to those reported previously for the E. coli and L. lactis NrdEF enzymes (17, 18).
FIG. 6.
Effect of ATP and dATP on the activity of the S. aureus NrdEF ribonucleotide reductase. Reduction of CDP to dCDP was determined in the standard assay mixture (see Materials and Methods) in the presence of increasing dATP (○) or increasing ATP (•) concentrations (A) and in the presence of 2.0 mM dATP and increasing ATP concentrations (B).
Concluding remarks.
The existence of multiple hydrogen donor systems in bacterial RNRs is well documented (29). S. aureus contains two well-characterized thiol redox systems, NrdH-thioredoxin reductase (NrdH/TrxB) (this work) and thioredoxin-thioredoxin reductase (TrxA/TrxB) (46). However, S. aureus and most other Gram-positive bacteria lack glutathione, the most common thiol in nature. Bacillithiol, a recently discovered low-molecular-weight antioxidant thiol abundant in Bacillus and S. aureus species, was proposed to serve as a substitute for glutathione in these bacteria (28). In this paper we show that S. aureus NrdH is an efficient reductant in vitro of disulfide bonds in low-molecular-weight substrates and proteins. Its activity in transferring electrons from NADPH via thioredoxin reductase to these substrates is comparable in efficiency to that of the homologous thioredoxin and thioredoxin reductase. Moreover, the S. aureus NrdH and thioredoxin proteins were equally as efficient as reductants of NrdEF. Thus, NrdH or thioredoxin can potentially function, with thioredoxin reductase, as the immediate reductant of the class Ib NrdEF RNR. These findings are consistent with the finding that S. aureus thioredoxin reductase is essential for aerobic growth (46). Since we show here that NrdH is dispensable for aerobic (and anaerobic) growth, thioredoxin is likely to be primarily responsible for the reduction of the class Ib RNR. Our conclusions are consistent with data from a recent study by Chaudhuri et al. (4), who performed a comprehensive mutational analysis of S. aureus SH1000 (the same strain used in this study) and showed that both thioredoxin and thioredoxin reductase are essential genes. Similarly, thioredoxin was found to be essential in Bacillus subtilis, which, like S. aureus, contains a class Ib RNR and lacks a glutathione redox system, based on the inability to isolate an insertional trxA mutant (36).
Given the existence of two apparently equally efficient NrdEF reductants, we speculated which of the two alternative reductants is used in vivo and under what physiological conditions. We suppose that there may exist conditions under which NrdH is preferentially required, rather than thioredoxin, but at present, these conditions are unknown. The situation resembles that found for E. coli, which contains class Ia and class Ib RNR systems, although the class Ib RNR is ordinarily not expressed in sufficient amounts to support growth when the class Ia RNR is inactive. However, when NrdEF is overexpressed (e.g., from a plasmid), it can support growth (12). In vitro studies of E. coli showed that NrdH reduces NrdEF, whereas thioredoxin (TrxA) does not (16). Because S. aureus has only the class Ib RNR, we supposed that NrdH reduces the NrdEF enzyme. However, and in contrast to E. coli, we find that S. aureus NrdH and TrxA both reduce NrdEF. Consequently, under ordinary conditions, we do not expect to see a phenotype for the S. aureus NrdH mutant. The molecular basis for this striking difference in the specificities of the E. coli and S. aureus NrdH proteins is unknown, although we present evidence that the two proteins differ in some important structural features. We suppose that NrdH may operate as a backup system when thioredoxin levels are low or when cells are exposed to stresses that activate its expression. It is noteworthy that in E. coli, S. Typhimurium, and L. lactis, nrdH is part of the class Ib RNR operon, whereas in S. aureus, nrdH is not part of the class Ib RNR operon and therefore not coordinately expressed with nrdEF. Thus, the expression and regulation of the NrdH, TrxA, and NrdEF proteins may be quite different. The possibility that NrdH may play a role in coping with oxidative stress led us to test the effect of oxidative agents on growth, but we failed to discern any significant effects on growth when NrdH was abolished. Further studies involving a broader range of physiological and environmental stresses may clarify a role for NrdH.
S. aureus NrdH is structurally different in some important aspects from previously characterized NrdH redoxins. Its redox-active site, C-P-P-C, differs from the canonical NrdH site, C-V/M-Q-C, and it lacks the highly conserved C-terminal [WF]SGFRP[DE] sequence motif that stabilizes an extended network of H-bonded interactions. The significance of these differences for NrdH function is unclear. If, as was proposed previously, NrdH is the probable ancestor of thioredoxins and glutaredoxins (40), S. aureus NrdH (and the archaeal NrdH redoxin-like proteins) forms a further division in the thioredoxin/glutaredoxin superfamily that emerged during evolution.
Supplementary Material
Acknowledgments
This research was funded in part by a grant from the NoE EuroPathoGenomics (EPG) program.
We thank Britt-Marie Sjoberg and Eduard Torrents for discussion and comment.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Cave, J. W., H. S. Cho, A. M. Batchelder, H. Yokota, R. Kim, and D. E. Wemmer. 2001. Solution nuclear magnetic resonance structure of a protein disulfide oxidoreductase from Methanococcus jannaschii. Protein Sci. 10:384-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaudhuri, R. R., A. G. Allen, P. J. Owen, G. Shalom, K. Stone, M. Harrison, T. A. Burgis, M. Lockyer, J. Garcia-Lara, S. J. Foster, S. J. Pleasance, S. E. Peters, D. J. Maskell, and I. G. Charles. 2009. Comprehensive identification of essential Staphylococcus aureus genes using transposon-mediated differential hybridisation (TMDH). BMC Genomics 10:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotruvo, J. A., Jr., and J. Stubbe. 2008. NrdI, a flavodoxin involved in maintenance of the diferric-tyrosyl radical cofactor in Escherichia coli class Ib ribonucleotide reductase. Proc. Natl. Acad. Sci. U. S. A. 105:14383-14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, I. W., A. Leaver-Fay, V. B. Chen, J. N. Block, G. J. Kapral, X. Wang, L. W. Murray, W. B. Arendall III, J. Snoeyink, J. S. Richardson, and D. C. Richardson. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35:W375-W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawes, S. S., D. F. Warner, L. Tsenova, J. Timm, J. D. McKinney, G. Kaplan, H. Rubin, and V. Mizrahi. 2003. Ribonucleotide reduction in Mycobacterium tuberculosis: function and expression of genes encoding class Ib and class II ribonucleotide reductases. Infect. Immun. 71:6124-6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fieschi, F., E. Torrents, L. Toulokhonova, A. Jordan, U. Hellman, J. Barbe, I. Gibert, M. Karlsson, and B. M. Sjoberg. 1998. The manganese-containing ribonucleotide reductase of Corynebacterium ammoniagenes is a class Ib enzyme. J. Biol. Chem. 273:4329-4337. [DOI] [PubMed] [Google Scholar]
- 10.Giachino, P., S. Engelmann, and M. Bischoff. 2001. Sigma(B) activity depends on RsbU in Staphylococcus aureus. J. Bacteriol. 183:1843-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gill, S. R., D. E. Fouts, G. L. Archer, E. F. Mongodin, R. T. Deboy, J. Ravel, I. T. Paulsen, J. F. Kolonay, L. Brinkac, M. Beanan, R. J. Dodson, S. C. Daugherty, R. Madupu, S. V. Angiuoli, A. S. Durkin, D. H. Haft, J. Vamathevan, H. Khouri, T. Utterback, C. Lee, G. Dimitrov, L. Jiang, H. Qin, J. Weidman, K. Tran, K. Kang, I. R. Hance, K. E. Nelson, and C. M. Fraser. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J. Bacteriol. 187:2426-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gon, S., M. J. Faulkner, and J. Beckwith. 2006. In vivo requirement for glutaredoxins and thioredoxins in the reduction of the ribonucleotide reductases of Escherichia coli. Antioxid. Redox Signal. 8:735-742. [DOI] [PubMed] [Google Scholar]
- 13.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 14.Holmgren, A. 1979. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254:9627-9632. [PubMed] [Google Scholar]
- 15.Horsburgh, M. J., J. L. Aish, I. J. White, L. Shaw, J. K. Lithgow, and S. J. Foster. 2002. SigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J. Bacteriol. 184:5457-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan, A., F. Aslund, E. Pontis, P. Reichard, and A. Holmgren. 1997. Characterization of Escherichia coli NrdH. A glutaredoxin-like protein with a thioredoxin-like activity profile. J. Biol. Chem. 272:18044-18050. [DOI] [PubMed] [Google Scholar]
- 17.Jordan, A., E. Pontis, F. Aslund, U. Hellman, I. Gibert, and P. Reichard. 1996. The ribonucleotide reductase system of Lactococcus lactis. Characterization of an NrdEF enzyme and a new electron transport protein. J. Biol. Chem. 271:8779-8785. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, A., E. Pontis, M. Atta, M. Krook, I. Gibert, J. Barbe, and P. Reichard. 1994. A second class I ribonucleotide reductase in Enterobacteriaceae: characterization of the Salmonella typhimurium enzyme. Proc. Natl. Acad. Sci. U. S. A. 91:12892-12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kreiswirth, B. N., S. Lofdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 20.Krivov, G. G., M. V. Shapovalov, and R. L. Dunbrack, Jr. 2009. Improved prediction of protein side-chain conformations with SCWRL4. Proteins 77:778-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Landau, M., I. Mayrose, Y. Rosenberg, F. Glaser, E. Martz, T. Pupko, and N. Ben-Tal. 2005. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 33:W299-W302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 23.Lovell, S. C., I. W. Davis, W. B. Arendall III, P. I. de Bakker, J. M. Word, M. G. Prisant, J. S. Richardson, and D. C. Richardson. 2003. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins 50:437-450. [DOI] [PubMed] [Google Scholar]
- 24.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 25.Luthman, M., and A. Holmgren. 1982. Glutaredoxin from calf thymus. Purification to homogeneity. J. Biol. Chem. 257:6686-6690. [PubMed] [Google Scholar]
- 26.Masalha, M., I. Borovok, R. Schreiber, Y. Aharonowitz, and G. Cohen. 2001. Analysis of transcription of the Staphylococcus aureus aerobic class Ib and anaerobic class III ribonucleotide reductase genes in response to oxygen. J. Bacteriol. 183:7260-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton, G. L., K. Arnold, M. S. Price, C. Sherrill, S. B. Delcardayre, Y. Aharonowitz, G. Cohen, J. Davies, R. C. Fahey, and C. Davis. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newton, G. L., M. Rawat, J. J. La Clair, V. K. Jothivasan, T. Budiarto, C. J. Hamilton, A. Claiborne, J. D. Helmann, and R. C. Fahey. 2009. Bacillithiol is an antioxidant thiol produced in bacilli. Nat. Chem. Biol. 5:625-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nordlund, P., and P. Reichard. 2006. Ribonucleotide reductases. Annu. Rev. Biochem. 75:681-706. [DOI] [PubMed] [Google Scholar]
- 30.Oehlmann, W., and G. Auling. 1999. Ribonucleotide reductase (RNR) of Corynebacterium glutamicum ATCC 13032—genetic characterization of a second class IV enzyme. Microbiology 145(Pt. 7):1595-1604. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petrey, D., and B. Honig. 2000. Free energy determinants of tertiary structure and the evaluation of protein models. Protein Sci. 9:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roca, I., E. Torrents, M. Sahlin, I. Gibert, and B. M. Sjoberg. 2008. NrdI essentiality for class Ib ribonucleotide reduction in Streptococcus pyogenes. J. Bacteriol. 190:4849-4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rychlewski, L., L. Jaroszewski, W. Li, and A. Godzik. 2000. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 9:232-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Scharf, C., S. Riethdorf, H. Ernst, S. Engelmann, U. Volker, and M. Hecker. 1998. Thioredoxin is an essential protein induced by multiple stresses in Bacillus subtilis. J. Bacteriol. 180:1869-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scotti, C., A. Valbuzzi, M. Perego, A. Galizzi, and A. M. Albertini. 1996. The Bacillus subtilis genes for ribonucleotide reductase are similar to the genes for the second class I NrdE/NrdF enzymes of Enterobacteriaceae. Microbiology 142(Pt. 11):2995-3004. [DOI] [PubMed] [Google Scholar]
- 38.Shatsky, M., R. Nussinov, and H. J. Wolfson. 2004. A method for simultaneous alignment of multiple protein structures. Proteins 56:143-156. [DOI] [PubMed] [Google Scholar]
- 39.Stehr, M., and Y. Lindqvist. 2004. NrdH-redoxin of Corynebacterium ammoniagenes forms a domain-swapped dimer. Proteins 55:613-619. [DOI] [PubMed] [Google Scholar]
- 40.Stehr, M., G. Schneider, F. Aslund, A. Holmgren, and Y. Lindqvist. 2001. Structural basis for the thioredoxin-like activity profile of the glutaredoxin-like NrdH-redoxin from Escherichia coli. J. Biol. Chem. 276:35836-35841. [DOI] [PubMed] [Google Scholar]
- 41.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 42.Thelander, L., B. R. Sjoberg, and S. Eriksson. 1978. Ribonucleoside diphosphate reductase (Escherichia coli). Methods Enzymol. 51:227-237. [DOI] [PubMed] [Google Scholar]
- 43.Torrents, E., A. Jordan, M. Karlsson, and I. Gibert. 2000. Occurrence of multiple ribonucleotide reductase classes in gamma-proteobacteria species. Curr. Microbiol. 41:346-351. [DOI] [PubMed] [Google Scholar]
- 44.Torrents, E., I. Roca, and I. Gibert. 2003. Corynebacterium ammoniagenes class Ib ribonucleotide reductase: transcriptional regulation of an atypical genomic organization in the nrd cluster. Microbiology 149:1011-1020. [DOI] [PubMed] [Google Scholar]
- 45.Torrents, E., M. Sahlin, D. Biglino, A. Graslund, and B. M. Sjoberg. 2005. Efficient growth inhibition of Bacillus anthracis by knocking out the ribonucleotide reductase tyrosyl radical. Proc. Natl. Acad. Sci. U. S. A. 102:17946-17951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uziel, O., I. Borovok, R. Schreiber, G. Cohen, and Y. Aharonowitz. 2004. Transcriptional regulation of the Staphylococcus aureus thioredoxin and thioredoxin reductase genes in response to oxygen and disulfide stress. J. Bacteriol. 186:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144(Pt. 11):3097-3104. [DOI] [PubMed] [Google Scholar]
- 48.Watson, S. P., M. O. Clements, and S. J. Foster. 1998. Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180:1750-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu, L., and J. Xie. 2010. Comparative genomics analysis of Mycobacterium NrdH-redoxins. Microb. Pathog. 48:97-102. [DOI] [PubMed] [Google Scholar]
- 50.Yang, F., S. C. Curran, L. S. Li, D. Avarbock, J. D. Graf, M. M. Chua, G. Lu, J. Salem, and H. Rubin. 1997. Characterization of two genes encoding the Mycobacterium tuberculosis ribonucleotide reductase small subunit. J. Bacteriol. 179:6408-6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.