Abstract
To cause disease, Clostridium difficile spores must germinate in the host gastrointestinal tract. Germination is initiated upon exposure to glycine and certain bile acids, e.g., taurocholate. Chenodeoxycholate, another bile acid, inhibits taurocholate-mediated germination. By applying Michaelis-Menten kinetic analysis to C. difficile spore germination, we found that chenodeoxycholate is a competitive inhibitor of taurocholate-mediated germination and appears to interact with the spores with greater apparent affinity than does taurocholate. We also report that several analogs of chenodeoxycholate are even more effective inhibitors. Some of these compounds resist 7α-dehydroxylation by Clostridium scindens, a core member of the normal human colonic microbiota, suggesting that they are more stable than chenodeoxycholate in the colonic environment.
Clostridium difficile is a Gram-positive, spore-forming, anaerobic bacterium that is pathogenic for both humans and animals (33, 44). Infections caused by C. difficile range from mild diarrhea to more life-threatening conditions, such as pseudomembranous colitis (33). In the classic case, prior antibiotic treatment that disrupts the normally protective colonic flora makes patients susceptible to C. difficile infection (CDI) (35, 53). Other antibiotics, such as vancomycin and metronidazole, are the most commonly used treatments for CDI (54). However, because these antibiotics also disrupt the colonic flora, 10 to 40% of patients whose symptoms have been ameliorated suffer from relapsing CDI (15, 24). The annual treatment-associated cost for CDI in the United States is estimated to be between $750 million and $3.2 billion (8, 9, 16, 31). Moreover, the number of fatal cases of CDI has been increasing rapidly (14, 39). Thus, there is an urgent need to find alternative therapies for CDI.
C. difficile infection likely is initiated by infection with the spore form of C. difficile (12). C. difficile elicits disease through the actions of two secreted toxins, TcdA and TcdB (48). TcdB was recently shown to be critical for pathogenesis in an animal model of disease (18). Since the toxins are produced by vegetative cells, not by spores (17), germination and outgrowth are prerequisites for pathogenesis.
Spore germination is triggered by the interaction of small molecules, called germinants, with receptors within the spore inner membrane. These germinants vary by bacterial species and can include ions, amino acids, sugars, nucleotides, surfactants, or combinations thereof (43). The recognition of germinants triggers irreversible germination, leading to Ca2+-dipicolinic acid release, the uptake of water, the degradation of the cortex, and, eventually, the outgrowth of the vegetative bacterium (43). The germination receptors that C. difficile uses to sense the environment have not been identified. Based on homology searches, C. difficile germination receptors must be very different from known germination receptors (42), but they appear to be proteinaceous (13).
Taurocholate, a primary bile acid, has been used for approximately 30 years by researchers and clinical microbiologists to increase colony formation by C. difficile spores from patient and environmental samples (3, 49, 51, 52). This suggested that C. difficile spores interact with bile acids along the gastrointestinal (GI) tract and that spores use a host-derived signal to initiate germination.
The liver synthesizes the two major primary bile acids, cholate and chenodeoxycholate (40). These compounds are modified by conjugation with either taurine (to give taurocholate or taurochenodeoxycholate) or glycine (producing glycocholate or glycochenodeoxycholate). Upon secretion into the digestive tract, bile aids in the absorption of fat and cholesterol; much of the secreted bile is actively absorbed and recycled back to the liver for reutilization (40). Though efficient, enterohepatic recirculation is not complete; bile enters the cecum of the large intestine at a concentration of approximately 2 mM (30).
In the cecum, bile is modified by the normal, benign colonic flora. First, bile salt hydrolases found on the surfaces of many bacterial species remove the conjugated amino acid, producing the deconjugated primary bile acids cholate and chenodeoxycholate (40). These deconjugated primary bile acids are further metabolized by only a few species of intestinal bacteria, including Clostridium scindens. C. scindens actively transports unconjugated primary bile acids into the cytoplasm, where they are 7α-dehydroxylated, converting cholate to deoxycholate and chenodeoxycholate to lithocholate (21, 40). The disruption of the colonic flora by antibiotic treatment abolishes 7α-dehydroxylation activity (41).
Building upon the work on Wilson and others (51, 52), we demonstrated that taurocholate and glycine, acting together, trigger the loss of the birefringence of C. difficile spores (45). All cholate derivatives (taurocholate, glycocholate, cholate, and deoxycholate) stimulate the germination of C. difficile spores (45). Recently it was shown that taurocholate binding is prerequisite to glycine binding (37). At physiologically relevant concentrations, chenodeoxycholate inhibits taurocholate-mediated germination (46) and also inhibits C. difficile vegetative growth, as does deoxycholate (45). In fact, C. difficile spores use the relative concentrations of the various bile acids as cues for germination within the host (10).
Since chenodeoxycholate is absorbed by the colonic epithelium and metabolized to lithocholate by the colonic flora (25, 40), the use of chenodeoxycholate as a therapy against C. difficile disease might be hindered by its absorption and conversion to lithocholate.
Here, we further characterize the interaction of C. difficile spores with various bile acids and demonstrate that chenodeoxycholate is a competitive inhibitor of taurocholate-mediated germination. Further, we identify chemical analogs of chenodeoxycholate that are more potent inhibitors of germination and that resist 7α-dehydroxylation by the colonic flora, potentially increasing their stability and effectiveness as inhibitors of C. difficile spore germination in the colonic environment.
MATERIALS AND METHODS
C. difficile and C. scindens growth conditions.
C. difficile UK1 (obtained from D. Gerding), an epidemic strain (REA type BI23), was isolated during a 2006 outbreak at Stoke-Mandeville Hospital in the United Kingdom. C. scindens is a core member of the gut microbiota that metabolizes primary bile acids to secondary bile acids (34, 50). C. difficile UK1 and C. scindens VPI12708 (obtained from P. Hylemon) were routinely grown on BHIS (BHI [brain heart infusion] plus 5 g/liter yeast extract plus 0.1% l-cysteine) agar medium in a Coy anaerobic chamber containing 5% CO2, 10% H2, and 85% N2.
C. difficile spore preparation.
To prepare spores, C. difficile UK1 was streaked on reduced BHIS agar medium and incubated under anaerobic conditions for 4 days. Plates then were removed from the chamber, and all surface growth (containing spores, vegetative cells, and debris) was scraped up and transferred to microcentrifuge tubes containing 1 ml sterile ice-cold water. After incubation overnight at 4°C to allow the release of the mature spores from mother cells, the suspension was washed five times in ice-cold sterile water. The washed pellets then were combined and resuspended in 3 ml ice-cold sterile water. The suspension was layered on top of a 10-ml bed volume of 50% (wt/vol) sucrose in water (17). The gradient was centrifuged in a swinging-bucket rotor at 3,200 × g for 20 min. During the centrifugation, vegetative cells and debris collected at the interface while spores migrated through the 50% sucrose bed and formed a pellet. After centrifugation, the spore pellet was washed five times to remove the sucrose and suspended in water. All spore preparations were >99% free of vegetative cells and debris, and all spores appeared phase bright.
Germination of C. difficile spores.
Purified spores were heat activated for 30 min at 60°C and placed on ice. Heat-activated spores were diluted in 1 ml BHIS only or BHIS supplemented with 2, 5, 10, 20, 50, or 100 mM taurocholate (Sigma-Aldrich T4009) with or without an inhibitor. Heat-activated spores in BHIS with 2% ethanol or 2% dimethylsulfoxide (DMSO) were used as controls to rule out the effects of organic solvent on germination. Germination was monitored spectrophotometrically at 600 nm. The ratio of the A600 at time X (Tx) to the A600 at time zero (T0) was plotted against time. Germination rates were determined using the slopes of the linear portions of the germination plots. Data are reported as the averages from three independent experiments with one standard deviation.
Chenodeoxycholate, ursodeoxycholate, and lithocholate were purchased from Acros Organics and dissolved in 100% ethanol at 100 mM. The chenodeoxycholate analogs were purchased from Steraloids Inc. and dissolved in DMSO at 10 mM.
7α-Dehydroxylation of bile acids and bile acid analogs.
C. scindens bile acid metabolism was induced in 18-h, exponential-phase cultures by incubation for 3 h after the addition of 100 μM cholate (50). Two hundred fifty microliters of the induced C. scindens culture then was diluted in 5 ml of BHIS medium supplemented with 100 μM (500 nmol) bile acid or bile acid analog. When analogs were being tested, cholate (100 μM [500 nmol]) was added as a positive control for 7α-dehydroxylation in the presence of the bile analog. After a 24-h incubation, NaOH was added to 1 M and the samples were incubated overnight at 65°C to hydrolyze esters. The solution was acidified with HCl and adjusted to 20% methanol. Hyodeoxycholate (HCA) was added as an internal standard for recovery, and the resulting mixture was purified over a C18 solid-phase extraction column (Thermo Sci. 60108-305) at a flow rate of 1 ml/min. The column was washed with 4 ml water and eluted with 3 ml 100% methanol. The de-esterified bile acids and analogs were dried at 60°C under a stream of forced air.
Bile acid labeling and HPLC separation.
We added 780 μl of 3 mM N,N-diisopropylethylamine and 390 μl of 3 mM 2-bromo-2-acetonaphthone, both dissolved in acetonitrile, to the tubes of dried, de-esterified bile acids and incubated the tubes overnight in the dark at 60°C. This reaction adds to all carboxyl groups a naphthylacylester that absorbs at 245 nm (29). The labeling compounds were added in excess. When monitored by high-performance liquid chromatography (HPLC), the labeling material was detected (data not shown), indicating that the labeling reaction proceeds to completion. The reaction mixture was dried in a SpeedVac, suspended in 60% acetonitrile-40% water (vol/vol) and subjected to reverse-phase high-performance liquid chromatography using a C18 column (4.6 by 250 mm; 5 μm C18; Phenomenex, Torrance, CA). Separation was carried out at a flow rate of 1 ml/min using 90 ml of 60% acetonitrile-40% water (vol/vol). Isopropanol then was introduced as a 30-ml gradient from 0 to 100% to elute lithocholate from the C18 column. The elution of the naphthylacylester derivatives of the bile acids was monitored at 245 nm. Peak area was determined using Breeze HPLC software (Waters, Inc.).
RESULTS
Kinetic analysis of C. difficile spore germination in the presence of taurocholate.
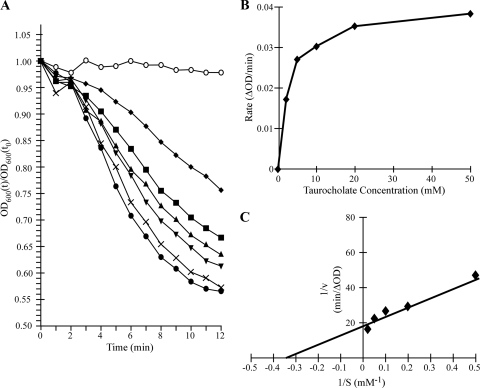
We have shown previously that taurocholate is a cogerminant with glycine for C. difficile spore germination (45). To further characterize the interaction of spores with taurocholate, we incubated purified C. difficile spores in glycine-containing rich medium (BHIS) with various concentrations of taurocholate. Mature spores are birefringent and appear bright under a phase-contrast microscope (designated phase bright). The initiation of germination can be measured spectrophotometrically by following the decrease in absorbance at 600 nm, corresponding to the transition from a phase-bright spore to a phase-dark spore (27). The loss of birefringence, one of the first measurable changes during germination, occurs within minutes after exposure to germinant (27). There was no decrease in A600 when spores in BHIS were incubated without taurocholate (Fig. 1A ) (45). The addition of increasing concentrations of taurocholate resulted in increased rates and extents of germination (Fig. 1A). To analyze the kinetics of germination, the slope of the linear range of absorbance decrease at each concentration of taurocholate was plotted against the taurocholate concentration (Fig. 1B). Increasing the concentration of taurocholate from 2 to 50 mM resulted in a rate curve that approached saturation (Fig. 1B).
FIG. 1.
Germination of Clostridium difficile spores with increasing concentrations of taurocholate. C. difficile spores were produced and purified as described in Materials and Methods. (A) Heat-activated, purified spores were suspended in BHIS medium containing no taurocholate (○), 2 mM taurocholate (⧫), 5 mM taurocholate (▪), 10 mM taurocholate (▴), 20 mM taurocholate (▾), 50 mM taurocholate (×), or 100 mM taurocholate (•). The ratio of the OD600 at the various time points to the OD600 at T0 is plotted versus time. (B) The linear portion of each curve in panel A was used to determine the maximum rate of germination under each condition. The maximum rate of germination under each condition was plotted versus taurocholate concentration. (C) The inverse rate (1/v [min/ΔOD600], where v = rate) versus the inverse taurocholate concentration (1/S [mM−1], where S = substrate concentration) was plotted. The linear best fit was generated and used to determine the apparent Km and Vmax for taurocholate-mediated germination.
Michaelis-Menten kinetic analysis of spore germination has been used to study spore germination in other pathogens (1, 36). Applying Michaelis-Menten kinetic analysis, we determined that C. difficile spores have an apparent Km for taurocholate of 2.8 mM and a Vmax of 0.053 optical density (OD) units/min (Fig. 1C). These results provide a quantitative measure of the interaction of taurocholate with the spores and indicate that the interaction occurs at a physiologically relevant bile acid concentration (30).
Chenodeoxycholate is a competitive inhibitor of C. difficile spore germination.
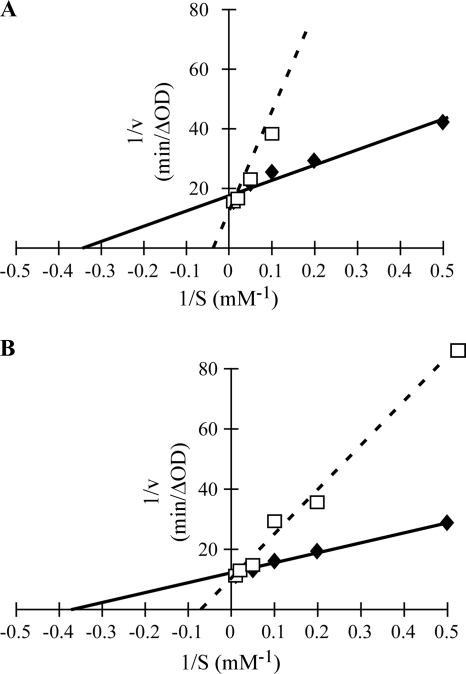
We have shown previously that chenodeoxycholate is an inhibitor of C. difficile spore germination (46). To determine the mechanism of inhibition, we incubated C. difficile spores in the presence of 2 mM chenodeoxycholate and increasing concentrations of taurocholate. As shown in Fig. 2 A, adding chenodeoxycholate increased the apparent Km for taurocholate but had almost no effect on the Vmax. We conclude that chenodeoxycholate is a competitive inhibitor of C. difficile spore germination, with an apparent Ki of approximately 370 μM (Table 1). These results indicate that C. difficile spores have a higher apparent affinity for chenodeoxycholate than for taurocholate.
FIG. 2.
Kinetics of C. difficile spore germination in the presence of inhibitors. (A) Heat-activated spores were suspended in BHIS medium containing 2, 5, 10, 20, 50, or 100 mM taurocholate (⧫) with or without 2 mM chenodeoxycholate (□). (B) Heat-activated spores were suspended in BHIS medium containing 2, 5, 10, 20, 50, or 100 mM taurocholate only (⧫) with or without 0.2 mM 5β-cholanic acid-3α,7α-diol-diacetate-methyl ester (37DAME) (□). The maximum rate of germination under each condition was determined as described for Fig. 1B, and the double-reciprocal plot was generated as described for Fig. 1C. The linear best fit was generated and used to determine the apparent Km for taurocholate, Vmax for germination, and Ki for chenodeoxycholate and 37DAME.
TABLE 1.
Inhibitor constants for chenodeoxycholate and chenodeoxycholate analogs
| Compound | Kia (μM) |
|---|---|
| CDCAb | 378 ± 55 |
| UCAb | 213 ± 80 |
| 6KLAb | 148 ± 60 |
| LCAc | 104 ± 12 |
| 3olAcc | 123 ± 41 |
| 37D3AMEc | 172 ± 96 |
| 37DAMEc | 93 ± 32 |
| 37DMEc | 44 ± 20 |
Ki values were determined using the equation Ki = [inhibitor]/(Kmtaurocholate,obs/Kmtaurocholate − 1) and are listed as the averages from three independent experiments with one standard deviation of the means.
Incubated with C. difficile spores at 2 mM.
Incubated with C. difficile spores at 0.2 mM.
Chenodeoxycholate analogs.
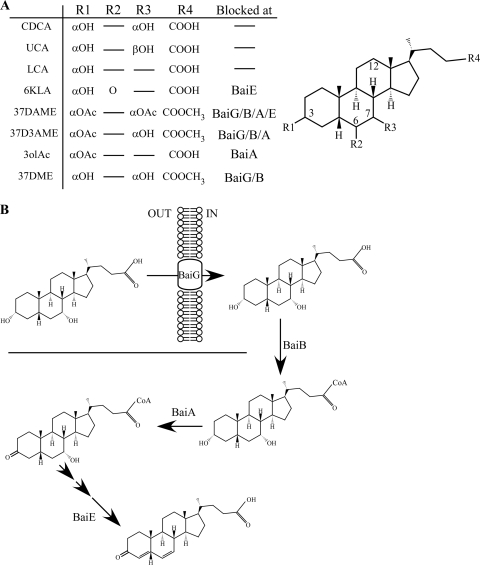
Chenodeoxycholate is metabolized by the colonic flora to lithocholate (40). The pathway for this 7α-dehydroxylation is known primarily through studies of C. scindens (5, 7, 20, 21, 40). C. scindens is a core member of the human colonic flora (34) and actively takes up unconjugated primary bile acids through the membrane protein BaiG (5, 21). In the cytoplasm, the first step in 7α-dehydroxylation is thought to be the ligation of the primary bile acid to coenzyme A (CoA) (20). Subsequent reactions target the 3α-hydroxyl group and finally the 7α-hydroxyl group (40). The modification of the primary bile acid carboxyl moiety likely would prevent the uptake of the bile acid and block its ability to be ligated to CoA, because uptake requires a deconjugated bile acid (2, 21) and ligation to CoA requires the presence of a free carboxyl group (20). Further, by altering the 3α- or the 7α-hydroxyl group, 7α-dehydroxylation may be blocked due to the inability of the bile acid to support the proper reaction chemistry. Using these criteria, we identified several chenodeoxycholate analogs (Fig. 3) that were predicted to resist 7α-dehydroxylation by the colonic flora, thereby potentially increasing their stability within the colonic environment. These compounds fall into two overlapping classes. First, 5β-cholanic acid-3α,7α-diol-3-acetate methyl ester (37D3AME), 5β-cholanic acid-3α,7α-diol-diacetate methyl ester (37DAME), and 5β-cholanic acid-3α,7α-diol methyl ester (37DME) are methyl esters and therefore were predicted to resist uptake by C. scindens BaiG or CoA ligation or both. The second class of analogs was predicted to be blocked in 7α-dehydroxylation because they are unable to support the proper reaction chemistry. These compounds, 5β-cholanic acid-3α-ol acetate (3olAc), 37D3AME, 37DAME, and 5β-cholanic acid-3α-ol-6-one (6KLA), have modifications at the 3, 7, or 6 position. 37D3AME and 37DAME belong to both classes of compounds.
FIG. 3.
Chemical analogs of chenodeoxycholate. (A) Analogs of chenodeoxycholate that are predicted to resist the 7α-dehydroxylating activity of the colonic flora are listed. Chenodeoxycholate (CDCA), ursodeoxycholate (UCA; the 7β-epimer of chenodeoxycholate), and lithocholate (LCA; the 7α-dehydroxylated form of chenodeoxycholate) also are listed. The step in the C. scindens 7α-dehydroxylation pathway at which each compound would be blocked is indicated. (B) A schematic representation of the C. scindens 7α-dehydroxylation pathway (40). Unconjugated CDCA is transported into the cytoplasm by the membrane protein BaiG. CoA is ligated through the action of BaiB. BaiA converts the 3α-hydroxyl to 3-keto. The 7α-hydroxyl is modified by BaiE.
Effects of chenodeoxycholate analogs on C. difficile spore germination.
We tested the chenodeoxycholate analogs (Fig. 3) individually for the inhibition of germination. Some compounds (ursodeoxycholate, lithocholate, 6KLA, 3olAc, and 37D3AME) had apparent Ki values similar to that of chenodeoxycholate, but other compounds (37DAME and 37DME) were more potent inhibitors than chenodeoxycholate (Table 1). Figure 2B shows the double-reciprocal plot for 37DAME. 37DAME was a competitive inhibitor of C. difficile spore germination, with an apparent Ki in this experiment of 81 μM.
37DME and 37DAME resist 7α-dehydroxylation by Clostridium scindens.
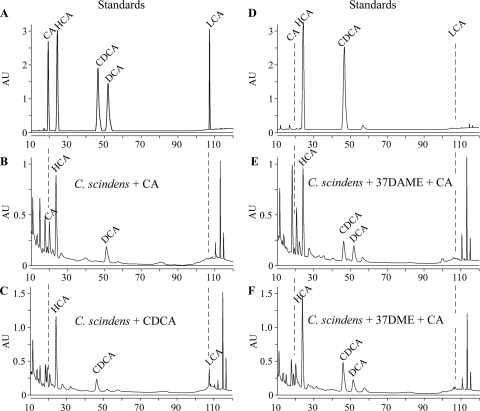
The modification of the carboxyl, the C-3 hydroxyl or the C-7 hydroxyl, or combinations thereof was predicted to prevent C. scindens-mediated 7α-dehydroxylation. To test this supposition, we incubated 37DME or 37DAME in the presence of C. scindens that had been induced for 7α-dehydroxylation by growth with 100 μM cholate. C. scindens has been shown to 7α-dehydroxylate primary, unconjugated bile acids. Cholate (CA) and chenodeoxycholate (CDCA), 500 nmol each, were used as positive controls for 7α-dehydroxylation. After incubation with C. scindens, bile acids were hydrolyzed in 1 M NaOH overnight at 65°C to generate free carboxyl groups that could be labeled with 2-bromo-2-acetonaphthone and detected by reverse-phase HPLC. The elution times of pure, hydrolyzed, labeled compounds are listed in Table 2 and illustrated in Fig. 4 A. Hyodeoxycholate (HCA; 500 nmol), which was added to every sample before purification as an internal control for extraction and labeling, eluted at approximately 24 min. As expected, the incubation of CA with C. scindens resulted in the appearance of a peak corresponding to deoxycholate (DCA) (Fig. 4B). By quantifying the peak area and comparing it to the area of a peak of known concentration (Fig. 4A), we determined that 108 nmol of DCA was formed from CA, corresponding to 48.6% conversion of CA to DCA (Table 3). Performing the same analysis for C. scindens incubated with CDCA, a peak corresponding to LCA was observed (Fig. 4C). This peak corresponded to 89 nmol of LCA or 49.2% conversion of CDCA to LCA (Table 3). Due to the labeling procedure, which labels all carboxyl groups, there are other peaks in the HPLC chromatograms that are not bile acids. 37DME and 37DAME (500 nmol each) were incubated with 7α-dehydroxylation-induced C. scindens in the presence of 500 nmol cholate. The ester hydrolysis of 37DME and 37DAME yields the parent CDCA compound (Fig. 3 and 4D). The incubation of 37DAME and cholate with C. scindens resulted in the conversion of cholate to deoxycholate (113 nmol; 40% conversion [Table 3]) but did not result in the formation of significant amounts of LCA (0.0025 nmol; 0.0025% conversion [Table 3]), the expected product of 7α-dehydroxylation and ester hydrolysis of 37DAME (Fig. 4E). Also, the incubation of 37DME and cholate with C. scindens resulted in the conversion of 113 nmol of CA to DCA (55.1% conversion) (Fig. 4F, Table 3) but essentially no conversion of 37DME to lithocholate (Fig. 4F, Table 3). These results indicate (i) that C. scindens is unable to 7α-dehydroxylate 37DME or 37DAME; (ii) that the modification of the C-3 hydroxyl, C-7 hydroxyl, or C-24 carboxyl prevents 7α-dehydroxylation; and (iii) that these compounds resist 7α-dehydroxylation and therefore should be more stable than chenodeoxycholate in the colonic environment.
TABLE 2.
HPLC retention times of bile acid standards
| Bile acid | Retention time (min) |
|---|---|
| Cholate | 19.3 |
| Chenodeoxycholate | 46.4 |
| Deoxycholate | 51.9 |
| Lithocholate | 107.2 |
| Hyodeoxycholate | 24.3 |
| 37DMEa | 46.4 |
| 37DAMEa | 46.4 |
The ester hydrolysis of 37DAME and 37DME yields chenodeoxycholate.
FIG. 4.
Metabolism of bile acids and bile acid analogs by C. scindens. Bile acids and bile acid analogs were incubated with C. scindens as described in Materials and Methods. (A) Pure cholate (CA), hyodeoxycholate (HCA), chenodeoxycholate (CDCA), deoxycholate (DCA), and lithocholate (LCA) were labeled with 2-bromo-2-acetonaphthone and separated by HPLC. (D) Pure 37DAME and 37DME were hydrolyzed, labeled, and separated by HPLC. C. scindens was incubated with cholate (B), chenodeoxycholate (C), 37DAME and cholate (E), or 37DME and cholate (F). Dashed lines indicate the expected mobilities of CA and LCA.
TABLE 3.
Quantification of HPLC peak area
| Compound(s) (500 nmolc each) incubated with C. scindens and compound formed | Amt eluted (nmol)d | % Recoverye | % Conversion |
|---|---|---|---|
| CA | |||
| CA | 114 | ||
| HCAa | 229 | 46 | |
| DCA | 108 | 48.6 | |
| CDCA | |||
| HCAa | 292 | 58 | |
| CDCA | 92 | ||
| LCA | 89 | 49.2 | |
| 37DAME + CA | |||
| CA | 177 | ||
| HCAa | 263 | 52 | |
| 37DAME (assayed as CDCA)b | 100 | ||
| DCA | 113 | 40.0 | |
| LCA | 0.0025 | 0.0025 | |
| 37DME + CA | |||
| CA | 110 | ||
| HCAa | 389 | 78 | |
| 37DME (assayed as CDCA)b | 219 | ||
| DCA | 135 | 55.1 | |
| LCA | 0.00096 | 0.00044 |
HCA was included as an internal extraction and labeling control.
Ester hydrolysis of 37DAME and 37DME yields chenodeoxycholate.
Indicates the total amount of compound labeled or incubated with C. scindens.
Indicates the total amount of compound purified.
Indicates the percent recovery of HCA during the purification process.
DISCUSSION
Bile acids are necessary to initiate the germination of C. difficile spores (Fig. 1) (45). By studying the kinetics of taurocholate-mediated germination, we found that C. difficile spores have an apparent Km of 2.8 mM for taurocholate (Fig. 1C). This is likely to be a physiologically relevant Km, because bile enters the cecum at approximately 2 mM (30). In the presence of chenodeoxycholate, the apparent affinity for taurocholate decreases by 5- to 8-fold (Fig. 2A). The receptors that C. difficile spores use to recognize potential germinants have not been identified. Based on the absence of homologs of receptors found in other Bacillus spp. and Clostridium spp. (42), it is likely that C. difficile receptors have unique properties. The analysis of the structure of cholate and chenodeoxycholate suggests that the 12α-hydroxyl group is important for the recognition of bile acids as germinants and as inhibitors of germination. The removal of the 12α-hydroxyl group of cholate converts the germinant to an inhibitor of germination (45, 46). Thus, the 12α-hydroxyl group may make an important contact with a germination receptor, triggering germination. In studying analogs of cholate and chenodeoxycholate, we hoped to define other side chains that are important for initiating germination or inhibiting germination. However, because all nonbiological compounds tested inhibited germination, albeit with different efficiencies, little structure/function analysis was possible. We can conclude that the 7α-hydroxyl group is not required either for germination or for the inhibition of germination, since lithocholate and 3olAc inhibit germination, whereas deoxycholate and cholate stimulate germination similarly (45). Further analysis of the C-3 position and combinations of C-3, C-7, and C-12 modifications could yield structure/function information.
The kinetic analysis suggests that chenodeoxycholate has an inhibitor constant (Ki) of approximately 370 μM (Table 1), indicating that spores interact with chenodeoxycholate more avidly than they interact with taurocholate. The higher apparent affinity of spores for chenodeoxycholate versus cholate has interesting consequences for the life cycle of the bacterium within the host. When C. difficile spores enter the small intestine, they encounter approximately equal concentrations of cholate derivatives and chenodeoxycholate derivatives (30). Under these conditions, C. difficile spores should be inhibited for germination, thereby preventing the production of vegetative bacteria in the aerobic, small intestinal environment (6).
In a normal, healthy host, the colonic microflora 7α-dehydroxylates chenodeoxycholate to lithocholate, also an inhibitor of germination. However, in an antibiotic-treated host, chenodeoxycholate is not 7α-dehydroxylated and has been shown to be absorbed by the colonic epithelium at a rate 10 times higher than that of cholate (25). Thus, in an antibiotic-treated individual, the ratio of cholate to chenodeoxycholate derivatives would favor germination.
These results also raise the possibility that chenodeoxycholate could be used to inhibit C. difficile spore germination in the GI tract. However, because chenodeoxycholate is metabolized by the colonic flora to lithocholate, its therapeutic use could be limited. Lithocholate also inhibits taurocholate-mediated germination (Table 1), but it is nearly insoluble, and secondary bile acids, such as lithocholate, have been implicated in colorectal carcinogenesis (23, 28, 38), precluding its use in therapeutics.
For the compounds in Fig. 3 to be effective inhibiters of C. difficile spore germination in the gut, they must resist uptake by the colonic epithelium and resist 7α-dehydroxylation by the normal flora. The farnesoid X receptor (FXR) is a nuclear receptor that is activated by bile acids in the cytoplasm of the eukaryotic cell (19, 32). Upon activation, FXR translocates to the nucleus, where it affects the transcription and upregulation of the bile acid binding proteins (BABPs) (11). BABPs are thought to aid in the transport of bile to the liver or protection of intestinal cells against high levels of bile acids (22, 47). Based on the crystal structures of FXR (26) and ileal-BABP (I-BABP) (4), 37DAME (Fig. 3) is unlikely to interact with either of these proteins. The acetyl group on C-3 of 37DAME would place a methyl group in the polar binding pocket of FXR (26). We predict that this would lead to an unproductive interaction that would not trigger FXR translocation to the nucleus. The interaction of 37DAME with I-BABP also should be less effective than that for natural bile acids. The acetylation of C-7 and the methyl ester of 37DAME should prevent the hydrogen bonding that seems to coordinate bile acids on I-BABP (4). On the other hand, 37DME might activate both of these proteins. Therefore, we predict that 37DAME is the more stable of the two analogs in the colon, as it may be unable to be trafficked and activate FXR signaling, though more work is needed to determine the validity of these predictions.
We have identified several chenodeoxycholate analogs that are predicted to resist the 7α-dehydroxylating activity of C. scindens (Fig. 3). Many of these compounds have modifications at the C-3, C-6, C-7, or carboxyl positions that would preclude them from being taken up by C. scindens or being ligated to CoA or supporting proper 7α-dehydroxylation reaction chemistry. Some of these compounds are more-potent inhibitors of germination than is CDCA (Table 1), and their ability to resist 7α-dehydroxylation was demonstrated experimentally (Fig. 4). If the behavior of C. scindens is typical of other 7α-dehydroxylating bacteria in the normal flora, some of these compounds might be useful in prophylaxis by providing a stable inhibitor of C. difficile spore germination in the presence of the flora before antibiotic treatment, or as therapy for relapsing CDI by preventing C. difficile spore germination after antibiotic treatment while the flora recovers from such treatment.
Acknowledgments
We thank Dale Gerding and Phillip Hylemon for the generous gifts of C. difficile UK1 and C. scindens VPI1208, respectively. We also thank Boris Belitsky, Laurent Bouillaut, Shonna McBride, and Michael Malamy for their comments and criticisms during the preparation of the manuscript.
This project was supported in part by funding from the National Institutes of Health under contract no. N01 AI30050 (S. Tzipori, principal investigator). J.A.S. acknowledges support through NIH Federal Training in Education and Critical Research Skills (TEACRS) fellowship K12 GM074869-02.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Akoachere, M., R. C. Squires, A. M. Nour, L. Angelov, J. Brojatsch, and E. Abel-Santos. 2007. Identification of an in vivo inhibitor of Bacillus anthracis spore germination. J. Biol. Chem. 282:12112-12118. [DOI] [PubMed] [Google Scholar]
- 2.Batta, A. K., G. Salen, R. Arora, S. Shefer, M. Batta, and A. Person. 1990. Side chain conjugation prevents bacterial 7-dehydroxylation of bile acids. J. Biol. Chem. 265:10925-10928. [PubMed] [Google Scholar]
- 3.Bliss, D. Z., S. Johnson, C. R. Clabots, K. Savik, and D. N. Gerding. 1997. Comparison of cycloserine-cefoxitin-fructose agar (CCFA) and taurocholate-CCFA for recovery of Clostridium difficile during surveillance of hospitalized patients. Diagn. Microbiol. Infect. Dis. 29:1-4. [DOI] [PubMed] [Google Scholar]
- 4.Capaldi, S., G. Saccomani, D. Fessas, M. Signorelli, M. Perduca, and H. L. Monaco. 2009. The x-ray structure of zebrafish (Danio rerio) ileal bile acid-binding protein reveals the presence of binding sites on the surface of the protein molecule. J. Mol. Biol. 385:99-116. [DOI] [PubMed] [Google Scholar]
- 5.Coleman, J. P., W. B. White, and P. B. Hylemon. 1987. Molecular cloning of bile acid 7-dehydroxylase from Eubacterium sp. strain VPI 12708. J. Bacteriol. 169:1516-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson, A. M., D. Trenchard, and A. Guz. 1965. Small bowel tonometry: assessment of small gut mucosal oxygen tension in dog and man. Nature 206:943-944. [DOI] [PubMed] [Google Scholar]
- 7.Dawson, J. A., D. H. Mallonee, I. Bjorkhem, and P. B. Hylemon. 1996. Expression and characterization of a C24 bile acid 7 alpha-dehydratase from Eubacterium sp. strain VPI 12708 in Escherichia coli. J. Lipid Res. 37:1258-1267. [PubMed] [Google Scholar]
- 8.Dubberke, E. R., K. A. Reske, M. A. Olsen, L. C. McDonald, and V. J. Fraser. 2008. Short- and long-term attributable costs of Clostridium difficile-associated disease in nonsurgical inpatients. Clin. Infect. Dis. 46:497-504. [DOI] [PubMed] [Google Scholar]
- 9.Ghantoji, S. S., K. Sail, D. R. Lairson, H. L. DuPont, and K. W. Garey. 2010. Economic healthcare costs of Clostridium difficile infection: a systematic review. J. Hosp. Infect. 74:309-318. [DOI] [PubMed] [Google Scholar]
- 10.Giel, J. L., J. A. Sorg, A. L. Sonenshein, and J. Zhu. 2010. Metabolism of bile salts in mice influences spore germination in Clostridium difficile. PLoS One 5:e8740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goodwin, B., and S. A. Kliewer. 2002. Nuclear receptors. I. Nuclear receptors and bile acid homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 282:G926-931. [DOI] [PubMed] [Google Scholar]
- 12.Jump, R. L. P., M. J. Pultz, and C. J. Donskey. 2007. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrob. Agents Chemother. 51:2883-2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kamiya, S., K. Yamakawa, H. Ogura, and S. Nakamura. 1989. Recovery of spores of Clostridium difficile altered by heat or alkali. J. Med. Microbiol. 28:217-221. [DOI] [PubMed] [Google Scholar]
- 14.Karas, J. A., D. A. Enoch, and S. H. Aliyu. A review of mortality due to Clostridium difficile infection. J. Infect. doi: 10.1016/j.jinf.2010.03.025. [DOI] [PubMed]
- 15.Kuijper, E. J., J. T. van Dissel, and M. H. Wilcox. 2007. Clostridium difficile: changing epidemiology and new treatment options. Curr. Opin. Infect. Dis. 20:376-383. [DOI] [PubMed] [Google Scholar]
- 16.Kyne, L., M. B. Hamel, R. Polavaram, and C. P. Kelly. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin. Infect. Dis. 34:346-353. [DOI] [PubMed] [Google Scholar]
- 17.Lawley, T. D., N. J. Croucher, L. Yu, S. Clare, M. Sebaihia, D. Goulding, D. J. Pickard, J. Parkhill, J. Choudhary, and G. Dougan. 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J. Bacteriol. 191:5377-5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyras, D., J. R. O'Connor, P. M. Howarth, S. P. Sambol, G. P. Carter, T. Phumoonna, R. Poon, V. Adams, G. Vedantam, S. Johnson, D. N. Gerding, and J. I. Rood. 2009. Toxin B is essential for virulence of Clostridium difficile. Nature doi: 10.1038/nature07822. [DOI] [PMC free article] [PubMed]
- 19.Makishima, M., A. Y. Okamoto, J. J. Repa, H. Tu, R. M. Learned, A. Luk, M. V. Hull, K. D. Lustig, D. J. Mangelsdorf, and B. Shan. 1999. Identification of a nuclear receptor for bile acids. Science 284:1362-1365. [DOI] [PubMed] [Google Scholar]
- 20.Mallonee, D. H., J. L. Adams, and P. B. Hylemon. 1992. The bile acid-inducible baiB gene from Eubacterium sp. strain VPI 12708 encodes a bile acid-coenzyme A ligase. J. Bacteriol. 174:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mallonee, D. H., and P. B. Hylemon. 1996. Sequencing and expression of a gene encoding a bile acid transporter from Eubacterium sp. strain VPI 12708. J. Bacteriol. 178:7053-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mara, G., R. Domenico, A. Michael, Z. Serena, P. Patrizia, R. Laura, T. Anna, and M. Henriette. 2008. Identification and functional characterization of the bile acid transport proteins in nonmammalian ileum and mammalian liver. Proteins 70:462-472. [DOI] [PubMed] [Google Scholar]
- 23.Martinez-Augustin, O., and F. Sanchez de Medina. 2008. Intestinal bile acid physiology and pathophysiology. World J. Gastroenterol. 14:5630-5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McFarland, L. 2008. Update on the changing epidemiology of Clostridium difficile-associated disease. Nat. Clin. Pract. Gstroentrol. Hepatol. 5:40-48. [DOI] [PubMed] [Google Scholar]
- 25.Mekhjian, H. S., S. F. Phillips, and A. F. Hofmann. 1979. Colonic absorption of unconjugated bile acids: perfusion studies in man. Dig. Dis. Sci. 24:545-550. [DOI] [PubMed] [Google Scholar]
- 26.Mi, L.-Z., S. Devarakonda, J. M. Harp, Q. Han, R. Pellicciari, T. M. Willson, S. Khorasanizadeh, and F. Rastinejad. 2003. Structural basis for bile acid binding and activation of the nuclear receptor FXR. Mol. Cell 11:1093-1100. [DOI] [PubMed] [Google Scholar]
- 27.Moir, A., and D. A. Smith. 1990. The genetics of bacterial spore germination. Annu. Rev. Microbiol. 44:531-553. [DOI] [PubMed] [Google Scholar]
- 28.Nagengast, F. M., M. J. Grubben, and I. P. van Munster. 1995. Role of bile acids in colorectal carcinogenesis. Eur. J. Cancer 31A:1067-1070. [DOI] [PubMed] [Google Scholar]
- 29.Nobilis, M., M. Pour, J. Kunes, J. Kopecky, J. Kvetina, Z. Svoboda, K. Sladkova, and J. Vortel. 2001. High-performance liquid chromatographic determination of ursodeoxycholic acid after solid phase extraction of blood serum and detection-oriented derivatization. J. Pharm. Biomed. Anal. 24:937-946. [DOI] [PubMed] [Google Scholar]
- 30.Northfield, T., and I. McColl. 1973. Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut 14:513-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O'Brien, J. A., B. J. Lahue, J. J. Caro, and D. M. Davidson. 2007. The emerging infectious challenge of Clostridium difficile-associated disease in Massachusetts hospitals: clinical and economic consequences. Infect. Control Hosp. Epidemiol. 28:1219-1227. [DOI] [PubMed] [Google Scholar]
- 32.Parks, D. J., S. G. Blanchard, R. K. Bledsoe, G. Chandra, T. G. Consler, S. A. Kliewer, J. B. Stimmel, T. M. Willson, A. M. Zavacki, D. D. Moore, and J. M. Lehmann. 1999. Bile acids: natural ligands for an orphan nuclear receptor. Science 284:1365-1368. [DOI] [PubMed] [Google Scholar]
- 33.Poutanen, S. M., and A. E. Simor. 2004. Clostridium difficile-associated diarrhea in adults. CMAJ 171:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin, J., R. Li, J. Raes, M. Arumugam, K. S. Burgdorf, C. Manichanh, T. Nielsen, N. Pons, F. Levenez, T. Yamada, D. R. Mende, J. Li, J. Xu, S. Li, D. Li, J. Cao, B. Wang, H. Liang, H. Zheng, Y. Xie, J. Tap, P. Lepage, M. Bertalan, J.-M. Batto, T. Hansen, D. Le Paslier, A. Linneberg, H. B. Nielsen, E. Pelletier, P. Renault, T. Sicheritz-Ponten, K. Turner, H. Zhu, C. Yu, S. Li, M. Jian, Y. Zhou, Y. Li, X. Zhang, S. Li, N. Qin, H. Yang, J. Wang, S. Brunak, J. Dore, F. Guarner, K. Kristiansen, O. Pedersen, J. Parkhill, J. Weissenbach, P. Bork, S. D. Ehrlich, and J. Wang. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raibaud, P., R. Ducluzeau, F. Dubos, S. Hudault, H. Bewa, and M. C. Muller. 1980. Implantation of bacteria from the digestive tract of man and various animals into gnotobiotic mice. Am. J. Clin. Nutr. 33:2440-2447. [DOI] [PubMed] [Google Scholar]
- 36.Ramirez, N., and E. Abel-Santos. 2010. Requirements for germination of Clostridium sordellii spores in vitro. J. Bacteriol. 192:418-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramirez, N., M. Liggins, and E. Abel-Santos. 2010. Kinetic evidence for the presence of putative germination receptors in C. difficile spores. J. Bacteriol. 192:4215-4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reddy, B. S. 1975. Role of bile metabolites in colon carcinogenesis. Animal models. Cancer 36:2401-2406. [DOI] [PubMed] [Google Scholar]
- 39.Redelings, M. D., F. Sorvillo, and L. Mascola. 2007. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg. Infect. Dis. 13:1417-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ridlon, J. M., D. Kang, and P. B. Hylemon. 2006. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47:241-259. [DOI] [PubMed] [Google Scholar]
- 41.Samuel, P., C. M. Holtzman, E. Meilman, and I. Sekowski. 1973. Effect of neomycin and other antibiotics on serum cholesterol levels and on 7α-dehydroxylation of bile acids by the fecal bacterial flora in man. Circ. Res. 33:393-402. [DOI] [PubMed] [Google Scholar]
- 42.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. G. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 43.Setlow, P. 2003. Spore germination. Curr. Opin. Microbiol. 6:550-556. [DOI] [PubMed] [Google Scholar]
- 44.Songer, J. G., and M. A. Anderson. 2006. Clostridium difficile: an important pathogen of food animals. Anaerobe 12:1-4. [DOI] [PubMed] [Google Scholar]
- 45.Sorg, J. A., and A. L. Sonenshein. 2008. Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190:2505-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorg, J. A., and A. L. Sonenshein. 2009. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J. Bacteriol. 191:1115-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tochtrop, G. P., K. Richter, C. Tang, J. J. Toner, D. F. Covey, and D. P. Cistola. 2002. Energetics by NMR: site-specific binding in a positively cooperative system. Proc. Natl. Acad. Sci. U. S. A. 99:1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weese, J. S., H. R. Staempfli, and J. F. Prescott. 2000. Isolation of environmental Clostridium difficile from a veterinary teaching hospital. J. Vet. Diagn. Investig. 12:449-452. [DOI] [PubMed] [Google Scholar]
- 50.White, B. A., R. L. Lipsky, R. J. Fricke, and P. B. Hylemon. 1980. Bile acid induction specificity of 7-alpha-dehydroxylase activity in an intestinal Eubacterium species. Steroids 35:103-109. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, K. H. 1983. Efficiency of various bile salt preparations for stimulation of Clostridium difficile spore germination. J. Clin. Microbiol. 18:1017-1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, K. H., M. J. Kennedy, and F. R. Fekety. 1982. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15:443-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilson, K. H., and F. Perini. 1988. Role of competition for nutrients in suppression of Clostridium difficile by the colonic microflora. Infect. Immun. 56:2610-2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zar, F., S. Bakkanagari, K. M. L. S. T. Moorthi, and M. Davis. 2007. A comparison of vancomycin and metronidazole for the treatment of Clostridium difficile-associated diarrhea, stratified by disease severity. Clin. Infect. Dis. 45:302-307. [DOI] [PubMed] [Google Scholar]