Abstract
The essential cell division protein FtsL is a substrate of the intramembrane protease RasP. Using heterologous coexpression experiments, we show here that the division protein DivIC stabilizes FtsL against RasP cleavage. Degradation seems to be initiated upon accessibility of a cytosolic substrate recognition motif.
Cell division in bacteria is a highly regulated process (1). The division site selection as well as assembly and disassembly of the divisome have to be strictly controlled (1, 4). Although the spatial control of the divisome is relatively well understood (2, 4, 14, 17), mechanisms governing the temporal control of division are still mainly elusive. Regulatory proteolysis was thought to be a potential modulatory mechanism (8, 9). The highly unstable division protein FtsL was shown to be rate limiting for division and would make an ideal candidate for a regulatory factor in the timing of bacterial cell division (7, 9). In Bacillus subtilis, FtsL is an essential protein of the membrane part of the divisome (5, 7, 8). It is necessary for the assembly of the membrane-spanning division proteins, and a knockout is lethal (8, 9, 12). We have previously reported that FtsL is a substrate of the intramembrane protease RasP (5).
These findings raised the question of whether RasP can regulate cell division by cleaving FtsL from the division complex. In order to mimic the situation in which FtsL is bound to at least one of its interaction partners, we used a heterologous coexpression system in which we synthesized FtsL and DivIC. It has been reported before that DivIC and FtsL are intimate binding partners in various organisms (6, 9, 15, 21, 22, 26) and that FtsL and DivIC (together with DivIB) can form complexes even in the absence of the other divisome components (6, 21). We therefore asked whether RasP is able to cleave FtsL in the presence of its major interaction partner DivIC, which would argue for the possibility that RasP could cleave FtsL within a mature divisome. In contrast, if interaction with DivIC could stabilize FtsL against RasP cleavage, this result would bring such a model into question. An alternative option for the role of RasP might be the removal of FtsL from the membrane. It has been shown that divisome disassembly and prevention of reassembly are crucial to prevent minicell formation close to the new cell poles (3, 16).
Construction of a coexpression system.
To test our hypothesis, we used an Escherichia coli-based coexpression system similar to one described before (5, 18). This approach is reasonable, since coexpression of B. subtilis FtsL and DivIC does not interfere with the E. coli cytokinetic machinery (22). For this purpose, the ftsL gene was amplified from chromosomal DNA of Bacillus subtilis 168 using primers ftslduet-for and ftslduet-rev. The fragment was cut with BamHI and EagI and cloned into MCS1 of the overexpression vector pACYCDuet-1. This resulted in plasmid pWB1 with the ftsL gene under the control of a T7 promoter and fused to a His tag. The yluC gene (coding for RasP) was amplified using primers ylucduet-for and ylucduet-rev. The resulting fragment was cloned with NdeI and KpnI into the MCS2 of pWB1, resulting in pWB2 with the yluC gene under the control of a T7 promoter and fused to an S tag. As a negative control, a second vector, pWB3, was constructed for coexpression of ftsL and yluC(E21A) genes coding for an inactive RasP mutant, RasP-E21A (RasP*). The mutation was introduced by using the modified forward primer ylucE21Aduet-for for yluC gene amplification.
The divIC gene was amplified using primers divICduet-for and divICduet-rev and cloned with NdeI and XhoI into the overexpression plasmid pETDUET-1. This resulted in pWB4 with the divIC gene under the control of a T7 promoter fused to an S tag.
For coexpression of ftsL, divIC, yluC, and yluC(E21A) genes in different combinations, the plasmids were transformed into E. coli BL21(DE3) (see Table S1 in the supplemental material). For expression cultures, 10 ml of LB medium was freshly inoculated with LB overnight cultures. Expression was induced at an optical density at 600 nm (OD600) of 0.7 by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.1 mM. Total cell extracts were analyzed by SDS-PAGE (24) and immunoblotting.
DivIC stabilizes FtsL against RasP cleavage.
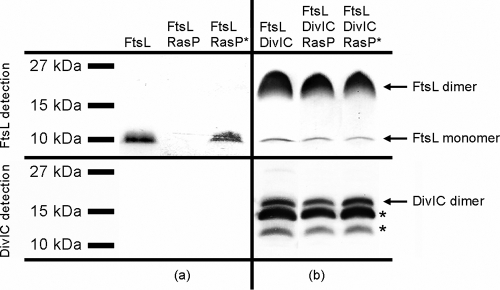
Coexpression of ftsL and yluC genes resulted in drastically decreased FtsL stability in comparison to coexpression of ftsL and yluC(E21A) genes (Fig. 1 a). FtsL was clearly degraded by RasP in the heterologous system as described before (5). It has to be noted that additional coexpression of the yluC or yluC(E21A) gene led to reduced expression levels of FtsL in general, probably due to the burden of expressing multiple recombinant membrane proteins. The intensity of this effect varied, probably depending on slightly varied growth conditions and overall expression between different experiments. When the ftsL gene was coexpressed with the divIC gene, the FtsL band was almost completely shifted to the molecular weight of a dimer (Fig. 1b). Such sodium dodecyl sulfate-resistant dimer formation has been reported for FtsL from E. coli as well (13). The amount of FtsL dimers exceeds the amount of FtsL monomer found in cells that did not express DivIC, suggesting that DivIC generally stabilized FtsL against E. coli endogenous proteases. This is in accordance with results of previous studies, which also showed that FtsL and DivIC can stabilize each other in an E. coli system (22). It was not clear whether the dimerization of FtsL was due to this stabilization and the higher protein levels or a specific effect of DivIC. Therefore, coexpression of ftsL and divIC genes was repeated but with significantly lower level of induction (see Fig. S1 in the supplemental material). At a lower concentration, most of FtsL was present as monomers, showing that the dimerization is strongly concentration dependent. DivIC itself was not completely stable in E. coli BL21. Antibody detection of the S tag showed only a DivIC dimer band (see Fig. S2a in the supplemental material), but the use of DivIC antibody revealed two distinct degradation bands (see Fig. S2b in the supplemental material).
FIG. 1.
Coexpression of ftsL, divIC, yluC, and yluC(E21A) genes. Shown are immunoblots of 10% Schägger SDS system gels. For FtsL detection, blots were treated with anti-PentaHis and anti-mouse IgG alkaline phosphatase (AP), and for DivIC detection, they were treated with anti-DivIC and anti-rabbit IgG AP. Proteins present in each strain after expression are noted on top. (a) Influence of active RasP and inactive RasP-E21A (RasP*) on FtsL levels. (b) Influence of DivIC on FtsL oligomerization and stability. Asterisks indicate DivIC degradation bands.
Additional coexpression of the yluC or yluC(E21A) gene had no effect on FtsL levels, showing that in the presence of DivIC, FtsL was not degraded by RasP (Fig. 1b). Obviously, DivIC is able to stabilize FtsL in general as well as against specific cleavage by RasP.
Importance of N-terminal domains for protein-protein interaction of FtsL and DivIC.
We wanted to investigate FtsL stabilization further. Another well-characterized substrate of RasP is the anti-sigma factor RsiW (10, 18, 19, 25). Sequence alignments between RsiW and FtsL revealed a putative substrate recognition motif for RasP cleavage in the N-terminal domain of FtsL (5). In vivo FtsL could be completely stabilized by either truncating or mutating this motif (5). FtsL and DivIC both consist of three domains, an extracellular domain, predicted to be a coiled-coil domain, a transmembrane helix, and the cytosolic N-terminal domain. It seems likely that during cell division, the recognition motif is buried within the cytosolic part of the divisome. This raised the question of whether a block of accessibility of the recognition motif is the main reason for FtsL stabilization in the FtsL-DivIC complex. To this end, we used an N-terminally truncated (ΔN) DivIC (DivICΔN) that should not be able to shield the N-terminal FtsL domain in the E. coli system. We tested whether the coexpression of ΔN-divIC genes could stabilize FtsL in the same manner as coexpression of divIC genes.
It was unclear if the N-terminal domains of FtsL and DivIC are essential for protein-protein interaction. Hence, interaction of the full-length and N-terminally truncated proteins was tested using a bacterial two-hybrid system (20). The various gene fragments were cloned with XbaI and KpnI into the plasmids pKT25 and pUT18C. From the resulting plasmids, ftsL, ΔN-ftsL, divIC, and ΔN-divIC genes were expressed C-terminally fused to the T25 or T18 part of adenylate cyclase (CyaA) of Bordetella pertussis. For complete plasmid and strain lists, see Tables S1 and S2 in the supplemental material. Plasmids were transformed into E. coli BTH101, and cells were spotted on LB plates supplemented with 100 μg ml−1 carbenicillin, 50 μg ml−1 kanamycin, 160 μg ml−1 X-galactosidase (X-Gal), and 1 mM IPTG. Incubation was done at 30°C for at least 24 h. The grown colonies were rediluted in LB medium, again spotted on supplemented LB plates, and incubated in the dark at 30°C for 30 h. Blue colonies indicate direct protein-protein interaction, and white colonies indicate no interaction. As a negative control, the empty plasmids were transformed, and the positive control was done with the leucine zipper of GCN4.
FtsL and DivIC both show self-interaction, as expected from the detected dimer bands on the coexpression blots (Fig. 1). Truncating the N terminus of FtsL, DivIC, or both did not completely abolish protein-protein-interaction between FtsL and DivIC (Fig. 2). However, the reduced intensity of the FtsLΔN/DivICΔN interaction compared to the FtsL/DivIC interaction suggests that the cytoplasmic domains of FtsL and DivIC at least contribute to dimer formation. This is interesting, because there is still a debate about the interaction of DivIC and FtsL (23, 26). However, self-interaction of FtsL is drastically impaired by the N-terminal truncation. Coexpression of full-length ftsL and ΔN-divIC genes should therefore still lead to protein complexes, albeit with better accessibility of the N-terminal FtsL domain.
FIG. 2.
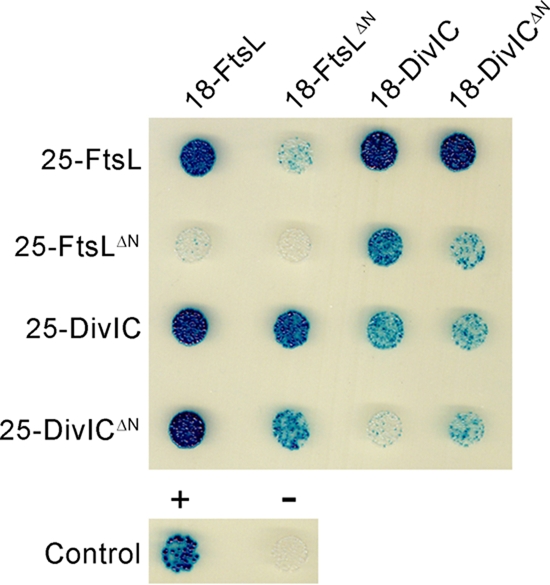
Protein-protein interactions of FtsL and DivIC in full-length and N-terminally truncated forms (FtsLΔN and DivICΔN) shown by a bacterial two hybrid assay. The fusion proteins to the T18 part of CyaA are noted on top, and the fusion proteins to the T25 part are noted on the left. Blue colonies indicate direct protein-protein interaction, and white colonies indicate no interaction.
Effect of the putative recognition motif and accessibility of the FtsL cytoplasmic domain for proteolysis.
The plasmids for expression of ΔN-ftsL (pWB5), ΔN-ftsL and yluC (pWB6), ΔN-ftsL and yluC(E21A) (pWB7), and ΔN-divIC (pWB8) genes were constructed in the same way as were the plasmids pWB1 to -3, using primers ΔN-ftslduet-for and ftslduet-rev and ΔN-divICduet-for and divICduet-rev. In addition, plasmids were constructed to coexpress yluC and yluC(E21A) genes and a version of the ftsL gene with the recognition motif mutated from KKRAS to KKAVA [ftsL(25B)] (5). Amplification of the ftsL(25B) gene was done using primers ftslduet-for and ftslduet-rev and chromosomal DNA of Bacillus subtilis 168 amyE:: ftsL(25B) ftsL::neo (5) as the template. Plasmids pWB17 to -19 were then constructed in the same way as were the plasmids pWB1 to -3.
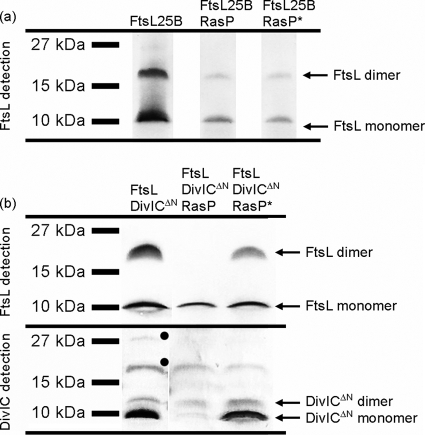
According to the in vivo data (5), expression of ΔN-ftsL and ftsL(25B) genes should result in stable protein levels not influenced by RasP. FtsLΔN was unstable in the E. coli system, and protein levels were below the detection limit (see Fig. S3 in the supplemental material). Only upon coexpression of divIC was FtsLΔN detected. This shows again that DivIC has a general stabilizing effect on FtsL. FtsL25B was stable in E. coli BL21 and present as monomers as well as dimers (Fig. 3 a). Coexpression of the yluC or yluC(E21A) gene showed that neither the FtsL25B monomers nor the dimers could be degraded by RasP.
FIG. 3.
Effect of mutation and accessibility of the putative recognition motif. (a) Stability of FtsL with a mutated recognition motif (FtsL25B). (b) Stability of FtsL in the presence of DivICΔN. Shown are immunoblots of 10% Schägger SDS system gels. For FtsL detection, blots were treated with anti-PentaHis and anti-mouse IgG AP, and for DivICΔN detection, they were treated with anti-DivIC and anti-rabbit IgG AP. Proteins present in each strain after expression are noted on top. Unspecific bands are marked with filled circles.
Expression of ΔN-divIC led to relatively low DivICΔN levels compared to full-length DivIC (Fig. 3b). As a result, significantly less of the full-length FtsL was shifted to the dimer band. Most important, FtsL was not stabilized against RasP activity. The protease seems to require only that the recognition motif be accessible to degrade FtsL. When the N-terminal domain was no longer shielded by the cytosolic DivIC domain, FtsL could be cleaved.
It should be noted that the amount of DivICΔN was decreased in the presence of active RasP. No other experiments ever indicated that DivIC is a substrate of RasP. It was more likely that the decrease in DivICΔN levels was a consequence of the FtsL degradation. While a stabilizing effect of FtsL on full-length DivIC was not observed, this might become noticeable at low DivICΔN concentrations. To test this idea, we coexpressed ΔN-divIC with ftsL(25B) and investigated the effect of RasP on DivICΔN for these cells (see Fig. S4 in the supplemental material). FtsL25B is not degraded by RasP, and as a result, DivICΔN indeed remained stable in the presence of RasP.
Conclusions.
In toto, we conclude that FtsL proteolysis by RasP is likely dependent on substrate recognition and can be prevented by mutation of the cytosolic recognition motif or by blocking its accessibility. FtsL is stabilized by DivIC. In this complex, FtsL cannot be cleaved by RasP, and DivIC, in turn, is relatively stabilized against proteolysis by endogenous E. coli proteases. A truncated DivIC form is not able to protect FtsL from RasP cleavage.
Our data are in good agreement with the current model for intramembrane proteolysis by site 2 proteases. Conclusions from a high-resolution structure of S2P from Methanocaldococcus jannaschii suggested that the diverse positions of cleavage sites for site 2 protease substrates are defined not by variations of the active site position but rather by recognition of a specific substrate motif (11). The same principle has recently been shown for rhomboids (27). The authors were able to show that a recognition motif is of utmost importance for substrate degradation and even determines the site of cleavage. In the case of FtsL, accessibility of the putative recognition motif seems to trigger degradation by the protease RasP. This puts into question the idea that RasP could directly cleave FtsL out of the division complex. An attractive model would be one in which FtsL is cleaved downstream of divisome disassembly. After cell division is completed, the divisome disassembles upon a yet-unknown signal, and the N-terminal domain of FtsL is no longer buried within the complex. RasP might then remove FtsL from the membrane to prevent reassembly. How the tight interaction between FtsL and DivIC is released after divisome disassembly, however, remains elusive.
Supplementary Material
Acknowledgments
We thank Richard Daniel (Newcastle University) for the kind gift of DivIC antibodies.
This study was financially supported by the Deutsche Forschungsgemeinschaft (SFB 635, project C6).
Footnotes
Published ahead of print on 19 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, D. W., and J. Errington. 2009. Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7:642-653. [DOI] [PubMed] [Google Scholar]
- 2.Barak, I., and A. J. Wilkinson. 2007. Division site recognition in Escherichia coli and Bacillus subtilis. FEMS Microbiol. Rev. 31:311-326. [DOI] [PubMed] [Google Scholar]
- 3.Bramkamp, M., R. Emmins, L. Weston, C. Donovan, R. A. Daniel, and J. Errington. 2008. A novel component of the division-site selection system of Bacillus subtilis and a new mode of action for the division inhibitor MinCD. Mol. Microbiol. 70:1556-1569. [DOI] [PubMed] [Google Scholar]
- 4.Bramkamp, M., and S. van Baarle. 2009. Division site selection in rod-shaped bacteria. Curr. Opin. Microbiol. 12:683-688. [DOI] [PubMed] [Google Scholar]
- 5.Bramkamp, M., L. Weston, R. A. Daniel, and J. Errington. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62:580-591. [DOI] [PubMed] [Google Scholar]
- 6.Buddelmeijer, N., and J. Beckwith. 2004. A complex of the Escherichia coli cell division proteins FtsL, FtsB and FtsQ forms independently of its localization to the septal region. Mol. Microbiol. 52:1315-1327. [DOI] [PubMed] [Google Scholar]
- 7.Daniel, R. A., and J. Errington. 2000. Intrinsic instability of the essential cell division protein FtsL of Bacillus subtilis and a role for DivIB protein in FtsL turnover. Mol. Microbiol. 36:278-289. [DOI] [PubMed] [Google Scholar]
- 8.Daniel, R. A., E. J. Harry, V. L. Katis, R. G. Wake, and J. Errington. 1998. Characterization of the essential cell division gene ftsL(yIID) of Bacillus subtilis and its role in the assembly of the division apparatus. Mol. Microbiol. 29:593-604. [DOI] [PubMed] [Google Scholar]
- 9.Daniel, R. A., M. F. Noirot-Gros, P. Noirot, and J. Errington. 2006. Multiple interactions between the transmembrane division proteins of Bacillus subtilis and the role of FtsL instability in divisome assembly. J. Bacteriol. 188:7396-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellermeier, C. D., and R. Losick. 2006. Evidence for a novel protease governing regulated intramembrane proteolysis and resistance to antimicrobial peptides in Bacillus subtilis. Genes Dev. 20:1911-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, L., H. Yan, Z. Wu, N. Yan, Z. Wang, P. D. Jeffrey, and Y. Shi. 2007. Structure of a site-2 protease family intramembrane metalloprotease. Science 318:1608-1612. [DOI] [PubMed] [Google Scholar]
- 12.Gamba, P., J. W. Veening, N. J. Saunders, L. W. Hamoen, and R. A. Daniel. 2009. Two-step assembly dynamics of the Bacillus subtilis divisome. J. Bacteriol. 191:4186-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghigo, J. M., and J. Beckwith. 2000. Cell division in Escherichia coli: role of FtsL domains in septal localization, function, and oligomerization. J. Bacteriol. 182:116-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goehring, N. W., and J. Beckwith. 2005. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr. Biol. 15:514-526. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez, M. D., and J. Beckwith. 2009. Divisome under construction: distinct domains of the small membrane protein FtsB are necessary for interaction with multiple cell division proteins. J. Bacteriol. 191:2815-2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory, J. A., E. C. Becker, and K. Pogliano. 2008. Bacillus subtilis MinC destabilizes FtsZ-rings at new cell poles and contributes to the timing of cell division. Genes Dev. 22:3475-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harry, E., L. Monahan, and L. Thompson. 2006. Bacterial cell division: the mechanism and its precison. Int. Rev. Cytol. 253:27-94. [DOI] [PubMed] [Google Scholar]
- 18.Heinrich, J., K. Hein, and T. Wiegert. 2009. Two proteolytic modules are involved in regulated intramembrane proteolysis of B. subtilis RsiW. Mol. Microbiol. 74:1412-1426. [DOI] [PubMed] [Google Scholar]
- 19.Heinrich, J., and T. Wiegert. 2006. YpdC determines site-1 degradation in regulated intramembrane proteolysis of the RsiW anti-sigma factor of Bacillus subtilis. Mol. Microbiol. 62:566-579. [DOI] [PubMed] [Google Scholar]
- 20.Karimova, G., N. Dautin, and D. Ladant. 2005. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J. Bacteriol. 187:2233-2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noirclerc-Savoye, M., A. Le Gouellec, C. Morlot, O. Dideberg, T. Vernet, and A. Zapun. 2005. In vitro reconstitution of a trimeric complex of DivIB, DivIC and FtsL, and their transient colocalization at the division site in Streptococcus pneumoniae. Mol. Microbiol. 55:413-424. [DOI] [PubMed] [Google Scholar]
- 22.Robichon, C., G. F. King, N. W. Goehring, and J. Beckwith. 2008. Artificial septal targeting of Bacillus subtilis cell division proteins in Escherichia coli: an interspecies approach to the study of protein-protein interactions in multiprotein complexes. J. Bacteriol. 190:6048-6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robson, S. A., K. A. Michie, J. P. Mackay, E. Harry, and G. F. King. 2002. The Bacillus subtilis cell division proteins FtsL and DivIC are intrinsically unstable and do not interact with one another in the absence of other septasomal components. Mol. Microbiol. 44:663-674. [DOI] [PubMed] [Google Scholar]
- 24.Schägger, H., and G. von Jagow. 1987. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166:368-379. [DOI] [PubMed] [Google Scholar]
- 25.Schöbel, S., S. Zellmeier, W. Schumann, and T. Wiegert. 2004. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52:1091-1105. [DOI] [PubMed] [Google Scholar]
- 26.Sievers, J., and J. Errington. 2000. The Bacillus subtilis cell division protein FtsL localizes to sites of septation and interacts with DivIC. Mol. Microbiol. 36:846-855. [DOI] [PubMed] [Google Scholar]
- 27.Strisovsky, K., H. J. Sharpe, and M. Freeman. 2009. Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36:1048-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.