Abstract
Long-chain and/or branched-chain polyamines are unique polycations found in thermophiles. Cytoplasmic polyamines were analyzed for cells cultivated at various growth temperatures in the hyperthermophilic archaeon Thermococcus kodakarensis. Spermidine [34] and N4-aminopropylspermine [3(3)43] were identified as major polyamines at 60°C, and the amounts of N4-aminopropylspermine [3(3)43] increased as the growth temperature rose. To identify genes involved in polyamine biosynthesis, a gene disruption study was performed. The open reading frames (ORFs) TK0240, TK0474, and TK0882, annotated as agmatine ureohydrolase genes, were disrupted. Only the TK0882 gene disruptant showed a growth defect at 85°C and 93°C, and the growth was partially retrieved by the addition of spermidine. In the TK0882 gene disruptant, agmatine and N1-aminopropylagmatine accumulated in the cytoplasm. Recombinant TK0882 was purified to homogeneity, and its ureohydrolase characteristics were examined. It possessed a 43-fold-higher kcat/Km value for N1-aminopropylagmatine than for agmatine, suggesting that TK0882 functions mainly as N1-aminopropylagmatine ureohydrolase to produce spermidine. TK0147, annotated as spermidine/spermine synthase, was also studied. The TK0147 gene disruptant showed a remarkable growth defect at 85°C and 93°C. Moreover, large amounts of agmatine but smaller amounts of putrescine accumulated in the disruptant. Purified recombinant TK0147 possessed a 78-fold-higher kcat/Km value for agmatine than for putrescine, suggesting that TK0147 functions primarily as an aminopropyl transferase to produce N1-aminopropylagmatine. In T. kodakarensis, spermidine is produced mainly from agmatine via N1-aminopropylagmatine. Furthermore, spermine and N4-aminopropylspermine were detected in the TK0147 disruptant, indicating that TK0147 does not function to produce spermine and long-chain polyamines.
Polyamines are positively charged aliphatic compounds. Putrescine [4], spermidine [34], and spermine [343] are common polyamines observed in various living organisms, from viruses to humans (16). Polyamines, which play important roles in cell proliferation and cell differentiation (19, 34), are thought to contribute to adaptation against various stresses (9, 26). In thermophilic microorganisms, polyamines contribute to growth under high-temperature conditions. Indeed, in the thermophilic bacterium Thermus thermophilus, a mutant strain lacking the enzyme related to polyamine biosynthesis shows defective growth at high temperatures (23). Furthermore, thermophilic archaea and bacteria possess long-chain and branched-chain polyamines such as N4-aminopropylspermidine [3(3)4], N4-aminopropylspermine [3(3)43], and N4-bis(aminopropyl)spermidine [3(3)(3)4], in addition to common polyamines (11, 13, 14). N4-aminopropylspermine was detected in the cells of thermophiles, such as Saccharococcus thermophilus, thermophilic Bacillus and Geobacillus spp. (Bacillus caldolyticus, B. caldotenax, B. smithii, Geobacillus stearothermophilus, and G. thermocatenulatus), Caldicellulosiruptor spp. (C. kristjanssonii and C. owensensis) and Calditerricola spp. (C. satsumensis and C. yamamurae) (10, 12, 22), but it was not detected in archaea. These unique polyamines are thought to support the growth of thermophilic microorganisms under high-temperature conditions. An in vitro study indicated that long-chain and branched-chain polyamines effectively stabilized DNA and RNA, respectively (32).
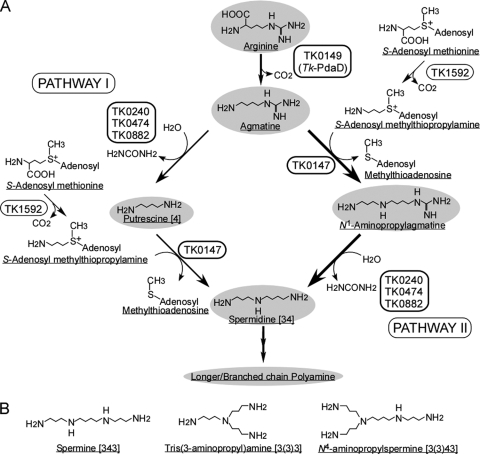
Polyamines are synthesized from amino acids such as arginine, ornithine, and methionine (26). In most eukaryotes, putrescine is synthesized directly from ornithine by ornithine decarboxylase (34). Plants and some bacteria possess additional or alternative putrescine biosynthesis pathways in which putrescine is synthesized from arginine via agmatine (18, 31, 35). In this pathway, agmatine is synthesized by arginine decarboxylase, and agmatine is converted to putrescine by agmatine ureohydrolase or a combination of agmatine iminohydrolase and N-carbamoylputrescine amidohydrolase. Longer polyamines are then produced by the addition of the aminopropyl group from decarboxylated S-adenosylmethionine. This pathway is shown on the left in Fig. 1 (pathway I). On the other hand, the thermophilic bacterium T. thermophilus possesses a unique polyamine-biosynthetic pathway (23) in which spermidine is synthesized from agmatine via N1-aminopropylagmatine by aminopropyl transferase followed by ureohydrolase, as shown on the right in Fig. 1 (pathway II).
FIG. 1.
Predicted biosynthetic pathway of polyamines in T. kodakarensis. (A) Predicted biosynthetic pathway. Pyruvoyl-dependent arginine decarboxylase proenzyme (TK0149), arginine/agmatine ureohydrolases (TK0240/TK0474/TK0882), aminopropyl transferase (TK0147), and pyruvoyl-dependent S-adenosylmethionine decarboxylase proenzyme (TK1592) are shown based on the genome analysis. (B) Structures of unique polyamines.
A sulfur-reducing hyperthermophilic archaeon, Thermococcus kodakarensis KOD1, was isolated from Kodakara Island, Kagoshima, Japan (1, 21). This archaeon grows at temperatures between 60°C and 100°C but optimally at 85°C. Under low- or high-temperature-stressed conditions, T. kodakarensis produces cold- or heat-inducible chaperones to adapt to unfavorable growth environments (4, 5, 30). The lipid composition of the membrane also changes depending on the growth shift (20). In addition to acting as such tolerance factors, polyamines have been suggested to play an important role in maintaining nucleosomes in high-temperature environments (15). A complete genome analysis of T. kodakarensis has been performed, and the pathway of polyamine biosynthesis has been predicted (Fig. 1) (6, 7). It has been speculated that putrescine is synthesized from arginine via agmatine by arginine decarboxylase (PdaDTk) and agmatine ureohydrolase. Long- and/or branched-chain polyamines are then produced by the addition of the aminopropyl group derived from decarboxylated S-adenosylmethionine. Previously, we revealed that PdaDTk catalyzed the first step of polyamine biosynthesis and was essential for cell growth (6). The strain DAD, which lacks the gene pdaDTk, does not grow in medium without agmatine. Archaeal cells are known to use agmatine to synthesize agmatidine, which is an agmatine-conjugated cytidine found at the anticodon wobble position of archaeal tRNAIle (17). Agmatine is important for agmatidine synthesis as well as long-chain polyamine. In the present study, we focused on the subsequent steps in polyamine biosynthesis, especially from agmatine to spermidine. T. kodakarensis possesses three agmatine ureohydrolase homologues (TK0240, TK0474, and TK0882); however, it is unclear which one is dominantly functional in T. kodakarensis cells. In a closely related genus, Pyrococcus, TK0474 and TK0882 orthologues have been identified, but the TK0240 orthologue is missing in Pyrococcus genomes. In Pyrococcus horikoshii, PH0083, which is an orthologue of TK0882, was shown to possess agmatine ureohydrolase activity (8). TK0882, hence, appears to possess agmatine ureohydrolase activity as well. It is unclear whether other agmatine ureohydrolase homologues (TK0240 and TK0474) are involved in polyamine synthesis and cell growth in T. kodakarensis. In addition to agmatine ureohydrolase, aminopropyl transferase plays a crucial role in the synthesis of polyamines. TK0147 was annotated first as spermidine synthase and shares sequence identity with aminopropyl transferase (PF0127) from Pyrococcus furiosus (3). It is therefore expected to harbor the function of aminopropyl transferase for long-chain-polyamine synthesis. Recombinant PF0127 showed broad amine acceptor specificity for agmatine, 1,3-diaminopropane (3), putrescine, cadaverine (5), sym-nor-spermidine (33), and spermidine. While maximal catalytic activity was observed with cadaverine, agmatine was most often preferred on the basis of the kcat/Km value (3), suggesting that pathway II is a dominant route for polyamine synthesis in P. furiosus. In the present study, various disruptants lacking genes for polyamine biosynthesis were constructed in order to understand the physiological roles of these enzymes in T. kodakarensis. The cell growth profiles and cytoplasmic polyamines of the wild type and the disruptants were analyzed and compared. Recombinant enzymes were also purified and characterized. The obtained results are expected to provide useful information regarding the specific roles of polyamines in thermophiles.
MATERIALS AND METHODS
Microorganisms, plasmids, and media.
T. kodakarensis KOD1 and its derivatives were cultivated anaerobically in a nutrient-rich medium (ASW-YT) with an addition of 2.0 g/liter of elemental sulfur or pyruvate or in a synthetic medium (ASW-AA) containing artificial seawater (ASW), amino acids, and elemental sulfur (29). For the solid medium, 1% Gelrite (Wako, Osaka, Japan) was added. Escherichia coli BL21-CodonPlus(DE3)-RIL (Stratagene, La Jolla, CA) and pET21a(+) (Novagen, Madison, WI) were used for gene expression. E. coli strains were routinely cultivated at 37°C in Luria-Bertani (LB) medium. Ampicillin (50 μg/ml) and/or chloramphenicol (25 μg/ml) was added to the medium when needed.
Polyamine analysis.
Polyamine analysis was carried out according to the reported procedure with slight modifications (23). T. kodakarensis strain KU216 (pyrF disruptant) and its derivatives were cultivated in an ASW-YT liquid medium with the addition of 2.0 g/liter of elemental sulfur at 60°C, 85°C, or 93°C until log phase and harvested (10,000 × g for 20 min). The cells were weighed and homogenized in 10% trichloroacetic acid of a one-third volume of cell suspension. Caldohexamine [33333] was added to the mixture (final concentration, 3 mM) as an internal standard to control for extraction and separation losses. The supernatant was obtained by centrifugation (10,000 × g for 20 min) and filtered with a 0.45-μm-pore-size filter (Millipore Corp., Bedford, MA). The supernatants (100 μl) containing 30 nmol caldohexamine were analyzed by high-performance liquid chromatography (HPLC) on a CK-10S column (6.0-mm inside diameter by 50 mm) of cation-exchange resin (GL Science, Tokyo, Japan). The column was equilibrated with elution buffer A [100 mM potassium citrate monohydrate, 1.7 M KCl, 650 mM 2-propanol, 2.4 mM polyoxyethylene(23)lauryl ether (Brij 35), and 65.0 ml of 1 N HCl per liter] at a flow rate of 1.0 ml/min at 70°C. The eluted polyamines were automatically mixed with detection buffer composed of 400 mM boric acid, 400 mM NaOH, 4.9 mM Brij 35, 7.5 mM o-phthalaldehyde, 171 mM ethanol, and 28 mM 2-mercaptoethanol at a flow rate of 0.5 ml/min at 70°C and detected with a fluorescence detector (GL-7453A) (GL Science). Standard samples of N1-aminopropylagmatine, N4-aminopropylspermine, caldohexamine, and homospermidine were synthesized by the method described previously (22-24). Other standard polyamines were kindly provided by Akira Shirahata and Yoshihiko Ikeguchi, Graduate School of Pharmaceutical Science, Josai University.
DNA manipulation and sequencing.
DNA manipulations were carried out by standard techniques, as described previously by Sambrook and Russell (27). Restriction enzymes and other modifying enzymes were purchased from Takara or Toyobo. Plasmid DNA was isolated with a Wizard Plus Minipreps DNA purification system (Promega, Madison, WI). GFX PCR DNA and gel band purification kit (GE Healthcare, Little Chalfont, United Kingdom) were used to recover DNA fragments from agarose gels after electrophoresis. DNA sequencing was performed with a BigDye-Terminator cycle-sequencing ready reaction kit, version 3.1, and a model 3130 capillary DNA sequencer (Applied Biosystems, Foster City, CA).
Construction of disruptants.
The theoretical background for specific gene disruption has been described previously (28). Vectors for disrupting the TK0240, TK0474, TK0882, and TK0147 genes through single-crossover homologous recombination were constructed as follows. DNA fragments containing the target gene together with its flanking regions (ca. 1,000 bp each) were amplified using T. kodakarensis genomic DNA as the template and the primer sets listed in Table 1 (TK0240-Fw and TK0240-Rv, TK0474-Fw and TK0474-Rv, TK0882-Fw and TK0882-Rv, and TK0147-Fw and TK0147-Rv). The amplified DNA was cloned into pUD2 (28). An inverse PCR was performed in order to amplify the entire plasmid, excluding the coding region of the target gene, using the primer sets listed in Table 1 (Inv-TK0240-Fw and Inv-TK0240-Rv, Inv-TK0474-Fw and Inv-TK0474-Rv, Inv-TK0882-Fw and Inv-TK0882-Rv, and Inv-TK0147-Fw and Inv-TK0147-Rv). By self-ligation, a plasmid containing the 5′- and 3′-flanking regions of the target gene immediately adjacent to one another was obtained. The appropriate constructions were confirmed by sequence determination. The plasmids obtained were used as donor DNA, and T. kodakarensis strain KU216 (pyrF disruptant) was used as the host. Strains with a host genotype and those with the intended disruptions were confirmed by PCR.
TABLE 1.
Strains and primers used in this study
| Strain or primer | Relevant characteristic(s) or sequence (5′ to 3′)a | Source or reference |
|---|---|---|
| Strains | ||
| E. coli TG1 | supE hsdΔ5 thi Δ(lac-proAB)/F′ traD36 proAB+lacIqlacZ ΔM15 | Stratagene |
| T. kodakarensis | ||
| KOD1 | Wild type | 1, 21 |
| KU216 | ΔpyrF | 28 |
| DAD | ΔpdaDTk | 6 |
| DUH2 | ΔTK0240 | This study |
| DUH4 | ΔTK0474 | This study |
| DUH8 | ΔTK0882 | This study |
| DUH24 | ΔTK0240 ΔTK0474 | This study |
| DUH48 | ΔTK0474 ΔTK0882 | This study |
| DUH28 | ΔTK0240 ΔTK0882 | This study |
| DUH248 | ΔTK0240 ΔTK0474 ΔTK0882 | This study |
| DAT | ΔTK0147 | This study |
| Primers | ||
| TK0240-Fw | 5′-AAAAGGATCCGACGGAAATTCGGGGCCAACTG-3′ | This study |
| TK0240-Rv | 5′-AAAAGGATCCCGAGGGCAGTTATCAGAACGCTC-3′ | This study |
| TK0474-Fw | 5′-AAAAGGATCCCCTGCCTCGAATGCTGGCGTG-3′ | This study |
| TK0474-Rv | 5′-AAAAGGATCCCTCGCCGACGACGGTTACATCGC-3′ | This study |
| TK0882-Fw | 5′-AAAAGGTACCGAGCAGAGCGTCACCTGAACAC-3′ | This study |
| TK0882-Rv | 5′-AAAAGGTACCGGTAGCCCATTGCCATGAGCG-3′ | This study |
| TK0147-Fw | 5′-AAGAATTCGGAGGAGGTTGAGATGAGCTGG-3′ | This study |
| TK0147-Rv | 5′-AAGAATTCGACCTCCCCCCGATACTTGGTG-3′ | This study |
| Inv-TK0240-Fw | 5′-GAGGATGTGGCCCTCCGCGAAGTTC-3′ | This study |
| Inv-TK0240-Rv | 5′-TCACCACACCCTCGTCCCGACCG-3′ | This study |
| Inv-TK0474-Fw | 5′-GTGAAGGGGCCAACCGAAGAAG-3′ | This study |
| Inv-TK0474-Rv | 5′-TCCTTCCACCAGCACAACATTGG-3′ | This study |
| Inv-TK0882-Fw | 5′-GCCTTCTGCGTTCTTTTTTCAG-3′ | This study |
| Inv-TK0882-Rv | 5′-TCTCTCACCTAAAGGCAAAAATAGAAGAGGG-3′ | This study |
| Inv-TK0147-Fw | 5′-ATTGGGCAACAAGAGAAAAGCTCTAAC-3′ | This study |
| Inv-TK0147-Rv | 5′-GAGTCTCACCACCGGTGGATACGATG-3′ | This study |
| TK0882-RT-Fw | 5′-GCTGGAGTTCCCGCTCGTAGAACC-3′ | This study |
| TK0882-RT-Rv | 5′-CAGCCGTCAATGCCGTCGGATCGC-3′ | This study |
| TK0147-RT-Fw | 5′-CGGCGGTGGAGACGGTGGAACGCTC-3′ | This study |
| TK0147-RT-Rv | 5′-GGCACGAAACCCTCTTCCAGATGCCGCGC-3′ | This study |
| 16SrDNA-Fw | 5′-GCCCCGAAACCCCCGGGCTACACGCGCGCT-3′ | This study |
| 16SrDNA-Rv | 5′-CGTATTCGCCGCGCGATGATGACACGCGGG-3′ | This study |
| TK0882-EX-Fw | 5′-AACATATGGAGTTCCTGTACACGTATG-3′ | This study |
| TK0882-EX-Rv | 5′-AAGGATCCTCAGCGCCCGAATTTTG-3′ | This study |
| TK0147-EX-Fw | 5′-AAAAAAACATATGGGATACAACGAGCAGGAG-3′ | This study |
| TK0147-EX-Rv | 5′-AAAGGATCCTTACTGCCCCTCTAGGAGC-3′ | This study |
Underlining indicates restriction enzyme sites.
Expression and purification of TK0882 protein.
The plasmid for overexpression of TK0882 (pET-TK0882) was constructed as follows. The TK0882 gene was amplified with T. kodakarensis genomic DNA as a template and two oligonucleotide primers, TK0882-EX-Fw and TK0882-EX-Rv (Table 1). The amplified fragment was digested with NdeI and BamHI and inserted into the NdeI and BamHI sites of pET21a(+). The constructed plasmid was designated pET-TK0882. E. coli BL21-CodonPlus(DE3)-RIL cells harboring pET-TK0882 were grown in LB medium at 37°C, and gene expression was induced with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at the mid-exponential growth phase with further incubation for 4 h. Cells were harvested by centrifugation (6,000 × g for 15 min at 4°C) and resuspended in 50 mM HEPES-NaOH buffer (pH 7.5). After sonication, the supernatant was obtained by centrifugation (25,000 × g for 30 min at 4°C). The supernatant was incubated at 80°C for 15 min and centrifuged (25,000 × g for 30 min at 4°C) again to obtain the supernatant. Ammonium sulfate was added to the soluble fraction to give 70% saturation. The precipitate collected by centrifugation (25,000 × g for 50 min at 4°C) was dissolved in 50 mM HEPES-NaOH buffer (pH 7.5) and dialyzed with the same buffer. The dialysate was applied to a HiTrap DEAE (5 ml) anion-exchange column (GE Healthcare). TK0882 was eluted with a linear gradient of NaCl (0 to 1.0 M) in 50 mM HEPES-NaOH buffer (pH 7.5), and fractions containing 250 to 350 mM NaCl were collected. The collection was then applied to a Superdex 200 HR 10/30 gel filtration column (GE Healthcare) with a mobile phase of 50 mM HEPES-NaOH buffer (pH 7.5) containing 150 mM NaCl. Protein concentration was determined with a protein assay kit (Bio-Rad, Hercules, CA) according to the instructions from the manufacturer, using bovine serum albumin as a standard.
Activity measurements of recombinant TK0882 protein.
To detect the ureohydrolase activity of TK0882, the products of the enzymatic reaction were analyzed by HPLC. The reaction mixture (200 μl) was composed of substrate (100 μM agmatine or 12.0 μM N1-aminopropylagmatine), 33 mM potassium citrate monohydrate, 563 mM KCl, 217 mM 2-propanol, 0.8 mM Brij 35, 10 mM NiCl2, and 21.7 ml of 1 N HCl per liter. Purified TK0882 (6.9 μg) in 100 mM HEPES-NaOH buffer (pH 7.5) was added and then incubated at 70°C for 60 min. Then, 100 μl of reaction mixture was filtered with a 0.45-μm filter and subjected to HPLC. HPLC analyses were performed as described for polyamine analysis. The quantities of enzyme products were calculated by measuring the peak area on the chromatogram. As standards, various amounts of putrescine or spermidine were subjected to HPLC, and peak areas on the chromatograms were measured.
Expression and purification of TK0147 protein.
The TK0147 gene was amplified with T. kodakarensis genomic DNA as a template and two oligonucleotide primers, TK0147-EX-Fw and TK0147-EX-Rv (Table 1). The amplified fragment was digested with NdeI and BamHI and inserted into the NdeI and BamHI sites of pET21a(+). The resulting plasmid was designated pET-TK0147. Expression of recombinant TK0147 in E. coli cells was carried out using the same conditions as TK0882 induction. Cells were harvested, disrupted, and heated at 80°C for 30 min to obtain a thermostable soluble fraction as well. Ammonium sulfate was added to the soluble fraction to give 80% saturation. The precipitate collected by centrifugation (25,000 × g for 50 min at 4°C) was dissolved in 20 mM Tris-HCl buffer (pH 8.0) and dialyzed with the same buffer. The dialysate was applied to a HiTrap Q (5 ml) anion-exchange column (GE Healthcare). TK0147 was eluted with a linear gradient of NaCl (0 to 1.0 M) in 20 mM Tris-HCl buffer (pH 8.0), and fractions containing 250 to 350 mM NaCl were collected. The collection was then applied to a Superdex 200 HR 10/30 gel filtration column with a mobile phase of 10 mM CHES-NaOH buffer (pH 9.0) containing 300 mM NaCl at a flow rate of 0.50 ml/min.
Activity measurements of recombinant TK0147 protein.
To detect the aminopropyl transferase activity of TK0147, the products of the enzymatic reaction were analyzed by HPLC. The reaction mixture (200 μl) was composed of 100 μM acceptor substrate (putrescine, agmatine, and spermidine), 100 μM donor substrate (S-adenosyl methylthiopropylamine), and purified TK0147 (15 μg) in 10 mM CHES-NaOH buffer (pH 9.0) and was incubated at 70°C for 5 min. Then, 100 μl of the reaction mixture was filtered through a 0.45-μm filter and subjected to HPLC. HPLC analyses were performed as described for polyamine analysis. The quantities of enzyme products were calculated by measuring the peak area on the chromatogram. As a standard, various amounts of spermidine or N1-aminopropylagmatine were subjected to HPLC, and peak areas on the chromatograms were measured.
RNA isolation and reverse transcription-PCR (RT-PCR).
Total RNA from T. kodakarensis was isolated from cells harvested at the logarithmic growth phase using RNeasy Midi kit (Qiagen, Tokyo, Japan). Each total RNA (0.25 μg) in 10 μl of a reaction mixture was used for reverse transcription at 55°C for 30 min with the reverse primer for each (TK0882RT-Rv and TK0147RT-Rv) (Table 1) designed on the basis of each gene and reverse transcriptase (Roche Diagnostics, Basel, Switzerland). PCR was then performed using the synthesized cDNA as a template with the primer pair for each (for TK0882, TK0882RT-Fw and TK0882RT-Rv; for TK0147, TK0147RT-Fw and TK0147-Rv) (Table 1). The PCR conditions were 20 cycles of 15 s at 94°C, 30 s at 60°C, and 45 s at 68°C. Three independent RT-PCRs were conducted to analyze the PCR products (TK0882; 787 bp, TK0147; 577 bp) by 1% (wt/vol) agarose gel electrophoresis. In order to confirm no DNA contamination in the prepared RNA, we performed PCR by specific primers (for TK0882, TK0882RT-Fw and TK0882RT-Rv; for TK0147, TK0147RT-Fw and TK0147-Rv) and confirmed no amplification in advance. As a control to ensure that the signal intensities reflected the initial levels of each transcript directly, RT-PCRs were performed with the same total RNAs and the 16S rRNA gene-specific primers (16SrDNA-Fw and 16SrDNA-Rv) (Table 1) under the PCR conditions described above, except for the reaction cycle (16 cycles) and annealing temperature (60°C). The resulting 174-bp fragments were analyzed in the same way.
RESULTS
Composition of intracellular polyamines in T. kodakarensis.
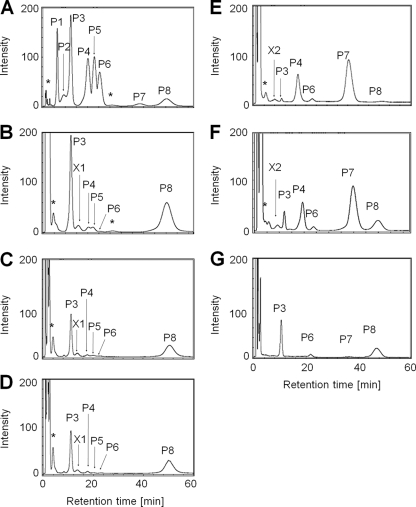
To obtain information on intracellular polyamines in T. kodakarensis, we analyzed polyamine compositions by HPLC as described in Materials and Methods. Intracellular polyamines were extracted from T. kodakarensis strain KU216 cells cultivated at 60°C, 85°C, or 93°C. Peaks were detected at the positions of agmatine, spermidine (P3 in Fig. 2), tris(3-aminopropyl)amine [3(3)3] (P5 in Fig. 2), spermine (P6 in Fig. 2) and N4-aminopropylspermine (P8 in Fig. 2). The peak of putrescine was hardly detected at the experimental temperatures. Moreover, an unknown signal, X1, was detected. It is noteworthy that the amount of spermidine decreased as the cell growth temperature increased, while the amount of N4-aminopropylspermine increased at elevated temperatures. At 60°C, 85°C, and 93°C, the spermidine amount was 1.14 μmol/g, 0.93 μmol/g, and 0.23 μmol/g in wet T. kodakarensis cells, respectively. It decreased as the growth temperature rose. In contrast, the N4-aminopropylspermine amount increased at elevated temperatures. The amount of N4-aminopropylspermine was 0.19 μmol/g at 60°C, 0.80 μmol/g at 85°C, and 0.73 μmol/g at 93°C.
FIG. 2.
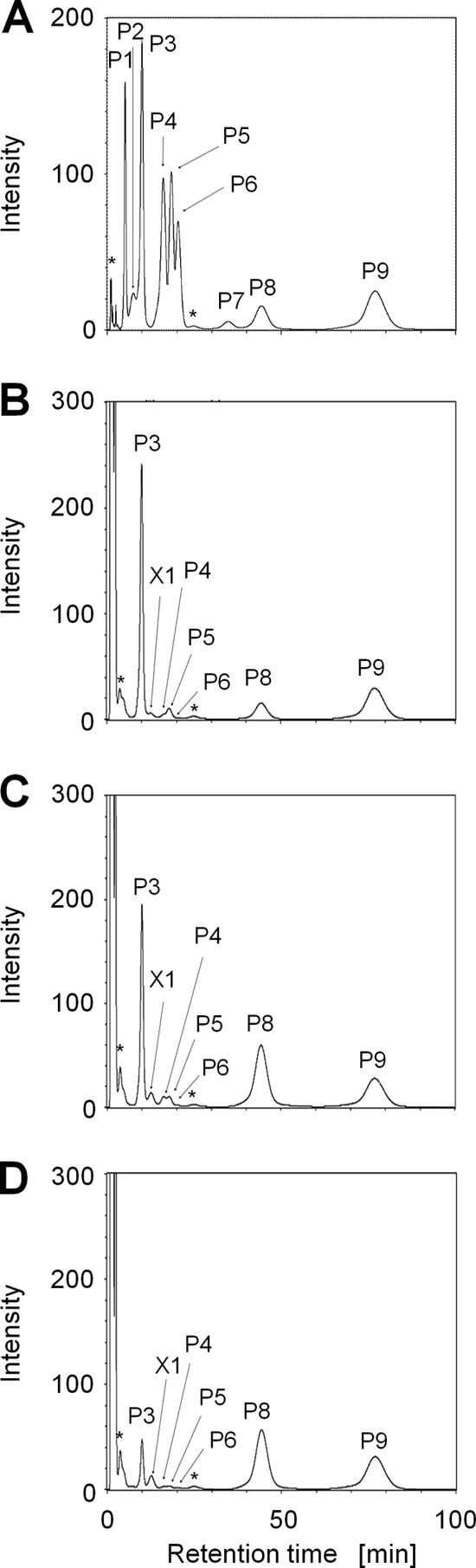
Intracellular polyamines in T. kodakarensis. Intracellular polyamines were extracted from T. kodakarensis KU216 cells grown at 60°C, 85°C, and 93°C. (A) Peak standards; (B) sample from 60°C; (C) sample from 85°C; (D) sample from 93°C. Abbreviations and used amounts are as follows: P1, putrescine (3 nmol); P2, S-adenosyl methylthiopropylamine (decarboxylated S-adenosyl methionine) (30 nmol); P3, spermidine (3 nmol); P4, agmatine (3 nmol); P5, tris(3-aminopropyl)amine (3 nmol); P6, spermine (3 nmol); P7, N1-aminopropylagmatine (3 nmol); P8, N4-aminopropylspermine (0.6 nmol); P9, caldohexamine (30 nmol). *, unexpected noise derived from buffer; X1, unknown peak.
Construction of TK0240, TK0474, and/or TK0882 gene disruptants and their growth characteristics.
A previous study revealed that PdaDTk (TK0149) catalyzed agmatine formation by the decarboxylation of arginine as the first step in polyamine biosynthesis. It is unclear which other enzymes are involved in the subsequent steps. In addition to arginine decarboxylase (PdaDTk), agmatine ureohydrolases (TK0240, TK0474, and TK0882), aminopropyl transferase (TK0147), and S-adenosylmethionine decarboxylase (TK1592) are considered to play crucial roles in polyamine synthesis, as summarized in Fig. 1. In some microorganisms, including T. thermophilus, a pathway from arginine to putrescine via ornithine has been proposed (25). In this pathway, arginine ureohydrolase and ornithine decarboxylase are required; however, in the T. kodakarensis genome, no homologue for ornithine decarboxylase has been identified. T. kodakarensis is therefore considered to synthesize polyamine without utilizing ornithine. We first focused on the pathway from agmatine to spermidine and constructed several single-gene disruptants of three agmatine ureohydrolase homologues to examine the effects of these homologues on cell growth. Disruptant strains lacking TK0240, TK0474, or TK0882 were designated DUH2 (ΔTK0240), DUH4 (ΔTK0474), and DUH8 (ΔTK0882), respectively. Double- and triple-gene disruptants were also constructed and designated DUH24 (ΔTK0240 ΔTK0474), DUH48 (ΔTK0474 ΔTK0882), DUH28 (ΔTK0240 ΔTK0882), and DUH248 (ΔTK0240 ΔTK0474 ΔTK0882) (Table 1). Disruption of the TK0240 or TK0474 gene did not affect the growth profiles at 85°C and 93°C; in contrast, disruption of the TK0882 gene led to a decreased growth rate at 85°C and a severe growth defect at 93°C (Fig. 3A and B). The growth profiles of multiple gene disruptants were also compared with that of a single TK0882 gene disruptant, strain DUH8. Multiple-gene disruptants lacking only the TK0882 gene showed deficient cell growth, as shown in Fig. 3. Hence, TK0882 seems important for cell growth, especially at high temperatures (Fig. 3C and D). Cytoplasmic polyamines in DUH8 and the enzyme characteristics of the TK0882 protein are discussed below.
FIG. 3.
Growth characteristics of T. kodakarensis lacking TK0240, TK0474, and/or TK0882 genes. (A and B) Growth curves of T. kodakarensis KU216 (ΔpyrF), DUH2 (ΔpyrF ΔTK0240), DUH4 (ΔpyrF ΔTK0474), and DUH8 (ΔpyrF ΔTK0882) grown at 85°C (A) and 93°C (B). (C and D) Growth curves of the disruptant strains DUH24 (ΔpyrF ΔTK0240 ΔTK0474), DUH28 (ΔpyrF ΔTK0240 ΔTK0882), DUH48 (ΔpyrF ΔTK0474 ΔTK0882), and DUH248 (ΔpyrF ΔTK0240 ΔTK0474 ΔTK0882) and original strain KU216 (ΔpyrF) grown at 85°C (C) and 93°C (D). OD660, optical density at 660 nm.
Composition of cytoplasmic polyamine in strains DUH2, DUH4, and DUH8.
Cytoplasmic polyamines were extracted from three disruptants (DUH2, DUH4, and DUH8), and their polyamine balances were analyzed. As shown in Fig. 4, no significant difference in polyamine contents was observed between DUH2 and KU216 or between DUH4 and KU216. However, the polyamine balance was changed in DUH8. As shown in Fig. 1, there are two predicted pathways for spermidine synthesis in T. kodakarensis, but it is unclear which the major one is. If pathway I is dominant, agmatine should accumulate in DUH8. If pathway II is dominant, N1-aminopropylagmatine should accumulate. The DUH8 cells cultivated at 85°C were harvested, and the cells obtained were used for the analysis. Both agmatine and N1-aminopropylagmatine were detected in DUH8, but spermidine, spermine, and N4-aminopropylspermidine were hardly detected (Fig. 4). It is noteworthy that both N1-aminopropylagmatine and agmatine were detected in large amounts, suggesting that both agmatine and N1-aminopropylagmatine are ureohydrolyzed by TK0882 in T. kodakarensis. In the thermophilic bacterium T. thermophilus, spermidine is synthesized from agmatine via N1-aminopropylagmatine (23). T. kodakarensis should produce spermidine via the same pathway. As shown in Fig. 4E and F, an unknown peak, X2, was detected in DUH8. The peak appeared close to peak P1 (putrescine), but X2 was distinguished from P1 when DUH8 cell extracts were fractionated with containing putrescine (data not shown). Further studies will be required to identify X1.
FIG. 4.
Intracellular polyamines of DUH2, DUH4, and DUH8. Intracellular polyamines of KU216, DUH2, DUH4, and DUH8 strains were extracted and analyzed by HPLC. Caldohexamine was added to the mixture as an internal control as well as an analysis of KU216 (Fig. 2). The peak of caldohexamine was eliminated from the chart. (A) Peak standards; (B) sample from KU216 cells cultivated at 85°C; (C) sample from DUH2 cells cultivated at 85°C; (D) sample from DUH4 cells cultivated at 85°C; (E) sample from DUH8 cells cultivated at 85°C; (F) sample from DUH8 cells cultivated at 85°C in the presence of spermidine; (G) sample from DUH8 cells cultivated at 93°C in the presence of spermidine. Peak abbreviations are the same as in Fig. 2. X1 and X2, unknown peaks.
Effect of polyamine addition on DUH8 cell growth and cytoplasmic polyamine balance.
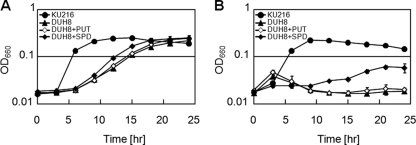
Disruption of the TK0882 gene led to a decreased growth rate at 85°C and a severe growth defect at 93°C. In the DUH8 strain, agmatine and N1-aminopropylagmatine accumulated in the cytoplasm. The growth deficiency of DUH8 was likely caused by inadequate long-chain-polyamine synthesis. In order to examine the effect of additional putrescine or spermidine on DUH8 growth, DUH8 cells were cultivated in a medium containing putrescine or spermidine. The growth rate of DUH8 was slightly increased, and cell growth was regained at 85°C and 93°C by the addition of spermidine only (Fig. 5). On the other hand, the growth defect was not eliminated by the addition of putrescine. Intracellular polyamines were extracted from DUH8 cells cultivated in the presence of spermidine at 85°C or 93°C and examined (Fig. 4F and G). N4-Aminopropylspermine (peak P8) was detected at both 85°C and 93°C. In contrast, N1-aminopropylagmatine (peak P7) was detected at 85°C but not at 93°C in DUH8.
FIG. 5.
Growth characteristics of DUH8 in the presence or absence of supplemental polyamine. Growth curves of KU216 and DUH8 were monitored at 85°C (A) and 93°C (B). OD660, optical density at 660 nm.
Examination of TK0882 function.
To confirm the agmatine ureohydrolase activity of TK0882, the recombinant form was expressed in E. coli cells, and TK0882 was purified. The thermostability of TK0882 was evaluated by circular dichroism (CD) spectrum analysis, and TK0882 was still not fully unfolded at 90°C (see Fig. S1 in the supplemental material). TK0882 seems functional even at 90°C. The ureohydrolase activity was examined by utilizing two substrates, agmatine and N1-aminopropylagmatine, at 70°C. As shown in Table 2, both substrates were accepted by TK0882, and putrescine and spermidine were produced from agmatine and N1-aminopropylagmatine, respectively. When the kinetic parameters were compared, TK0882 showed a higher kcat/Km value for N1-aminopropylagmatine (165 ± 37 M−1·s−1) than for agmatine (3.87 ± 0.38 M−1·s−1), suggesting that TK0882 functions mainly as N1-aminopropylagmatine ureohydrolase to produce spermidine. TK0882 accepts N1-aminopropylagmatine more efficiently, because it possesses a 54-fold-lower Km value for N1-aminopropylagmatine (6.42 μM) than for agmatine (486 μM), as shown in Table 2. The enzymatic activities for both substrates increased as the reaction temperature increased. When we examined the activity at 70°C, 80°C, and 90°C, the highest activity was detected at 90°C (data not shown).
TABLE 2.
Kinetic parameters of ureohydrolase activity of TK0882
| Substrate | Concn (μM) | Km (μM) | kcat (s−1) (10−3) | kcat/Km (M−1·s−1) |
|---|---|---|---|---|
| Agmatine | 50-1,000 | 486 ± 46.1 | 1.86 ± 0.06 | 3.87 ± 0.38 |
| N1-Aminopropyl agmatine | 4.82-24.1 | 6.42 ± 2.34 | 1.01 ± 0.11 | 165 ± 37 |
Construction of the TK0147 disruptant and growth characteristics.
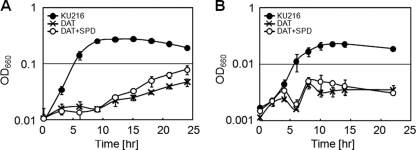
TK0147 was first annotated as spermidine synthase by genome analysis. Based on the sequence similarity, aminopropyl transferase was expected to catalyze the aminopropylation of polyamines. TK0147 is the only candidate for aminopropyl transferase among all annotated genes in the genome. Furthermore, TK0882 has been shown to function as an aminopropylagmatine ureohydrolase to produce spermidine. Hence, we speculated that TK0147 catalyzes aminopropyl chain transfer from decarboxylated S-adenosylmethionine (S-adenosyl methylthiopropylamine) to agmatine. If this is the case, then knockout of the TK0147 gene would influence the polyamine balance in the cytoplasm, and agmatine would accumulate in the TK0147 disruptant. To examine the role of TK0147, we constructed a TK0147 gene disruptant, designated strain DAT (disruptant of aminopropyl transferase), and examined DAT cell growth. As shown in Fig. 6, the DAT strain showed decreased growth at 85°C and 93°C. Interestingly, the growth rate of DAT was slightly restored at 85°C by the addition of spermidine (Fig. 6A). However, unlike that of DUH8, the growth of DAT at 93°C was not restored even when spermidine was added (Fig. 6B). These results suggest that spermidine is incorporated and contributes to cell growth at 85°C but not at 93°C. TK0147 may transfer an aminopropyl group to produce the longer-chain and/or branched-chain polyamines required for cell growth at 93°C. To examine the role of TK0147, cytoplasmic polyamines of DAT were analyzed.
FIG. 6.
Growth profiles of strain DAT. Growth curves of KU216 and DAT were monitored at 85°C (A) and 93°C (B). OD660, optical density at 660 nm.
Composition of cytoplasmic polyamine in strain DAT.
DAT cells were cultivated at 85°C in a medium with or without spermidine and harvested by centrifugation. The cells obtained were used for the analysis. Agmatine, spermidine, spermine, putrescine, and N4-aminopropylspermine were detected in DAT cells (Fig. 7C). It is noteworthy that larger amounts of agmatine accumulated in DAT than in KU216 (Fig. 4B and C). In T. kodakarensis, TK0147 is likely to function as N1-aminopropylagmatine synthase rather than as spermidine synthase. Small amounts of spermidine, spermine, and N4-aminopropylspermine were also produced in DAT, suggesting that an unidentified enzyme which functions as aminopropyl transferase exists in vivo. By the addition of spermidine, the growth of DAT at 85°C was partially restored, as shown in Fig. 6A. To examine what kinds of polyamines are synthesized under the stated condition, cytoplasmic polyamines were extracted from DAT cells cultivated in the presence of spermidine and analyzed. The peak P8 of N4-aminopropylspermine was detected, as shown in Fig. 7D, indicating that the synthesis of long- and branched-chain polyamines is catalyzed by an unknown aminopropyl transferase besides TK0147.
FIG. 7.
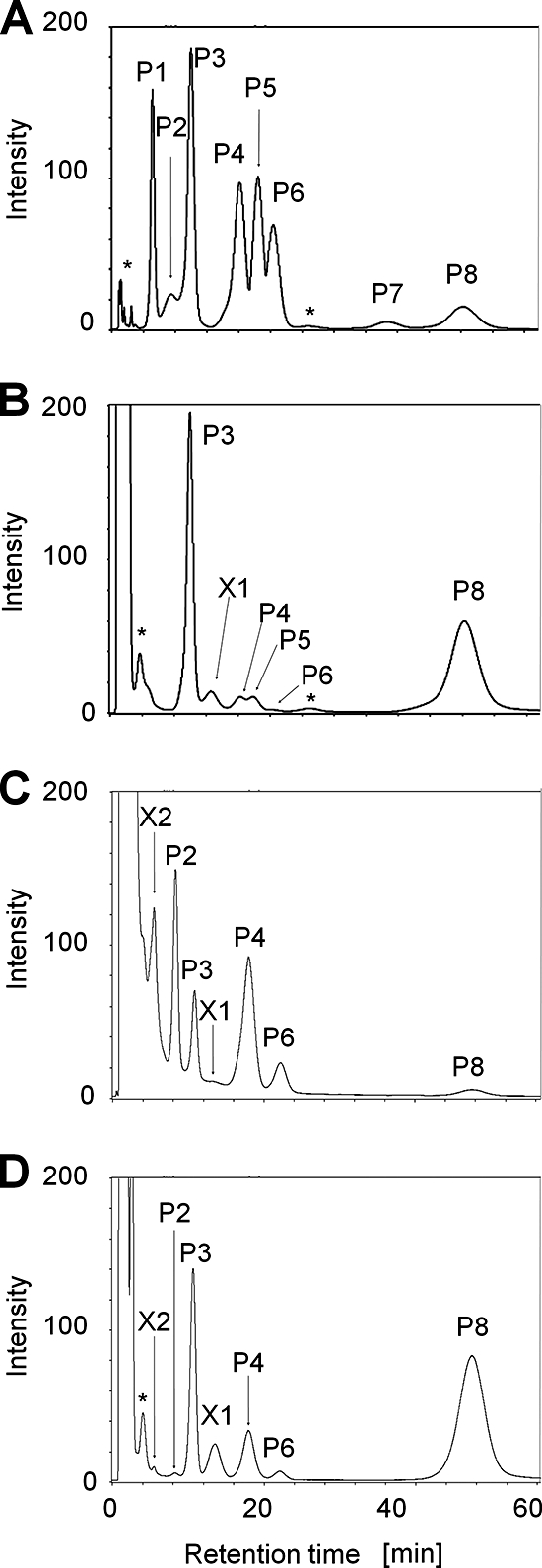
Intracellular polyamines of strain DAT. Intracellular polyamines of DAT were extracted and analyzed by HPLC. Caldohexamine was added to the mixture as an internal control as well as an analysis of KU216 (Fig. 2). Peak of caldohexamine is eliminated from the chart. (A) Peak standards; (B) sample from KU216 cells cultivated at 85°C; (C) sample from DAT cells cultivated at 85°C; (D) sample from DAT cells cultivated at 85°C in the presence of spermidine. Peak abbreviations are the same as in Fig. 2. X1 and X2, unknown peaks.
Examination of TK0147 function.
To confirm the aminopropyl transferase activity of TK0147, the recombinant form was expressed in E. coli cells, and TK0147 was purified. The aminopropyl transferase activity was examined utilizing three acceptor substrates, agmatine, cadaverine, and putrescine, in addition to decarboxylated S-adenosylmethionine (S-adenosyl methylthiopropylamine), as an aminopropyl chain donor. Because decarboxylated S-adenosylmethionine is spontaneously degraded at temperatures higher than 70°C, an enzyme assay was carried out at 70°C. As shown in Table 3, all acceptor substrates were utilized, and N1-aminopropylagmatine and spermidine were produced from agmatine and putrescine, respectively. When the kcat/Km values were compared, TK0147 showed higher aminopropyl transferase activity for agmatine (22.5 ×103 M−1·s−1) than for putrescine (0.29 ×103 M−1·s−1) and cadaverine (0.29 ×103 M−1·s−1). TK0147 showed a lower Km for agmatine (12.2 μM) than for putrescine (1,053 μM), indicating that TK0147 catalyzes the aminopropyl chain transfer to agmatine more efficiently than that to putrescine. Activity for cadaverine was also detected, but the activity was less than that for agmatine. Hardly any activity for spermidine (spermine synthase activity) was obtained, because the signal peak was very small. TK0147 mainly functions as agmatine aminopropyl transferase to produce N1-aminopropylagmatine rather than spermidine/spermine synthase. It is noteworthy that spermine and N4-aminopropylspermine were produced in DAT, showing the involvement of an unknown aminopropyl transferase in addition to TK0147 in the synthesis of a long- and branched-chain polyamine in T. kodakarensis.
TABLE 3.
Kinetic parameters of aminopropyl transferase activity of TK0147
| Substrate | Concn (μM) | Km (μM) | kcat (s−1) | kcat/Km (M−1·s−1) (103) |
|---|---|---|---|---|
| Putrescine | 100-1,400 | 1,053 ± 101 | 0.30 ± 0.03 | 0.29 ± 0.01 |
| Agmatine | 1-80 | 12.2 ± 1.41 | 0.28 ± 0.07 | 22.5 ± 3.9 |
| Cadaverine | 10-100 | 80.7 ± 36.9 | 0.02 ± 0.007 | 0.29 ± 0.06 |
Expression profiles of TK0882 and TK0147.
As reported above, both TK0882 and TK0147 seem to contribute to high-temperature adaptation. To examine the expression level of these two genes at various growth temperatures, we performed quantitative RT-PCR analysis using the total RNA from T. kodakarensis cells grown at 60°C, 85°C, and 93°C. As shown in Fig. 8, signals for TK0882 and TK0147 transcripts were detected at all examined temperatures by RT-PCR. However, both mRNA levels slightly decreased with an increase of the growth temperature. At 93°C, both transcripts were detected, suggesting that the level was sufficient to produce functional amounts in the cytoplasm.
FIG. 8.
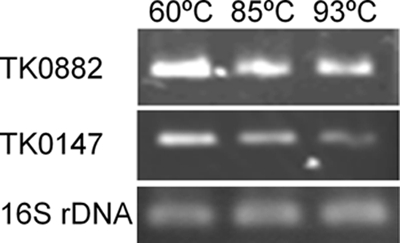
Expression profile of TK0882 and TK0147 genes. Transcription levels of TK0882 and TK0147 genes at several growth temperatures were examined. RT-PCR was performed by using total RNA isolated from cells grown at 60°C, 85°C, and 93°C in logarithmic phase. To ensure that the intensities directly reflected the initial levels of each transcript, the levels of 16S rRNA were examined as an internal control.
DISCUSSION
In T. kodakarensis, N4-aminopropylspermine was identified in addition to conventional polyamines such as putrescine and spermidine. Long-chain and branched-chain polyamines are known to stabilize DNA and RNA as thermotolerance factors (32). In T. kodakarensis, they are considered to be involved in thermoadaptation at elevated growth temperatures. Under the present HPLC conditions, tris(3-aminopropyl)amine can barely be distinguished from N4-aminopropylspermidine, thermine [333], and thermospermine [334]. It is unclear whether these polyamines are produced in T. kodakarensis. It has been assumed that unique branched-chain polyamines are synthesized from spermidine. According to the genomic analysis in T. kodakarensis, two kinds of synthetic pathways from agmatine to spermidine were predicted, as shown in Fig. 1. One is the pathway via putrescine (pathway I), and the other is that via N1-aminopropylagmatine (pathway II). HPLC analysis revealed that putrescine was detected in the cytoplasm, suggesting the production of spermidine by pathway I. Furthermore, N1-aminopropylagmatine as well as agmatine was detected in strain DUH8, lacking the TK0882 gene, indicating that spermidine is synthesized mainly from agmatine by pathway II. Pathway II has been observed in the thermophilic bacterium T. thermophilus (23). In the present study, the same pathway was shown to be achievable in the hyperthermophilic archaeon T. kodakarensis. In P. furiosus, PF0127 was shown to have aminopropyl transferase activity. Agmatine was most often preferred on the basis of the kcat/Km value (3), suggesting that pathway II is also achievable in P. furiosus. Pathway II has not been detected in mesophilic microorganisms; it seems to be unique to thermophilic microorganisms. As for agmatine ureohydrolase homologues, T. kodakarensis possesses TK0240 and TK0474 genes in addition to the TK0882 gene. Their transcriptional levels were examined by RT-PCR, showing that both TK0240 and TK0474 were also transcribed at all temperatures examined (data not shown). However, TK0240 and TK0474 were not involved in polyamine synthesis, because their disruptions did not cause any change in polyamine contents in vivo. These genes are considered to play roles other than that of polyamine synthesis.
N4-aminopropylspermidine was found in Pyrococcus spp. (P. furiosus and P. woesei) and Thermococcus spp. (T. celer, T. litoralis, and T. stetteri), closely related hyperthermophiles belonging to the order Thermococcales (11, 14). In T. kodakarensis, N4-aminopropylspermidine may be synthesized as well. Under the present HPLC conditions, tris(3-aminopropyl)amine was hardly distinguished from N4-aminopropylspermidine. If N4-aminopropylspermidine were produced, N4-aminopropylspermine could be produced from spermidine via N4-aminopropylspermidine. To confirm the possibility, further precise analysis is needed. In the present analyses, an unknown signal, X1, was detected. We speculated that it was homospermidine [44], aminopropylcadaverine [35], or spermidine due to its retention time. In order to examine this issue, we compared it with homospermidine [44], aminopropylcadaverine [35], and spermidine. However, peak X1 did not match any standard peaks (see Fig. S2 in the supplemental material), and X1 therefore remains an unknown compound.
In P. horikoshii, PH0083, which is an orthologue of TK0882, has been characterized as agmatine ureohydrolase (8). PH0083 also catalyzed the hydrolysis of agmatine but not of arginine. Our kinetic analyses clearly indicate that TK0882 mainly functions as N1-aminopropylagmatine ureohydrolase to produce spermidine. Kinetic information on agmatine ureohydrolases is limited. The kcat/Km value of TK0882 (3.87 M−1·s−1 for agmatine, 165 M−1·s−1 for N1-aminopropylagmatine) was low compared with that for agmatine ureohydrolase from rodent brain (320 M−1·s−1 for agmatine) (33).
The cell growth of DUH8 was partially retrieved at 85°C and 93°C by the addition of spermidine. On the other hand, the growth defect was not eliminated by the addition of putrescine. Putrescine may be barely incorporated into cells due to the lack of a transport system. No obvious homologue involved in polyamine transport has been identified in the T. kodakarensis genome (6). Another possibility would be poor aminopropyl transferase activity for putrescine. When we analyzed intracellular polyamines from DUH8 cells cultivated in the presence of spermidine at 85°C or 93°C (Fig. 4F and G), N4-aminopropylspermine (peak P8) was detected at both 85°C and 93°C in DUH8. However, N1-aminopropylagmatine (peak P7) was detected at 85°C but not at 93°C, suggesting that aminopropyl transferase, which functions mainly at 93°C, is involved in N4-aminopropylspermine synthesis.
The results of our gene disruption study reveal that TK0147 functions as N1-aminopropylagmatine synthase and spermidine synthase. In vitro results also support this idea: recombinant TK0147 catalyzed N1-aminopropylagmatine synthesis from agmatine as well as spermidine synthesis from putrescine. However, TK0147 did not catalyze spermine synthesis from spermidine. In addition, spermine was produced in the DAT strain. These results show that spermine is synthesized not by TK0147 but by an unidentified aminopropyl transferase in vivo. When we tried to list aminopropyl transferase candidates on the basis of sequence similarity, TK0147 was the only suitable candidate identified. The unidentified aminopropyl transferase may possess low structural similarity to TK0147. In strain DAT (ΔTK0147), the amount of N4-aminopropylspermine was increased by the addition of spermidine, indicating that the synthesis of long-chain and branched-chain polyamines is also catalyzed in the absence of TK0147. We are not sure how many aminopropyl transferases T. kodakarensis possesses. Several unidentified aminopropyl transferases may independently catalyze aminopropyl transfer from putrescine to spermidine, spermidine to spermine, and spermine to N4-aminopropylspermine.
Aminopropyl transferases from various bacteria and archaea have been studied. PF0127 in P. furiosus is one of the characterized enzymes. PF0127 showed broad amine acceptor specificity for agmatine, 1,3-diaminopropane, putrescine, cadaverine, and sym-norspermidine, but agmatine was the most preferable on the basis of the kcat/Km value (134 × 102 M−1·s−1 for agmatine, 5.50 × 102 M−1·s−1 for cadaverine, 23 M−1·s−1 for putrescine, and 32 M−1·s−1 for 1,3-diaminopropane) (3). The higher activity for agmatine is explained by the crystal structure, which shows that agmatine is easily accommodated in the binding site pocket and that several additional hydrogen bonds are formed between the agmatine amide group and residues Glu7, Tyr9, Val51, and Asp162 of PF0127 (3). These residues are conserved in TK0147 as Glu12, Tyr14, Val56, and Asp169, respectively. Agmatine acts as an aminopropyl acceptor in the TK0147 reaction as well as in the PF0127 reaction. However, TK0147 does not accept cadaverine efficiently, on the basis of the Km value (12.2 μM for agmatine, 80.7 μM for cadaverine, and 1,053 μM for putrescine) and the kcat/Km value (22.5 × 103 M−1·s−1 for agmatine, 0.29 × 103 M−1·s−1 for cadaverine, and 0.29 ×103 M−1·s−1 for putrescine). These two enzymes appear to possess different enzyme properties, although they show high amino acid sequence similarity (67% identity). In Sulfolobus solfataricus, orthologous aminopropyl transferase does not accept cadaverine as a substrate (2). In the present study, cadaverine and aminopropylcadaverine were not identified at all examined temperatures in T. kodakarensis. It is unclear why cadaverine is accepted by PF0127. In Pyrococcus spp., aminopropylcadaverine might be synthesized under some stress conditions, as speculated in a previous study (3).
Supplementary Material
Acknowledgments
This study was supported by grants from the Japan Society for the Promotion of Science (JSPS) KAKENHI (20580090), Grant-in-Aid for Creative Scientific Research (project no. 18GS0421), and Support Project to Assist Private Universities in Developing Bases for Research by Ministry of Education, Culture, Sports, Science and Technology.
We thank Masaru Niitsu, Josai University, for technical advice on performing HPLC analyses and Akira Shirahata and Fumihiko Ikeguchi, Josai University, for kindly providing standard polyamines. We thank Prakarn Krathinthong and Toshiyuki Moriya for technical assistance.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Atomi, H., T. Fukui, T. Kanai, M. Morikawa, and T. Imanaka. 2004. Description of Thermococcus kodakaraensis sp. nov., a well studied hyperthermophilic archaeon previously reported as Pyrococcus sp. KOD1. Archaea 1:263-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cacciapuoti, G., M. Porcelli, M. Carteni-Farina, A. Gambacorta, and V. Zappia. 1986. Purification and characterization of propylamine transferase from Sulfolobus solfataricus, an extreme thermophilic archaebacterium. Eur. J. Biochem. 161:263-271. [DOI] [PubMed] [Google Scholar]
- 3.Cacciapuoti, G., M. Porcelli, M. A. Moretti, F. Sorrentino, L. Concilio, V. Zappia, Z. J. Liu, W. Tempel, F. Schubot, J. P. Rose, B. C. Wang, P. S. Brereton, F. E. Jenney, and M. W. Adams. 2007. The first agmatine/cadaverine aminopropyl transferase: biochemical and structural characterization of an enzyme involved in polyamine biosynthesis in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 189:6057-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danno, A., W. Fukuda, M. Yoshida, R. Aki, T. Tanaka, T. Kanai, T. Imanaka, and S. Fujiwara. 2008. Expression profiles and physiological roles of two types of prefoldins from the hyperthermophilic archaeon Thermococcus kodakaraensis. J. Mol. Biol. 382:298-311. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara, S., R. Aki, M. Yoshida, H. Higashibata, T. Imanaka, and W. Fukuda. 2008. Expression profiles and physiological roles of two types of molecular chaperonins from the hyperthermophilic archaeon Thermococcus kodakarensis. Appl. Environ. Microbiol. 74:7306-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukuda, W., N. Morimoto, T. Imanaka, and S. Fujiwara. 2008. Agmatine is essential for the cell growth of Thermococcus kodakaraensis. FEMS Microbiol. Lett. 287:113-120. [DOI] [PubMed] [Google Scholar]
- 7.Fukui, T., H. Atomi, T. Kanai, R. Matsumi, S. Fujiwara, and T. Imanaka. 2005. Complete genome sequence of the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1 and comparison with Pyrococcus genomes. Genome Res. 15:352-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goda, S., H. Sakuraba, Y. Kawarabayasi, and T. Ohshima. 2005. The first archaeal agmatinase from anaerobic hyperthermophilic archaeon Pyrococcus horikoshii: cloning, expression, and characterization. Biochim. Biophys. Acta 1748:110-115. [DOI] [PubMed] [Google Scholar]
- 9.Groppa, M. D., and M. P. Benavides. 2008. Polyamines and abiotic stress: recent advances. Amino Acids 34:35-45. [DOI] [PubMed] [Google Scholar]
- 10.Hamana, K., H. Hamana, M. Niitsu, K. Samejima, T. Sakane, and A. Yokota. 1993. Tertiary and quaternary branched polyamines distributed in thermophilic Saccharococcus and Bacillus. Microbios 75:23-32. [PubMed] [Google Scholar]
- 11.Hamana, K., and T. Itoh. 2001. Polyamines of the hyperthermophilic archaebacteria belonging to the genera Thermococcus and Methanothermus and two new genera Caldivirga and Palaeococcus. Microbios 104:105-114. [PubMed] [Google Scholar]
- 12.Hamana, K., M. Niitsu, K. Samejima, and T. Itoh. 2001. Polyamines of the thermophilic eubacteria belonging to the genera Thermosipho, Thermaerobacter and Caldicellulosiruptor. Microbios 104:177-185. [PubMed] [Google Scholar]
- 13.Hamana, K., M. Niitsu, K. Samejima, T. Itoh, H. Hamana, and T. Shiozawa. 1998. Polyamines of the thermophilic eubacteria belonging to the genera Thermotoga, Thermodesulfovibrio, Thermoleophilum, Thermus, Rhodothermus and Meiothermus, and the thermophilic archaebacteria belonging to the genera Aeropyrum, Picrophilus, Methanobacterium and Methanococcus. Microbios 94:7-21. [Google Scholar]
- 14.Hamana, K., T. Tanaka, R. Hosoya, M. Niitsu, and T. Itoh. 2003. Cellular polyamines of the acidophilic, thermophilic and thermoacidophilic archaebacteria, Acidilobus, Ferroplasma, Pyrobaculum, Pyrococcus, Staphylothermus, Thermococcus, Thermodiscus and Vulcanisaeta. J. Gen. Appl. Microbiol. 49:287-293. [DOI] [PubMed] [Google Scholar]
- 15.Higashibata, H., S. Fujiwara, S. Ezaki, M. Takagi, K. Fukui, and T. Imanaka. 2000. Effect of polyamines on histone-induced DNA compaction of hyperthermophilic archaea. J. Biosci. Bioeng. 89:103-106. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi, K., and K. Kashiwagi. 1999. Polyamine transport in bacteria and yeast. Biochem. J. 344(Pt. 3):633-642. [PMC free article] [PubMed] [Google Scholar]
- 17.Ikeuchi, Y., S. Kimura, T. Numata, D. Nakamura, T. Yokogawa, T. Ogata, T. Wada, and T. Suzuki. 2010. Agmatine-conjugated cytidine in a tRNA anticodon is essential for AUA decoding in archaea. Nat. Chem. Biol. 6:277-282. [DOI] [PubMed] [Google Scholar]
- 18.Imai, A., T. Matsuyama, Y. Hanzawa, T. Akiyama, M. Tamaoki, H. Saji, Y. Shirano, T. Kato, H. Hayashi, D. Shibata, S. Tabata, Y. Komeda, and T. Takahashi. 2004. Spermidine synthase genes are essential for survival of Arabidopsis. Plant Physiol. 135:1565-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Janne, J., L. Alhonen, T. A. Keinanen, M. Pietila, A. Uimari, E. Pirinen, M. T. Hyvonen, and A. Jarvinen. 2005. Animal disease models generated by genetic engineering of polyamine metabolism. J. Cell Mol. Med. 9:865-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuno, Y., A. Sugai, H. Higashibata, W. Fukuda, K. Ueda, I. Uda, I. Sato, T. Itoh, T. Imanaka, and S. Fujiwara. 2009. Effect of growth temperature and growth phase on the lipid composition of the archaeal membrane from Thermococcus kodakaraensis. Biosci. Biotechnol. Biochem. 73:104-108. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa, M., Y. Izawa, N. Rashid, T. Hoaki, and T. Imanaka. 1994. Purification and characterization of a thermostable thiol protease from a newly isolated hyperthermophilic Pyrococcus sp. Appl. Environ. Microbiol. 60:4559-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moriya, T., T. Hikota, I. Yumoto, T. Ito, Y. Terui, A. Yamagishi, and T. Oshima. 16 April 2010, posting date. Calditerricola satsumensis gen. nov., sp. nov. and C. yamamurae sp. nov., extreme thermophiles isolated from a high temperature compost. Int. J. Syst. Evol. Microbiol. doi: 10.1099/ijs.0.018416-0. [Epub ahead of print.] [DOI] [PubMed]
- 23.Ohnuma, M., Y. Terui, M. Tamakoshi, H. Mitome, M. Niitsu, K. Samejima, E. Kawashima, and T. Oshima. 2005. N1-aminopropylagmatine, a new polyamine produced as a key intermediate in polyamine biosynthesis of an extreme thermophile, Thermus thermophilus. J. Biol. Chem. 280:30073-30082. [DOI] [PubMed] [Google Scholar]
- 24.Oshima, T. 1983. Novel polyamines in Thermus thermophilus: isolation, identification, and chemical synthesis. Methods Enzymol. 94:401-411. [Google Scholar]
- 25.Pantazaki, A. A., C. G. Anagnostopoulos, E. E. Lioliou, and D. A. Kyriakidis. 1999. Characterization of ornithine decarboxylase and regulation by its antizyme in Thermus thermophilus. Mol. Cell Biochem. 195:55-64. [DOI] [PubMed] [Google Scholar]
- 26.Rhee, H. J., E. J. Kim, and J. K. Lee. 2007. Physiological polyamines: simple primordial stress molecules. J. Cell Mol. Med. 11:685-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 28.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2005. Improved and versatile transformation system allowing multiple genetic manipulations of the hyperthermophilic archaeon Thermococcus kodakaraensis. Appl. Environ. Microbiol. 71:3889-3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sato, T., T. Fukui, H. Atomi, and T. Imanaka. 2003. Targeted gene disruption by homologous recombination in the hyperthermophilic archaeon Thermococcus kodakaraensis KOD1. J. Bacteriol. 185:210-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shimada, Y., W. Fukuda, Y. Akada, M. Ishida, J. Nakayama, T. Imanaka, and S. Fujiwara. 2009. Property of cold inducible DEAD-box RNA helicase in hyperthermophilic archaea. Biochem. Biophys. Res. Commun. 389:622-627. [DOI] [PubMed] [Google Scholar]
- 31.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terui, Y., M. Ohnuma, K. Hiraga, E. Kawashima, and T. Oshima. 2005. Stabilization of nucleic acids by unusual polyamines produced by an extreme thermophile, Thermus thermophilus. Biochem. J. 388:427-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uribe, E., M. Salas, S. Enriquez, M. S. Orellana, and N. Carvajal. 2007. Cloning and functional expression of a rodent brain cDNA encoding a novel protein with agmatinase activity, but not belonging to the arginase family. Arch. Biochem. Biophys. 461:146-150. [DOI] [PubMed] [Google Scholar]
- 34.Wallace, H. M., A. V. Fraser, and A. Hughes. 2003. A perspective of polyamine metabolism. Biochem. J. 376:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang, Z., and C. D. Lu. 2007. Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 189:3945-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.