Abstract
The nitrogen assimilation control protein (NAC) is a LysR-type transcriptional regulator (LTTR) that is made under conditions of nitrogen-limited growth. NAC's synthesis is entirely dependent on phosphorylated NtrC from the two-component Ntr system and requires the unusual sigma factor σ54 for transcription of the nac gene. NAC activates the transcription of σ70-dependent genes whose products provide the cell with ammonia or glutamate. NAC represses genes whose products use ammonia and also represses its own transcription. In addition, NAC also subtly adjusts other cellular functions to keep pace with the supply of biosynthetically available nitrogen.
NITROGEN REGULATION
In 1952, Boris Magasanik noticed that a histidine auxotroph of Klebsiella pneumoniae (then known as Aerobacter aerogenes) required 30 times more histidine for growth in minimal medium (rich in ammonium salts) when inositol was supplied as the carbon source than it did when glucose was supplied (76). This led to the double assumption that there must be an enzymatic pathway to catabolize histidine and that the expression of this pathway must be repressed by glucose. A series of publications in the 1950s proved both of these conclusions correct and demonstrated both the histidine utilization enzymes encoded by the hut operons (36, 43, 64) and their repression by glucose (40, 52, 53). In pursuing the repression, Magasanik coined the term catabolite repression to describe the absence of the enzymes responsible for catabolizing a poorer carbon source when a more effective carbon source is being catabolized (41). Histidine can be used as the sole source of carbon, but growth is slower than that with glucose as the carbon source. Inositol is also a poor carbon source, so less repression of the histidine degrading enzymes is seen when inositol replaces glucose. Histidine can also serve as a nitrogen source for K. pneumoniae when a poor carbon source (like inositol) is present, with two of its nitrogen atoms available for incorporation into cellular material (40, 43). Thus, it was predicted that K. pneumoniae would be able to use histidine as a sole nitrogen source only when glucose was absent. However, surprisingly, K. pneumoniae grows well in glucose minimal medium with histidine as the nitrogen source, and the enzymes of histidine degradation are derepressed under these conditions (53).
In 1956, Neidhardt and Magasanik (53) showed that the hut enzymes could be derepressed in glucose minimal medium, but only if ammonium was absent. They showed that either nitrogen limitation (N limitation) or carbon limitation would allow expression of the hut enzymes. Moreover, the derepression in response to N limitation did not result from elimination of catabolite repression. In fact, in glucose minimal medium, N limitation leads to an increased catabolite repression of traditional indicators like β-galactosidase and tryptophanase (63). This phenomenon was termed “relief of catabolite repression” and appeared to be specific for pathways that yielded ammonium or glutamate and to be independent of the effects of CRP-cAMP.
But the exploration reached a dead end at that point. There was no system of genetic exchange characterized for K. pneumoniae at that time. And the two enterobacteria with good genetic systems, Escherichia coli and Salmonella enterica (then called S. typhimurium), were unsuitable. E. coli lacks the hut operons and cannot catabolize histidine under any growth condition. S. enterica has a hut system, but in S. enterica the catabolite repression of hut is not relieved by nitrogen limitation (8, 45), and thus, S. enterica cannot use histidine as the sole source of nitrogen when glucose is the carbon source. So, in the absence of good genetics, the question of how hut could be derepressed by nitrogen limitation lay unanswered for a dozen or more years.
THE Ntr SYSTEM
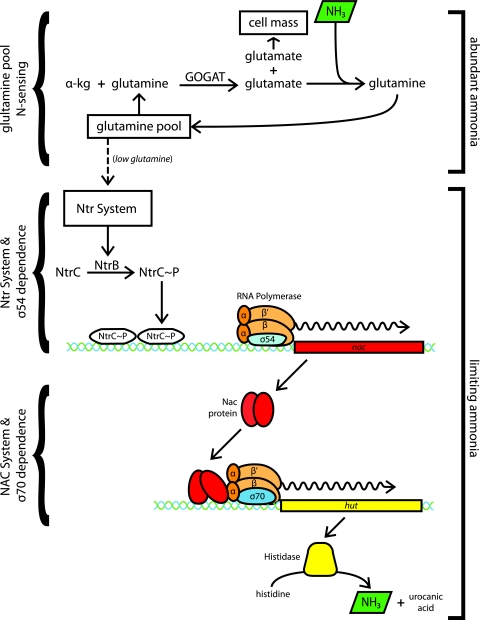
The discovery of a transducing phage for K. pneumoniae (38) and the subsequent adaptation of the coliphage P1 tools for use with K. pneumoniae (17) allowed a reopening of the study of this nitrogen regulatory effect (63). A study of K. pneumoniae glutamine auxotrophs and their revertants (7, 62) led ultimately to the discovery of the Ntr system. The Ntr system has been reviewed extensively elsewhere (42, 61), and a detailed discussion of it is beyond the scope of this review. A few basic features of the Ntr system (Fig. 1) are pertinent to this discussion. (i) The Ntr system is a two-component regulatory system in which the transcriptional regulator, NtrC, is phosphorylated to its active form by NtrB under conditions of N-limited growth (30, 56). (ii) The signal for nitrogen limitation is a low intracellular pool of glutamine (3, 25). (iii) NtrC∼P activates transcription by RNA polymerase bearing the unusual sigma factor σ54 (23, 24), which recognizes an unusual, GC-rich promoter sequence (22) that is not recognized by σ70.
FIG. 1.
A cartoon illustrating the nitrogen regulation of the histidine utilization (hut) genes of K. pneumoniae. Step one: under conditions of ammonia limitation, the intracellular pool of glutamine is filled by the action of glutamine synthetase and depleted by the action of glutamate synthase (GOGAT). The glutamine pool is also depleted by the biosynthetic needs of the cell, both directly and by incorporation of glutamate into cellular material. Step two: when the intracellular pool of glutamine is low, the glnD and glnB products (not shown) signal NtrB to phosphorylate (activate) NtrC to NtrC∼P. NtrC∼P activates transcription of the nac gene by RNA polymerase bearing the unusual sigma factor σ54. Step three: the NAC protein activates transcription of the hutUH operon by RNA polymerase bearing the “housekeeping” sigma factor σ70. The products of the hut operons cleave histidine to generate ammonia, which is then assimilated into glutamine, restoring the cycle. (Urocanic acid, the other product of histidine cleavage, is further degraded to glutamate by the remaining hut-encoded enzymes.)
K. pneumoniae is a “nitrogen generalist,” capable of using a wide range of organic and inorganic compounds as its sole source of nitrogen (75). The metabolism of nearly all of these alternative N sources is controlled in response to N limitation, and this N regulation requires the Ntr system in almost every case. This regulation by the σ54-dependent Ntr system can be direct, as it is for the glnA gene, or indirect, as it is for the hut (histidine utilization) operon.
DISCOVERY OF NAC
As part of a study of nitrogen fixation in a related Klebsiella strain, Valentine and his coworkers isolated the first true nitrogen regulatory mutation, which they called asm-1 (51). This mutation was in the gltBD operon, which encodes GOGAT (glutamate synthase), one of the two enzymes that allow de novo synthesis of glutamate from α-ketoglutarate (44, 74). These gltBD mutants were unable to activate expression of the nif (nitrogen fixation) or hut operon in response to nitrogen limitation (51). Similar gltB mutants that could not activate hut or put expression were then identified in K. pneumoniae (7). These mutants were still able to activate hut in response to carbon limitation, and thus, the defect seemed to be specific to N regulation. The reason for the inability to activate hut expression was not known at that time (21), but revertants that could activate hut expression were easily obtained, and these were constitutively active for hut expression, even in the presence of excess ammonia (7). The mutations responsible for the constitutive expression of hut were mapped to the ntrB gene (69), tightly linked to glnA, the structural gene for glutamine synthetase (73). Unexpectedly, these revertants were unable to grow in glucose minimal medium with ammonia as the sole source of nitrogen (7). This inability was caused by a repression of the gdhA gene, encoding the enzyme glutamate dehydrogenase, the only enzyme other than GOGAT that can effect a net assimilation of ammonia into glutamate. Once again, revertants that could grow with ammonia as the sole nitrogen source were easily isolated. These revertants were constitutive for gdhA expression and constitutively repressed for hut expression (4). The mutations responsible for some of the revertants were located in ntrC, again tightly linked to the glnA locus, but the majority defined a new locus called nac.
Under conditions of N limitation, K. pneumoniae nac mutants are still able to activate expression of glnA and a variety of other N-regulated genes (e.g., nif, nas, a catabolic asparaginase gene, and a tryptophan permease gene) but are not able to activate expression of several others, e.g., hut, put, and ure (4, 37). Even when the Ntr system was constitutively active (due to an ntrB mutation), nac mutants could not activate this subset of nitrogen-regulated genes. The glutamate dehydrogenase (GDH) gene, gdhA, also fell into the class of NAC-regulated genes, but in the opposite direction. GDH was expressed at high levels in the absence or presence of ammonia in nac mutants (4, 37). Operons whose regulation did not require NAC (glnA-ntrB-ntrC and nifLA) had σ54-dependent promoters, as expected for Ntr-regulated genes (22, 42). However, operons whose regulation did require NAC (hutUH and gdhA) had σ70-dependent promoters (28, 54), which should not be recognized by the Ntr system. Studies with nac-lacZ fusions showed that nac was itself regulated by the Ntr system (37). This was confirmed by in vitro transcription using purified components of the Ntr system (11). Finally, experiments with mutants that put the nac gene under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter proved that nac expression was both necessary and sufficient for activation of hutUH and repression of gdhA expression in vivo (72).
Thus, the basic picture of N regulation in K. pneumoniae (Fig. 1) was understood (1): in the absence of the preferred N source (ammonia), the glutamine pool falls and leads to activation of the Ntr system and the consequent phosphorylation of the NtrC protein. NtrC∼P activates expression of a number of genes, all of which are dependent on σ54. One of those genes is nac. NAC in turn activates or represses expression of a number of operons, whose expression is dependent on the normal sigma factor, σ70. One of those operons is hutUH.
SCOPE OF NAC REGULON
A survey of related enterobacteria showed that E. coli has a nac gene but S. enterica does not. The location of nac between two direct repeats of an asparaginyl tRNA gene explains the origin of the nac-1 mutation of K. pneumoniae and the lack of a nac gene in S. enterica. In both cases, there is only one of those Asn-tRNA genes, and the genes between them are either missing or located elsewhere on the chromosome (20). This explains why the catabolite repression of hut in S. enterica is not relieved by N limitation even though it has a functioning Ntr system.
Genetic and physiological studies of individual catabolic pathways led to an initial list of 8 operons regulated by NAC in K. pneumoniae (Table 1). A comparable survey for E. coli identified 4 operons. Zimmer et al. (78) used an E. coli microarray to examine the scope of the NAC regulon and found 9 operons whose regulation depends on NAC. As expected, each of the operons whose products are known is involved in N metabolism. We used chromatin immunoprecipitation (ChIP) to identify potential NAC-regulated genes in K. pneumoniae and found at least 89 unique DNA regions that bound NAC (16). Primer extension assays with 16 of these genes showed that 15 required NAC for their regulation. (The 16th gave no signal under the growth conditions tested.) Of the genes for which a function was known or could be inferred, about half were clearly related to N metabolism. The remaining genes affect the mobilization of carbon sources, the synthesis of macromolecules, and cell division. In other words, the NAC regulon of K. pneumoniae is large and broad, that of E. coli is much smaller and may be limited to nitrogen metabolism, and S. enterica lacks a nac gene. However, it should be noted that the hutUH operon of S. enterica retains the sites needed for regulation by NAC. If its hutUH operon is transferred to K. pneumoniae, it is regulated by nitrogen (63). Conversely, transfer of a cloned nac gene from K. pneumoniae to S. enterica allows relief of catabolite repression in response to N limitation (5).
TABLE 1.
Evidence for NAC regulation of gene expression
| Category and operon | Function | Reference(s) for evidence |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
In vivo |
In vitro |
||||||||
| Enzyme assaya | lacZ fusionb | Primer extensionc | NBS mutantd | EMSAe | DNase I footprintf | Transcriptiong | ChIPh | ||
| Activated by NAC | |||||||||
| hut | Histidine → NH3 + glutamate + formamide | 4, 3 | 60 | 19 | 19 | 19 | |||
| put | Proline permease | 37 | 9 | 9 | 19 | ||||
| ure | Urea → NH3 − CO2 | 37 | 33 | 16, 33 | 15, 33 | 15, 68 | 19 | 16 | |
| cod | Cytosine → NH3 + uracil | 50 | 50 | 50 | 50, 66 | 50 | 50 | 16 | |
| dad | Alanine → NH3 + pyruvate | 26 | 26 | 26 | 16 | ||||
| folA | Dihydrofolate reductase (thymine synthesis) | 16 | 16 | ||||||
| gyrA | DNA gyrase | 16 | 16 | ||||||
| ldcA | l,d-Carboxypeptidase (murein integrity) | 16 | 16 | ||||||
| oppA | Oligopeptide transport | 16 | 16 | ||||||
| rpmI | Ribosomal protein L35 | 16 | 16 | ||||||
| ybjI | (Putative lipoprotein) | 16 | 16 | ||||||
| yceO | (Biofilm and acid stress) | 16 | 16 | ||||||
| Repressed by NAC | |||||||||
| gdhA | NH3 + αkgi → glutamate | 4, 37 | 20 | 28 | 20 | 20, 29, 68 | 20 | 16 | |
| gltB | Glutamine + αkgi → glutamate | 4, 21 | |||||||
| nac | Transcription regulation | 37 | 11 | 12, 66, 67 | 12, 67 | 12 | |||
| argP | Transcription regulation | 16 | 16 | ||||||
| holE | DNA replication | 16 | 16 | ||||||
| secY | Protein secretion | 16 | 16 | ||||||
| glgX | Glycogen debranching | 16 | 16 | ||||||
| ygdQ | (Putative transport protein) | 16 | 16 | ||||||
| yobA | (Unknown) | 16 | 16 | ||||||
An enzyme encoded by the operon was assayed in a nac+ and nac mutant background.
A promoterless lacZ gene was fused to the promoter of the operon, and β-galactosidase expression was measured in a nac+ and nac mutant background.
Primer extension was performed using RNA extracted from cells grown in excess or limiting nitrogen or from nac+ and nac mutant cells grown in limiting nitrogen.
A mutation was introduced into the NAC-binding site (NBS) as determined by DNase I footprinting. The enzyme activities of the mutant and wild type were compared under conditions of NAC-regulated expression.
EMSAs were performed with purified NAC and DNA fragments containing the NBS.
DNase I protection of the NBS by purified NAC.
In vitro transcription with purified components.
Chromatin immunoprecipitation using purified anti-NAC antibody.
αkg, α-ketoglutarate.
The differences in the size of K. pneumoniae's NAC regulon and that of E. coli or S. enterica probably reflect the different ecology of K. pneumoniae. K. pneumoniae is at best a minor component of the human gut and, despite its fearsome species name, is merely an opportunistic pathogen. It contrast to E. coli and S. enterica, K. pneumoniae survives and thrives in soil and aquatic environments, where nutrient limitation, especially N limitation, would favor an expanded repertoire of pathways that could be expressed when needed. It is telling that several of the genes that are regulated by NAC in K. pneumoniae are either not regulated (26) or weakly regulated (48) in E. coli. In at least one case (26), this loss in E. coli resulted from a single base pair change in an otherwise recognizable activation consensus sequence. Thus, it appears that the E. coli set of NAC-regulated genes is shrinking while that of K. pneumoniae may be expanding. S. enterica has lost NAC entirely but still contains remnants of a NAC regulon in that hutUp of S. enterica carries all the information for regulation by NAC. Of course, it is possible that S. enterica acquired hut recently from a NAC-containing source. However, the location of hut, between the gal and bio operons, is identical in K. pneumoniae and S. enterica, arguing that hut is ancestral to both. Although NAC has allowed K. pneumoniae to coopt many operons into the N limitation response, NAC is not the only path to a rich metabolic diversity of nitrogen-yielding pathways. The pseudomonads can use a very large number of N sources, and yet no true homolog of NAC has been found (77).
One final observation about the E. coli nac gene deserves comment. Although E. coli nac appears to complement K. pneumoniae nac mutants completely (48), there is a surprising divergence of sequence between these two homologous genes. Many E. coli proteins are 90 to 95% identical to their K. pneumoniae homologs in their amino acid sequence. The E. coli and K. pneumoniae NAC proteins are only 79% identical in amino acid sequence (48). However, the E. coli and K. pneumoniae NAC proteins are more than 90% identical in the N-terminal one-third of the protein, the portion that is sufficient for binding to DNA and activating transcription (49, 68). This divergence in the C-terminal domain of NAC has its explanation in the structure of NAC.
STRUCTURE
NAC is a LysR-type transcriptional regulator with strong similarity to other LysR-type transcriptional regulator (LTTRs). For example, the N-terminal domain of NAC (amino acids 1 to 100) is 39% identical in amino acid sequence to OxyR from E. coli and 15% identical in the C-terminal domain (71). So, it is not surprising that NAC shares many of the properties common to most LTTRs (39). NAC is a tetramer in solutions at concentrations above 7 μM (68) and dissociates into dimers at lower concentrations, consistent with the general “dimer of dimers” structure of LTTRs. Like most LTTRs, NAC is quite insoluble, though the K. pneumoniae NAC (but not the E. coli NAC) is soluble in high salt (19). This differential solubility provided a simple protocol for purification of the K. pneumoniae NAC so that both native NAC and C-terminally His-tagged NAC can be isolated with ease. Finally, NAC regulates expression of its own gene (see below), though there is no oppositely directed gene that NAC activates (20, 37), as is the case with many LTTRs whose regulons are more limited (39, 70).
However, NAC differs from the generic LTTR (39, 70) in several key ways. NAC dimers activate transcription at most of the well-characterized promoters (68). Most LTTRs use tetramers to activate transcription. Most LTTRs bind a physiologically relevant coeffector (or undergo a covalent modification) to regulate their function in response to environmental signals. NAC's function is regulated solely at the level of synthesis. NAC is active whether purified from cells grown under N excess or N limitation (19). NAC's activity in vitro is unchanged by likely candidates for coeffectors such as α-ketoglutarate, glutamate, glutamine, and ammonia (19). If NAC is produced in vivo from an IPTG-inducible promoter, NAC-dependent gene expression responds to IPTG and not to the nitrogen status of the cell (72). Taken together, these in vivo and in vitro data argue that the NAC polypeptide (as a dimer or as a tetramer) is necessary and sufficient for regulation of NAC-dependent genes.
The C-terminal domain of most LTTRs contains the sites of coeffector binding, for formation of tetramers from dimers, and for conformational rearrangement of the dimers within the tetramer in response to coeffector binding (39, 70). Thus, it is not surprising that this domain is dispensable for many (though not all) of NAC's functions. Substitution mutants of NAC (NACL111K and NACL125R) have been isolated that cannot form tetramers and remain as dimers (68). These mutants activate transcription of hut and ure about as well as wild-type NAC (NACWT). In addition, truncation mutants of NAC with only the 125, 100, or even 86 N-terminal amino acids have been isolated. These are dimers in solution and activate as well as NACWT wherever tested (49, 68). But the C-terminal domain does have a function: tetramer formation, which is essential for the repression of gdhA (20, 49, 68) and is also part of the regulation of codBA (50) and nac (12, 66) as described below.
NAC-ACTIVATED PROMOTERS
The hutUH promoter, hutUp.
At hutUp, a dimer of NAC (68) binds to a site centered at −64 relative to the transcription start site (19) and activates transcription of hutUp by RNA polymerase bearing σ70 (19). The promoter activated by NAC is the same promoter that is used for basal expression of hutUp in the absence of activators. This same promoter is also activated by CRP-cAMP (58), which has binding sites centered at −81.5 and −41.5 relative to the transcription start site (58). In all three cases (basal expression, NAC-mediated activation, and CRP-cAMP-mediated activation), RNA polymerase bearing σ70 is used (19, 55, 58). The common question of whether the two positive activators of hutUp, NAC and CRP-cAMP, would act synergistically or antagonistically is moot, since these two activators would never be present in the same cell at the same time.
Several lines of evidence establish that the dimer is the active form of NAC at hutUp. The extent of the mobility shift in an electrophoretic mobility shift assay (EMSA) is consistent with a dimer, as is the 28-bp size of the DNase I footprint (19, 68). In addition, NAC mutants that cannot form tetramers (NACL111K) are fully active at hutUp (29, 68). Truncated versions of NAC (NAC100ter and NAC86ter) containing only the 100 or 86 N-terminal amino acids of NAC are fully active at hutUp in vivo (49, 68). Thus, the N-terminal domain of NAC is sufficient for NAC activation at hutUp. The C-terminal domain plays no known role at hutUp.
The nature of the interaction of NAC with RNA polymerase is not known. Genetic analysis of a related LTTR, CysB, identified a positive control (PC) mutant (Y27G, affecting amino acid 27) that still binds normally to DNA but fails to activate transcription of the cysP operon (34). Those studies showed that mutations affecting amino acids K271 and E273 of the alpha subunit of RNA polymerase also showed defects in activation by CysB (35). In addition, a two-hybrid assay showed an interaction between a domain of CysB and the alpha subunit of RNA polymerase that is not found with the Y27G substitution of CysB (35). This has suggested a necessary interaction between Y27 of CysB and the “273 determinant” of the alpha subunit as the key to transcriptional activation. A comparable PC mutant of NAC, NACH26D, has been isolated and shown to have a PC phenotype (67), so there is reason to suspect that many of the features of activation by LTTRs may be generalizable. However, it seems likely that H26 of NAC plays a more complex role in activation (15, 67).
NACH26A is wild type for activation of hutUp, and several other drastic substitution mutants (e.g., NACH26K and NACH26Y) also retain a substantial ability to activate hutUp (67). NAC activates ureDp from a site centered at −47, overlapping the RNA polymerase binding site and on the opposite face of the DNA helix from the site at hutUp (33), and yet NACH26D shows a positive-control phenotype at this promoter as well (67). It seems unlikely that H26 could make the same kinds of contact with RNA polymerase in both cases. So, we favor a model where H26 plays an important role in allowing NAC to undergo the DNA-mediated conformational change that is required for allowing it to activate transcription.
The interaction of NAC with its DNA site is better understood. NAC's 28-bp DNase I footprint is centered on a 15-bp symmetric sequence, ATA-n9-TAT (19). Genetic analysis of the 5 or 6 bp flanking this consensus suggests that they do not provide specific information for binding or activation (60). A more careful analysis of the 15-bp core, including both mutational analysis and comparison with other known NAC-binding sites (NBS), suggests that the critical nucleotides in this more detailed “activation consensus” are ATAA-n5-TnGTAT (19, 60). The asymmetry of this sequence is critical to NAC's ability to activate hutUp expression (60). Inverting the 15-bp site from ATA ACA AAA TTG TAT (the wild type) to ATA CAA TTT TGT TAT leaves NAC's binding and DNase I footprint intact but severely reduces activation. A site with symmetric hexamers and based on the promoter-proximal half-site (ATA CAA AAA TTG TAT) bound NAC weakly but still showed activation proportional to the reduced binding. A symmetric site based on the promoter distal half-site (ATA ACA AAA TGT TAT) bound NAC better than the wild-type site but was severely defective for activation.
At hutUp, binding of NAC is not sufficient for activation. The sequence of key nucleotides determines whether a bound NAC dimer will activate transcription or not (60). In other words, the DNA binding site is an allosteric effector for NAC. NAC bound to some sites is in an inactive conformation; NAC bound to other sites is active. This agrees with a pattern seen in other LTTRs where the dimers within the tetramers play different roles in mediating activation of transcription (39, 70). One of the dimer-binding sites used by such a tetramer functions in binding but lacks some information that allows a bound protein to activate transcription. The other dimer-binding site used by this tetramer carries information necessary for activation as well as binding.
The ureDABCEFG promoter, ureDp.
The ureDABCEFG operon of K. pneumoniae encodes an N-regulated urease that provides the cell with ammonia from urea (13). The ureA, ureB, and ureC genes encode the subunits of the apoenzyme, and the other ure genes are involved in the insertion of a required nickel atom to generate an active holoenzyme (32). In contrast to the well-studied urease from Proteus spp., whose formation requires the presence of urea, the urease from K. pneumoniae is regulated solely by N limitation (46). The ureDp region contains a weakly regulated promoter (ureDp2), which I shall ignore here, and the NAC-dependent ureDp1 promoter (33).
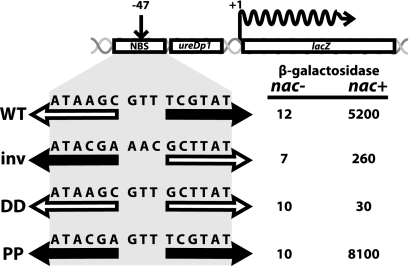
Transcription from ureDp1 is the most tightly regulated of all the characterized NAC-dependent promoters. In fact, in the absence of NAC, β-galactosidase expression from a ureDp1-lacZ fusion (Fig. 2) was barely detectable, making ureDp1 at least 300- to 500-fold inducible by NAC (33). NAC-mediated activation of ureDp1 shares several features with that of hutUp. A dimer of NAC binds to a region near the promoter and protects a 28-bp region of DNA with an “activation consensus” site (ATAA-N5-TnGTAT) at its center, the same activation consensus found at hutUp (19). NACL111K, NAC86ter, and NAC100ter, which cannot form tetramers, activate ureDp1 about as well as NACWT (68). The asymmetry of the NBS is important (15). Inverting the site nearly abolishes the ability of NAC to activate ureDp1 (Fig. 2). A symmetric site based on the promoter-proximal half-site is even more effective than the native site. In contrast, a symmetric site based on the promoter-distal half-site is totally inactive, in spite of the fact that all four types of sites bind NAC well (15).
FIG. 2.
Sequence specificity and directionality of the NAC-binding site (NBS) within the ureD promoter. The upper drawing illustrates the position of the NBS at −47 relative to the start of transcription of a fusion of a promoterless lacZ gene to the nitrogen-regulated ureDp1 promoter. The wild-type (WT) sequence of the NBS yielded 433 times as much β-galactosidase expression in a nac+ strain as in a nac mutant strain. Inverting the 15-bp core of the NBS (inv) greatly reduced the ability of NAC to activate expression of the fusion. An alteration of the NBS that increased the symmetry of the core by making an inverted repeat based on the promoter-distal half-site (DD) virtually eliminated the ability of NAC to activate expression of the fusion. The converse alteration that increased symmetry based on the promoter-proximal half-site (PP) made the effect of NAC on expression even greater than that in the wild type. All four NBS sites shown here bound NAC well in vitro.
However, the ureDp1 promoter differs from hutUp in several important features. The NBS at ureDp1 is centered at −47 relative to the start of transcription (33), and its DNase I footprint extends from −37 to −60 (19), overlapping the RNA polymerase binding site and on a different side of the helix (relative to RNA polymerase) than the NBS at hutUp. In addition to being the most NAC-dependent of the NAC-regulated promoters, ureDp1 is also the most NAC sensitive. When nac expression was titrated by varying the IPTG concentrations for an IPTG-inducible nac gene construct, we found that urease expression was fully induced at NAC levels that did not induce histidase at all (72). This was also evident from unpublished EMSA experiments with mixtures of ureDp and hutUp fragments. The ureDp fragments were shifted at lower concentrations of NAC than were the hutUp fragments. It bears repeating that most of the NAC-regulated promoters we have studied show considerable basal (NAC-independent) expression, and NAC-mediated activation results in a 3- to 10-fold increase in expression (37). For ureDp1, there is no detectable expression in the absence of NAC. Unfortunately, early (unsuccessful) attempts to show NAC-mediated activation of ureDp were not resumed (19). Thus, hutUp from K. pneumoniae and codBp from E. coli remain the only promoters whose NAC regulation has been confirmed in vitro with purified components.
THE codBA OPERON
K. pneumoniae has two cytosine deaminase genes, one of which is part of a codBXA operon that encodes a putative cytosine permease (codB), the cytosine deaminase (codA), and a gene of unknown function (codX). The other cytosine deaminase (codA2) is a standalone gene (50). Both of these appear to be regulated by NAC. Experiments using sensitivity to 5-fluorocytosine as an indicator of codA expression suggest that cytosine deaminase expression in K. pneumoniae is activated in a NAC-dependent manner (50). Cloned codBXA from K. pneumoniae was transferred to an E. coli cod deletion mutant, and the cytosine deaminase expression of the resulting strain was derepressed about 5-fold in response to N limitation (50). Further explorations of NAC-mediated regulation of cytosine deaminase expression were done using the better-characterized codBA operon from E. coli.
Plasmid-borne cod-lac fusions show a NAC-dependent derepression of about 3- to 5-fold. This recapitulates the 3-fold derepression of cytosine deaminase expression by N limitation (50). In vitro transcription from a supercoiled plasmid containing the codB promoter showed a NAC-dependent activation of the codBp-driven transcript (50). Thus, NAC is both necessary and sufficient for activation of codBp in response to N limitation.
Both hutUp and ureDp are activated by NAC dimers. However, both EMSA and DNase I footprinting have shown that NAC binds as a tetramer at codBp (50). In contrast to the 26- to 28-bp footprint seen at hutUp or ureDp, the NAC footprint at codBp is about 56 bp, with a single hypersensitive site in the middle (50). The promoter-proximal half of the footprinted region contains a standard NBS that is able to bind NAC dimers in the absence of the rest of the region. The promoter-distal half of the footprinted region cannot bind a NAC dimer on its own, but the presence of an ATA triplet (half of a NBS consensus) is necessary for tetramer binding to the complete site. The length of the DNase I footprint, the mobility in gel shift assays, and the minimal amount of hypersensitivity all suggest that the NAC tetramer bound at codBp has a conformation similar to that of a typical LTTR in its active form, the form with the two dimers binding the DNA with little or no space between them (66).
Mutational analysis showed that the promoter-proximal NBS at codBp is essential for activation (50). However, deletion of the promoter-distal NBS fragment within the region, resulting in dimer binding as determined by gel shifts, only slightly reduced the ability of NAC to activate a cod-lac fusion on a plasmid (50). Thus, although a NAC tetramer appears to be more effective, it appears that a NAC dimer can also activate codBp.
The regulation of codBA is complex. Cytosine deaminase provides the cell with ammonia (for general biosynthesis) but also with uracil (on the salvage pathway that leads to UTP and CTP). Expression of codBA is regulated by PurR in response to purine levels, and this regulation appears to be independent of NAC (31). Expression of codBA is also regulated by the stringent response, with a strong repression mediated by ppGpp and DksA (14). In response to severe N limitation, DksA-mediated repression and NAC-mediated activation counteract each other. In a DksA deletion, codBA expression is high and NAC provides little or no further activation. Thus, NAC's role appears to allow the cells to extract ammonia from cytosine, even when the stringent response signals that there is no need for uracil (i.e., pyrimidines) for RNA synthesis.
THE lac OPERON
The wild-type lacZYA operon of E. coli is not activated by N limitation. In fact, it is subject to an especially strong catabolite repression under these conditions (63). When the NBS from the K. pneumoniae hut operon (NBShutU) was inserted into the lacZp region, β-galactosidase expression was derepressed about 13-fold by N limitation (59). Primer extension analysis showed that most of the increased expression came from the principal lacZ promoter, lacZp1 (59). However, a second minor lacZ promoter, lacZp2, was also activated. NBShutU in this construct was centered at −64 and −42 relative to the start of transcription from lacZp1 and lacZp2, respectively. When the NBShutU was moved 5 bp farther from lacZp, another minor promoter, lacZp3, was activated, but lacZp1 and lacZp2 were not. When NBShutU was moved another 5 bp away, only lacZp2 was activated. A pattern emerged from these data, with NAC activating when NBShutU was 42, 52, 54, or 64 bp from the start of transcription but not when at 47, 49, 59, or 74 bp from the start of transcription. In other words, NAC activated lacZp when NBShutU was on the correct side of the DNA helix and not too far away (59). It is curious that the NBS at ureDp1 is located at −47, and yet ureDp1 is the most responsive of all the NAC-dependent promoters.
This suggested that there may be nothing special to distinguish those promoters that are activated by NAC from those that are not. This also suggested that the position of the NBS is variable but not infinitely so. The NBS from the gdhA promoter, where NAC represses transcription, contains the “repression consensus” of ATA-n9-GAT (see below), which differs slightly from the activation consensus discussed thus far. When NBSgdhA was used instead of NBShutU, no activation was seen (59). Thus, inverting an NBS or using an NBS from a repression site eliminates NAC activation, suggesting that the information needed for activation of transcription includes the sequence, direction, and location of the NBS.
THE dadAB OPERON
The dadAB operon (dadAX in E. coli) is responsible for l-alanine catabolism in K. pneumoniae (26). The dadB gene encodes an alanine racemase, and dadA encodes the small subunit of d-amino acid dehydrogenase. The regulation of dadAB in K. pneumoniae is complex. It is both induced (in the presence of alanine) and repressed (in the absence of alanine) by the leucine-responsive protein, Lrp (27). As a result, cells grown with alanine added to glucose minimal medium have 25 to 30 times as much racemase and dehydrogenase as cells grown in glucose minimal medium without alanine. The dadAB operon is also weakly activated (about 2-fold) by CRP-cAMP (26). Neither the E. coli nor the S. enterica dad operon shows regulation by nitrogen (26). However, the K. pneumoniae dad operon is activated by NAC in response to N-limited growth.
Studying the nitrogen regulation of dadAB in K. pneumoniae was initially complicated by the fact that alanine (both the l and d isomers) strongly inhibits glutamine synthetase activity (2, 26). The resulting glutamine starvation leads to derepression of the entire Ntr system, including NAC expression (Fig. 1). By using nac and ntr mutants and adding glutamine to the growth medium, it was possible to identify the role of NAC in regulating dadAB expression (26). In the absence of alanine (i.e., dad repressed by Lrp), artificial induction of nac raised dad expression about 5-fold. In the presence of alanine, (i.e., dad activated by Lrp), constitutive expression of the Ntr system (and NAC) raised dad expression about 3-fold. Thus, the addition of alanine to glucose minimal medium has three physiological effects. (i) It lifts the Lrp-mediated repression of dad. (ii) It brings about the Lrp-mediated activation of dad. (iii) It induces the Ntr system, resulting in NAC-mediated activation of dad. Taken together, these effects result in a 50-fold increase in dad expression.
NAC-dependent gel shifts of the dadAp region and inspection of the DNA sequence of the dadAp region suggest that NAC binds to dadAp as a dimer (26), though DNase I footprints have not yet confirmed this. The NAC-activated transcript has the same start site as the Lrp-activated one (26), and there is an NBS consensus (ATAA-n5-CnGTAT) that differs by only 1 bp from the activation consensus and is centered at −44 relative to this start site. The dadAB regions of K. pneumoniae and E. coli are nearly identical from −56 to +23 except for the region of the putative NBS. In particular, ATA-n9-TAT of K. pneumoniae is replaced by ATA-n9-CAT (26). This explains why NAC fails to bind to the E. coli dadAp region in vitro and why there is no activation of E. coli dad expression in vivo.
THE put OPERON
The putP and putA genes of K. pneumoniae are a pair of divergently transcribed genes that encode a proline permease and a bifunctional proline oxidase/pyrroline-5-carboxylic acid dehydrogenase, respectively (10). Proline oxidase expression is elevated by N limitation (63), and this elevation is NAC dependent (37). NAC binds to and protects a 25-bp site in the regulatory region between putP and putA (19). This site contains the NBS of ATA-n9-TAT, and genetic analysis showed that these are important for NAC binding both in vivo and in vitro (9). Primer extension studies show strong derepression of the putP transcript in response to N limitation (10) and suggest that the regulation of proline oxidase expression from putA may be indirect. In addition to its two enzymatic functions, the PutA product also represses transcription of the put genes in the absence of proline (47). Thus, an increase in proline transport (caused by NAC) might be sufficient to cause derepression of putA expression.
OTHER ACTIVATED GENES
ChIP experiments using anti-NAC antibody identified 89 unique DNA regions that are candidates for NAC-mediated regulation (16). Regulation of 15 of these was tested by comparing primer extension products from nac+ and nac mutant strains grown under N limitation. Nine of these 15 had a stronger signal in the nac+ strain than in the nac mutant (i.e., they were activated by NAC). These nine were folA, gyrA, ldcA, mltE, oppA, rpmI, ureD, ybjP, and yceO. Although not characterized in detail, these are likely candidates for NAC-activated genes. The remaining six genes appear to be repressed by NAC (see below).
GENES REPRESSED BY NAC
Like most other LTTRs, NAC represses its own synthesis, but NAC also represses the expression of several other operons and uses a variety of mechanisms to exert that repression. A slightly different NBS consensus seems to be used in these sites. Instead of the ATA-n9-TAT sequence seen within the activation consensus NBS, the NBS consensus for repression seems to be ATA-n9-GAT or its equivalent, ATC-n9-TAT (20, 68).
Glutamate dehydrogenase (gdhA gene).
The gdhA gene of K. pneumoniae is activated by ArgP, which binds to the gdhAp region and footprints a region from −50 to −100 relative to the transcriptional start site (18). A NAC dimer binds to a NBS and footprints a region from −75 to −100, overlapping the ArgP-binding site (20). Since ArgP and NAC share a binding site, the presence of NAC eliminates ArgP-mediated activation and reduces gdhA expression about 3-fold (18). The presence of lysine in the growth medium inhibits ArgP-mediated activation of gdhA to the same extent as NAC's binding to the region upstream of the gdhA promoter (28). It is clear that a NAC dimer is sufficient for this effect, since NACL111K, which cannot tetramerize, can exert this effect, sometimes called “weak repression” (68).
NAC also exerts a much stronger repression of gdhAp expression, but this “strong repression” requires that NAC be able to form a tetramer (68). The strong repression requires the dimer-binding site centered at −89 (mentioned above), but it also requires a second dimer-binding site centered at +57, about 147 bp away from the other NBS (20). Moreover, it is important that the two dimer-binding sites be on the correct side of the DNA helix. Inserting 5 extra base pairs between them reduces the effect of NAC, but inserting 10 bp between them leaves strong repression largely intact (20). The best model to explain these data is that a tetramer of NAC with one dimer bound at −89 and another at +57 causes a DNA loop to form and this loop interferes with the ability of RNA polymerase to transcribe from gdhAp.
Glutamate synthase (gltBD operon).
The gltBD operon of K. pneumoniae codes for the two subunits of glutamate synthase (commonly called GOGAT). GOGAT activity is repressed as much as 17-fold under conditions of severe N limitation (21), though somewhat less under less severe limitation (4, 21, 37). This repression is NAC dependent (4, 48). A similar NAC-dependent repression of GOGAT formation (about 8-fold) is seen in E. coli (21, 48). The transcription start site for gltBp is known in E. coli (57), and the strong DNA sequence conservation in this region suggests that it is the same in K. pneumoniae (21), though this has not been confirmed. Preliminary experiments from this laboratory suggest that NAC binds to two sites in the gltBp region, one of which could overlap the −35 region of gltBp and the other of which lies about 270 bp farther upstream. Each of these regions contains a sequence that resembles or matches a known NBS consensus (centered at about −60 and −305 relative to the transcription start site). Little else is known about the nature of this regulation in K. pneumoniae, but the presence of two NAC-binding sites might suggest a mechanism similar to that seen at gdhA. Alternatively, NAC might directly affect RNA polymerase binding, it might interfere with the essential role of Lrp in activating gltBp, or it might act through none of these mechanisms. Clearly this important operon deserves further study.
The nac gene.
Like most LTTRs, NAC downregulates its own synthesis (5, 12, 37). This autoregulation is important, since NAC is always in its active form and overproduction of NAC severely inhibits growth (6). Most LTTRs limit transcription of their own gene by a simple repression mechanism. However, NAC's autoregulation is more complex. The nac gene is transcribed by RNA polymerase bearing the unusual sigma factor σ54 (11, 37). This transcription absolutely requires the presence of an activator (NtrC∼P) bound to two enhancer sites at −155 and −135 relative to the transcription start site (11). NAC is one of the very few negative regulators of σ54-dependent transcription and acts by binding as a tetramer to a site between the enhancer and the promoter (12). DNase I footprints show that NAC protects a region from −58 to −130, occupying almost all the space between bound NtrC∼P and the RNA polymerase (11, 68). NAC does not interfere with the binding of NtrC∼P or RNA polymerase (11), but it does introduce a significant bend (about 113 degrees) into the DNA (12, 68). Because the NtrC∼P-dependent enhancer is so close to the promoter, the flexibility of the “linker DNA” is limited, and anything that interferes with this flexibility will impede the ability of NtrC∼P to activate the polymerase. The severe bend introduced by NAC has precisely this effect (12).
The NAC-binding region at the nac promoter is composed of two NBSs (66), one of which contains a match to the repression consensus (ATC-n9-TAT, which is equivalent to ATA-n9-GAT) and the other of which has a one-base mismatch (ATA-n9-GcT). Each of these sites is capable of binding a dimer of NAC (though not a tetramer) in the absence of the other. Furthermore, the nontetramerizing mutant NACL111K fills both sites but bends the DNA much less than the tetramer (66, 68). Curiously, NACL111K blocks NtrC∼P-mediated activation nearly as well as NACWT (29). Perhaps the lesser bend induced by a NAC dimer (65) is the reason for this effect. However, it is more likely that filling the space between the bound NtrC∼P and RNA polymerase with two dimers of NACL111K restricts the flexibility of the DNA enough to prevent the interaction of NtrC∼P with the RNA polymerase (12). Strictly speaking, the regulation of nac by NAC is an antiactivation rather than a repression.
FLEXIBILITY OF NAC TETRAMER CONFORMATIONS
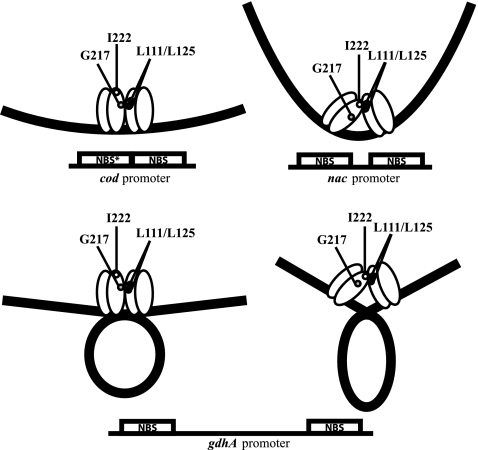
The three sites where NAC has been shown to function as a tetramer (Fig. 3) differ in the position of the two dimer-binding sites that make up the complete tetramer-binding site (66). At cod, the two dimer-binding sites are closely apposed (50), at nac, the two sites are separated by 10 bp (11, 68), and at gdh, the two sites are separated by 148 bp (20). The architectures of the first two types (cod and nac) resemble sites recognized by the activated and nonactivated forms of most LTTRs (39, 70). Since NAC recognizes both sites without any coeffector signal to change its conformation (66), there was a question of whether NAC did in fact have two different conformations. Using the structures of OxyR and CbnR as a guide, mutants of NAC were generated that should have been unable to assume one or the other predicted conformation (67). One such mutant readily bound cod but not nac; the other readily bound nac but not cod. Thus, NACWT can assume either conformation and can change from one to the other depending only on the spacing of the DNA site presented. In other words, the DNA site is the allosteric effector that determines the conformation of the NAC tetramer (compact and not bending the DNA, as at cod, or extended with a significant DNA bend, as at nac).
FIG. 3.
NAC tetramers bind to three different kinds of sites. NAC tetramers appear to require an interaction between the L111/L125 region of one dimer with either the I222 region or the G217 region of another dimer. The NAC mutants NACL111K and NACL125R are dimers under all conditions and cannot form tetramers at any known site. The NAC mutants NACI222R and NACG217R bind as tetramers at some sites, but the double mutant NACI222R,G217 is a dimer. When two NBSs are adjacent to each other as they are at the cod promoter, NAC assumes a compact conformation that depends on an interaction between the L111/L125 region and a region near I222. NACI222R can still form tetramers at some sites (nac) but binds poorly at cod. When two NBSs are separated by one turn of the DNA helix, NAC assumes an extended conformation that depends on an interaction between the L111/L125 region and a region near G217. NACG217R can still form tetramers at some sites (cod) but not at sites separated by one turn of the helix (nac). When the NBSs are separated by many turns of the helix (as at gdh), both the compact and extended conformations of NAC can bind as a tetramer by forming a DNA loop between the two dimers of the tetramer. Both NACI222R and NACG217R can bind and repress gdh, but the double mutant NACI222R,G217R cannot. The asterisk in one of the NBSs at cod indicates that this is a weak NBS that cannot independently bind a NAC dimer; all other NBSs here can bind a dimer of NAC in the absence of the other NBS.
When the distance between the two dimer-binding sites is large enough to allow a DNA loop to form (as it is at gdhAp and perhaps also at gltBp), either conformation of NAC should be able to function. In fact, the cod-specific mutants and the nac-specific mutants can each still form tetramers and repress gdh expression. However, the double mutant cannot bind as a tetramer at either cod or nac and does not form the tetramers that repress gdhAp (67).
Recall that the DNA sequence of the dimer-binding sites at hut and ure also serves as an allosteric effector. Binding of a NAC dimer to an NBS is not sufficient for activation of transcription. A specific set of nucleotides in the promoter-proximal half of the NBS is required for the bound NAC to be active at hut, ure, or even lacZ. But this allosteric effect is different from the spacing effects that influence tetramer conformation. However, the question of whether the dimer effects influence the tetramer conformation remains open.
OVERVIEW
NAC is one of the most versatile regulators in bacteria. NAC is promiscuous in that at least four different DNA sequences have been shown to be recognized by the helix-turn-helix of a NAC monomer. As a result, the family of genes regulated by NAC is exceptionally large for an LTTR. It is clear that many of those genes have functions other than nitrogen metabolism, suggesting that NAC provides K. pneumoniae with the means to coopt genes or operons into the nitrogen regulon. Moreover, NAC's regulon overlaps the regulons of other global regulators. For example, CAP-cAMP regulates hut and put, DksA regulates cod, Lrp regulates dad and gltB, and NtrC regulates nac.
The degeneracy of the NBS sequence (there are three or four NBSs in pUC19! [19]) suggests that NAC will bind to nearly any AT-rich DNA at some level. This may explain why NAC regulates its own expression and why overproduction of NAC is so detrimental to K. pneumoniae. This also makes it easy for evolution to add NAC activation to an operon. A weak binding of NAC to a promoter can become stronger and can acquire the “activation sequence” without too may constraints on the precise location of the NBS or on its sequence. Acquiring repression is even easier.
OPEN QUESTIONS
It has been more than half a century since Magasanik began to ask how N limitation could relieve the catabolite repression of histidase formation in K. pneumoniae. The properties of NAC, produced under the control of the Ntr system, have provided the answer. But two pressing questions remain, and both require an analysis of NAC structure for their answers. The first is how the sequence of the NBS can convert NAC from a bound (but inactive) dimer to its activating form. Cocrystals of NAC complexes with activating and nonactivating NBSs will be necessary to answer this question. The second obvious question pertains to all LTTRs, not only NAC. What is the nature of the interaction of NAC with RNA polymerase that results in increased transcription? Findings for the positive-control mutants of CysB, GcvA, and NAC have been interpreted to suggest that amino acid Y27 of CysB (equivalent to H26 of NAC and F31 of GcvA) makes a direct contact with the “273 determinant” of the alpha-subunit C-terminal domain (α-CTD) of RNA polymerase. However, it is unlikely that amino acid H26 of NAC could make the same contact with the α-CTD at both hutUp, where NAC lies centered at −64, and ureDp, where NAC lies centered at −47. It seems more likely that H26 is important in transmitting a conformation signal from the DNA-binding (helix-turn-helix) region of NAC to the rest of the N-terminal domain of NAC (the N-terminal 86 amino acids). The effect of the H26D substitution remains a mystery. Do NACH26D dimers bend DNA differently than does NACWT? Does NAC interact with different subunits of RNA polymerase at hutUp and ureDp? The fact that very short fragments of NAC (with as few as 86 amino acids) are fully functional makes NAC a good candidate for solving this mystery.
Acknowledgments
This minireview is dedicated to Boris Magasanik on the occasion of his 90th birthday, in recognition of his more than 5 decades spent unraveling the intricacies of microbial nitrogen metabolism.
This work was supported by Public Health Service grant GM47156 from the National Institutes of Health.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Bender, R. A. 1991. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol. Microbiol. 5:2575-2580. [DOI] [PubMed] [Google Scholar]
- 2.Bender, R. A., K. A. Janssen, A. D. Resnick, M. Blumenberg, F. Foor, and B. Magasanik. 1977. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J. Bacteriol. 129:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bender, R. A., and B. Magasanik. 1977. Regulatory mutations in the Klebsiella aerogenes structural gene for glutamine synthetase. J. Bacteriol. 132:100-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bender, R. A., P. M. Snyder, R. Bueno, M. Quinto, and B. Magasanik. 1983. Nitrogen regulation system of Klebsiella aerogenes: the nac gene. J. Bacteriol. 156:444-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Best, E. A., and R. A. Bender. 1990. Cloning of the Klebsiella aerogenes nac gene, which encodes a factor required for nitrogen regulation of the histidine utilization (hut) operons in Salmonella typhimurium. J. Bacteriol. 172:7043-7048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blauwkamp, T. A., and A. J. Ninfa. 2002. Nac-mediated repression of the serA promoter of Escherichia coli. Mol. Microbiol. 45:351-363. [DOI] [PubMed] [Google Scholar]
- 7.Brenchley, J. E., M. J. Prival, and B. Magasanik. 1973. Regulation of the synthesis of enzymes responsible for glutamate formation in Klebsiella aerogenes. J. Biol. Chem. 248:6122-6128. [PubMed] [Google Scholar]
- 8.Brill, W. J., and B. Magasanik. 1969. Genetic and metabolic control of histidase and urocanase in Salmonella typhimurium, strain 15-59. J. Biol. Chem. 244:5392-5402. [PubMed] [Google Scholar]
- 9.Chen, L. M., T. J. Goss, R. A. Bender, S. Swift, and S. Maloy. 1998. Genetic analysis, using P22 challenge phage, of the nitrogen activator protein DNA-binding site in the Klebsiella aerogenes put operon. J. Bacteriol. 180:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, L. M., and S. Maloy. 1991. Regulation of proline utilization in enteric bacteria: cloning and characterization of the Klebsiella put control region. J. Bacteriol. 173:783-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng, J., T. J. Goss, R. A. Bender, and A. J. Ninfa. 1995. Activation of transcription initiation from the nac promoter of Klebsiella aerogenes. J. Bacteriol. 177:5523-5534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng, J., T. J. Goss, R. A. Bender, and A. J. Ninfa. 1995. Repression of the Klebsiella aerogenes nac promoter. J. Bacteriol. 177:5535-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedrich, B., and B. Magasanik. 1977. Urease of Klebsiella aerogenes: control of its synthesis by glutamine synthetase. J. Bacteriol. 131:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frisch, R. L. 2009. The role of the nitrogen assimilation control protein (NAC) in the response of Klebsiella pneumoniae to nitrogen limitation, Ph.D. thesis. University of Michigan, Ann Arbor, MI.
- 15.Frisch, R. L., and R. A. Bender. 2010. Properties of the NAC (nitrogen assimilation control protein)-binding site within the ureD promoter of Klebsiella pneumoniae. J. Bacteriol. 192:4821-4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frisch, R. L., and R. A. Bender. 2010. Expanded role for the nitrogen assimilation control protein in the response of Klebsiella pneumoniae to nitrogen stress. J. Bacteriol. 192:4812-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg, R. B., R. A. Bender, and S. L. Streicher. 1974. Direct selection for P1-sensitive mutants of enteric bacteria. J. Bacteriol. 118:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goss, T. J. 2008. The ArgP protein stimulates the Klebsiella pneumoniae gdhA promoter in a lysine-sensitive manner. J. Bacteriol. 190:4351-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss, T. J., and R. A. Bender. 1995. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J. Bacteriol. 177:3546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goss, T. J., B. K. Janes, and R. A. Bender. 2002. Repression of glutamate dehydrogenase formation in Klebsiella aerogenes requires two binding sites for the nitrogen assimilation control protein, NAC. J. Bacteriol. 184:6966-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goss, T. J., A. Perez-Matos, and R. A. Bender. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183:6607-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gussin, G. N., C. W. Ronson, and F. M. Ausubel. 1986. Regulation of nitrogen fixation genes. Annu. Rev. Genet. 20:567-591. [DOI] [PubMed] [Google Scholar]
- 23.Hirschman, J., P. K. Wong, K. Sei, J. Keener, and S. Kustu. 1985. Products of nitrogen regulatory genes ntrA and ntrC of enteric bacteria activate glnA transcription in vitro: evidence that the ntrA product is a sigma factor. Proc. Natl. Acad. Sci. U. S. A. 82:7525-7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt, T. P., and B. Magasanik. 1985. Transcription of glnA by purified Escherichia coli components: core RNA polymerase and the products of glnF, glnG, and glnL. Proc. Natl. Acad. Sci. U. S. A. 82:8453-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ikeda, T. P., A. E. Shauger, and S. Kustu. 1996. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259:589-607. [DOI] [PubMed] [Google Scholar]
- 26.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janes, B. K., and R. A. Bender. 1999. Two roles for the leucine-responsive regulatory protein in expression of the alanine catabolic operon (dadAB) in Klebsiella aerogenes. J. Bacteriol. 181:1054-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janes, B. K., P. J. Pomposiello, A. Perez-Matos, D. J. Najarian, T. J. Goss, and R. A. Bender. 2001. Growth inhibition caused by overexpression of the structural gene for glutamate dehydrogenase (gdhA) from Klebsiella aerogenes. J. Bacteriol. 183:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janes, B. K., C. J. Rosario, and R. A. Bender. 2003. Isolation of a negative control mutant of the nitrogen assimilation control protein, NAC, in Klebsiella aerogenes. J. Bacteriol. 185:688-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keener, J., and S. Kustu. 1988. Protein kinase and phosphoprotein phosphatase activities of nitrogen regulatory proteins NTRB and NTRC of enteric bacteria: roles of the conserved amino-terminal domain of NTRC. Proc. Natl. Acad. Sci. U. S. A. 85:4976-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilstrup, M., L. M. Meng, J. Neuhard, and P. Nygaard. 1989. Genetic evidence for a repressor of synthesis of cytosine deaminase and purine biosynthesis enzymes in Escherichia coli. J. Bacteriol. 171:2124-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. H., S. B. Mulrooney, M. J. Renner, Y. Markowicz, and R. P. Hausinger. 1992. Klebsiella aerogenes urease gene cluster: sequence of ureD and demonstration that four accessory genes (ureD, ureE, ureF, and ureG) are involved in nickel metallocenter biosynthesis. J. Bacteriol. 174:4324-4330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Q., and R. A. Bender. 2007. Complex regulation of urease formation from the two promoters of the ure operon of Klebsiella pneumoniae. J. Bacteriol. 189:7593-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 35.Lochowska, A., R. Iwanicka-Nowicka, J. Zaim, M. Witkowska-Zimny, K. Bolewska, and M. M. Hryniewicz. 2004. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol. Microbiol. 53:791-806. [DOI] [PubMed] [Google Scholar]
- 36.Lund, P., and B. Magasanik. 1965. N-formimino-L-glutamate formiminohydrolase of Aerobacter aerogenes. J. Biol. Chem. 240:4316-4319. [PubMed] [Google Scholar]
- 37.Macaluso, A., E. A. Best, and R. A. Bender. 1990. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J. Bacteriol. 172:7249-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.MacPhee, D. G., I. W. Sutherland, and J. F. Wilkinson. 1969. Transduction in Klebsiella. Nature 221:475-476. [DOI] [PubMed] [Google Scholar]
- 39.Maddocks, S. E., and P. C. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 40.Magasanik, B. 1955. The metabolic control of histidine assimilation and dissimilation in Aerobacter aerogenes. J. Biol. Chem. 213:557-569. [PubMed] [Google Scholar]
- 41.Magasanik, B. 1961. Catabolite repression. Cold Spring Harb. Symp. Quant Biol. 26:249-256. [DOI] [PubMed] [Google Scholar]
- 42.Magasanik, B. 1996. Regulation of nitrogen utilization, p. 1344-1356. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 43.Magasanik, B., and H. R. Bowser. 1955. The degradation of histidine by Aerobacter aerogenes. J. Biol. Chem. 213:571-580. [PubMed] [Google Scholar]
- 44.Meers, J. L., D. W. Tempest, and C. M. Brown. 1970. Glutamine(amide):2-oxoglutarate amino transferase oxido-reductase (NADP); an enzyme involved in the synthesis of glutamate by some bacteria. J. Gen. Microbiol. 64:187-194. [DOI] [PubMed] [Google Scholar]
- 45.Meiss, H. K., W. J. Brill, and B. Magasanik. 1969. Genetic control of histidine degradation in Salmonella typhimurium, strain LT-2. J. Biol. Chem. 244:5382-5391. [PubMed] [Google Scholar]
- 46.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muro-Pastor, A. M., P. Ostrovsky, and S. Maloy. 1997. Regulation of gene expression by repressor localization: biochemical evidence that membrane and DNA binding by the PutA protein are mutually exclusive. J. Bacteriol. 179:2788-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muse, W. B., and R. A. Bender. 1998. The nac (nitrogen assimilation control) gene from Escherichia coli. J. Bacteriol. 180:1166-1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muse, W. B., and R. A. Bender. 1999. The amino-terminal 100 residues of the nitrogen assimilation control protein (NAC) encode all known properties of NAC from Klebsiella aerogenes and Escherichia coli. J. Bacteriol. 181:934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muse, W. B., C. J. Rosario, and R. A. Bender. 2003. Nitrogen regulation of the codBA (cytosine deaminase) operon from Escherichia coli by the nitrogen assimilation control protein, NAC. J. Bacteriol. 185:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagatani, H., M. Shimizu, and R. C. Valentine. 1971. The mechanism of ammonia assimilation in nitrogen fixing bacteria. Arch. Mikrobiol. 79:164-175. [DOI] [PubMed] [Google Scholar]
- 52.Neidhardt, F. C., and B. Magasanik. 1957. Effect of mixtures of substrates on the biosynthesis of inducible enzymes in Aerobacter aerogenes. J. Bacteriol. 73:260-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neidhardt, F. C., and B. Magasanik. 1957. Reversal of the glucose inhibition of histidase biosynthesis in Aerobacter aerogenes. J. Bacteriol. 73:253-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nieuwkoop, A. J., S. A. Baldauf, M. E. Hudspeth, and R. A. Bender. 1988. Bidirectional promoter in the hut(P) region of the histidine utilization (hut) operons from Klebsiella aerogenes. J. Bacteriol. 170:2240-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nieuwkoop, A. J., S. A. Boylan, and R. A. Bender. 1984. Regulation of hutUH operon expression by the catabolite gene activator protein-cyclic AMP complex in Klebsiella aerogenes. J. Bacteriol. 159:934-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ninfa, A. J., and B. Magasanik. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 83:5909-5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Oliver, G., G. Gosset, R. Sanchez-Pescador, E. Lozoya, L. M. Ku, N. Flores, B. Becerril, F. Valle, and F. Bolivar. 1987. Determination of the nucleotide sequence for the glutamate synthase structural genes of Escherichia coli K-12. Gene 60:1-11. [DOI] [PubMed] [Google Scholar]
- 58.Osuna, R., S. A. Boylan, and R. A. Bender. 1991. In vitro transcription of the histidine utilization (hutUH) operon from Klebsiella aerogenes. J. Bacteriol. 173:116-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pomposiello, P. J., and R. A. Bender. 1995. Activation of the Escherichia coli lacZ promoter by the Klebsiella aerogenes nitrogen assimilation control protein (NAC), a LysR family transcription factor. J. Bacteriol. 177:4820-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pomposiello, P. J., B. K. Janes, and R. A. Bender. 1998. Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein. J. Bacteriol. 180:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Porter, R. I., A. K. North, and S. Kustu. 1995. Mechanism of transcriptional activation by NtrC, p. 147-158. In J. A. Hoch and T. J. Silhavy (ed.), Two-component signal transduction. American Society for Microbiology, Washington, DC.
- 62.Prival, M. J., J. E. Brenchley, and B. Magasanik. 1973. Glutamine synthetase and the regulation of histidase formation in Klebsiella aerogenes. J. Biol. Chem. 248:4334-4344. [PubMed] [Google Scholar]
- 63.Prival, M. J., and B. Magasanik. 1971. Resistance to catabolite repression of histidase and proline oxidase during nitrogen-limited growth of Klebsiella aerogenes. J. Biol. Chem. 246:6288-6296. [PubMed] [Google Scholar]
- 64.Revel, H. R., and B. Magasanik. 1958. The enzymatic degradation of urocanic acid. J. Biol. Chem. 233:930-935. [PubMed] [Google Scholar]
- 65.Rosario, C. J. 2005. The importance of tetramer formation by the nitrogen assimilation control protein (NAC) for DNA binding and repression at the gdhA promoter in Klebsiella pneumoniae. Ph.D. thesis. University of Michigan, Ann Arbor, MI.
- 66.Rosario, C. J., R. L. Frisch, and R. A. Bender. 2010. The LysR-type nitrogen assimilation control protein forms complexes with both long and short DNA binding sites in the absence of coeffectors. J. Bacteriol. 192:4827-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rosario, C. J., B. K. Janes, and R. A. Bender. 2010. Genetic analysis of the nitrogen assimilation control protein from Klebsiella pneumoniae. J. Bacteriol. 192:4834-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rosario, C. J., and R. A. Bender. 2005. Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae. J. Bacteriol. 187:8291-8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rothman, N., D. Rothstein, F. Foor, and B. Magasanik. 1982. Role of glnA-linked genes in regulation of glutamine synthetase and histidase formation in Klebsiella aerogenes. J. Bacteriol. 150:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 71.Schwacha, A., and R. A. Bender. 1993. The nac (nitrogen assimilation control) gene from Klebsiella aerogenes. J. Bacteriol. 175:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Streicher, S. L., R. A. Bender, and B. Magasanik. 1975. Genetic control of glutamine synthetase in Klebsiella aerogenes. J. Bacteriol. 121:320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tempest, D. W., J. L. Meers, and C. M. Brown. 1970. Synthesis of glutamate in Aerobacter aerogenes by a hitherto unknown route. Biochem. J. 117:405-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tyler, B. 1978. Regulation of the assimilation of nitrogen compounds. Annu. Rev. Biochem. 47:1127-1162. [DOI] [PubMed] [Google Scholar]
- 76.Ushiba, D., and B. Magasanik. 1952. Effects of auxotrophic mutations on the adaptation to inositol degradation in Aerobacter aerogenes. Proc. Soc. Exp. Biol. Med. 80:626-632. [DOI] [PubMed] [Google Scholar]
- 77.Zhang, X. X., and P. B. Rainey. 2008. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 178:185-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. U. S. A. 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]