Abstract
The nitrogen assimilation control protein (NAC) of Klebsiella pneumoniae is a LysR-type transcriptional regulator that activates transcription when bound to a DNA site (ATAA-N5-TnGTAT) centered at a variety of distances from the start of transcription. The NAC-binding site from the hutU promoter (NBShutU) is centered at −64 relative to the start of transcription but can activate the lacZ promoter from sites at −64, −54, −52, and −42 but not from sites at −47 or −59. However, the NBSs from the ureD promoter (ureDp) and codB promoter (codBp) are centered at −47 and −59, respectively, and NAC is fully functional at these promoters. Therefore, we compared the activities of the NBShutU and NBSureD within the context of ureDp as well as within codBp. The NBShutU functioned at both of these sites. The NBSureD has the same asymmetric core as the NBShutU. Inverting the NBSureD abolished more than 99% of NAC's ability to activate ureDp. The key to the activation lies in the TnG segment of the TnGTAT half of the NBSureD. Changing TnG to GnT, TnT, or GnG drastically reduced ureDp activation (to 0.5%, 6%, or 15% of wild-type activation, respectively). The function of the NBSureD, like that of the NBShutU, requires that the TnGTAT half of the NBS be on the promoter-proximal (downstream) side of the NBS. Taken together, our data suggest that the positional specificity of an NBS is dependent on the promoter in question and is more flexible than previously thought, allowing considerable latitude both in distance and on the face of the DNA helix for the NBS relative to that of RNA polymerase.
The LysR-type transcriptional regulators (LTTRs) represent the largest family of regulatory proteins in all of the bacterial world (29). The nitrogen assimilation control protein (NAC) is unusual among the LTTRs in several important ways: it regulates scores of genes with diverse functions rather than regulating just a few genes with a specific function (14, 25), it functions as a dimer rather than as a tetramer at many of the genes whose expression it activates (8, 9, 24), and it requires no coeffector to assume its active conformation (22, 26), using the DNA sequence of the binding site to determine activity (21). The features of the dimer-binding sites that allow a NAC dimer to bind and activate are not well understood. The NAC-binding site at the hutU promoter (NBShutU) is the best characterized of the NBSs (21). The NBShutU is centered on the sequence T-N11-A, typical of most LTTRs (25). A more specific 15-bp “core sequence,” ATA-N9-TAT, has been recognized as a consensus for sites where NAC dimers activate transcription (7). A comparison of the four well-studied promoters where a dimer of NAC activates expression (hutUp, putPp, ureDp, and dadAp) suggested an “activation consensus” of ATAA-N5-TnGTAT (1, 7, 8). Genetic analysis of the NBShutU showed that its 15-bp core contained all the information needed for NAC to bind and activate hut expression (20). The promoter-proximal half of the activation consensus (TnGTAT) was crucial for the NBShutU to function in activating hutUp (21). The promoter-distal half of the NBShutU appeared to be much less effective in allowing activation by NAC but contributed to the binding of NAC. Thus, the asymmetry in the activation consensus was important for NAC's function at hutUp.
Within the hutUp region, the NBShutU is centered at −64 and situated such that a NAC dimer bound there could potentially interact with the C-terminal domain of the α subunit (αCTD) of RNA polymerase. However, the NBShutU is also quite versatile, and its ability to activate transcription is not specific to the hutU promoter. The NBShutU was able to activate all three promoters in the lacZ promoter region and was able to do so from a variety of distances, including from positions −64, −54, −52, and −42 relative to the start of transcription (20). But this flexibility was limited. It did not activate expression when the distance was −74 from the start of transcription, nor when the NBShutU was on the opposite face of the helix at position −69, −59, −49, or −47 (20). Thus, it was surprising when we discovered that NAC not only activates ureDp from an NBS centered at −47 but does so very effectively (9). Moreover, NAC activates the Escherichia coli codB promoter from an NBS centered at −59, though less effectively (19). This was even more surprising given a positive-control mutant (NACH26D), which by analogy to other LTTRs (10, 30) was initially thought to define the interaction region of LTTRs with the α subunit of RNA polymerase. However, NACH26D fails to activate both hutUp and ureDp (23), and it seems unlikely that amino acid H26 makes the same contact with α from such very different positions on the DNA. That led us to examine whether there was something unique to the NBSureD or the ureDp that differed from their hut counterparts. Therefore, we tested whether the NBShutU could replace the NBSureD, whether the asymmetry of the NBSureD was important at ureDp, and, if so, whether it was the promoter-proximal half of the NBS (the TnGTAT portion) that was needed for NAC to be able to activate ure operon expression. We also tested whether the NBShutU would function in the context of the codBp. The binding of NAC at the wild-type codBp is complex, with an NBScodB (ATAT-N5-AnATAT) that differs significantly from the activation consensus defined above and a second half-site located just upstream of the NBScodB. As a result, NAC binds to wild-type codBp as a tetramer, but deletion of the upstream half-site results in binding of a dimer of NAC to the NBScodB and a somewhat weaker activation of codBp.
MATERIALS AND METHODS
Strains and bacterial growth.
Klebsiella pneumoniae strains used in this study were derived from strain W70 (13). Cells cultured under nitrogen-limiting conditions were grown in W4 salts containing 0.4% (wt/vol) glucose and 0.2% (wt/vol) monosodium glutamate (12). For general use as rich medium for cloning and propagation of cells, L broth was used (16). For selection of alleles and plasmids, the following antibiotics were used at the indicated concentrations: ampicillin (100 μg/ml), kanamycin sulfate (50 μg/ml), streptomycin sulfate (50 μg/ml), and tetracycline hydrochloride (25 μg/ml). Solid media were supplemented with 1.5% Bacto agar.
Genetic techniques.
DNA manipulation was carried out as described by Maniatis et al. (15). Mutant promoters were amplified via PCR using wild-type promoter templates and primers containing the desired mutations. In this study, the 15-bp core NAC-binding site plus 6 bp upstream and downstream were used as the NAC-binding site. The sequences of the sites in the study (with the 15-bp core underlined) are as follows: for NBSureD, GATGACATAAGCGTTTCGTATGACCGG, for NBScodB, CTCATTCATATAAAAAATATATTTCCCC, and for NBShutU, CGCAATATAACAAAATTGTATCATTTC. In each case, the 27-bp sequence was inserted such that the 15-bp core was in the same position in the mutant promoter as the native 15-bp core was in the wild-type promoter. The architecture of the transcriptional ureDp-lacZ fusion has been described previously (9). This fusion contains a segment of ure DNA from −77 to +107 relative to the start of transcription and fuses the promoterless lacZ gene after 64 bp of the ureD sequence (9). The transcriptional codBp fusion (19) contains a segment of cod DNA from −120 to +67 and fuses the promoterless lacZ gene after 76 bp of the codB coding sequence (19). The DNA sequences of primers used in this study are available upon request. PCR products were cloned into the EcoRI and BamHI sites of pRS415 (27). The DNA sequence of each cloned fragment was determined to ensure that no unwanted mutations had been introduced. In order to integrate the fusions into the chromosome of K. pneumoniae, the fusions were subcloned into a pir-dependent vector, pCB1583, based on pKAS46 (6, 28). Fusions were integrated into the rbs landing pad as previously described (6). Transduction of the nac-2 allele, an in-frame deletion of the NAC open reading frame, with the aph-1 cassette replacing the deleted material (2) was performed utilizing P1vir grown on KC5447 as previously described (5).
NAC purification.
K. pneumoniae NAC was purified from E. coli cells as described previously (7). Purity was monitored via SDS-PAGE and staining by the method of Fairbanks et al. (3). Purified protein was quantified by the method of Lowry et al. and compared to a bovine serum albumin (BSA) standard (11). Protein was diluted 1:1 in glycerol and stored at −20°C.
Electrophoretic mobility shift assay (EMSA).
Gel mobility shifts were performed with purified NAC as described previously (7). Briefly, 6 μl (0.08 pmol) of purified PCR-amplified fragments of ureDp, codBp, or mutant promoter regions was mixed with 4 μl of buffer 6 (50% glycerol, 125 mM NaCl, 50 mM NaH2PO4 [pH 7.0], 1.25 mM MgCl2, 0.5 mM 2-mercaptoethanol, 1 mg/ml BSA) containing 0, 0.07, 0.14, or 0.28 pmol of purified NAC. Reaction mixtures were incubated at room temperature for 20 min, and then 1 μl of 10× loading buffer (25% Ficoll, 100 mM Tris-HCl, 10 mM EDTA, 0.05% [wt/vol] cresol red, and 0.05% [wt/vol] orange G, pH 7.4) was added to each reaction mixture. A 10-μl portion of each reaction mixture was loaded on a prerun gel (5% polyacrylamide buffered with 0.5× Tris-borate-EDTA [TBE]). Bound and unbound species were separated by electrophoresis at 10 V/cm for 60 min. Gels were stained with ethidium bromide (40 μg/ml) and then destained with water. The mobilities of the dimer-bound and tetramer-bound promoter fragments were determined by comparison to known shifts of ureDp (dimer) and codBp (tetramer), as shown previously (19, 24).
β-Galactosidase assay.
Liquid cultures of 10 ml were grown in 125-ml sidearm flasks at 30°C and 250 rpm to mid-log phase (50 Klett units, ca. 1.2 × 108 CFU/ml). Cells were washed once with one culture volume of cold 1% KCl. Cells were resuspended in 1/10 culture volume of cold 1% KCl. Assays were performed on whole cells permeabilized by detergent as previously described (12). Three different volumes of 10× cells were assayed for each culture, and each strain was cultured at least three independent times. Values for specific activity are reported as nanomoles of product formed per minute per milligram of total protein (as determined by the method of Lowry) at 30°C (11).
RESULTS
NBShutU functions in the ureD and codB promoters.
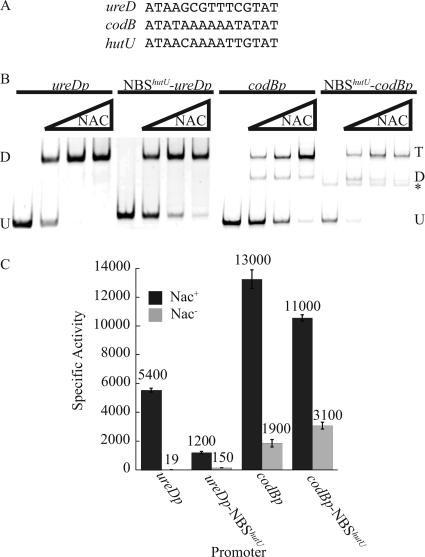
Both the ureD and codB promoters are activated by NAC, even though the NBSureD is centered at −47 and the NBScodB is centered at −59, positions where the NBShutU appeared to be ineffective in the context of the lac promoters (20). The 27-bp NBShutU used here has been defined by DNase I footprinting (7) and contains the 15-bp activation consensus (ATAA-N5-TnGTAT) and 6 bp of DNA sequence on either side of that 15-bp core. The NBShutU was used to replace the native NBS within the context of the ureD or codB promoters. In both cases, the 27-bp replacement was aligned such that the 15-bp core of the inserted NBShutU was in the same location as the cores of the native NBSureD and NBScodB, centered at −47 and −59 bp, respectively, relative to the start of transcription of the reporter. A transcriptional fusion of ureDp to a promoterless lacZ gene was used to measure ureDp activity. This fusion, described previously (9), carries a segment of ure DNA from −77 to +107 relative to the start of ureDp transcription. Thus, the lacZ fusion occurred after 64 bp of ureD sequence. The comparable reporter for codBp described previously (19) carries cod DNA from −120 to +67, and thus the lacZ fusion occurred after 76 bp of codB sequence. Each of these fusions was inserted into the K. pneumoniae chromosome in single copy at the rbs locus using “landing pad technology” (6). The 15-bp cores of the NBShutU and the NBSureD are prefect matches to the activation consensus, but the core of the NBScodB differs in three positions from the consensus (Fig. 1A). Nevertheless, the NBScodB still contains the critical ATA-N9-TAT motif.
FIG. 1.
Functional equivalence of the NAC-binding sites. (A) Alignment of the core, 15-nucleotide NAC-binding sites from the ureD, codB, and hutU promoters. (B) Electrophoretic mobility shift assay (EMSA) of the ureDp, NBShutU-ureDp, codBp, and NBShutU-codBp fragments mixed with buffer 6 and increasing concentrations of NAC. U indicates the mobility of the unbound DNA, D indicates the band corresponding to a dimer of NAC associated with the DNA, T indicates the band corresponding to a tetramer of NAC associated with the DNA, and * indicates a PCR artifact in the NBShutU-codBp mobility shifts that is not NAC reactive. (C) β-Galactosidase activities of ureDp, NBShutU-ureDp, codBp, and NBShutU-codBp as lacZ fusions integrated into the Nac+ or Nac− background and grown under nitrogen-limiting conditions. Bars indicate standard errors.
NAC formed a stable interaction, as assayed by electrophoretic mobility shift assay (EMSA), with ureDp containing the NBShutU and codBp containing the NBShutU (Fig. 1B). NAC interacted with the NBShutU as a dimer within the context of ureDp and as a tetramer within the context of codBp, determined as described in Materials and Methods. This is not surprising, since NAC associates with the native ureDp as a dimer and the native codBp as a tetramer (19, 22, 24). Since NAC was able to associate with the NBShutU in both the ureDp and codBp contexts, the ability of NAC to activate transcription from these constructs was tested.
Nac+ and Nac− derivatives of the reporter strains were grown under nitrogen-limiting conditions and assayed for β-galactosidase activity (Fig. 1C). Transcription of all four promoters was dependent on NAC. The NAC-mediated activation of the ureD promoter containing the NBShutU was similar to the 8-fold activation of the native hutUp by NAC (21). The NBShutU was also able to activate transcription when positioned at −59 in the context of the codB promoter. In this case, the maximal activation was similar to the maximal activation seen for the wild-type codB promoter rather than the wild-type hutUp. Clearly, the positions of −47 and −59 are not forbidden for the NBShutU, at least in the context of the NAC-activated promoters, ureDp and codBp, respectively. The basal level of activity from both the ureD and codB promoters containing the NBShutU was higher than that of the wild-type ureD or codB promoter, and this will be addressed in Discussion.
The two halves of the NBSureD are functionally different.
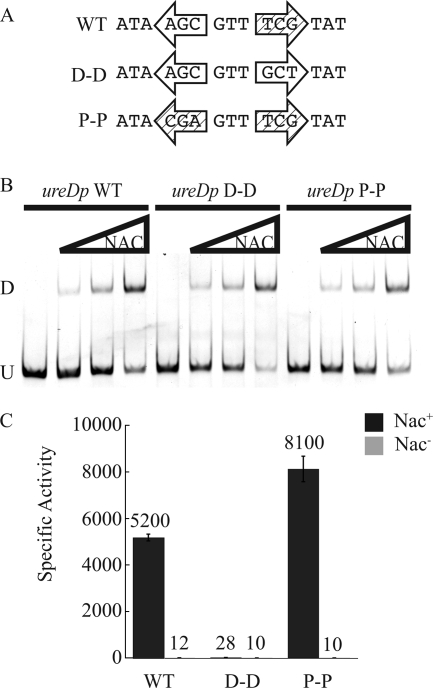
Despite the presence of some symmetry within the NBShutU (ATA-N9-TAT), the more complete activation sequence of the NBShutU is asymmetric (ATAA-N5-TnGTAT) and this asymmetry is important for NAC activation of the hutU promoter (21). Within the context of hutUp, the promoter-distal half of the NBShutU (containing the hexanucleotide ATAAnn) is important for strong binding of NAC and the promoter-proximal half of the site (the hexanucleotide TnGTAT) is important for activation of transcription by NAC (21). We reasoned that the NBSureD, located closer to the promoter and on the opposite face of the DNA helix, might display different requirements. The role of each half of the NBSureD was examined by creating mutant constructs with either two distal or two proximal half-sites (D-D and P-P, respectively) (Fig. 2A). The promoter-distal half of the site (shown in the open arrow) was used to replace the promoter-proximal half of the site (shown in the cross-hatched arrow) to create the construct referred to as distal-distal (D-D). The opposite replacement was made to create the proximal-proximal construct (P-P). NAC bound to promoters containing the D-D and P-P NBS mutants in a manner similar to that of the wild-type ureD promoter (Fig. 2B). This suggested that unlike the NBShutU, both halves of the NBSureD were sufficient for strong NAC binding, perhaps reflecting a role for the 6 bp of flanking sequence.
FIG. 2.
Asymmetry of the ureD promoter NAC-binding site. (A) Alignment of the core, 15-nucleotide NAC-binding sites of wild-type ureD NAC (WT) and the proximal-proximal (P-P) and distal-distal (D-D) mutants. The proximal half-site is shown as a cross-hatched arrow, and the distal half-site is an open arrow. (B) EMSA of the ureDp, D-D, and P-P fragments mixed with buffer 6 and increasing concentrations of NAC. U indicates the mobility of the unbound DNA, and D indicates the band corresponding to a dimer of NAC associated with the DNA. (C) β-Galactosidase activities of ureDp and the D-D and P-P mutants as lacZ fusions integrated into the Nac+ or Nac− background and grown under nitrogen-limiting conditions. Bars indicate standard errors.
Since both the D-D and P-P constructs bound NAC well, we next tested their ability to function in NAC-mediated activation of ureDp. Fusions of ureD promoters containing the P-P or D-D mutant NBSs to lacZ were integrated in single copy into the K. pneumoniae chromosome in Nac+ and Nac− strain backgrounds. NAC was severely defective in activating transcription from the promoter containing the D-D mutant NBS (Fig. 2C) even though NAC bound well to this site (Fig. 2B). In contrast, the NAC-mediated activation of the promoter containing the P-P mutant was even stronger than the wild-type ureD promoter (Fig. 2C). These data suggested that at least one TnGTAT-containing half-site is required for NAC transcriptional activation from the NBSureD.
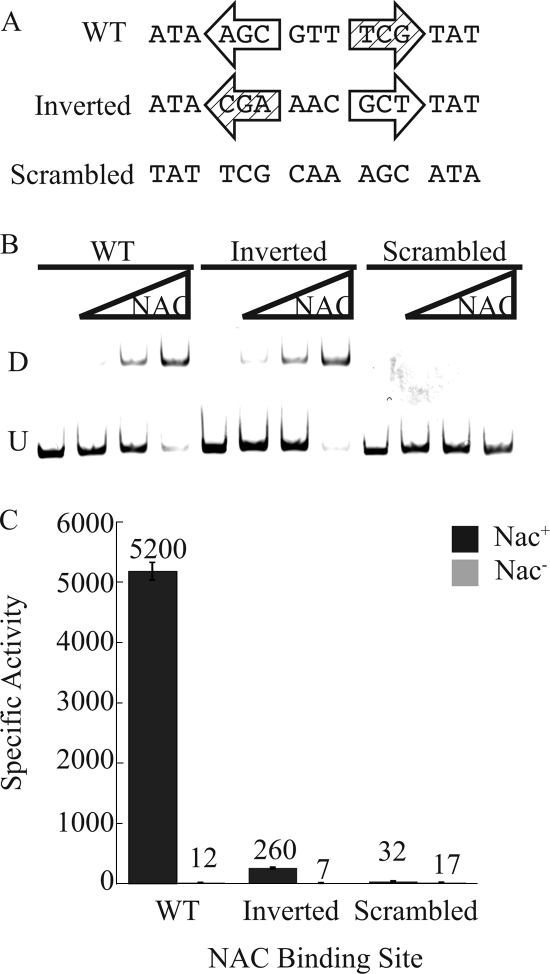
NBSureD is directional.
We next asked if the orientation of the TnGTAT half of the NBSureD was important. To test this, we constructed ureD promoters containing an inverted NBSureD or a scrambled NBSureD (Fig. 3A). As expected, both the inverted NBS and the wild-type NBS bound NAC well and the scrambled NBS failed to bind NAC (Fig. 3B). Single-copy promoter-lacZ fusions with either the inverted or scrambled NAC-binding site were integrated into the K. pneumoniae chromosome. Not surprisingly, the scrambled site, to which NAC did not bind in vitro, demonstrated little or no NAC-mediated transcriptional activation (Fig. 3C). The inverted site, to which NAC bound well in vitro, exhibited only 5% as much activation as the wild-type NBS (Fig. 3C). This suggested that NBSureD requires the TnGTAT-containing half of the NBS to be on the promoter-proximal side of the NBS in order for NAC to activate transcription effectively.
FIG. 3.
Directionality of the ureD promoter NBS. (A) Alignment of the wild type and the inverted and scrambled mutants containing ureD promoter NAC-binding sites. The distal half-site is shown as an open arrow, and the proximal half-site is a cross-hatched arrow. (B) EMSA of the wild-type and the inverted and scrambled ureDp fragments. U indicates the mobility of the unbound DNA, and D indicates the band corresponding to a dimer of NAC associated with the DNA. (C) β-Galactosidase activities of the wild type and the inverted and scrambled mutants containing ureDp as lacZ fusions integrated into the Nac+ or Nac− background and grown under nitrogen-limiting conditions. Bars indicate standard errors.
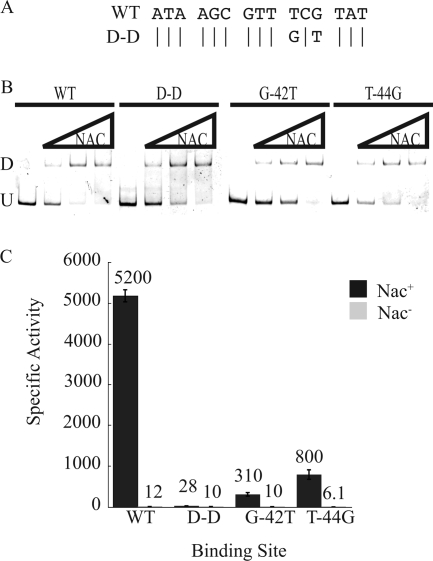
The TnG sequence is critical for NAC activation.
The D-D mutant, which failed in activation despite strong binding, contained only two changes from the wild-type NBSureD. The two changes occurred in the 10th and 12th nucleotides of the 15-bp core. These nucleotides correspond to nucleotides −44 and −42 with respect to the start of transcription (Fig. 4A). Since the D-D construct had lost virtually all NAC-mediated transcriptional control of the ureD promoter, the role of each of these changes, T−44G and G−42T, was assessed individually.
FIG. 4.
Role of T−44 and G−42 in NBSureD. (A) Alignment of the wild-type and D-D ureDp NAC-binding sites. (B) EMSA of the wild-type and the D-D-, G−42T-, and T−44G-containing ureD promoter fragments. U indicates the mobility of the unbound DNA, and D indicates the band corresponding to a dimer of NAC associated with the DNA. (C) β-Galactosidase activities of the wild-type and the D-D-, G−42T-, and T−44G-containing ureDp as lacZ fusions integrated into the Nac+ or Nac− background and grown under nitrogen-limiting conditions. Bars indicate standard errors.
EMSA was performed with ureD promoters containing the D-D, T−44G, and G−42T NBSureD mutants. NAC stably interacted with both single mutants (Fig. 4B). Single-copy promoter-lacZ fusions of the mutant promoters integrated into the K. pneumoniae chromosome were assayed in Nac+ and Nac− backgrounds (Fig. 4C). The promoter that contained the G−42T mutation had NAC-mediated activation that was only 6% of that of wild-type ureDp (Fig. 4C). The promoter that contained the T−44G mutation was slightly better, with 15% of the NAC-mediated activation of wild-type ureDp (Fig. 4C). These data suggest that nucleotides T−44 and G−42 work synergistically to elicit NAC-mediated activation of the ureD promoter.
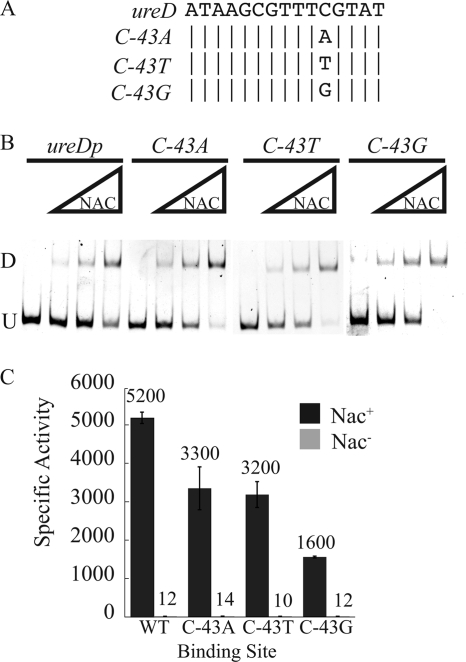
The 10th and 12th nucleotides of the NBSureD core were critical for NAC-mediated activation of the promoter but were nonessential for NAC binding to the promoter. This raised the question of whether the 11th nucleotide of the core, C−43, which remained unchanged in both the P-P and D-D constructs, plays a role in NAC-mediated transcriptional activation of the ureD promoter. The role of C−43 in the NAC-mediated transcriptional activation of the ureD promoter was examined by changing C−43 to the other three nucleotides, creating C−43A, C−43T, and C−43G constructs (Fig. 5A). The ability of these three constructs to interact with NAC in vitro and in vivo was examined to determine the role of C−43 in both NAC binding and NAC-mediated transcriptional activation under nitrogen-limiting conditions.
FIG. 5.
Role of C−43 in NBSureD. (A) Alignment of the wild-type and the C−43A, C−43T, and C−43G ureDp NAC-binding sites. (B) EMSA of the wild-type and the C−43A-, C−43T-, and C−43G-containing ureD promoter fragments. U indicates the mobility of the unbound DNA, and D indicates the band corresponding to a dimer of NAC associated with the DNA. (C) β-Galactosidase activities of the wild-type and the C−43A-, C−43T-, and C−43G-containing ureDp as lacZ fusions integrated into the Nac+ or Nac− background and grown under nitrogen-limiting conditions. Bars indicate standard errors.
NAC bound all three of the C−43 substitution mutants in vitro (Fig. 5B). Single-copy integrants of the mutant promoters fused to lacZ were assayed in the Nac+ and Nac− backgrounds (Fig. 5C). All three of the promoters containing NBSs with C−43 substitution mutations showed less activation in response to NAC than the wild-type ureD promoter. The C−43A and C−43T mutants had slightly less NAC-mediated activation, 65% and 61% of the wild-type ureDp activity, respectively (Fig. 5C). The promoter containing the C−43G mutant had activation that was 30% of that of the wild-type ureD promoter.
DISCUSSION
Previous data had suggested that NAC-mediated activation of transcription required that NAC be located on the same face of the DNA helix as RNA polymerase but that the distance from the RNA polymerase was somewhat flexible (20). The NBShutU brought the lac promoters of E. coli under the control of NAC when placed at −64, −54, and −42, all positions on the same face of the helix. Transcriptional activation by NAC at the closer distances was weaker than that seen from −64, suggesting that NBSs located close to the promoter are less effective (20). The NBShutU failed to bring the lac promoters under the control of the nitrogen regulon when located at −69, −59, and −47, all positions on the opposite face of the helix (20). Interestingly, the NBSureD is located at the “weaker” spacing and on the “forbidden” face of the helix and demonstrates strong NAC-dependent transcriptional activation (9). The data presented here show that the NBShutU was able to activate the ureD promoter even at −47, a position where it failed to activate the lac promoter. In addition, the NBShutU activated the codB promoter even when it was at −59, a position from which it failed to activate at the lac promoter. Thus, there appears to be considerable flexibility in how NAC activates transcription.
We noted that many constructs with altered NBSs had elevated levels of basal activity compared to those of the wild-type promoters. The ureDp is an extremely poor promoter in the absence of NAC. Visual inspection of the −10 and −35 regions fails to detect any good match to the consensus sequences. The replacement of the NBSureD with the NBShutU changed the nucleotide composition as close to the promoter as position −34, and these changes, increasing the A/T richness of the −35 region, might improve the overall strength of an extremely poor promoter. Likewise, replacement of the NBScodB with the NBShutU increased the A/T richness of the −60 region of the codB promoter, perhaps improving the binding of the αCTD, which again might improve the overall strength of a promoter. These changes appear to have increased the basal level of expression from these promoters. Nevertheless, NAC was still able to activate expression from these promoters.
The NBShutU contains two functionally important regions: the promoter-distal half of the site (containing the sequence ATAA), which is important for NAC binding affinity, and the promoter-proximal half of the site (TnGTAT), which is important for NAC transcriptional activation of the hutU promoter (21). In order for NAC to yield strong activation, the TnGTAT half of the site must be located in the proper orientation (facing the promoter), suggesting that for an NBS positioned at −64, the activation of transcription by NAC occurs in a directional manner (21). The NBSureD shows the same nonequivalent halves of the NBS as does the NBShutU. Only the TnGTAT half of the site allows activation, and this half must be present on the side nearest the start of transcription. So it appears that the NBSureD, like the NBShutU, induces a conformation change in the bound NAC that allows NAC to activate transcription by RNA polymerase.
Studies with the positive-control (PC) mutant of CysB, CysBY27A, have been interpreted to suggest that amino acid Y27 of CysB interacts with a region of RNA polymerase near amino acid E273 of the α subunit (10). An analogous PC mutant of NAC, NACH26D, has been isolated. NACH26D is able to bind DNA and repress transcription but cannot activate either hutUp or ureDp (23). NAC can activate hutUp from an NBS at −64 and ureDp from an NBS at −47, on the face of the DNA helix opposite RNA polymerase. Thus, it seems unlikely that amino acid H26 of NAC could contact similar regions of RNA polymerase (e.g., the E273 region of the α subunit) in both cases. Moreover, the crystal structure of a related LTTR (17) suggests that Y27 of CysB and H26 of NAC lie at the edge of the first helix in the DNA-binding helix-turn-helix domain and are likely to lie close to the DNA. Thus, it appears that H26 of NAC may play a role other than making direct contact with the α subunit of RNA polymerase. We suggest that H26 may interfere with the conformational change in NAC that is induced by the TnGTAT sequence and that is required for NAC's ability to activate transcription. But this raises the broader question of how NAC activates transcription. Perhaps NAC interacts with RNA polymerase at hutUp and ureDp in fundamentally different ways. For example, NAC might interact with α at hutUp and with a different subunit of RNA polymerase at ureDp. This might help to explain how NAC can activate so many different promoters (4) and do so from so many different distances relative to the start of transcription (9, 19, 20). Alternatively, NAC might distort the DNA at hutUp and ureDp in ways that allow rather poor promoter sequences to function more effectively. It is worth noting that hutUp has a rather poor −10 region (TATATAT). The −10 region of the NAC-dependent ureDp is even worse, and it has no recognizable −35 sequence at all. NAC dimers are known to bend the NBSnac about 42° (24), and the NBShutU an estimated 23° (18). So a model involving DNA distortion is certainly plausible. The answers to these questions will require a structure for a cocrystal of NAC bound to its NBS. What is clear, however, is that NAC is an unusually versatile regulator that can function as a dimer or tetramer, on the same face of the DNA helix as RNA polymerase or on the opposite face, and as an activator or a repressor and whose ability to activate transcription depends on the sequence of the NBS that it uses for binding.
Acknowledgments
This work was supported by Public Health Service grant GM47156 from the National Institutes of Health to R.A.B.
Footnotes
Published ahead of print on 9 July 2010.
REFERENCES
- 1.Chen, L. M., T. J. Goss, R. A. Bender, S. Swift, and S. Maloy. 1998. Genetic analysis, using P22 challenge phage, of the nitrogen activator protein DNA-binding site in the Klebsiella aerogenes put operon. J. Bacteriol. 180:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairbanks, G., T. L. Steck, and D. F. Wallach. 1971. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry 10:2606-2617. [DOI] [PubMed] [Google Scholar]
- 4.Frisch, R. L., and R. A. Bender. 2010. Expanded role for the nitrogen assimilation control protein in the response of Klebsiella pneumoniae to nitrogen stress. J. Bacteriol. 192:4812-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg, R. B., R. A. Bender, and S. L. Streicher. 1974. Direct selection for P1-sensitive mutants of enteric bacteria. J. Bacteriol. 118:810-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goss, T. J. 2008. The ArgP protein stimulates the Klebsiella pneumoniae gdhA promoter in a lysine-sensitive manner. J. Bacteriol. 190:4351-4359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goss, T. J., and R. A. Bender. 1995. The nitrogen assimilation control protein, NAC, is a DNA binding transcription activator in Klebsiella aerogenes. J. Bacteriol. 177:3546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janes, B. K., and R. A. Bender. 1998. Alanine catabolism in Klebsiella aerogenes: molecular characterization of the dadAB operon and its regulation by the nitrogen assimilation control protein. J. Bacteriol. 180:563-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu, Q., and R. A. Bender. 2007. Complex regulation of urease formation from the two promoters of the ure operon of Klebsiella pneumoniae. J. Bacteriol. 189:7593-7599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lochowska, A., R. Iwanicka-Nowicka, J. Zaim, M. Witkowska-Zimny, K. Bolewska, and M. M. Hryniewicz. 2004. Identification of activating region (AR) of Escherichia coli LysR-type transcription factor CysB and CysB contact site on RNA polymerase alpha subunit at the cysP promoter. Mol. Microbiol. 53:791-806. [DOI] [PubMed] [Google Scholar]
- 11.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 12.Macaluso, A., E. A. Best, and R. A. Bender. 1990. Role of the nac gene product in the nitrogen regulation of some NTR-regulated operons of Klebsiella aerogenes. J. Bacteriol. 172:7249-7255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacPhee, D. G., I. W. Sutherland, and J. F. Wilkinson. 1969. Transduction in Klebsiella. Nature 221:475-476. [DOI] [PubMed] [Google Scholar]
- 14.Maddocks, S. E., and P. C. Oyston. 2008. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154:3609-3623. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis, T., J. Sambrook, and E. F. Fritsch. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 16.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 17.Muraoka, S., R. Okumura, N. Ogawa, T. Nonaka, K. Miyashita, and T. Senda. 2003. Crystal structure of a full-length LysR-type transcriptional regulator, CbnR: unusual combination of two subunit forms and molecular bases for causing and changing DNA bend. J. Mol. Biol. 328:555-566. [DOI] [PubMed] [Google Scholar]
- 18.Muse, W. B. 1996. The nac gene of E. coli and its role in nitrogen regulation. Ph.D. thesis. University of Michigan, Ann Arbor, MI.
- 19.Muse, W. B., C. J. Rosario, and R. A. Bender. 2003. Nitrogen regulation of the codBA (cytosine deaminase) operon from Escherichia coli by the nitrogen assimilation control protein, NAC. J. Bacteriol. 185:2920-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomposiello, P. J., and R. A. Bender. 1995. Activation of the Escherichia coli lacZ promoter by the Klebsiella aerogenes nitrogen assimilation control protein (NAC), a LysR family transcription factor. J. Bacteriol. 177:4820-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pomposiello, P. J., B. K. Janes, and R. A. Bender. 1998. Two roles for the DNA recognition site of the Klebsiella aerogenes nitrogen assimilation control protein. J. Bacteriol. 180:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosario, C. J., R. L. Frisch, and R. A. Bender. 2010. The LysR-type nitrogen assimilation control protein forms complexes with both long and short DNA binding sites in the absence of coeffectors. J. Bacteriol. 192:4827-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosario, C. J., B. K. Janes, and R. A. Bender. 2010. Genetic analysis of the nitrogen assimilation control protein from Klebsiella pneumoniae. J. Bacteriol. 192:4834-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosario, C. J., and R. A. Bender. 2005. Importance of tetramer formation by the nitrogen assimilation control protein for strong repression of glutamate dehydrogenase formation in Klebsiella pneumoniae. J. Bacteriol. 187:8291-8299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 26.Schwacha, A., and R. A. Bender. 1993. The product of the Klebsiella aerogenes nac (nitrogen assimilation control) gene is sufficient for activation of the hut operons and repression of the gdh operon. J. Bacteriol. 175:2116-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simons, R. W., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 28.Skorupski, K., and R. K. Taylor. 1996. Positive selection vectors for allelic exchange. Gene 169:47-52. [DOI] [PubMed] [Google Scholar]
- 29.Sonnhammer, E. L., S. R. Eddy, and R. Durbin. 1997. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28:405-420. [DOI] [PubMed] [Google Scholar]
- 30.Tao, K., C. Zou, N. Fujita, and A. Ishihama. 1995. Mapping of the OxyR protein contact site in the C-terminal region of RNA polymerase alpha subunit. J. Bacteriol. 177:6740-6744. [DOI] [PMC free article] [PubMed] [Google Scholar]