Abstract
Chlamydiae are a group of obligate intracellular bacteria comprising several important human pathogens. Inside the eukaryotic cell, chlamydiae remain within a host-derived vesicular compartment, termed the inclusion. They modify the inclusion membrane through insertion of unique proteins, which are involved in interaction with and manipulation of the host cell. Among chlamydiae, inclusion membrane proteins have been exclusively found in members of the family Chlamydiaceae, which predominantly infect mammalian and avian hosts. Here, the presence of inclusion membrane proteins in Protochlamydia amoebophila UWE25, a chlamydial endosymbiont of free-living amoebae, is reported. A genome-wide screening for secondary structure motifs resulted in the identification of 23 putative inclusion membrane proteins for this organism. Immunofluorescence analysis demonstrated that five of these proteins were expressed, and four of them could be localized to a halo surrounding the intracellular bacteria. Colocalization studies showed an almost complete overlap of the signals obtained for the four putative inclusion membrane proteins, and immuno-transmission electron microscopy unambiguously demonstrated their location in the inclusion membrane. The presence of inclusion membrane proteins (designated IncA, IncQ, IncR, and IncS) in P. amoebophila shows that this strategy for host cell interaction is conserved among the chlamydiae and is used by chlamydial symbionts and pathogens alike.
Chlamydiae were once considered a small group of very closely related bacteria which form an evolutionarily well-separated lineage in the tree of life. These obligate intracellular bacteria infect a broad spectrum of at least 60 eukaryotic host organisms including both invertebrates and vertebrates. Until recently, the family Chlamydiaceae was the only described chlamydial family, which is exclusively comprised of human and animal pathogens. It includes Chlamydia trachomatis, the most prevalent sexually transmitted bacteria worldwide, with over 90 million new cases of infection per year (56), and the world's leading cause of preventable blindness (57). The other major human pathogen among the chlamydiae, Chlamydophila pneumoniae (basonym, Chlamydia pneumoniae) (15), causes community-acquired pneumonia and is also implicated in several chronic diseases such as arteriosclerosis and multiple sclerosis (33, 38). The discovery of several chlamydia-like endosymbionts in amoebae in the late 1990s represented the first evidence for a much larger diversity of chlamydiae in the environment (3, 7, 17, 27). Since then, numerous new chlamydial organisms were discovered, which led to the description of several new families (9, 25, 30).
A characteristic feature of all chlamydiae is the developmental cycle, which includes morphologically and physiologically distinct developmental forms (1, 37). Infection of host cells starts with the attachment of the infectious form of the chlamydiae, the elementary body (EB), to a host cell and uptake by a phagocytosis-like mechanism. Once inside the eukaryotic cell, the chlamydiae remain surrounded by the host-derived membrane in a nonacidic compartment termed the inclusion (18). Inside the inclusion, the EB differentiates into a reticulate body (RB), which is metabolically active and divides by binary fission. After a phase of bacterial replication, the RBs differentiate into EBs and leave the host cell by exocytosis of the inclusion or by lysis of the host cell (29), and a new infectious cycle begins.
The chlamydial inclusion rapidly excludes early or late endosomal or late endosomal/lysosomal markers (18, 47) and acquires properties associated with the Golgi apparatus and host multivesicular bodies (6). The inclusion acquires lipid components necessary for its expansion during the chlamydial replicative phase by hijacking intracellular membrane transport vesicles from the host (19). These processes are dependent on chlamydial proteins as inhibition of bacterial protein synthesis blocks lipid accumulation in the inclusion membrane, and the inclusion fuses with lysosomes (48). Furthermore, as chlamydiae are metabolically impaired, they need to import a variety of metabolites from their host cells. Microinjection experiments demonstrated that no diffusion through the inclusion membrane above 520 Da takes place (23). Import of host metabolites is thus achieved either by (chlamydial) transport proteins or by fusion of the inclusion with vesicles from the host intracellular transport pathways (19).
Chlamydial proteins likely to be responsible for these processes are the inclusion membrane proteins (Incs), which are type III effectors located in the inclusion membrane (42, 44, 51). This family of proteins is encoded in all sequenced chlamydial genomes (26, 39). Known Inc proteins do not share a sequence motif and do not show distinct sequence similarities. However, most of them share a large bi-lobed hydrophobic domain (5) possibly responsible for anchoring the Incs in the inclusion membrane by formation of a hairpin-like structure (11). Even though up to 70 to 90 Inc proteins are predicted for chlamydial genomes (42, 54), only a few of these proteins have been characterized beyond their localization to the inclusion membrane (24, 31).
The first recognized Inc protein was Chlamydophila caviae (formerly Chlamydia caviae) IncA (44), and this protein as well as its homologue in C. trachomatis still are the most extensively characterized Incs. Its C terminus points to the host cell cytoplasm (43), and C. trachomatis IncA is responsible for fusion of the chlamydial inclusions (11, 20, 45, 52). This is mediated by interaction between IncA proteins located on the surface of the inclusions. IncA proteins encode a SNARE-like motif (11-12), which enables them to form homodimers leading to membrane fusion in a similar manner as described for fusion of intracellular membrane transport vesicles in eukaryotic cells (14). Functional characterization of a limited number of additional Inc proteins identified their interactions with the host protein 14-3-3β (49) and Rab GTPases (10), interference with host cell cytokinesis (2), and inhibition of NF-κB activation (57). Detailed characterization of Inc protein function is limited by the lack of a chlamydial genetic transformation system.
Inc proteins are chlamydia specific, and, due to the lack of any other available chlamydial genome sequence until recently (55), have been described only for members of the Chlamydiaceae. The genome sequence of Protochlamydia amoebophila UWE25 (26), a chlamydia-like symbiont of acanthamoebae and member of the family Parachlamydiaceae, provided the first opportunity to search for Inc proteins outside the Chlamydiaceae (11, 26, 42). The identification of Inc proteins in the Parachlamydiaceae will shed new light onto the evolution of this unusual group of proteins. The symbiont P. amoebophila is of special interest with respect to Inc proteins as this organism is found mostly in inclusions containing single bacteria that apparently divide as the bacteria multiply (8). In the present study, 23 novel putative inclusion membrane proteins from P. amoebophila were identified by in silico genomic analysis, 15 of which were found to be unique to this amoeba symbiont. Expression and subcellular location for four P. amoebophila Inc proteins were demonstrated by immunofluorescence and immuno-transmission electron microscopy during chlamydial multiplication in amoeba host cells.
MATERIALS AND METHODS
Organisms and cultivation.
Uninfected or P. amoebophila UWE25-infected Acanthamoeba castellanii strains Neff and UWC1 were grown in TSY medium (30 g/liter Trypticase soy broth, 10g/liter yeast extract, pH 7.3) at 20°C. Culture medium was changed every 3 to 4 days. Escherichia coli BL21(DE3) (Invitrogen) was grown in standard LB medium.
Purification of P. amoebophila EBs.
Unsynchronized A. castellanii Neff cultures infected with P. amoebophila were harvested and washed, and amoebal cells were disrupted by at least two cycles of freezing (dry ice/ethanol bath) and rapid thawing (55°C water bath). An equal volume of glass beads was added to the lysed cells, followed by vortexing for 3 min. The mixture was centrifuged at 3,400 × g for 10 min at 4°C, and the pellet was discarded. The supernatant was centrifuged at 50,000 × g for 40 min at 4°C. The pellet, mainly containing P. amoebophila elementary bodies, was resuspended in sucrose phosphate-buffered glutamic acid (SPG) buffer (750 g/liter sucrose, 5.2 g/liter KH2PO4, 23 g/liter NaHPO4·7 H2O, 7.5 g of glutamic acid). The SPG wash was repeated, and the pellet was resuspended in SPG buffer. This material was filtered several times through a 0.45-μm-pore-size syringe to disperse individual EBs. Purified EBs were stored in SPG buffer at −80°C until further use.
In silico identification of putative Inc proteins.
All P. amoebophila proteins lacking similarity to functionally characterized proteins (annotated as conserved hypothetical proteins or as hypothetical proteins [26]) and without a signal peptide (as determined by SignalP prediction) were analyzed using a Kyte-Doolittle hydrophilicity profile (window size of seven) through the MacVector software suite (40). Hydrophilicity plots were analyzed manually, and all proteins with a large dominant bi-lobed hydrophobic domain (5) were regarded as putative Inc proteins. Coiled-coil structures were determined by MARCOIL (13 [http://www.isrec.isb-sib.ch/webmarcoil/webmarcoilC1.html]). Amino acid sequence identity values were determined by pairwise alignment (35 [http://pir.georgetown.edu/]).
Cloning, protein expression, and purification of five putative Incs.
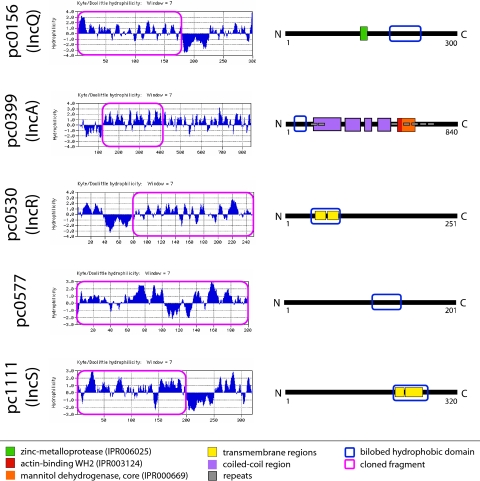
Fragments of genes encoding the predicted Inc proteins pc0156, pc0399, pc0530, pc0577, and pc1111 were cloned into the expression vector pET16b, which adds an N-terminal His tag to the expressed proteins (Novagen). The fragments used for cloning generally excluded the bi-lobed hydrophobic domain and included amino acids 1 to 180 for pc0156, 120 to 402 for pc0399, 78 to 251 for pc0530, 2 to 201 for pc0577, and 1 to 199 for pc1111 (Fig. 1). The primers used for amplification included the restriction enzyme sites for NdeI in the forward and BamHI in the reverse primer. Amplified sequences were cloned into the pET16b vector, and the resulting constructs were validated by sequencing. The plasmids were transformed into E. coli BL21(DE3) (Invitrogen). Heterologous expression of each recombinant protein was achieved at room temperature following induction with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). Expressed proteins were purified with HisTrap purification columns (HiTrap HP; GE Healthcare Amersham) according to recommendations of the manufacturer. The identity of the purified proteins was determined by one-dimensional (1D) gel electrophoresis in combination with mass spectrometry (data not shown).
FIG. 1.
Characteristics of five putative P. amoebophila inclusion proteins. Kyte-Doolittle hydrophilicity profiles of proteins (left) and a schematic overview of predicted domains as determined by InterPro, MARCOIL, and RADAR (right) are shown. The sequence length in amino acids is indicated.
Antisera and IgY preparation.
For antibody production P. amoebophila EBs were purified by ultracentrifugation on a gastrografin gradient. The absence of host cell debris in the EB fraction was verified by staining nucleic acids with 4′,6′-diamidino-2-phenylindole (DAPI) and membranes with 3,3′-dihexyloxacarbocyanine iodide (DiOC6; Invitrogen) (3; also data not shown). Protein amount was determined with a BCA Protein Assay Kit (Pierce Biotechnology), and purified EBs were used for immunization of two rabbits and two chickens (Eurogentec).
Heterologously expressed and purified proteins were dialyzed against buffer of 1× phosphate-buffered saline (PBS; 130 mM NaCl, 10 mM Nax·PO4, pH 7.2 to 7.4) containing up to 0.05% sodium dodecyl sulfate and used for immunization of animals (Eurogentec). Animals were chosen as follows: one rabbit and one guinea pig each for pc0530 and pc0577 and one rabbit and one chicken each for pc0156, pc0399, and pc1111. IgY from egg yolks was purified with an Eggcellent IgY Purification Kit (Pierce Biotechnology) for purified proteins or with HiTrap IgY Purification HP columns (GE Healthcare Amersham) for chickens immunized with whole EBs. For all obtained antiserum and egg yolk samples, preimmune serum and egg yolk collections from the respective animals were obtained and tested by immunofluorescence analysis of A. castellanii cultures infected with P. amoebophila. Preimmune serum yielded no signals in immunofluorescence experiments. All serum and egg yolk fractions obtained after immunization were also tested by immunofluorescence in uninfected A. castellanii cultures as a negative control to check for signals caused by amoebal proteins. Background reactivity against amoebae was observed for most of the animals. To remove antibodies binding to amoebal proteins, all antiserum and IgY fractions were adsorbed with amoeba cells prior to immunofluorescence experiments. For this, amoebae were lysed by at least three freeze-thaw steps, and the resulting lysate was incubated with the serum and IgY fractions in a 1:1 mixture overnight and then centrifuged at 12,300 × g for 2 min at 4°C to remove cell debris with bound antibodies.
Western blotting.
Lysates of E. coli cells expressing the respective putative Inc proteins were subjected to 1D gel electrophoresis and transferred onto a polyvinylidene difluoride membrane (Hybond-P; GE Healthcare Amersham) by semidry blotting (TE77 semidry transfer unit; GE Healthcare Amersham) in transfer buffer (14.4 g of glycine, 3.03 g of Tris-HCl, 20% methanol, pH 8.3) for 1.5 h as recommended by the manufacturer. Primary antibodies against putative inclusion membrane proteins and corresponding secondary antibodies labeled with horseradish peroxidase (dianova, Germany) were incubated each for 1 h at room temperature, respectively. Signal detection was performed by a Western Lightning Chemiluminescence Plus kit (Perkin Elmer), and X-ray films were exposed in the dark.
Immunofluorescence.
Acanthamoeba host cells containing P. amoebophila were fixed with either methanol for 10 min at room temperature or with 4% paraformaldehyde (PFA) in 1× PBS for 1 h at room temperature. After the fixative was removed, cells were covered with 1× PBS and either used immediately for immunofluorescence or stored at 4°C for up to 3 days. PFA-fixed cells were permeabilized prior to immunofluorescence via incubation in 0.05% Tween 20 in 1× PBS for 25 min at room temperature. After removal of Tween, the cells were covered with 10 mM Tris-HCl (pH 6.5) containing 5 μg/ml lysozyme (L1667; Sigma) and incubated for 1 h at room temperature. Fixed cells were incubated with blocking solution (2% bovine serum albumin [BSA] in 1× PBS) for 10 min at room temperature. Then, blocking solution was replaced with the respective dilution of primary antibody in blocking solution. After incubation for 1 h at room temperature, cells were washed three times with 1× PBS, and secondary antibodies (dianova) diluted in blocking solution were added. Incubation was performed for 1 h in the dark at room temperature. Secondary antibodies used in this study were labeled with either one of the hydrophilic sulfoindocyanine fluorescent dyes Cy2, Cy3, and Cy5 or fluorescein isothiocyanate (FITC). Afterwards, the cells were washed three times with 1× PBS and embedded in Mowiol, which was prepared by stirring 2.4 g of Mowiol 4-88 from Sigma, 6 g of glycerin, 6 ml of H2O, and 12 ml of 0.2 M Tris-HCl, pH 8.5, at 50 to 60°C for several hours; the solution was then centrifuged at 7,700 × g for 15 min at room temperature to remove air bubbles and nondissolved matter, and the supernatant was stored at −20°C until further use. Embedded samples were examined by confocal laser scanning microscopy (LSM 510 Meta; Carl Zeiss).
Immuno-electron microscopy.
A. castellanii UWC1 host cells infected with P. amoebophila were processed for immunogold labeling by either of two different methods (using either cryosections or Lowicryl HM20 embedding). For cryosections, cells were fixed with 2% paraformaldehyde-0.1% glutaraldehyde in 100 mM HEPES buffer (pH 7.2) for 2 h 20 min at 20°C, washed twice with 100 mM HEPES, and then encapsulated in 12% gelatin and processed as described previously (50). Cryosections were cut using a Leica Ultracut UC7 cryo-ultramicrotome (Leica Microsystems, Austria). Semithin sections were mounted onto glass slides pretreated with BioBond (British Biosystems International, United Kingdom). Ultrathin sections were mounted onto carbon-coated formvar-pilioform films on 200-mesh nickel electron microscopy (EM) grids (Gilder, Netherlands).
Alternatively, high-pressure freezing and low-temperature freeze-substitution (FS) were used to prepare thin sections (41). Cells were grown on sapphire discs (1.2 mm), dipped into 20% BSA, and then snap-frozen in liquid nitrogen at a pressure of 2.1 × 108 Pa using a Leica Empact high-pressure freezing device (Leica Microsystems). Frozen cells were transferred to the chamber of the Leica EM AFS2 freeze-substitution device (Leica Microsystems). Cells were simultaneously fixed and substituted in FS medical-grade methanol solution containing 0.5% uranyl acetate (UA) for 96 h at −88°C, after which the temperature was slowly increased to −54°C at a rate of 2°C per h and maintained for 8 h. The temperature was raised to −24°C for optimal action of the UA for 5 h and returned to −40°C for a further 2 h. At −40°C the samples were washed in methanol three times for 10 min. The freeze-substitution solution was removed and replaced by Lowicryl HM20 resin-methanol mixture at 1:2 for 15 min, 1:1 for 30 min, and 2:1 for 90 min, followed by two changes of pure fresh Lowicryl HM20 resin (Chemische Werke Lowi, Waldkraiburg, Germany) for 90 min each, and maintained in pure resin for a further 20 h. Polymerization with UV light proceeded at −40°C for 2 days. Ultrathin sections were cut using a Leica Supercut UCT ultramicrotome (Leica Microsystems).
Immunolabeling of ultrathin sections was performed at room temperature as described previously (50). The primary antibody dilution was determined empirically and optimized by testing immunofluorescence labeling of semithin cryosections. Goat-anti rabbit or goat-anti chicken antibodies conjugated with Ultrasmall gold (Aurion, Netherlands) were used as secondary antibodies at a 1:40 dilution. Ultrasmall gold labeling was silver enhanced using an R-Gent SE-EM kit (Aurion, Netherlands) according to the manufacturer's instructions. Cryosections were contrasted with 1% uranyl acetate-1% methyl cellulose solution for 10 min. Lowicryl HM20 sections were contrasted with 0.5% uranyl acetate for 20 min and lead citrate for 7 min. All grids were examined in a Zeiss 902 transmission electron microscope at 80 kV accelerating voltage.
Statistical evaluation of immuno-electron microscopy staining.
A systematic, uniform random sampling of the whole section was performed for statistical analysis of immunolabeled sections (32). A series of either 10 images at a magnification of ×12,000 or 20 images at a magnification of ×20,000 was captured at defined intervals to record at least 100 gold particles. Point counting (using 2-cm point intervals on transparent film overlay onto printed images) of defined cell compartments (host cytoplasm, inclusion, bacteria, and mitochondria) was used to estimate relative proportions of cell compartment areas. Then, the numbers of gold particles found within each compartment were counted manually. The relative labeling index (RLI) summed for all images was used to compare summed expected gold particles to the actual number of gold particles counted (34). Compartments showing an RLI above 1 were assessed using a chi-square equation (null hypothesis equals random distribution of gold label), and the value was compared to a chi-square table (3 degrees of freedom) to assess the probability of random gold labeling for the compartment of interest.
RESULTS
Twenty-three putative inclusion membrane proteins in P. amoebophila.
The genome-wide survey for proteins of P. amoebophila UWE25 with a bi-lobed hydrophobic domain revealed 23 putative Inc proteins (Table 1). Only three of the 23 candidate Inc proteins shared unambiguous sequence similarity with proteins from members of the Chlamydiaceae (Table 2). We further analyzed the candidate Inc proteins of P. amoebophila, most of which were annotated as hypothetical or unknown proteins, by BLAST and used patterns and domains identified by PEDANT and InterPro (4, 16) to gain indications for their possible function. Based on this analysis, five putative Inc proteins described below were selected for further investigation of their expression and subcellular locations during intracellular development of P. amoebophila.
TABLE 1.
Inclusion membrane proteins and putative inclusion membrane proteins of P. amoebophila
| Gene IDa | Protein | Description | Length (aa)b | Mass (Da) | pI |
|---|---|---|---|---|---|
| pc0063 | Hypothetical protein | 295 | 33,395.3 | 8.74 | |
| pc0156* | IncQ | Hypothetical protein | 903 | 35,051.9 | 8.88 |
| pc0164 | Unknown protein | 154 | 17,372 | 9.80 | |
| pc0184 | Hypothetical protein | 435 | 50,097.4 | 5.35 | |
| pc0399* | IncA | Similar to inclusion protein IncA | 840 | 95,891 | 5.61 |
| pc0508 | Unknown protein | 125 | 14,497.7 | 5.45 | |
| pc0530* | IncR | Unknown protein | 251 | 28,197.7 | 8.88 |
| pc0577 | Unknown protein | 201 | 22,807.8 | 4.98 | |
| pc0579 | Unknown protein | 201 | 22,769.7 | 4.74 | |
| pc0699 | Unknown protein | 447 | 51,630.6 | 8.79 | |
| pc0726 | Unknown protein | 164 | 18,084.9 | 5.61 | |
| pc0791 | Hypothetical protein | 483 | 54,440.4 | 6.98 | |
| pc0922 | Unknown protein | 502 | 60,035.6 | 9.09 | |
| pc1111* | IncS | Unknown protein | 320 | 35,575.8 | 6.33 |
| pc1114 | Unknown protein | 320 | 37,096 | 9.38 | |
| pc1290 | Unknown protein | 287 | 31,730.7 | 6.30 | |
| pc1422 | Unknown protein | 101 | 11,840.4 | 9.70 | |
| pc1540 | Unknown protein | 856 | 98,885.7 | 8.93 | |
| pc1549 | Unknown protein | 234 | 26,937.5 | 6.21 | |
| pc1730 | Unknown protein | 257 | 29,399.2 | 8.61 | |
| pc1737 | Conserved hypothetical protein | 342 | 37,286.4 | 6.98 | |
| pc1857 | Conserved hypothetical protein | 343 | 40,463.1 | 5.17 | |
| pc1910 | Unknown protein | 318 | 35,939 | 7.87 |
Proteins analyzed in this study are in bold letters; proteins localized to the inclusion are indicated by asterisks. The protein pc0399 only shows weak similarity (E-value of >0.001) to IncA proteins of the Chlamydiaceae.
aa, amino acids.
TABLE 2.
Conservation of putative P. amoebophila inclusion membrane proteins among the Chlamydiaceae
| P. amoebophila UWE25 protein | Homologue (% identity)a |
|||||
|---|---|---|---|---|---|---|
| C. trachomatis D/UW-3/CX | C. muridarum Nigg | C. pneumoniae CWL029 | C. caviae GPIC | C. abortus S26/3 | C. felis Fe/C-56 | |
| pc0156 (IncQ) | CT642 (32) | TC0010 (31) | Cpn0770 (32) | CCA00987 (32) | CAB957 (32) | CF0026 (31) |
| pc0184 | CT616 (24) | TC0906 (22) | Cpn0755 (28) | CCA01002 (23) | CAB972 (25) | CF0010 (24) |
| pc1857 | CT484 (43) | TC0771 (42) | Cpn0602 (42) | CCA00138 (47) | CAB137 (46) | CF0868 (42) |
Homologues were determined by BLASTp at the NCBI site against the nonredundant protein sequences database (46); cutoff was set at an E-value of 1E-10. The protein pc0399 only shows weak similarity (E-value of > 0.001) to IncA proteins of the Chlamydiaceae and is therefore not listed. C. muridarum, Chlamydia muridarum; C. abortus, Chlamydophila abortus; C. felis, Chlamydophila felis.
The putative Inc protein pc0399, previously identified as a distant IncA homologue by Delevoye and coworkers (11), is notably longer than the C. trachomatis IncA (840 versus 273 amino acids). The amino acid sequence of pc0399 contains a bi-lobed hydrophobic domain at the N terminus and a coiled-coil structure (Fig. 1), both of which are found in other IncA molecules. The large coiled-coil domains of pc0399 range from amino acids 140 to 276, 294 to 370, 386 to 423, and 453 to 516. In addition, several repeats can be found in this region (amino acids 126 to 154 and 169 to 196; determined by the RADAR program at http://www.ebi.ac.uk/Radar/ [21]) and in the C-terminal part of the protein, where two repeats show similarity to a repeat region of several Entamoeba histolytica hypothetical proteins (amino acids 518 to 546, 577 to 604, 613 to 643, 648 to 678, and 685 to 723). However, none of the tandem repeats described for some Chlamydiaceae IncA are present (58). The protein pc0399 also contains an actin-binding motif and the catalytic core of mannitol dehydrogenases (Fig. 1). In contrast to pc0399, the Chlamydiaceae IncA coiled-coil region is much shorter and contains no long sequence repeats.
Another putative P. amoebophila Inc protein we selected for further analysis is pc0156, which shares similarity with proteins in other Chlamydiaceae. This protein is a putative zinc metalloprotease (IPR006025) (Fig. 1) which might serve as a chlamydial virulence factor (36). It is intriguing that among the Chlamydiaceae homologues of pc0156 (Table 2), the P. amoebophila protein is the only protein that contains a bi-lobed hydrophobic domain.
A feature observed for several Chlamydiaceae Incs is their occurrence in different isoforms originating from gene duplications (42). The only putative Incs found in P. amoebophila that might have arisen from gene duplication are pc0577 and pc0579, which share 86% amino acid sequence identity. Therefore, we chose pc0577 as one of the candidate Inc proteins to be analyzed further. The putative Inc proteins pc0530 and pc1111 do not share any significant sequence similarity with proteins from other organisms, nor were any conserved patterns or motifs recognized. This points at still unknown functions of these proteins, and they were therefore also subjected to further analyses (Table 1; Fig. 1).
Expression of putative Inc proteins during intracellular multiplication of P. amoebophila.
Rabbits, guinea pigs, and chickens were immunized with purified fragments (excluding the hydrophobic domain) of the putative P. amoebophila Incs pc0156, pc0399, pc0530, and pc1111 and the full-length protein pc0577 (Fig. 1). To analyze the specificity of the obtained sera and egg yolks, Western blot analysis of E. coli expressing the respective proteins was performed, resulting in a strong, specific detection of all respective proteins; the antibodies against pc0530 and pc1111 also showed some nonspecific binding to E. coli proteins (data not shown).
In contrast to members of the Chlamydiaceae, P. amoebophila is able to establish a long-term relationship with its host, in which both bacteria and amoebae multiply in a synchronized manner. We therefore investigated the expression of the five selected putative Inc proteins during coexistence of P. amoebophila within its Acanthamoeba host by immunofluorescence analysis using an unsynchronized culture containing EBs, intermediate bodies, and RBs of the chlamydial symbiont (Fig. 2). For this, anti-Inc antibodies were used simultaneously with antibodies against the immunodominant components of the outer membrane of P. amoebophila (anti-P. amoebophila), which were obtained by immunization of animals with whole EBs but also detect P. amoebophila RBs (data not shown). Immunofluorescence analysis resulted in distinct signals for all antibodies (Fig. 3). No differences between methanol and PFA fixation could be observed (data not shown).
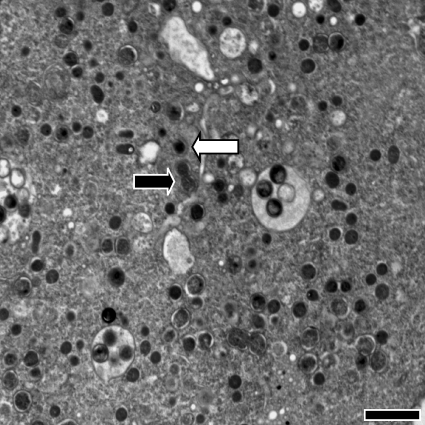
FIG. 2.
An Acanthamoeba trophozoite infected with P. amoebophila in a continuously grown culture. Both developmental forms, EBs and RBs, are present in one amoeba host cell in comparable ratios. The black arrow indicates a typical RB, and the white arrow indicates a typical EB. Bar, 2 μm.
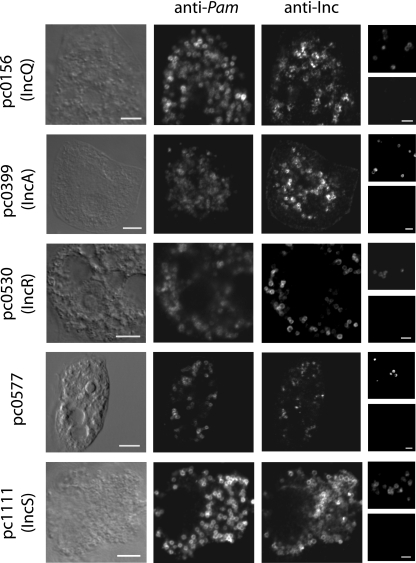
FIG. 3.
Localization of putative Inc proteins of P. amoebophila by immunofluorescence and confocal laser scanning microscopy. The first three columns represent identical microscopic fields (bar, 5 μm). Images are as follows: first column, the amoeba host in transmitted light mode; second column, intracellular P. amoebophila (EBs and RBs) visualized by anti-P. amoebophila (anti-Pam) antibodies; third column, immunofluorescence derived from antibodies against the five candidate inclusion proteins as indicated on the left. Note that with P. amoebophila each inclusion contains only a single bacterial cell, and the inclusion membrane is closely attached to the P. amoebophila cell surface; the halo-like appearance of the fluorescence signals derived from anti-Inc antibodies and the overlap with those from anti-P. amoebophila antibodies demonstrate that the putative Inc proteins are located either in the cell envelope of P. amoebophila or in the inclusion membrane. The fourth column shows two identical microscopic fields of purified P. amoebophila cells stained with anti-P. amoebophila antibodies (top image) and the respective anti-Inc antibodies (bottom image; bar, 2 μm). The absence of fluorescence signals with the anti-Inc antibodies suggests that the putative Inc proteins of P. amoebophila are not located in the cell envelope of P. amoebophila.
The putative Inc proteins pc0156, pc0399, pc0530, and pc1111 were located in a halo-like structure around the bacterial cytoplasm of P. amoebophila EBs and RBs, which represents either the chlamydial inner or outer membrane or the inclusion membrane (Fig. 3). As P. amoebophila lives in single-cell inclusions in its host cell, the inclusion membrane is directly adjacent to the bacterial outer membrane (8, 17). To test for the presence of the candidate Incs in the P. amoebophila cell envelope, purified P. amoebophila EBs were analyzed by immunofluorescence. While purified EBs could be readily visualized with anti-P. amoebophila antibodies, they were not detected with antibodies against the putative Inc proteins pc0156, pc0399, pc0530, or pc1111, which indicates that these proteins are not located in the P. amoebophila EB cell envelope (Fig. 3). The antiserum against protein pc0577 was the only serum tested that did not show a clear and consistent halo-like signal, and the observed signal suggests that the protein is localized in the bacterial cytoplasm (Fig. 3).
Colocalization experiments with antibodies against the four candidate Inc proteins resulting in halo-like signals around the P. amoebophila cells showed an almost complete overlap between the different putative Inc proteins (shown for pc1111 and pc0530 in Fig. 4). In addition, almost all cells positive for polyclonal antibodies raised against outer membrane proteins of P. amoebophila also showed signals with the candidate Inc-specific antibodies in asynchronous cultures (Fig. 4). This suggests that the Inc proteins pc0156, pc0399, pc0530, and pc1111 are continuously expressed during the intracellular life of P. amoebophila.
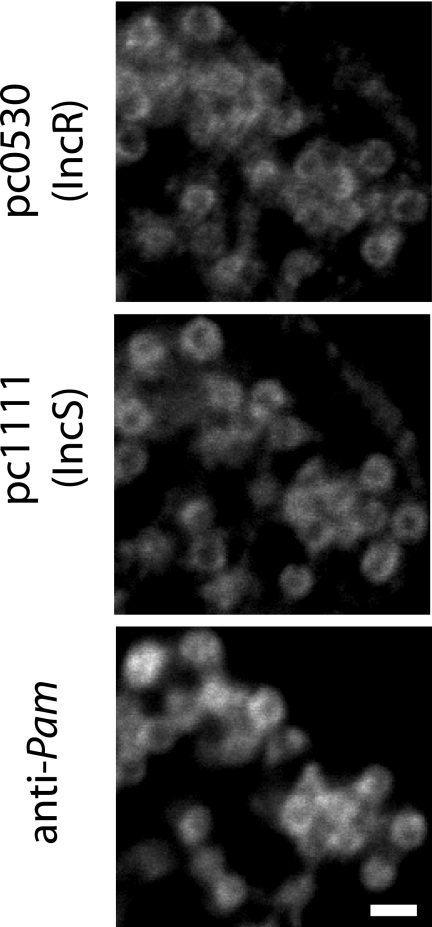
FIG. 4.
Colocalization of Inc proteins of P. amoebophila. Immunofluorescence images of identical microscopic fields are shown. Antibodies against the two Inc proteins pc530 (IncR) and pc1111 (IncS) were used in combination with an antibody against immunodominant components of P. amoebophila. The overlay shows that almost all inclusions are labeled by both anti-Inc antibodies. Bar, 1 μm.
Localization of four putative Inc proteins in the inclusion membrane.
To further explore the origin of the halo-like signals observed in the immunofluorescence experiments with antibodies against the putative Inc proteins pc0156, pc0399, pc0530, and pc1111 and to investigate the subcellular location of these proteins, we performed immuno-electron microscopy. For this purpose, the protein-specific polyclonal antibodies and appropriate gold-labeled secondary antibodies were used to detect the proteins on either cryosections or ultrathin sections of low-temperature HM20 resin.
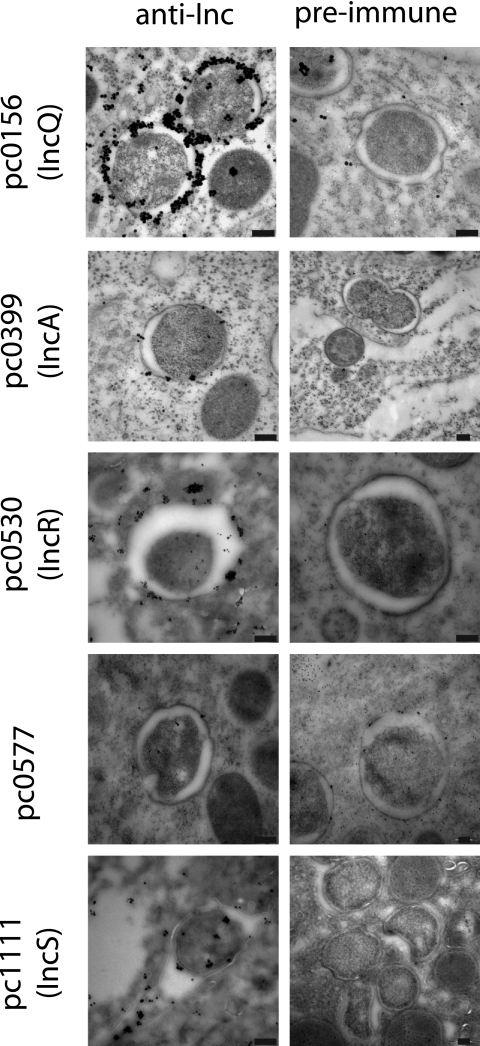
Immuno-electron microscopy clearly visualized the proteins pc0156, pc0399, pc0530, and pc1111 while all control experiments using the respective preimmune serum resulted in no signal (Fig. 5). For the candidate inclusion proteins pc0156 and pc0399, gold labels were clearly visible and limited to the inclusion membrane enclosing P. amoebophila within the host cytoplasm. The proteins pc1111 and pc0530 could also be detected at the inclusion membrane but were in addition located within the bacteria and in the cell envelope. Some gold labels can also be seen in the inclusion lumen (Fig. 5, pc1111) or at places where a possible connection between the bacteria and the inclusion membrane exists (Fig. 5, pc0530). The antibodies targeting the protein pc0577 did not yield a clear signal in immuno-electron microscopy, perhaps indicating that the epitopes targeted by these antibodies were not recovered by this technique.
FIG. 5.
Localization of Inc proteins of P. amoebophila by immuno-transmission electron microscopy. Images shown in the left column were produced using antibodies against the four candidate inclusion proteins, as indicated; for the images in the right column, the corresponding preimmune serum was used as a control. Bar, 200 nm. Gold labeling was performed on cryosections for pc1111 (IncS) and on Lowicryl HM20 resin sections for all other proteins. An unambiguous association of pc0156 (IncQ), pc0399 (IncA), pc0530 (IncR), and pc1111 (IncS) with the inclusion membrane is demonstrated. No association with the inclusion was observed for pc0577.
The observed location of the putative Inc proteins was further confirmed by stereological analysis of the obtained gold labeling, where between 120 and 2,600 gold particles were counted for each anti-Inc antibody. Their locations were categorized into four different compartments: P. amoebophila cells, the inclusion (including the lumen as well as the membrane), the amoeba cytoplasm, and amoeba mitochondria. Despite the small size of the chlamydial inclusion compared, e.g., with the amoeba cytoplasm, stereology identified a significant association (P < 0.0005) of the antibodies targeting the four Inc proteins, pc0156, pc0399, pc0530, and pc1111, with the inclusion (Table 3). As expected, no association with the inclusion membrane was observed for the protein pc0577, which is consistent with the observed cytoplasmic signal with these antibodies in the immunofluorescence experiments.
TABLE 3.
Significance of the subcellular localization of immunogold labeling for putative inclusion membrane proteins based on stereological analysis
| Gene ID | Protein | Association of the immunogold particles witha: |
|||||
|---|---|---|---|---|---|---|---|
|
P. amoebophila |
Inclusion |
Amoeba cytoplasm |
Mitochondria |
||||
| RLI | P value | RLI | P value | RLI | RLI | ||
| pc0156 | IncQ | 7.6 | <0.0005 | 2.37 | <0.0005 | 0.28 | 0.3 |
| pc0399 | IncA | 1.79 | <0.0005 | 6.32 | <0.0005 | 0.29 | 0.6 |
| pc0530 | IncR | 4.0 | <0.0005 | 1.2 | <0.0025 | 0.7 | 0.6 |
| pc0577 | 2.51 | <0.01 | 1.3 | NS | 0.99 | 0.45 | |
| pc1111 | IncS | 2.76 | <0.0005 | 1.72 | <0.0005 | 0.77 | 0.31 |
P values are based on a chi-square test; statistical tests were not performed if the relative labeling index (RLI) was <1. NS, not significant.
DISCUSSION
Inc proteins are one of the most enigmatic features of the chlamydiae (54), and similar proteins have only rarely been identified in other bacteria (28, 53). Located directly at the interface between bacteria and host cell, the inclusion membrane is a central mediator for interaction with and manipulation of the host cell by the chlamydiae. The inclusion membrane might also contribute to host specificity and play a crucial role in the uptake of nutrients from the host cell (42). In this study we raised the question of whether this unique group of proteins is also used by the chlamydia-related amoeba symbiont P. amoebophila, which does not share various other classic chlamydial virulence factors, such as, e.g., the polymorphic membrane proteins or the translocated actin-recruiting protein, with the Chlamydiaceae (22). In addition, in contrast to the Chlamydiaceae, P. amoebophila remains in single-cell inclusions and stably coexists with its host. Despite these differences, we could, however, identify 23 putative Inc proteins in the genome of P. amoebophila based on their hydrophilicity profiles (Table 1). Comparative sequence analysis demonstrated a low degree of conservation of Inc proteins between Chlamydiaceae and Parachlamydiaceae. Only four P. amoebophila Inc candidates showed considerable (pc0156, pc0184, and pc1857) or only partial and low (pc0399) sequence similarities to Chlamydiaceae proteins (Table 2). Two of these four proteins, pc0156 and pc0399, were further analyzed in this study, as well as three proteins without homologues in the Chlamydiaceae. Immunofluorescence analysis of pc0156, pc0399, pc0530, and pc1111 (Fig. 3) showed a halo-like signal, which demonstrated a location in the inclusion membrane or the P. amoebophila cell envelope. Further investigations by fluorescence and immuno-electron microscopy unambiguously demonstrated the location of pc0156, pc0399, pc0530, and pc1111 in the inclusion membrane (Fig. 3 and 5 and Table 3). Two of these proteins (pc1111 and pc0530) were present not only in the inclusion membrane but also on the outside of the P. amoebophila cells (pc1111) or at connections formed between the inclusion membrane and the bacterial outer membrane (pc0530), which could indicate a means of nutrient transport between the bacteria and the amoeba host. The names IncA (pc0399), IncQ (pc0156), IncR (pc0530), and IncS (pc1111) are suggested for these inclusion membrane proteins of P. amoebophila.
Although the function of most Inc proteins remains cryptic, the presence of an IncA protein (pc0399) in P. amoebophila with similarity to Chlamydiaceae IncA, which is the key player in fusion of inclusions (11, 20, 45, 52), seems remarkable as the differences between these proteins might account for the absence of fusion of P. amoebophila inclusions. In contrast to Chlamydiaceae IncA, the protochlamydial IncA (pc0399) encodes an actin-binding motif (Fig. 1). This might suggest an IncA-mediated interaction of the P. amoebophila inclusion with the host cell cytoskeleton. The specific functions of the putative P. amoebophila inclusion proteins IncQ, IncR, and IncS (pc0156, pc0530, and pc1111, respectively) are unknown, and no specific indications can be attributed based on their sequences. The analysis of their interaction partners, which might be of bacterial and/or eukaryotic origin, could help to provide deeper insights into their role during establishment and maintenance of the P. amoebophila inclusion.
In summary, we demonstrated that inclusion proteins are not specific for the Chlamydiaceae but are also present in the amoeba symbiont P. amoebophila. The occurrence of these unique proteins among only distantly related chlamydial groups indicates that pathogens and symbionts share similar mechanisms for host cell interaction and suggests that at least some inclusion proteins like IncA might be ancient and might have been present in the chlamydial ancestor, which lived roughly 700 million years ago (26). The expansion of this group of proteins in the Chlamydiaceae and the low number of shared Inc homologues among all known chlamydiae indicate that Inc proteins are also important components determining host specificity and the pathogenic lifestyle in mammalian cells, further supporting the notion of inclusion proteins as important chlamydial virulence factors.
Acknowledgments
This work was supported by Austrian Science Fund (FWF) grant Y277-B03, a grant from the University of Vienna in the framework of the University Research Focus “Molecular interactions between intracellular bacteria and their eukaryotic host cells,” and by grants from the National Institutes of Health (AI48769 and AI031448).
We thank Sara Weeks and Aishu Ramaswamy for technical assistance, Sven Poppert for providing antibodies, and Siegfried Reipert and Daniela Gruber for support with electron microscopy.
Footnotes
Published ahead of print on 30 July 2010.
REFERENCES
- 1.Abdelrahman, Y. M., and R. J. Belland. 2005. The chlamydial developmental cycle. FEMS Microbiol. Rev. 29:949-959. [DOI] [PubMed] [Google Scholar]
- 2.Alzhanov, D. T., S. K. Weeks, J. R. Burnett, and D. D. Rockey. 2009. Cytokinesis is blocked in mammalian cells transfected with Chlamydia trachomatis gene CT223. BMC Microbiol. 9:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R., N. Springer, W. Schonhuber, W. Ludwig, E. N. Schmid, K. D. Muller, and R. Michel. 1997. Obligate intracellular bacterial parasites of acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Apweiler, R., T. K. Attwood, A. Bairoch, A. Bateman, E. Birney, M. Biswas, P. Bucher, L. Cerutti, F. Corpet, M. D. R. Croning, R. Durbin, L. Falquet, W. Fleischmann, J. Gouzy, H. Hermjakob, N. Hulo, I. Jonassen, D. Kahn, A. Kanapin, Y. Karavidopoulou, R. Lopez, B. Marx, N. J. Mulder, T. M. Oinn, M. Pagni, F. Servant, C. J. A. Sigrist, and E. M. Zdobnov. 2001. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bannantine, J. P., R. S. Griffiths, W. Viratyosin, W. J. Brown, and D. D. Rockey. 2000. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2:35-47. [DOI] [PubMed] [Google Scholar]
- 6.Beatty, W. L. 2008. Late endocytic multivesicular bodies intersect the chlamydial inclusion in the absence of CD63. Infect. Immun. 76:2872-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 349:925-926. [DOI] [PubMed] [Google Scholar]
- 8.Collingro, A., E. R. Toenshoff, M. W. Taylor, T. R. Fritsche, M. Wagner, and M. Horn. 2005. “Candidatus Protochlamydia amoebophila,” an endosymbiont of Acanthamoeba spp. Int. J. Syst. Evol. Microbiol. 55:1863-1866. [DOI] [PubMed] [Google Scholar]
- 9.Corsaro, D., M. Valassina, and D. Venditti. 2003. Increasing diversity within Chlamydiae. Crit. Rev. Microbiol. 29:37-78. [DOI] [PubMed] [Google Scholar]
- 10.Cortes, C., K. A. Rzomp, A. Tvinnereim, M. A. Scidmore, and B. Wizel. 2007. Chlamydia pneumoniae inclusion membrane protein Cpn0585 interacts with multiple Rab GTPases. Infect. Immun. 75:5586-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delevoye, C., M. Nilges, A. Dautry-Varsat, and A. Subtil. 2004. Conservation of the biochemical properties of IncA from Chlamydia trachomatis and Chlamydia caviae: oligomerization of IncA mediates interaction between facing membranes. J. Biol. Chem. 279:46896-46906. [DOI] [PubMed] [Google Scholar]
- 12.Delevoye, C., M. Nilges, P. Dehoux, F. Paumet, S. Perrinet, A. Dautry-Varsat, and A. Subtil. 2008. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4:e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delorenzi, M., and T. Speed. 2002. An HMM model for coiled-coil domains and a comparison with PSSM-based predictions. Bioinformatics 18:617-625. [DOI] [PubMed] [Google Scholar]
- 14.Dietrich, L. E., C. Boeddinghaus, T. J. LaGrassa, and C. Ungermann. 2003. Control of eukaryotic membrane fusion by N-terminal domains of SNARE proteins. Biochim. Biophys. Acta 1641:111-119. [DOI] [PubMed] [Google Scholar]
- 15.Everett, K. D., R. M. Bush, and A. A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 16.Frishman, D., K. Albermann, J. Hani, K. Heumann, A. Metanomski, A. Zollner, and H. W. Mewes. 2001. Functional and structural genomics using PEDANT. Bioinformatics 17:44-57. [DOI] [PubMed] [Google Scholar]
- 17.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K. H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackstadt, T., E. R. Fischer, M. A. Scidmore, D. D. Rockey, and R. A. Heinzen. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol. 5:288-293. [DOI] [PubMed] [Google Scholar]
- 19.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 20.Hackstadt, T., M. A. Scidmore-Carlson, E. I. Shaw, and E. R. Fischer. 1999. The Chlamydia trachomatis IncA protein is required for homotypic vesicle fusion. Cell Microbiol. 1:119-130. [DOI] [PubMed] [Google Scholar]
- 21.Heger, A., and L. Holm. 2000. Rapid automatic detection and alignment of repeats in protein sequences. Proteins 41:224-237. [DOI] [PubMed] [Google Scholar]
- 22.Heinz, E., P. Tischler, T. Rattei, G. Myers, M. Wagner, and M. Horn. 2009. Comprehensive in silico prediction and analysis of chlamydial outer membrane proteins reflects evolution and life style of the Chlamydiae. BMC Genomics 10:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinzen, R. A., and T. Hackstadt. 1997. The Chlamydia trachomatis parasitophorous vacuolar membrane is not passively permeable to low-molecular-weight compounds. Infect. Immun. 65:1088-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heuer, D., C. Kneip, A. P. Maurer, and T. F. Meyer. 2007. Tackling the intractable: approaching the genetics of Chlamydiales. Int. J. Med. Microbiol. 297:569-576. [DOI] [PubMed] [Google Scholar]
- 25.Horn, M. 2008. Chlamydiae as symbionts in eukaryotes. Annu. Rev. Microbiol. 62:113-131. [DOI] [PubMed] [Google Scholar]
- 26.Horn, M., A. Collingro, S. Schmitz-Esser, C. L. Beier, U. Purkhold, B. Fartmann, P. Brandt, G. J. Nyakatura, M. Droege, D. Frishman, T. Rattei, H. W. Mewes, and M. Wagner. 2004. Illuminating the evolutionary history of chlamydiae. Science 304:728-730. [DOI] [PubMed] [Google Scholar]
- 27.Horn, M., M. Wagner, K. D. Muller, E. N. Schmid, T. R. Fritsche, K. H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 28.Huang, B., M. J. Troese, S. Ye, J. T. Sims, N. L. Galloway, D. L. Borjesson, and J. A. Carlyon. 2010. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect. Immun. 78:1864-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hybiske, K., and R. S. Stephens. 2007. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U. S. A. 104:11430-11435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuo, C.-C., M. Horn, and R. S. Stephens. The order Chlamydiales, p. 522-523. In B. Hedlund, N. R. Krieg, W. Ludwig, B. J. Paster, J. T. Staley, N. Ward, and W. B. Whitman (ed.), Bergey's manual of systematic bacteriology, in press. Springer, New York, NY.
- 31.Li, Z., C. Chen, D. Chen, Y. Wu, Y. Zhong, and G. Zhong. 2008. Characterization of fifty putative inclusion membrane proteins encoded in the Chlamydia trachomatis genome. Infect. Immun. 76:2746-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucocq, J. M., A. Habermann, S. Watt, J. M. Backer, T. M. Mayhew, and G. Griffiths. 2004. A rapid method for assessing the distribution of gold labeling on thin sections. J. Histochem. Cytochem. 52:991-1000. [DOI] [PubMed] [Google Scholar]
- 33.Mahony, J. B., B. K. Coombes, and M. A. Chernesky. 2003. Chlamydia and Chlamydophila, p. 991-1004. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 8 ed., vol. 1. ASM Press, Washington, DC. [Google Scholar]
- 34.Mayhew, T. M., J. M. Lucocq, and G. Griffiths. 2002. Relative labelling index: a novel stereological approach to test for non-random immunogold labelling of organelles and membranes on transmission electron microscopy thin sections. J. Microsc. 205:153-164. [DOI] [PubMed] [Google Scholar]
- 35.McGarvey, P. B., H. Huang, W. C. Barker, B. C. Orcutt, J. S. Garavelli, G. Y. Srinivasarao, L.-S. L. Yeh, C. Xiao, and C. H. Wu. 2000. PIR: a new resource for bioinformatics. Bioinformatics 16:290-291. [DOI] [PubMed] [Google Scholar]
- 36.Miyoshi, S., and S. Shinoda. 2000. Microbial metalloproteases and pathogenesis. Microbes Infect. 2:91-98. [DOI] [PubMed] [Google Scholar]
- 37.Moulder, J. W. 1964. The psittacosis group as bacteria. John Wiley, New York, NY.
- 38.Mussa, F. F., H. Chai, X. Wang, Q. Yao, A. B. Lumsden, and C. Chen. 2006. Chlamydia pneumoniae and vascular disease: an update. J. Vasc. Surg. 43:1301-1307. [DOI] [PubMed] [Google Scholar]
- 39.Peters, J., D. P. Wilson, G. Myers, P. Timms, and P. M. Bavoil. 2007. Type III secretion a la Chlamydia. Trends Microbiol. 15:241-251. [DOI] [PubMed] [Google Scholar]
- 40.Rastogi, P. A. 2000. MacVector. Integrated sequence analysis for the Macintosh. Methods Mol. Biol. 132:47-69. [DOI] [PubMed] [Google Scholar]
- 41.Reipert, S., and G. Wiche. 2008. High-pressure freezing and low-temperature fixation of cell monolayers grown on sapphire coverslips. Methods Cell Biol. 88:165-180. [DOI] [PubMed] [Google Scholar]
- 42.Rockey, D. D., and D. T. Alzhanov. 2006. Proteins in the chlamydial inclusion membrane. Horizon Bioscience, Norfolk, United Kingdom.
- 43.Rockey, D. D., D. Grosenbach, D. E. Hruby, M. G. Peacock, R. A. Heinzen, and T. Hackstadt. 1997. Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion. Mol. Microbiol. 24:217-228. [DOI] [PubMed] [Google Scholar]
- 44.Rockey, D. D., R. A. Heinzen, and T. Hackstadt. 1995. Cloning and characterization of a Chlamydia psittaci gene coding for a protein localized in the inclusion membrane of infected cells. Mol. Microbiol. 15:617-626. [DOI] [PubMed] [Google Scholar]
- 45.Rockey, D. D., W. Viratyosin, J. P. Bannantine, R. J. Suchland, and W. E. Stamm. 2002. Diversity within inc genes of clinical Chlamydia trachomatis variant isolates that occupy non-fusogenic inclusions. Microbiology 148:2497-2505. [DOI] [PubMed] [Google Scholar]
- 46.Sayers, E. W., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. DiCuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, I. Mizrachi, J. Ostell, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, E. Yaschenko, and J. Ye. 2009. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 37:D5-D15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 1996. Sphingolipids and glycoproteins are differentially trafficked to the Chlamydia trachomatis inclusion. J. Cell Biol. 134:363-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 50.Slot, J. W., and H. J. Geuze. 2007. Cryosectioning and immunolabeling. Nat. Protoc. 2:2480-2491. [DOI] [PubMed] [Google Scholar]
- 51.Subtil, A., C. Parsot, and A. Dautry-Varsat. 2001. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol. Microbiol. 39:792-800. [DOI] [PubMed] [Google Scholar]
- 52.Suchland, R. J., D. D. Rockey, J. P. Bannantine, and W. E. Stamm. 2000. Isolates of Chlamydia trachomatis that occupy nonfusogenic inclusions lack IncA, a protein localized to the inclusion membrane. Infect. Immun. 68:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Teng, C. H., R. U. Palaniappan, and Y. F. Chang. 2003. Cloning and characterization of an Ehrlichia canis gene encoding a protein localized to the morula membrane. Infect. Immun. 71:2218-2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toh, H., K. Miura, M. Shirai, and M. Hattori. 2003. In silico inference of inclusion membrane protein family in obligate intracellular parasites chlamydiae. DNA Res. 10:9-17. [DOI] [PubMed] [Google Scholar]
- 55.Wolf, K., G. V. Plano, and K. A. Fields. 2009. A protein secreted by the respiratory pathogen Chlamydia pneumoniae impairs IL-17 signaling via interaction with human Act1. Cell Microbiol. 11:769-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.World Health Organization. 2001. Global prevalence and incidence of curable sexually transmitted infections. WHO/CDS/CDR/EDC/2001.10. World Health Organization, Geneva, Switzerland.
- 57.World Health Organization. 2008. Priority eye diseases. World Health Organization, Geneva, Switzerland. http://www.who.int/blindness/causes/priority/en/index2.html.
- 58.Yousef Mohamad, K., A. Rekiki, G. Myers, P. M. Bavoil, and A. Rodolakis. 2008. Identification and characterisation of coding tandem repeat variants in incA gene of Chlamydophila pecorum. Vet. Res. 39:56. [DOI] [PubMed] [Google Scholar]