Abstract
Phytochromes are biliprotein photoreceptors that are found in plants, bacteria, and fungi. Prototypical phytochromes have a Pr ground state that absorbs in the red spectral range and is converted by light into the Pfr form, which absorbs longer-wavelength, far-red light. Recently, some bacterial phytochromes have been described that undergo dark conversion of Pr to Pfr and thus have a Pfr ground state. We show here that such so-called bathy phytochromes are widely distributed among bacteria that belong to the order Rhizobiales. We measured in vivo spectral properties and the direction of dark conversion for species which have either one or two phytochrome genes. Agrobacterium tumefaciens C58 contains one bathy phytochrome and a second phytochrome which undergoes dark conversion of Pfr to Pr in vivo. The related species Agrobacterium vitis S4 contains also one bathy phytochrome and another phytochrome with novel spectral properties. Rhizobium leguminosarum 3841, Rhizobium etli CIAT652, and Azorhizobium caulinodans ORS571 contain a single phytochrome of the bathy type, whereas Xanthobacter autotrophicus Py2 contains a single phytochrome with dark conversion of Pfr to Pr. We propose that bathy phytochromes are adaptations to the light regime in the soil. Most bacterial phytochromes are light-regulated histidine kinases, some of which have a C-terminal response regulator subunit on the same protein. According to our phylogenetic studies, the group of phytochromes with this domain arrangement has evolved from a bathy phytochrome progenitor.
Phytochromes are biological photoreceptors that were discovered in plants, where they control development throughout the life cycle in manifold ways (21, 33). Today, a large number of homologs are known also from cyanobacteria, other bacteria, and fungi, which are termed cyanobacterial phytochromes (Cphs), bacteriophytochromes (BphPs), and fungal phytochromes (Fphs), respectively (20, 24). The chromophore is autocatalytically assembled within the N-terminal part of the protein, the photosensory core module (PCM), which contains the PAS, GAF, and PHY domains (30). Typically, phytochromes are converted by light between two spectrally different forms, the red-absorbing Pr and the far-red-absorbing Pfr forms. Photoconversion is initiated by an isomerization of the covalently bound bilin chromophore (32).
Plant and cyanobacterial phytochromes incorporate phytochromobilin (PΦB) and phycocyanobilin (PCB) as natural chromophores, respectively, which are covalently bound to Cys residues in the GAF domains. All characterized phytochromes that belong to these groups have a Pr ground state. Plant phytochromes can undergo dark conversion of Pfr to Pr (5), whereas the Pfr form of typical cyanobacterial phytochromes is stable in darkness (26).
Bacteriophytochromes utilize biliverdin (BV) instead as a natural chromophore (1), which is covalently attached to a Cys residue in the N terminus of the PAS domain (26). Since the conjugated system of BV is longer than that of PΦB or PCB, the absorption maxima of bacteriophytochromes are found at higher wavelengths than those of cyanobacterial or plant homologs.
With the discovery of a bacterial phytochrome from Bradyrhizobium sp. strain ORS278, termed BrBphP1, the first phytochrome with a Pfr ground state and dark conversion from Pr to Pfr was found (10). Thereafter, five more phytochromes with dark conversion of Pr to Pfr were described: Rhodopseudomonas palustris BphP1 (RpBphP1) from strain CEA001, RpBphP5, and RpBphP6 from strain CGA009 (11); Agrobacterium tumefaciens Agp2 (or AtBphP2) from strain C58 (18); and Pseudomonas aeruginosa BphP1 (PaBphP1) (40). These phytochromes are now termed bathy phytochromes because the absorption maxima of their ground states are bathochromically (to longer wavelengths) shifted compared to those of all other phytochromes.
Moreover, some other bacterial phytochromes with unusual properties have been described. In the Ppr from Rhodospirillum centenum, a photoactive yellow protein (PYP) domain is fused to the N terminus of a phytochrome homolog. The phytochrome part of Ppr assembles with BV to form a Pr adduct. However, irradiation does not result in the formation of Pfr but in a bleaching of the Pr spectrum (23). The BV adduct of RpBphP3 from R. palustris, which has a Pr ground state, photoconverts to the so-called Pnr form with a blue-shifted absorption maximum (12). RpBphP4 from R. palustris strains Ha2 and BisB5 and Bradyrhizobium BphP3 (BrBphP3) from Bradyrhizobium BTAi1, both with a Pr ground state, photoconvert into a long-lived MetaR form (8, 42). MetaRa and MetaRc are intermediates in the photoconversion from Pr to Pfr of prototypical phytochromes (3). BphP3 from the Bradyrhizobium strain ORS 278 is an exception among bacteriophytochromes as it binds PCB as a natural chromophore. This phytochrome adopts a so-called Po (P-orange) ground state with an absorbance maximum in the orange range (11, 15). Upon irradiation, this phytochrome converts into the Pr form. RpBphP4 from R. palustris CGA009 lacks the biliverdin binding cysteine and does not bind a chromophore (42).
With the rapidly growing number of bacterial genome sequences, many new bacterial phytochromes are being discovered. Thus, a large and increasing number of newly identified phytochromes remain spectroscopically uncharacterized. We established an in vivo photometry approach which allowed the rapid acquisition of spectral information about phytochromes from intact bacterial cells. In the beginning period of plant phytochrome research, in vivo photometry was extensively applied (4, 6, 29, 34). This method, in fact, allowed the identification of phytochromes for the first time in plant tissues (6), which led to the purification of phytochromes from plant extracts (37). Here, we apply in vivo photometry for the first time to organisms outside the plant kingdom. This method is especially useful for studying species with single phytochrome genes. The approach is also helpful for comparing properties of native phytochromes in vivo and of their recombinant proteins in vitro.
In the present study, we concentrate on nonphotosynthetic species of the order Rhizobiales which belongs to the Alphaproteobacteria. The family Rhizobiaceae comprises plant-interacting soil bacteria. A. tumefaciens and Agrobacterium vitis can transfer genes into plants to induce plant tumors, whereas many other Rhizobiaceae can live as plant symbionts in nodules of stems or roots in which they assimilate molecular nitrogen to produce NH4+, which is used by the plant for synthesis of amino acids and other nitrogen-containing molecules. A. tumefaciens C58 contains two phytochromes, termed Agp1 (or AtBphP1) and Agp2 (or AtBphP2), that have been characterized as recombinant proteins (14, 18, 26, 35) and whose spectral activities have been measured in extracts of wild-type and knockout mutants (31). A large number of phytochromes from photosynthetic Bradyrhizobium and Rhodopseudomonas species, which also belong to the order Rhizobiales, have been characterized as recombinant proteins (11), some of which have already been noted above.
It turned out that most of our analyzed phytochromes undergo dark conversion of Pr to Pfr and thus belong to the group of bathy phytochromes. Such phytochromes, which absorb at around 750 nm, clearly dominate among Rhizobiales. We propose that this specific property reflects an adaptation to the light regime in the soil. Our studies also suggest that bacterial phytochromes with a C-terminal response regulator have evolved from a bathy phytochrome progenitor.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains of Rhizobiales were grown at 28°C. The M1 growth medium (5 g of peptone, 3 g of meat extract per liter, pH 7.0) was used for A. tumefaciens strain C58 and its Δagp1, Δagp2, and Δagp1 Δagp2 mutant strains (31) and for Azorhizobium caulinodans ORS 571. A. vitis S4 was grown in YEP medium (5 g of meat extract, 1 g of peptone, 1 g of yeast extract, 5 g of sucrose, and 2 mmol of MgCl2 per liter, pH 7.0). Rhizobium leguminosarum 3841, Rhizobium etli CIAT 652, and Xanthobacter autotrophicus Py2 were cultivated in TY medium (5 g of tryptone, 3 g of yeast extract, 0.7 g of CaCl2 per liter). For the Δagp1 and Δagp2 mutant strains, 300 μg/ml spectinomycin was added to the growth medium; for the Δagp1 Δagp2 double mutant, 300 μg/ml spectinomycin and 100 μg/ml gentamicin were added. Escherichia coli strains DH5α and BL21 were usually grown at 37°C in either LB or RBP (10 g of tryptone, 5 g of yeast extract, 10 g of Nacl, 0.2% glucose, 2.3 g of KH2PO4, 12 g of K2HPO4 per liter [pH 7.0]) medium. The expression clones carrying the plasmids pETavi3496 and pETavi9083 (see below) were grown in medium supplemented with 100 μg/ml ampicillin (Amp).
Cloning of bacteriophytochrome genes.
Avp1 and Avp2 were amplified from genomic DNA of A. vitis S4 using Phusion polymerase (New England Biolabs) and the primer pair avp1_5′ (CATATGGGCAATGACTACACCGATG) and avp1_3′ (GCTCACTAAACCCGTCGCACACCACCACCACCACCACTGAGCGGCCGC) and the pair avp2_5′ (CATATGCCTACGCTGCCTTCGAAAGCC) and avp2_3′ (CTCGAGGCTGGTGGTGGCGTCCGGTTG), respectively, with a PCR program consisting of 98°C for 60 s, followed by 35 cycles of 98°C for 10 s, 58°C for 30 s, and 72°C for 80 s, with a final step at 72°C for 60s. Products were A-tailed with Taq polymerase (Sigma) and ligated with pGEM T Easy (Promega), yielding pGEMavi9083 and pGEMavi3496, respectively. Plasmids were amplified in E. coli DH5α and digested with NotI and EcoRI, in the case of pGEMavi9083, and with NdeI and XhoI, in the case of pGEMavi3496. The fragments were ligated with identically digested pET21b (Novagen). The obtained expression plasmids pETavi9083 and pETavi3496 contain the genes for Avp1 and Avp2, respectively, downstream of a Shine-Dalgarno sequence and with additional codons for a C-terminal His6 tag. The plasmids were then transformed into the E. coli BL21 expression strain.
Recombinant protein expression.
For protein expression, 2-liter E. coli cultures were grown at 37° to an optical density at 600 nm (OD600) of 0.6. Specific Avp1 or Avp2 expression was induced by the addition of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG). Thereafter, the bacterial cultures were shaken at 10°C for 5 days. Cells were collected by centrifugation (5,000 × g for 10 min at 4°C), washed with TES buffer (50 mM Tris-Cl, 5 mM EDTA, 300 mM NaCl, pH 7.8), centrifuged again, and suspended in 30 ml of extraction buffer (50 mM Tris-Cl, 10 mM EDTA, 300 mM NaCl, 1 mM dithiothreitol [DTT], 100 mM cholic acid, pH 7.8). Cells were disrupted with a French pressure cell at 1,000 lb/in2. Insoluble material was removed by centrifugation (20,000 × g for 20 min at 4°C).
Protein purification.
Protein purification was done principally as described earlier (27). Soluble protein of the extract was precipitated overnight at 4°C by the addition of ammonium sulfate (AMS) from a 3.3 M stock solution to a final concentration of 1.65 M, followed by centrifugation at 10,000 × g for 20 min at 4°C. The pellet after AMS precipitation was resuspended in 30 ml of low-IMI buffer (50 mM Tris-Cl, 300 mM NaCl, 10 mM imidazole, 100 mg/liter cholic acid, 10 mM β-mercaptoethanol, pH 7.8). After centrifugation at 25,000 × g for 20 min at 4°C, the supernatant was loaded on a Ni2+ affinity column (30 ml of Ni2+-nitrilotriacetic acid [NTA] agarose matrix; Qiagen). The column was washed with 30 column volumes of low-IMI buffer. Bound proteins were subsequently eluted with high-IMI buffer (50 mM Tris-Cl, 300 mM NaCl, 200 mM imidazole, 100 mg/liter cholic acid, 10 mM β-mercaptoethanol, pH 7.8). The eluate was concentrated by AMS precipitation and resuspended in TES buffer supplemented with 1 mM DTT. Samples were frozen in liquid nitrogen and stored at −80°C. For analysis, samples were mixed with a 1.5-fold molar excess of biliverdin (Fisher Scientific) under a green safety light (555 nm). After self-assembly in the dark, excess chromophore was removed by desalting columns (NAP columns; GE Healthcare).
Spectral characterization of bacteriophytochromes.
For in vivo spectrophotometric measurements, we used a Jasco V550 photometer equipped with an Ulbricht sphere (Jasco ISN-723; 60 mm) and a custom-built computer-controlled irradiation device. Bacteria were grown in a 1-liter culture in darkness to an OD600 of 0.5 to 0.7. Thereafter, bacteria were kept either in darkness or under green safelight. Cells were collected by centrifugation (6,000 × g for 10 min at 4°C), and the cell pellet was washed with 45 ml of TES buffer, pelleted again, shock frozen in liquid nitrogen, and stored at −80°C until further use. For the measurements, samples were thawed at room temperature and transferred into a plastic cuvette with a 4-mm by 10-mm cross section, and the measuring light path was 4 mm. The cuvette was placed in front of the Ulbricht sphere in such a way that the window was completely covered by the bacterial sediment. A standard measuring series consists of four different irradiation programs according to the following scheme (FR and R stand for far-red and red actinic light irradiations, respectively, and D stands for dark incubation; the superscript indicates the scan number): (i) for dark conversion after FR, FR 1 D 2 FR 3 D 4 FR 5 D 6 FR 7; (ii) for photoconversion I, D 1 FR 2 R 3 FR 4 R 5 FR 6 R 7; (iii) for dark conversion after R, R 1 D 2 R 3 D 4 R 5 D 6 R 7; (iv) for photoconversion II, D 1 R 2 FR 3 R 4 FR 5 R 6 FR 7.
Spectral scans were performed at a scan speed of 200 nm/min with a 2-nm bandwidth. Actinic FR and R irradiations were made with 780-nm and 655-nm light-emitting diodes, respectively. The intensity of FR at the upper position of the cuvette was 1,000 μmol cm−2 s−1, and the duration was 60 s. The R intensity was 40 μmol cm−2 s−1, and the duration was 240 s. The dark incubation period during dark conversion measuring series was 1 h, unless otherwise noted.
The differences between the second and third, fourth and fifth, and sixth and seventh absorbance spectra were calculated for each measuring series, and the average difference spectrum was calculated from these three differences.
Nonscattering samples with extracted, recombinant proteins were measured either in the same way or in the standard photometer setup, i.e., without the Ulbricht sphere. In this case, the scan speed was set to 1,000 nm/min, and the bandwidth was adjusted to 1 nm.
Computer analysis.
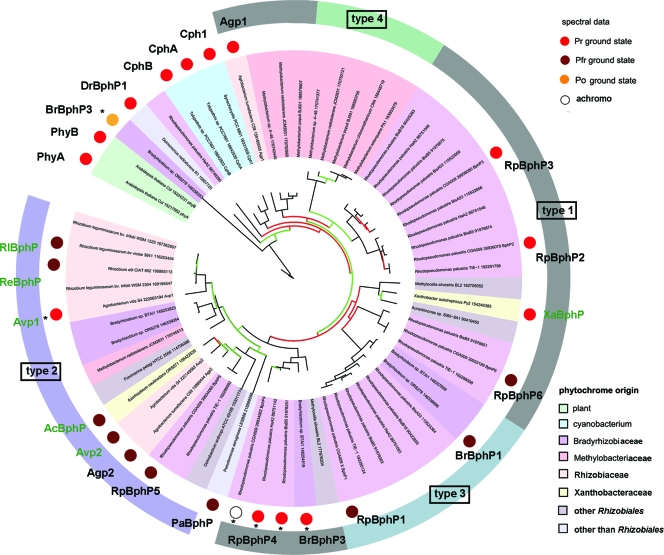
Database searches for bacteriophytochrome protein sequences were performed with NCBI BLAST by using the sequence of A. tumefaciens Agp1 (gi 159185005) as a query. The results were restricted to 49 sequenced Rhizobiales strains (see Table SA1 in the supplemental material). An alignment of the selected protein sequences, together with additional representative phytochrome sequences (R. palustris CGA009 RpBphP1 mutated pseudogene, Bradyrhizobium ORS 271 BrBphP3, Arabidopsis thaliana PhyA and PhyB, Deinococcus radiodurans R1 BphP [DrBphP], P. aeruginosa PaBphP, Synechocystis sp. strain PCC 6801 Cph1, and Tolypothrix sp. strain PCC 7601 CphA and CphB) was performed with ClustalX2 and the default parameter settings. The part comprising the GAF and PHY domains, equivalent to amino acids 337 to 756 of Agp1, was chosen for construction of a phylogenetic tree with PhyML, using the maximum-likelihood algorithm and default parameters (13). The resulting data were illustrated using ITOL (28) and further graphically processed. Protein domain classification was analyzed by PFAM (version 24.0).
RESULTS
Computer analysis.
Using the protein sequence of A. tumefaciens Agp1 as a query, we screened 49 Rhizobiales genomes (see Table SA1 in the supplemental material) for phytochrome homologs. We found 93 homologous protein sequences, which were further analyzed by PFAM (version 24.0) for the presence of PAS, GAF, and PHY domains. Whereas 38 proteins were homologous only to the C-terminal histidine kinase part of Agp1, 55 sequences of 25 strains were found to contain a GAF and a PHY domain, which constitute the photosensory core module (PCM). These proteins are thus supposed to be true bacterial phytochromes (see Table SA1). Eleven strains of nine species contain only a single phytochrome (Fig. 1). With the exception of two Methylobacterium strains, all of these bacteria are nonphotosynthetic. The photosynthetic bacteria that belong to the genera Bradyrhizobium and Rhodopseudomonas contain between two and five phytochrome genes.
FIG. 1.
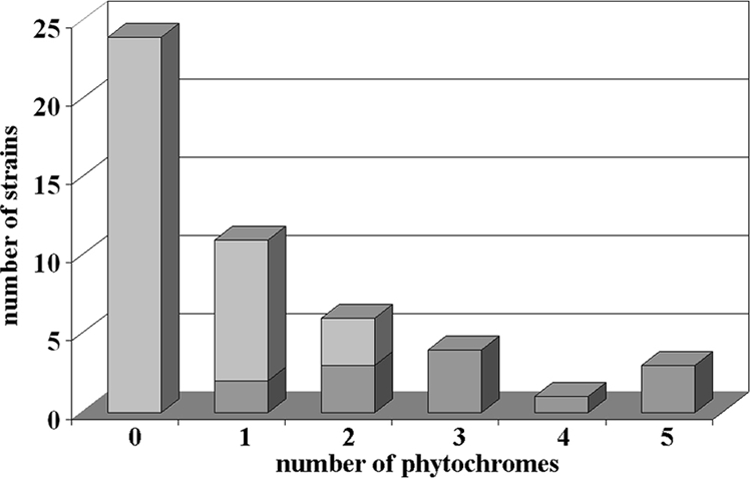
BphP genes in sequenced strains of Rhizobiales. The graph shows the number of strains carrying a given number of BphPs. Dark gray, photosynthetic; light gray, nonphotosynthetic.
The domain arrangement of all selected phytochromes was analyzed by PFAM and SMART, and a phylogenetic tree was constructed based on the sequences of the GAF and PHY domains. In addition to these 55 phytochrome sequences that were (re)identified in Rhizobiales, 10 other phytochromes from other bacteria and plants were included in the phylogenetic analysis.
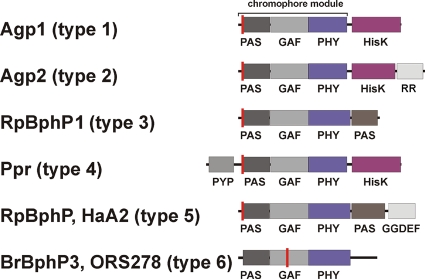
The rhizobial phytochromes show six different types of protein domain arrangement, which we designate type 1 through type 6. In the type 1 phytochromes, a histidine kinase is located C-terminal of the PCM. This is the most common domain arrangement of bacterial phytochromes, and 24 of the 55 rhizobial phytochrome homologs belong to this group. The domain arrangement of type 2 phytochromes is comparable to that of type 1, but these proteins have an additional C-terminal response regulator, which might serve as substrate of the histidine kinase. In all 15 of the type 2 proteins, the histidine kinase is of the HWE type, a newly characterized subgroup of histidine kinases with canonical amino acids (19). Interestingly, all other phytochromes analyzed here have non-HWE histidine kinases or other C-terminal domains, suggesting that type 2 phytochromes have a common evolutionary origin. This view is supported by our phylogeny analysis (Fig. 2) and by an earlier phylogenetic study (11), which was based on a different subset of bacterial phytochromes. The type 3 phytochromes have no histidine kinase but a C-terminal PAS domain. We found this type only in the photosynthetic bacteria Bradyrhizobium and Rhodopseudomonas. These phytochromes cluster also in one group (Fig. 2) (11). The type 4 phytochromes are comparable with the type 1 as they bear a C-terminal histidine kinase. However, their characteristic feature is an additional N-terminal photoactive yellow protein domain. A PYP-phytochrome chimeric protein of this type, Ppr from Rhodospirillum centenum, which stands outside the Rhizobiales, has been described before (16, 23). Although the PCM of Ppr incorporates a bilin chromophore, the spectral properties of this holoprotein are different from those of typical phytochromes. We found five phytochromes of this type in the genus Methylobacterium. All type 4 phytochromes cluster in one branch of the phylogenetic tree (Fig. 2). The domain arrangements of type 5 and 6 phytochromes (Fig. 3) are found in single phytochromes only and will not be further considered here.
FIG. 2.
Phylogenetic tree of Rhizobiales BphPs and additional phytochromes. Branches of the tree are given in black, green, and red for bootstrap values of >75%, >50%, and <50%, respectively. Colors underlying the strain names indicate taxonomic groups according to the color key at the bottom of the figure. The ground states of spectrally characterized phytochromes are marked by large colored dots according to the key at the top of the figure. Common abbreviations are also given for these phytochromes. The following phytochromes were characterized earlier (reference): Agp1 (26), Agp2 (18), Bradyrhizobium ORS278 BrBphP1 (10), Bradyrhizobium ORS278 BrBphP3 (15), Bradyrhizobium strain BTAi1 BrBphP3 (11), Cph1 (27), CphA (17), CphB (17), DrBphP1 (7), PaBphP (40), PhyA (36), PhyB (36), RpBphP1 (11), RpBphP2 (12), RpBphP3 (8), R. palustris CGA009/HaA2/BisB5 RpBphP4 (42), RpBphP5 (11), and RpBphP6 (11). The domain arrangements of type 1 through type 4 Rhizobiales phytochromes are indicated by the colored circle segments around the tree. Asterisks indicate phytochromes with unusual spectral characteristics.
FIG. 3.
Domain arrangements of BphPs from examined strains (Agp1 and Agp2 from A. tumefaciens C58; RpBphP [gi 86749290] and RpBphP1 from R. palustris HaA2 and CGA009, respectively; BrBphP3 from Bradyrhizobium sp. ORS278; and Ppr from R. centenum SW). Domain arrangements were drawn according to PFAM results. The indicated domains are as follows: PAS, Per-Arnt-Sim; GAF, cGMP-specific and -regulated cyclic nucleotide phosphodiesterase, adenylyl cyclase, and E. coli transcription factor FhlA; PHY, phytochrome-specific; HisK, histidine kinase; and RR, response regulator. The chromophore binding site is indicated by the red line.
All selected rhizobial phytochromes with the exception of two have a conserved Cys residue in the PAS domain, which serves as BV binding site (25, 26). The two exceptions are the above-mentioned BphP3 from Bradyrhizobium ORS 278, which binds PCB as a natural chromophore (15), and RpBphP4 from R. palustris CGA009, which does not incorporate a chromophore (42).
For our in vivo spectroscopic studies, we selected two species with two phytochromes, A. tumefaciens C58 and A. vitis S4, and five other species which have a single phytochrome only, R. leguminosarum 3841, R. etli CIAT 652, A. caulinodans ORS 571, X. autotrophicus Py2, and Methylobacterium extorquens PA1.
Spectroscopy with A. tumefaciens.
A. tumefaciens C58 served as a standard for our measurements. Both phytochromes, Agp1 and Agp2, of this soil bacterium have extensively been studied as recombinant proteins, and the spectral activities of both phytochromes have been measured in cell extracts (31). Moreover, knockout mutants are available (31) which allow the spectral characterization of single phytochromes in vivo.
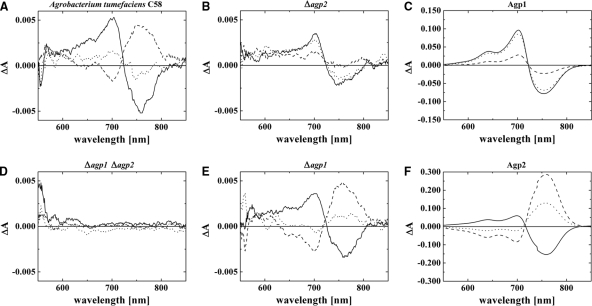
After having optimized the in vivo measuring parameters with respect to cuvette dimensions, repetition rate, duration and strength of irradiation, and other settings (see Materials and Methods), we obtained typical difference spectra with A. tumefaciens wild-type cells by subtracting absorption spectra taken after red (R) from those taken after far-red (FR) irradiation (Fig. 4). The peak-to-peak amplitude was ca. 0.01 ΔΔA, (where A is absorption), and the maxima and minima were found at 700 nm and 750 nm, respectively. Similar difference spectra with lower amplitudes were also measured in Δagp2 and Δagp1 knockout mutants, and the difference spectrum of the Δagp1 Δagp2 double knockout had no apparent phytochrome signature (Fig. 4). These results show that Agp1 and Agp2 are the only photoactive compounds in A. tumefaciens cells that are detected under the present measuring conditions and that both proteins are present in the cells at roughly equal concentrations, as has been postulated before (31). These observations made us confident that phytochromes can be detected in other species by the same in vivo approach as well, given that the content of screening bacterial pigments is not too high.
FIG. 4.
Difference spectra of bacterial cell sediments of A. tumefaciens C58 wild type (WT) (A) and agp1 agp2 (D), agp2 (B), agp1 (E) knockout mutants and recombinant proteins Agp1 (C) and Agp2 (F). Photoconversion (solid lines), difference spectra obtained from subtracting absorbance spectra after R from spectra after FR; dark conversion after FR (dotted lines), spectra after 1 h dark of incubation minus spectra after subsequent FR (a positive signal around 700 nm indicates Pfr to Pr dark conversion); dark conversion after R (dashed lines), spectra after 1 h of dark incubation minus spectra after R (a positive signal around 750 nm indicates Pr to Pfr dark conversion). All samples were measured with an Ulbricht sphere; details are given in the Materials and Methods section.
The A. tumefaciens wild-type and mutant cells were also used for measuring the direction of dark conversion in the living cells. As noted above, three principles are realized among known phytochromes: dark conversion of Pfr to Pr (normal phytochromes), dark conversion of Pr to Pfr (bathy phytochromes), and a stable dark state (normal phytochromes). Recombinant Agp1 and Agp2 follow the first and second principles, respectively. For in vivo Pfr-to-Pr dark conversion measurements, the samples were initially irradiated with R. Thereafter, the cells were kept for 1 h in darkness. After an absorption spectrum was measured, the samples were again irradiated with R and measured. The procedure was typically repeated three times, and the average difference spectrum was calculated from the spectra before and after the R treatments (see also Materials and Methods). To measure the dark conversion of Pr to Pfr, the same procedure was performed except that R was replaced by FR. The shapes and amplitudes of the obtained difference spectra give clear information about the extent and the direction of dark conversion during 1 h. In wild-type A. tumefaciens cells, both directions of dark conversion were found, whereas in the agp1 and agp2 mutants, dark conversion only of Pr to Pfr and of Pfr to Pr, respectively, was observed. Thus, the direction of dark conversion in the A. tumefaciens cells correlates with the direction of dark conversion of recombinant proteins (Fig. 4).
Besides the overall consistency between in vivo and in vitro measurements, the in vivo spectra of Agp2 in the agp1 mutant differ from those of recombinant Agp2 with respect to the absorbance changes at 700 nm and 750 nm (Fig. 4E and F). The difference spectrum of Agp2 in vivo exhibits a balanced ΔA700/ΔA750 absorbance change ratio, whereas the ΔA700/ ΔA750 absorbance change ratio of recombinant Agp2 is much lower than 1 (about 0.3 in the photoconversion difference spectrum). The same difference has already been observed between Agp2 measured in cell extracts and purified recombinant Agp2 (22). We also noted that the photoconversion (Fig. 4F, solid line) and dark conversion (Fig. 4F, dashed line) difference spectra of recombinant Agp2, but not those of Agp2 in vivo, are dissimilar. In the former, the ΔA700/ΔA750 absorbance change ratio was lower for dark conversion than for photoconversion.
Spectroscopy with A. vitis.
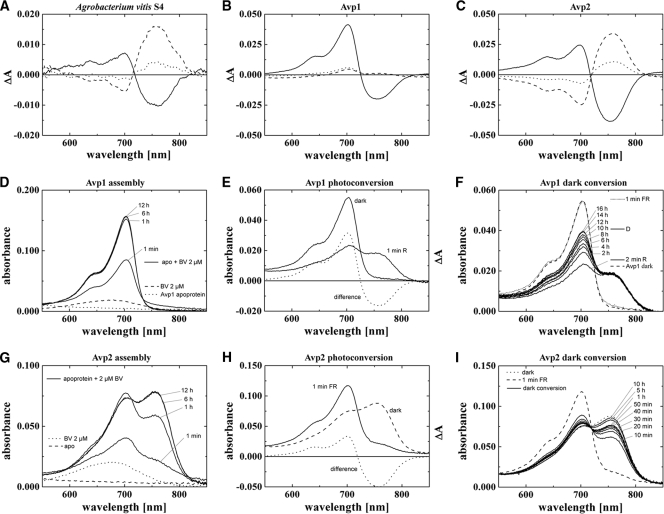
The second species of the present study with two phytochrome genes was A. vitis S4, a bacterium which is closely related to A. tumefaciens. Both A. vitis phytochromes, which we have termed Avp1 and Avp2, have a C-terminal response regulator and belong to the type 2 group. We would like to note here that another closely related species, the nonpathogenic Agrobacterium radiobacter, contains no phytochrome gene at all (see Table SA1 in the supplemental material).
We measured a ΔΔA peak-to-peak value of ca. 0.02 in A. vitis cell pellets, a signal which is about twice as high as that of A. tumefaciens. The positions of maxima and minima (at around 700 nm and 750 nm) are also well in accordance with a phytochrome which uses BV as a chromophore. Remarkably, difference spectra for photoconversion and even more for dark conversion exhibit a low ΔA700/ΔA750 absorbance change ratio. As opposed to the case in A. tumefaciens, dark conversion only of Pr to Pfr was detected, and no dark conversion of Pfr to Pr was observed.
Thus, at least one of the A. vitis phytochromes is a bathy phytochrome. The other one might share the same dark conversion properties or might be expressed at low concentrations or have other spectral features that remain undetected by the in vivo approach.
We decided to concentrate on the characterization of recombinant, poly-His-tagged Avp1 and Avp2 to address this question. The purification of both proteins from E. coli proved difficult, however. Although expression levels of Avp1 and Avp2 were reasonably high, a predominant portion of the proteins remained insoluble. By lowering the temperature during recombinant expression, we could, however, obtain sufficient amounts of soluble proteins. After Ni2+ affinity chromatography, it was possible to follow chromophore assembly photometrically, to measure absorbance spectra of ground states and photoproducts, and to follow dark conversion.
As shown in Fig. 5, recombinant Avp2 undergoes dark conversion of Pr to Pfr during or immediately after chromophore assembly and after Pfr-to-Pr photoconversion. Thus, Avp2 is a typical bathy phytochrome. It resembles Agp2 of A. tumefaciens although assembly kinetics and dark conversion kinetics of Avp2 are slower than those of Agp2. The difference spectra of recombinant Avp2 match well with the in vivo difference spectra of A. vitis although the more balanced ΔA700/ΔA750 absorbance change ratio in the difference spectrum for dark conversion after FR of recombinant Avp2 contrasts with the in vivo measurement.
FIG. 5.
A. vitis S4 phytochrome spectra. (A) In vivo difference spectra of A. vitis bacterial cell sediment. (B and C) Difference spectra of recombinant Avp1 and Avp2, measured with the Ulbricht sphere. (D to F) Absorption and difference spectra of recombinant Avp1 during assembly (D), photoconversion (E), and dark conversion (E) as measured with standard setup (without the Ulbricht sphere). (G to I) Absorption and difference spectra of recombinant Avp2 during assembly (G), photoconversion (H), and dark conversion (I) as measured with a standard setup. For panels A to C solid lines indicate difference spectra obtained from subtracting absorption spectra after R from absorption spectra after FR, dotted lines indicate spectra after 1 h of dark incubation subtracted from spectra after FR (a positive signal around 700 nm indicates Pfr-to-Pr dark conversion), and dashed lines indicate spectra after 1 h of dark incubation subtracted from spectra after R (a positive signal around 750 nm indicates Pr-to-Pfr dark conversion). Symbols in panels D to I are identified on the figure.
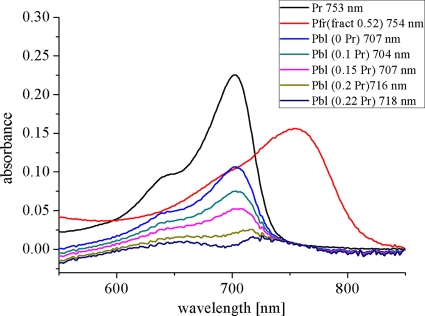
A different feature was found for recombinant Avp1. This phytochrome assembles in the Pr form, with a maximum at 703 nm. The rate of assembly is again slow: Avp1 requires ca. 6 h for completion of assembly (Fig. 5D). Irradiation of the Avp1 adduct results in the formation of Pfr, which absorbs with a maximum at 750 nm. However, photoconversion into Pfr seems incomplete as the light-induced decrease at 700 nm is larger than the concomitant increase at 750 nm (Fig. 5E); i.e., a high ΔA700/ΔA750 nm absorbance change ratio was observed in the difference spectrum. This observation suggests that in addition to Pr and Pfr, a third species is present in the irradiated sample. Measurements during dark incubation corroborated this assumption: the Pfr form of recombinant Avp1 is stable in darkness, as the absorbance in the 750-nm range remained unchanged over the observation time of 16 h. However, the Pr signal increased steadily during 16 h (Fig. 5F). This increase must come from a third species that converts to Pr in darkness. This species is termed Pbl (bleached phytochrome) here. The three spectra of the dark-assembled, red-irradiated, and dark-incubated Avp1 samples may be used for empirical estimations on the contents and spectral shapes of the photoproducts (see reference 2) because dark-assembled Avp1 contains only Pr, and the sample after irradiation with red light and an 18-h dark incubation contains only Pr and Pfr. If the fractions of Pr, Pfr, and Pbl were exactly known, the spectra of pure Pbl and Pfr could be exactly calculated. Based on the available data, it was not possible to obtain accurate values for each fraction, but it was possible to estimate fraction ranges. According to our analysis, the fraction of Pfr in the R-irradiated sample lies between 0.5 and 0.55, and the fraction of Pr directly after R is 0.22 or lower. The Pfr spectra that are calculated under these assumptions are comparable with (calculated) Pfr spectra of, e.g., Agp1. The spectrum of Pbl has either a Pr-like shape in the red region (above 600 nm), or it does not absorb at all in this wavelength range (Fig. 6). In either case the Pbl extinction coefficient is significantly lower than that of Pr or Pfr. We noted similarities of our Pbl photoproduct with that of R. centenum Ppr. This PYP-phytochrome chimeric protein assembles with BV and forms a Pr-like adduct, which becomes bleached upon irradiation. The Ppr photoproduct also undergoes dark conversion into Pr (23). However, unlike Avp1, Ppr does not form Pfr. Most of our calculated Pbl spectra (Fig. 6) are also reminiscent of MetaRc spectra (3) and photoproduct spectra of Agp1 mutants which are arrested in their photocycle (41). We thus propose that Pbl could be a photocycle intermediate, probably with a deprotonated chromophore, which is unable to undergo conversion into Pfr.
FIG. 6.
Avp1 spectra. The black line shows the Pr spectrum of the dark-assembled sample, the red line gives the calculated Pfr spectrum under the assumption that the Pfr fraction in the red-irradiated sample after 18 h of dark incubation is 0.52. The other spectra are calculated for Pbl under the assumption that the Pfr fraction in the R-treated sample is always 0.52, and the Pr fraction is 0, 0.1, 0.15, 0.2, or 0.22, as indicated on the figure. The positions of the absorption maxima are also given on the figure.
In vivo spectroscopy with Rhizobiales containing single phytochromes.
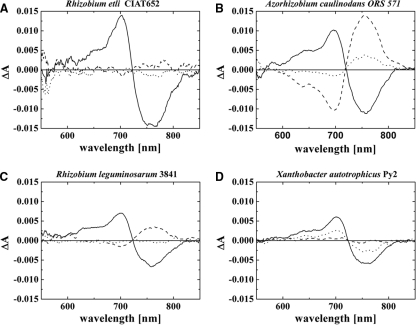
We also performed spectral in vivo measurements with cells of R. etli CIAT 652 (Fig. 7A), R. leguminosarum 3841 (Fig. 7B), A. caulinodans ORS 571 (Fig. 7C), X. autotrophicus Py2 (Fig. 7D), and M. extorquens PA1. As noted above, each of these bacteria contains a single phytochrome gene. These phytochromes are denominated ReBphP, RlBphP, AcBphP, XaBphP, and MeBphP, respectively. We detected a phytochrome signal in all strains with the exception of M. extorquens (data not shown). All difference spectra of FR irradiated minus R irradiated samples had maxima and minima around 700 nm and 750 nm, respectively. The highest signal of all strains was obtained for R. etli CIAT 652, which had a peak-to-peak value above 0.02 ΔΔA. The peak-to-peak values of the other strains were between 0.01 and 0.02 ΔΔA.
FIG. 7.
Difference spectra of bacterial cell sediments of R. etli, A. caulinodans, R. leguminosarum, and X. autotrophicus are shown. Solid lines, difference spectra obtained from subtracting absorbance spectra after far-red irradiation (780 nm) from absorption spectra after red irradiation (655 nm); dotted lines, spectra after 1 h of dark incubation subtracted from spectra after far-red irradiation (a positive signal around 700 nm indicates Pfr-to-Pr dark conversion); dashed lines, spectra after 1 h of dark incubation subtracted from spectra after red irradiation (a positive signal around 750 nm indicates Pr-to-Pfr dark conversion).
Phytochromes of A. caulinodans and R. leguminosarum revealed a dark conversion of Pr to Pfr although dark conversion of the latter species was not completed within the 1-h dark period. Thus, AcBphP and RlBhpP are thereby directly assigned as bathy phytochromes. Dark conversion of XaBphP proceeds from Pfr to Pr; this phytochrome may be classified as normal or Agp1-like. In R. etli, a very weak Pr-to-Pfr dark conversion was observed during the 1-h dark incubation period. However, when an FR-treated sample was incubated for 16 h in darkness, subsequent FR irradiation induced clear photoconversion. In addition, we found that FR induced photoconversion of completely dark-cultivated R. etli samples. This indicates that ReBphP undergoes a very slow dark conversion of Pr to Pfr. Hence, we classify ReBphP also as a bathy phytochrome. The ΔΔA values that were obtained by the initial FR irradiation of completely dark-adapted samples or 16-h dark-adapted samples were ca. 0.5-fold lower than ΔΔA values obtained by subsequent R to FR photoconversion. Thus, residual Pr is present in dark-adapted samples. This Pr could be freshly synthesized phytochrome. Bathy phytochromes are assembled in the Pr form, which converts to Pfr during subsequent dark incubation. If the rate of dark conversion is slower than the rate of synthesis, it is to be expected that a significant fraction of phytochrome is in the Pr form.
DISCUSSION
We have shown that phytochrome difference spectra can be directly measured in bacterial cells. The methodology involves concentration of bacterial cells by centrifugation, transfer of the cell pellet into a measuring cuvette of appropriate dimensions, UV-visible light spectroscopy with an Ulbricht sphere which partially compensates for scattering losses, and the use of strong actinic light in order to induce maximal photoconversion. By this approach, one can easily obtain spectral information of phytochromes in vivo as well as information on the direction and kinetics of dark conversion within the cell.
A. tumefaciens.
C58, which has two phytochromes, served as a control for our studies. Both phytochromes are well characterized as recombinant proteins, and A. tumefaciens knockout mutants allowed measurements on strains in which only one phytochrome was expressed. In vivo spectral properties and dark conversion of Agp1 and Agp2 matched well with those of cell extracts and the recombinant proteins: Agp1 and Agp2 undergo dark conversion of Pfr to Pr and Pr to Pfr in vivo, respectively. We have previously reported that spectral properties of recombinant Agp2 are specifically modified by compounds of the cell extract (22, 31). Our in vivo difference spectra are well in accordance with those of Agp2 in cell extracts and more dissimilar from those of the pure recombinant protein.
A. vitis.
S4 contains also two phytochromes, Avp1 and Avp2. Despite the close relationship between the two Agrobacterium species, the phytochrome systems differ. In A. vitis we observed only Pr-to-Pfr dark conversion and not conversion in the reverse direction. The bathy feature corresponds to the properties of recombinant Avp2. According to our studies on recombinant Avp1, we consider this phytochrome as a new type of phytochrome with a Pr ground state and two photoproducts, Pfr and Pbl. Whereas Pfr of recombinant Avp1 was found to be stable in darkness, the Pbl form undergoes dark conversion into the Pr state. We did not observe a Pbl-to-Pr dark conversion in our in vivo measurements. This is most probably due to the interference with the dark conversion of Pr to Pfr of Avp2. The net dark conversion of Avp2 after R proceeds from Pr to Pfr, i.e., in the same direction as after FR (Fig. 5C), because R also produces a significant amount of Pr. The weak dark conversion of Avp1 after R (Fig. 5B) is thus not seen in the in vivo measurements. The photoconversion ΔA700/ΔA750 absorbance change ratios of recombinant Avp1 (high), recombinant Avp2 (low), and the in vivo measurements (balanced) (Fig. 5A to C, solid lines) suggest that both phytochromes are present in A. vitis cells and that both contribute to the difference spectrum. Please note that the absorbance change ratios of dark conversion after FR differ between recombinant Avp2 and the in vivo measurements as well.
The spectral in vivo analysis of knockout mutants might show whether Avp1 has also unusual spectral characteristics in vivo.
Altogether, five phytochromes (Agp2, Avp2, ReBphP, RlBphP, and AcBphP) characterized in the present study belong to the bathy phytochromes, whereas only two (Agp1 and XaBphP) showed a normal dark conversion of Pfr to Pr, and one (Avp1) revealed unusual spectral properties. With the inclusion of all spectrally characterized Rhizobiales phytochromes, a similar picture emerges: in this group there are nine bathy phytochromes, seven phytochromes with a Pfr-to-Pr dark conversion, and three to four phytochromes with unusual spectral properties. Thus, the bathy phytochromes obviously dominate among the Rhizobiales, and they are most frequently represented in plant-interacting nodule-forming soil bacteria. Indeed, it is expected that the light regime in the soil is rich in long-wavelength far-red light. First, the green canopy filters almost all visible light, whereas long-wavelength red and infrared light is only weakly absorbed by photosynthetic pigments (9, 38). Second, long-wavelength red light penetrates deeper into the soil than shorter wavelengths, an effect which is independent of the soil type (38, 43). Third, plant stems and plant roots function as light vessels that guide light into the soil (39). In this case, far-red light in the range of 750 nm has the highest transmission rates. This effect might be especially important for bacteria that undergo symbiosis with plant roots.
Origin of bathy phytochromes.
With the increased number of bathy phytochromes, we can now shine more light on the evolutionary origin of this particular subgroup. In the phylogenetic tree which comprises phytochromes of Rhizobiales together with representatives of other groups, we have indicated all 10 bathy phytochromes (Fig. 2, brown dots). Seven of these, PaBphP, RpBphP5, Agp2, Avp2, AcBphP1, ReBphP, and RlBphP, are found within a cluster of 16 phytochromes which comprise all 15 phytochromes with a type 2 domain arrangement and 1 phytochrome, PaBphP, with a type 1 domain arrangement. This last phytochrome is from a species that does not belong to the Rhizobiales. It has been included in our analyses because its photochemical and structural details are well known (40, 44). However, we assume that the grouping of PaBphP together with the type 2 phytochromes does not reflect a close relationship but is the consequence of a convergent evolution or domain rearrangement in the evolution of PaBphP because of the dissimilar C-terminal moieties (HWE histidine kinases with a response regulator in the type 2 and classical histidine kinase without a response regulator in type 1 phytochromes). The only exception among the type 2 cluster with respect to dark conversion is the unusual A. vitis phytochrome Avp1. The dark conversion properties of the other members of the type 2 cluster are as yet unknown. It is very likely that the common ancestor of the type 2 phytochromes was a bathy phytochrome and that Avp1 has undergone a reversion into a phytochrome with a Pr ground state.
There are then also three other Rhizobiales bathy phytochromes outside the type 2 cluster, BrBphP1, RpBphP1, and RpBphP6. These phytochromes are found in a separate cluster of our phylogenetic tree which comprises phytochromes with the type 1 (RpBphP6) and type 3 domain arrangements. Dark conversion of other phytochromes of this cluster has as yet not been analyzed. Also these phytochromes might have originated from a common bathy phytochrome ancestor.
Supplementary Material
Acknowledgments
We thank Léon Otten (IBMP, University of Strasbourg), Victor Zuniga (Center for Genomic Sciences, UNAM), Dirk Janssen (University of Groningen), Peter Young (Department of Biology, University of York), and Hiroshi Oyaizu (Biotechnology Research Center, University of Tokyo) for kindly providing bacterial strains. We thank Sybille Wörner, Kai Pfannebecker, and Chaoran Li for their help with culturing and measuring bacteria.
This work was supported by the Deutsche Forschungsgemeinschaft.
Footnotes
Published ahead of print on 30 July 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Bhoo, S. H., S. J. Davis, J. Walker, B. Karniol, and R. D. Vierstra. 2001. Bacteriophytochromes are photochromic histidine kinases using a biliverdin chromophore. Nature 414:776-779. [DOI] [PubMed] [Google Scholar]
- 2.Borucki, B., and T. Lamparter. 2009. A polarity probe for monitoring light induced structural changes at the entrance of the chromophore pocket in a bacterial phytochrome. J. Biol. Chem. 38:26005-26016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borucki, B., D. von Stetten, S. Seibeck, T. Lamparter, N. Michael, M. A. Mroginski, H. Otto, D. H. Murgida, M. P. Heyn, and P. Hildebrandt. 2005. Light-induced proton release of phytochrome is coupled to the transient deprotonation of the tetrapyrrole chromophore. J. Biol. Chem. 280:34358-34364. [DOI] [PubMed] [Google Scholar]
- 4.Butler, W. L. 1964. Absorption spectroscopy in vivo—theory and application. Annu. Rev. Plant Physiol. 15:451-470. [Google Scholar]
- 5.Butler, W. L., H. C. Lane, and H. W. Siegelman. 1963. Nonphotochemical transformations of phytochrome in vivo. Plant Physiol. 38:514-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler, W. L., K. H. Norris, H. W. Siegelman, and S. B. Hendricks. 1959. Detection, assay, and preliminary purification of the pigment controlling photoresponsive development of plants. Proc. Natl. Acad. Sci. U. S. A. 45:1703-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis, S. J., A. V. Vener, and R. D. Vierstra. 1999. Bacteriophytochromes: phytochrome-like photoreceptors from nonphotosynthetic eubacteria. Science 286:2517-2520. [DOI] [PubMed] [Google Scholar]
- 8.Evans, K., A. P. Fordham-Skelton, H. Mistry, C. D. Reynolds, A. M. Lawless, and M. Z. Papiz. 2005. A bacteriophytochrome regulates the synthesis of LH4 complexes in Rhodopseudomonas palustris. Photosynth. Res. 85:169-180. [DOI] [PubMed] [Google Scholar]
- 9.Franklin, K. A., and G. C. Whitelam. 2005. Phytochromes and shade-avoidance responses in plants. Ann. Bot. 96:169-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giraud, E., J. Fardoux, N. Fourrier, L. Hannibal, B. Genty, P. Bouyer, B. Dreyfus, and A. Vermeglio. 2002. Bacteriophytochrome controls photosystem synthesis in anoxygenic bacteria. Nature 417:202-205. [DOI] [PubMed] [Google Scholar]
- 11.Giraud, E., and A. Vermeglio. 2008. Bacteriophytochromes in anoxygenic photosynthetic bacteria. Photosynth. Res. 97:141-153. [DOI] [PubMed] [Google Scholar]
- 12.Giraud, E., L. Vuillet, L. Hannibal, J. Fardoux, S. Zappa, J. M. Adriano, C. Berthomieu, P. Bouyer, D. Pignol, and A. Verméglio. 2005. A new type of bacteriophytochrome acts in tandem with a classical bacteriophytochrome to control the antennae synthesis in Rhodopseudomonas palustris. J. Biol. Chem. 280:32389-32397. [DOI] [PubMed] [Google Scholar]
- 13.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PhyML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acid Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inomata, K., S. Noack, M. A. S. Hammam, H. Khawn, H. Kinoshita, Y. Murata, N. Michael, P. Scheerer, N. Krauß, and T. Lamparter. 2006. Assembly of synthetic locked chromophores with Agrobacterium phytochromes Agp1 and Agp2. J. Biol. Chem. 281:28162-28173. [DOI] [PubMed] [Google Scholar]
- 15.Jaubert, M., J. Lavergne, J. Fardoux, L. Hannibal, L. Vuillet, J. M. Adriano, P. Bouyer, D. Pignol, E. Giraud, and A. Vermeglio. 2007. A singular bacteriophytochrome acquired by lateral gene transfer. J. Biol. Chem. 282:7320-7328. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, Z. Y., L. R. Swem, B. G. Rushing, S. Devanathan, G. Tollin, and C. E. Bauer. 1999. Bacterial photoreceptor with similarity to photoactive yellow protein and plant phytochromes. Science 285:406-409. [DOI] [PubMed] [Google Scholar]
- 17.Jorissen, H. J., B. Quest, A. Remberg, T. Coursin, S. E. Braslavsky, K. Schaffner, N. Tandeau de Marsac, and W. Gärtner. 2002. Two independent, light-sensing two-component systems in a filamentous cyanobacterium. Eur. J. Biochem. 269:2662-2671. [DOI] [PubMed] [Google Scholar]
- 18.Karniol, B., and R. D. Vierstra. 2003. The pair of bacteriophytochromes from Agrobacterium tumefaciens are histidine kinases with opposing photobiological properties. Proc. Natl. Acad. Sci. U. S. A. 100:2807-2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karniol, B., and R. D. Vierstra. 2004. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J. Bacteriol. 186:445-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karniol, B., J. R. Wagner, J. M. Walker, and R. D. Vierstra. 2005. Phylogenetic analysis of the phytochrome superfamily reveals distinct microbial subfamilies of photoreceptors. Biochem. J. 392:103-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kendrick, R. E., and G. H. M. Kronenberg. 1994. Photomorphogenesis in plants, 2nd ed. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 22.Krieger, A., I. Molina, I. Oberpichler, N. Michael, and T. Lamparter. 2008. Spectral properties of phytochrome Agp2 from Agrobacterium tumefaciens are specifically modified by a compound of the cell extract. J. Photochem. Photobiol. B 93:22. [DOI] [PubMed] [Google Scholar]
- 23.Kyndt, J. A., T. E. Meyer, and M. A. Cusanovich. 2004. Photoactive yellow protein, bacteriophytochrome, and sensory rhodopsin in purple phototrophic bacteria. Photochem. Photobiol. Sci. 3:519-530. [DOI] [PubMed] [Google Scholar]
- 24.Lamparter, T. 2004. Evolution of cyanobacterial and plant phytochromes. FEBS Lett. 573:1-5. [DOI] [PubMed] [Google Scholar]
- 25.Lamparter, T., M. Carrascal, N. Michael, E. Martinez, G. Rottwinkel, and J. Abian. 2004. The biliverdin chromophore binds covalently to a conserved cysteine residue in the N terminus of Agrobacterium phytochrome Agp1. Biochemistry 43:3659-3669. [DOI] [PubMed] [Google Scholar]
- 26.Lamparter, T., N. Michael, F. Mittmann, and B. Esteban. 2002. Phytochrome from Agrobacterium tumefaciens has unusual spectral properties and reveals an N-terminal chromophore attachment site. Proc. Natl. Acad. Sci. U. S. A. 99:11628-11633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamparter, T., F. Mittmann, W. Gärtner, T. Börner, E. Hartmann, and J. Hughes. 1997. Characterization of recombinant phytochrome from the cyanobacterium Synechocystis. Proc. Natl. Acad. Sci. U. S. A. 94:11792-11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic, I., and P. Bork. 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23:127-128. [DOI] [PubMed] [Google Scholar]
- 29.Mancinelli, A. L. 1988. Phytochrome photoconversion in vivo. Comparison between measured and predicted values. Plant Physiol. 86:749-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montgomery, B. L., and J. C. Lagarias. 2002. Phytochrome ancestry: sensors of bilins and light. Trends Plant Sci. 7:357-366. [DOI] [PubMed] [Google Scholar]
- 31.Oberpichler, I., I. Molina, O. Neubauer, and T. Lamparter. 2006. Phytochromes from Agrobacterium tumefaciens: difference spectroscopy with extracts of wild type and knockout mutants. FEBS Lett. 580:437-442. [DOI] [PubMed] [Google Scholar]
- 32.Rockwell, N. C., Y. S. Su, and J. C. Lagarias. 2006. Phytochrome structure and signaling mechanisms. Annu. Rev. Plant Biol. 57:837-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sage, L. C. 1992. Pigment of the imagination: a history of phytochrome research. Academic Press, New York, NY.
- 34.Schäfer, E., B. Marchal, and D. Marme. 1972. In vivo measurements of the phytochrome photostationary state in far red light. Photochem. Photobiol. 15:457-464. [Google Scholar]
- 35.Scheerer, P., N. Michael, J. H. Park, S. Nagano, H. W. Choe, K. Inomata, B. Borucki, N. Krauss, and T. Lamparter. 2010. Light-induced conformational changes of the chromophore and the protein in phytochromes: bacterial phytochromes as model systems. Chemphyschem 11:1090-1105. [DOI] [PubMed] [Google Scholar]
- 36.Sharrock, R. A., and P. H. Quail. 1989. Novel phytochrome sequences in Arabidopsis thaliana: structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3:1745-1757. [DOI] [PubMed] [Google Scholar]
- 37.Siegelman, H. W., and E. M. Firer. 1964. Purification of phytochrome from oat seedlings. Biochemistry 3:418-423. [DOI] [PubMed] [Google Scholar]
- 38.Smith, H. 1982. Light quality, photoperception, and plant strategy. Annu. Rev. Plant Physiol. 33:481-518. [Google Scholar]
- 39.Sun, Q., K. Yoda, and H. Suzuki. 2005. Internal axial light conduction in the stems and roots of herbaceous plants. J. Exp. Bot. 56:191-203. [DOI] [PubMed] [Google Scholar]
- 40.Tasler, R., T. Moises, and N. Frankenberg-Dinkel. 2005. Biochemical and spectroscopic characterization of the bacterial phytochrome of Pseudomonas aeruginosa. FEBS J. 272:1927-1936. [DOI] [PubMed] [Google Scholar]
- 41.von Stetten, D., S. Seibeck, N. Michael, P. Scheerer, M. A. Mroginski, D. H. Murgida, N. Krauß, M. P. Heyn, P. Hildebrandt, B. Borucki, and T. Lamparter. 2007. Highly conserved residues D197 and H250 in Agp1 phytochrome control the proton affinity of the chromophore and Pfr formation. J. Biol. Chem. 282:2116-2123. [DOI] [PubMed] [Google Scholar]
- 42.Vuillet, L., M. Kojadinovic, S. Zappa, M. Jaubert, J. M. Adriano, J. Fardoux, L. Hannibal, D. Pignol, A. Vermeglio, and E. Giraud. 2007. Evolution of a bacteriophytochrome from light to redox sensor. EMBO J. 26:3322-3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woolley, J. T., and E. W. Stoller. 1978. Light penetration and light-induced seed germination in soil. Plant Physiol. 61:597-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang, X., J. Kuk, and K. Moffat. 2008. Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:14715-14720. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.